Abstract
The Arabidopsis thaliana leucine-rich repeat receptor kinase FLAGELLIN SENSING2 (FLS2) is required for the recognition of bacterial flagellin in innate immunity. Recently, FLS2 was proposed to act as a multispecific receptor recognizing unrelated exogenous and endogenous peptide ligands, including CLAVATA3 (CLV3), a key regulator of shoot meristem stem cell production. Here, we report experimental evidence demonstrating that FLS2 does not recognize CLV3 and that the shoot apical meristem is immune to bacteria independently of CLV3 perception.
Plants use surface-localized transmembrane receptor-like kinases (RLKs) to regulate intercellular communication during development and in response to environmental challenges (De Smet et al., 2009; Lehti-Shiu et al., 2009). Arabidopsis thaliana and rice (Oryza sativa) are predicted to contain 443 and 786 transmembrane RLKs, respectively (Lehti-Shiu et al., 2009). The expansion of the RLK family suggests that plants have diversified their surveillance systems to ensure specificity of ligand–RLK interactions. As a result, RLKs are thought to be highly specific for a single ligand or ligand family. The exact biological role for most plant RLKs is still unknown, and only a handful plant RLKs have been matched with a corresponding ligand. Even then, proof of direct binding is often lacking (Butenko et al., 2009). In addition, it is clear that some RLKs may not bind ligands directly, but rather function as regulatory proteins for other RLKs (Li, 2010). The difficulty in connecting RLK-ligand binding activity with downstream biological responses is illustrated by the tomato (Solanum lycopersicum) leucine-rich repeat (LRR)-RLK BRASSINOSTEROID INSENSITIVE1 (BRI1), which acts as the receptor for brassinosteroids to control growth and development. BRI1 was proposed to bind systemin, a Solanaceae-specific endogenous defense elicitor peptide that is produced in response to insect attack and wounding (Montoya et al., 2002; Scheer and Ryan, 2002; Scheer et al., 2003). Later studies found that BRI1 was dispensable for systemin-induced responses, despite conferring systemin binding (Holton et al., 2007, 2008; Lanfermeijer et al., 2008; Malinowski et al., 2009). However, it appears that some systemin-induced responses in Solanum pimpinellifolium are BRI1 dependent (e.g., root elongation), suggesting an interaction between the brassinosteroid and systemin pathways (Holton et al., 2007). Therefore, understanding the mechanisms and specificity of RLK–ligand interactions is critical for deciphering the molecular language of cell-to-cell communication in plants and their interactions with the environment.
FLS2 is a well-documented receptor for the bacterial pathogen-associated molecular pattern (PAMP) flagellin (or its derived peptide flg22) leading to the activation of antibacterial innate immune responses (Gómez-Gómez and Boller, 2000; Zipfel et al., 2004; Chinchilla et al., 2006). Flagellin is a major protein component of bacterial flagella and is found in diverse pathogenic species, including Pseudomonas syringae. In response to flg22 binding, FLS2 rapidly heterooligomerizes with the regulatory LRR-RLK BRI1-ASSOCIATED KINASE1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE3 (BAK1/SERK3) and other SERK class RLKs, resulting in phosphorylation of the complex and activation of downstream signaling (Chinchilla et al., 2007; Heese et al., 2007; Schulze et al., 2010; Roux et al., 2011; Schwessinger et al., 2011). Downstream responses induced by FLS2 activation include ion fluxes, rapid production of reactive oxygen species, transient activation of mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases, and induction of defense-related gene expression (Segonzac and Zipfel, 2011).
Surprisingly, Arabidopsis FLS2 was also recently reported to bind the endogenous peptide CLAVATA3 (CLV3p) that normally regulates maintenance of the shoot apical meristem (SAM) stem cell niche during development (Lee et al., 2011). CLV3 function in the SAM is mediated by the cooperative activity of several FLS2-unrelated LRR-containing receptors, including the LRR-RLK CLV1 (Wang and Fiers, 2010). CLV3 has been demonstrated to bind directly to the extracellular domain of CLV1, consistent with the genetic requirement for this RLK in CLV3 function in planta (Ogawa et al., 2008). Activation of CLV3-dependent RLKs is thought to restrict meristem size mainly by inhibiting the expression of the homeodomain protein WUSCHEL in a feedback loop fashion (De Smet et al., 2009; Katsir et al., 2011).
Flg22 and mature CLV3p sequences are highly divergent. Despite this, Lee et al. (2011) provided evidence that CLV3 triggers FLS2-dependent immune responses and thereby restricts bacterial infection in the SAM. Lee et al. (2011) found that treatment of Arabidopsis mesophyll protoplasts and seedlings with synthetic CLV3p induces similar responses to flg22, including FLS2-BAK1 complex formation, MAPK activation, induction of early defense marker genes (Flg22-induced receptor-like kinase1 [FRK1], WRKY DNA-binding protein29 [WRKY29], and WRKY30), and enhanced resistance to the pathogenic bacterium P. syringae pv tomato (Pto) DC3000. This result is surprising given the sequence divergence between the two ligands. In particular, the extremely high concentrations of CLV3p required to induce FLS2-dependent responses (Lee et al., 2011) are unlikely to be physiologically relevant. Micromolar concentrations of CLV3p are required to trigger FLS2-dependent function. By contrast, flg22 is active at nanomolar levels. Micromolar concentrations of CLV3p also terminate SAM growth (Ohyama et al., 2009; Kinoshita et al., 2010). It is therefore unlikely that CLV3p levels in growing wild-type plants are sufficient to trigger FLS2 in the SAM.
The claim that FLS2 could be a bona fide receptor for CLV3p is based on data that suggest that: (1) radiolabeled CLV3p binds to FLS2 upon FLS2 overexpression in Arabidopsis fls2 protoplasts with a dissociation constant (Kd) of 34.7 nM, (2) the flg22 antagonist flg22Δ2 competes with CLV3p for FLS2 binding and CLV3p-induced FLS2-BAK1 association, and (3) two mutants in the FLS2 LRR region (fls2-24 and LRR23b) are impaired in both flg22- and CLV3p-induced MAPK activation and flg22 and CLV3p binding (Lee et al., 2011). The finding that unlabeled CLV3p, flg22, and flg22Δ2 could compete similarly with radiolabeled CLV3p is not consistent with the divergent concentrations required to induce biological defense responses between the different peptides (micromolar versus nanomolar range) (Lee et al., 2011). Despite this, the authors also suggest that FLS2–CLV3 interactions are highly specific, as the CLV3p-related peptide CLE40p does not induce FLS2-dependent responses or bind to FLS2 (Lee et al., 2011). CLE40p and CLV3p differ by only three amino acids, yet CLV3p and flg22, which can outcompete each other for binding, do not share any sequence. It is therefore surprising that FLS2 has the ability to distinguish between CLV3p and CLE40p, which even CLV1 cannot do. On a similar note, there is a great diversity of CLE peptides expressed throughout development in tissues overlapping with FLS2 (Robatzek et al., 2006; Jun et al., 2010). Many of them can functionally complement CLV3p (Ni and Clark, 2006), yet none of them seem capable of activating FLS2, as wild-type plants do not show constitutive defense responses.
Lee et al. (2011) also performed infection assays and suggested that Pto DC3000 replication in SAM tissue is restricted by both CLV3 and FLS2. Pto DC3000 infects aboveground tissues of plants by entering through stomata, hydathodes, and wounds. Once inside, Pto DC3000 replicates in intercellular spaces (Alfano and Collmer, 1996). The vegetative SAM lacks both stomata and hydathodes and is protected from damage by overlying leaf primordia. In addition, SAM cells are tightly packed with no intercellular spaces. It is therefore not clear how Pto DC3000 would enter the SAM or where it would replicate once inside. Consistent with this, the number of green fluorescent protein (GFP)-labeled Pto DC3000 bacteria supposed to be inside the SAM only increases by an estimated 1.5- to twofold over 2 d of infection in fls2 and clv3 plants as assayed by confocal imaging (see Figure 14E in Lee et al., 2011) or by approximately eightfold within 4 d of infection as assayed by quantitative PCR (see Figure 15 in Lee et al., 2011). Pto DC3000 is a highly invasive pathogen and typically grows 100- to 10,000-fold during the same time frame in leaf tissue (Alfano and Collmer, 1996). Thus, the increase in GFP-positive signals seen by Lee et al. (2011) in susceptible SAM tissue is comparable to changes in bacterial numbers seen during nonpathogenic infections or during an active resistance response but does not correspond to a typical compatible infection. All of these considerations have led us to question the biological relevance of the Lee et al. (2011) experiments.
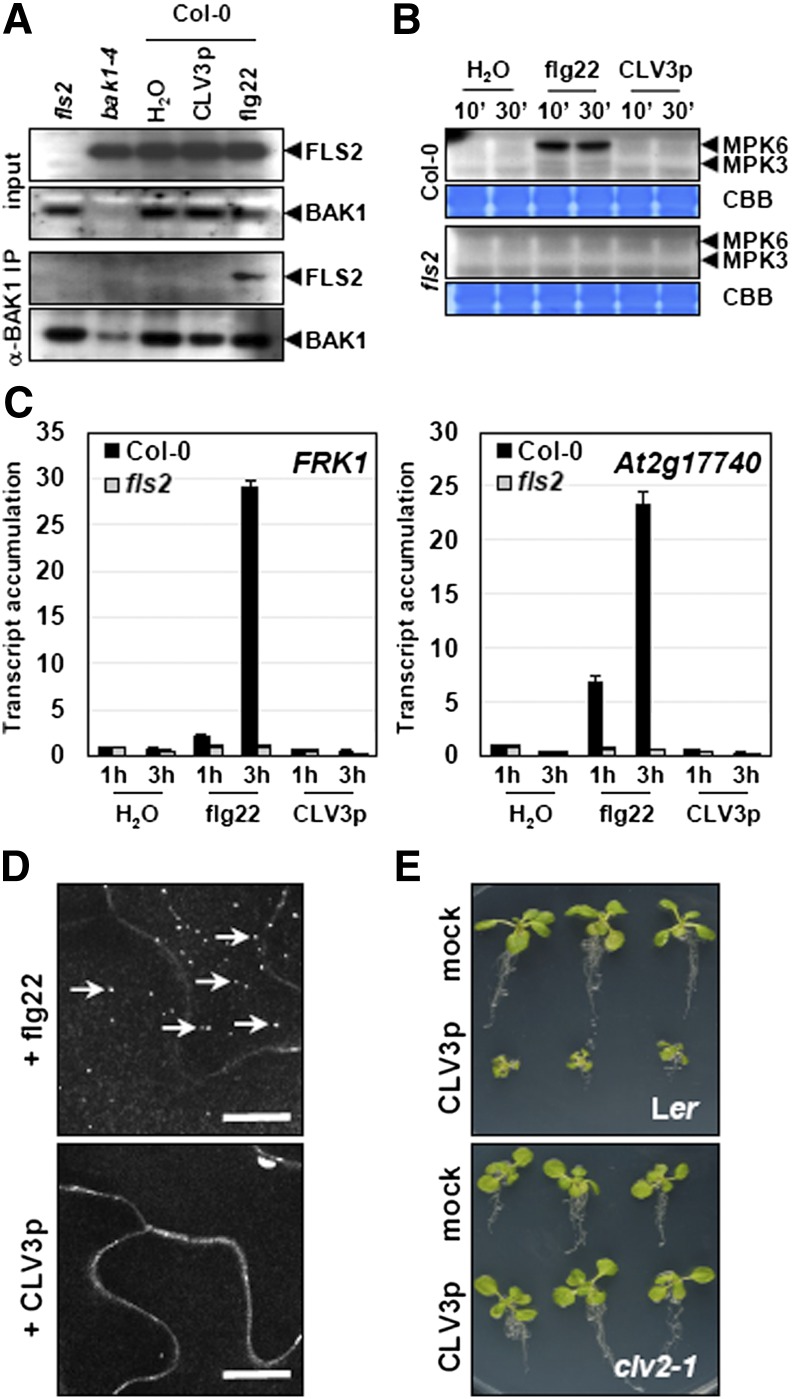
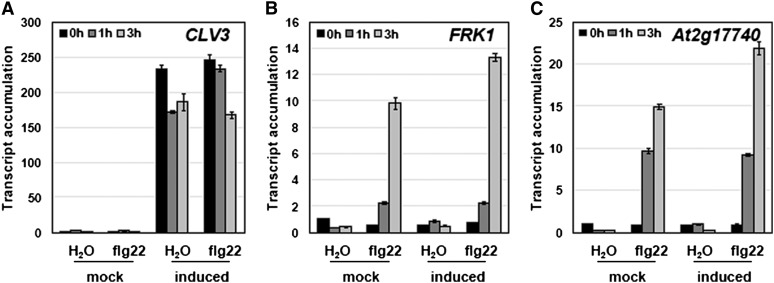
Consequently, we sought to confirm some of the results reported by Lee et al. (2011) from Arabidopsis mesophyll protoplasts, seedlings, and SAM-enriched tissues by studying typical defense responses in Arabidopsis seedlings. We found that treatment of Arabidopsis seedlings with 10 μM biologically active synthetic CLV3p did not induce rapid complex formation between FLS2 and BAK1 (Figure 1A), activation of MAPKs (Figure 1B), or increased transcript accumulation of the early immune marker genes FRK1 and At2g17740 (Figure 1C). By contrast, all of these responses were readily induced by 100 nM flg22 (Figures 1A to 1C). As an additional assay not used previously by Lee et al. (2011), we tested if CLV3p could trigger FLS2 endocytosis in a manner similar to flg22 (Robatzek et al., 2006; Figure 1D). Again, no effect following treatment with 10 μM CLV3p could be observed (Figure 1D). Distinct synthetic CLV3p forms are classically used. The arabinosylated and hydroxyprolinated 13–amino acid CLV3p corresponds to the native mature form and is considered to be the most active (Ohyama et al., 2009). Lee et al. (2011) used a nonarabinosylated and hydroxyprolinated 12–amino acid CLV3p form (Kondo et al., 2006), while we used for our study a nonmodified (nonarabinosylated and nonhydroxyprolinated) 13–amino acid CLV3p form. Despite these differences, the relevance of the hydroxyprolination and arabinosylation modifications is still under investigation (Kondo et al., 2006; Song et al., 2012). We confirmed that the synthetic CLV3p used here was biologically active, as it inhibited the shoot and root growth of Arabidopsis seedlings in a CLV2-dependent manner (Figure 1E), and that concentrations as high as 10 μM (10-fold higher than the level of CLV3p used by Lee et al. [2011] to invoke immune responses) did not induce immune responses in our experiments (Figures 1A to 1D). Furthermore, we tested if the native mature CLV3p expressed transgenically in Arabidopsis in an inducible manner (as described in Nimchuk et al., 2011) could elicit early immune marker gene expression. As observed with the exogenous treatment with synthetic unmodified CLV3p (Figure 1), dexamethasone-induced expression of CLV3 (Figure 2A) did not lead to increased transcript accumulation of FRK1 and At2g17740 (Figures 2B and 2C). Together, our results argue that CLV3p, including CLV3p in its native form as expressed from a transgene, does not confer FLS2-dependent defense responses in vivo.
Figure 1.
CLV3p Does Not Induce PAMP-Triggered Responses.
(A) CLV3p does not induce FLS2/BAK1 complex. Arabidopsis seedlings were treated with water, 100 nM flg22, or 10 μM CLV3p for 10 min. Total proteins (input) were extracted and submitted to immunoprecipitation with anti-BAK1 antibodies. FLS2 protein presence was revealed by protein immunoblot with anti-FLS2 antibodies.
(B) CLV3p does not induce MAPKs activation. Arabidopsis seedlings were treated with water, 1 μM flg22, or 10 μM CLV3p for 10 and 30 min. Activated MAPKs were detected by protein immunoblots with anti-pTEpY antibodies. Coomassie blue (CBB) staining of the membranes is shown to assess equal loading.
(C) CLV3p does not induce PAMP marker gene expression. Arabidopsis seedlings were treated with water, 100 nM flg22, or 10 μM CLV3p for 1 and 3 h. Accumulation of marker gene transcript was assessed by qRT-PCR and is presented as relative to the Col-0 accumulation in the water sample.
(D) CLV3p does not induce internalization of FLS2. Arabidopsis Col-0 transgenic line expressing FLS2-GFP was treated with 1 μM flg22 or 1 μM CLV3p for 30 min. Confocal images show cross sections of cotyledon leaf epidermal cells. Arrows in the top panel indicate FLS2-GFP containing vesicles. Bars = 5 μm.
(E) CLV3p used in this study is active. Arabidopsis seedlings (Ler and clv2-1) were grown on medium supplemented or not with 10 μM CLV3p. Picture were taken 14 d after germination.
Figure 2.
Endogenous CLV3p Does Not Induce PAMP Marker Gene Expression.
Fourteen-day-old Arabidopsis clv3-2/Dex:CLV3 seedlings were treated with 0.01% ethanol (mock) or 30 μM dexamethasone (induced) for 16 h then with water or 100 nM flg22 for 1 or 3 h. Accumulation of the transgene CLV3 (A) and the marker genes FRK1 (B) and At2g17740 (C) transcript was assessed by qRT-PCR and is presented as relative to the accumulation in the untreated sample (0 h).
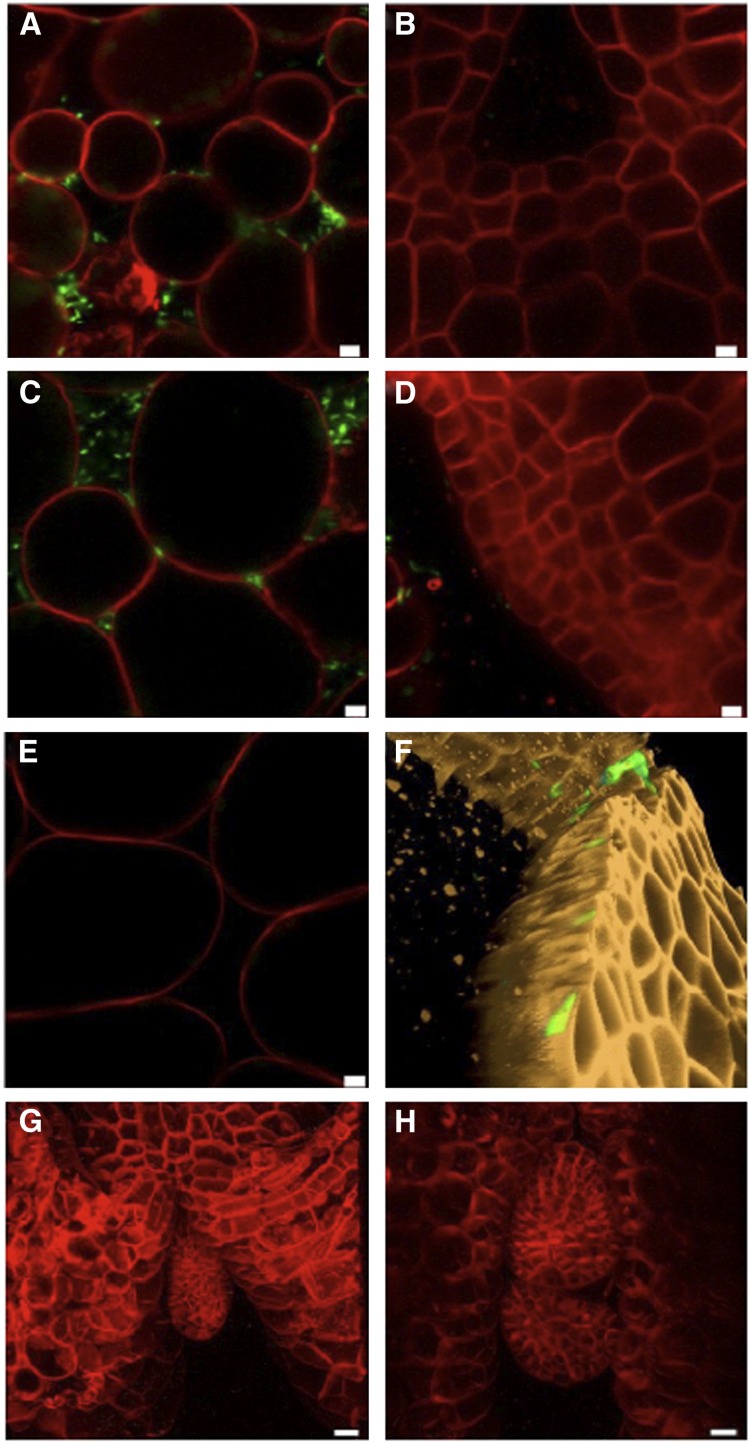
To test if the presence of endogenous CLV3p indeed restricts bacterial infection, we inoculated 2-d-old wild-type Landsberg erecta-0 (Ler-0) and clv3-2 Arabidopsis seedlings with GFP-labeled Pto DC3000, as per Lee et al. (2011), and analyzed tissues by live imaging using confocal microscopy 4 d later. We used stable transgenic lines expressing a plasma membrane–localized dtTOMATO fusion to positively identify cell and tissues structures in seedlings. This construct does not interfere with Pto DC3000 infection (Figure 3). While SAMs were enlarged and slightly domed in clv3-2 seedlings, SAMs in Ler-0 seedlings were generally flatter and small, consistent with previous analysis (Autran et al., 2002). As a control for the efficiency of the infection, we always analyzed cotyledon tissues. As shown in Figures 3A and 3C, Pto DC3000-GFP was readily detected in the apoplastic spaces of Ler-0 and clv3-2 cotyledons. By contrast, no Pto DC3000-GFP could be detected in noninoculated Ler-0 cotyledons (Figure 3E) or in dissected Ler-0 and clv3-2 SAMs from inoculated seedlings (Figures 3B and 3D). Notably, no apoplastic space was observed in SAMs, suggesting that the niche in which Pto DC3000-GFP grows is absent from this tissue (Figures 3B, 3D, and 3F). An Amira three-dimensional reconstruction of a clv3-2 SAM revealed that the only GFP signal occasionally detected is from the SAM surface (Figure 3F), most likely reflecting epiphytic bacteria remaining after washing seedlings that were submerged in the inoculation medium. While our analysis was performed after careful dissection of meristems, we also attempted to use the squash method reported by Lee et al. (2011) to visualize SAMs after infection. In our hands, we only observed young leaf primordia using this method, not SAMs (Figures 3G and 3H).
Figure 3.
Pto DC3000 Does Not Infect SAM Tissues.
(A) Confocal image of Pto DC3000 infection of the intracellular spaces between palisade mesophyll cells of a 6-d-old Ler-0 cotyledon. Pto DC3000, green; Arabidopsis cell outlines (PM-dtTomato), red.
(B) Image of the SAM from the same seedling as in (A); note the lack of intracellular spaces and bacteria.
(C) Imaging as in (A), clv3-2 genotype.
(D) Image of the SAM from the same seedling as in (C).
(E) Imaging controls of uninfected clv3-2 cotyledon. (A) to (E) were imaged at the same resolution using identical imaging settings.
(F) Three-dimensional reconstruction of a clv3-2 SAM showing GFP-positive bacteria restricted to the surface of the SAM.
(G) and (H) Imaging of the apex of 6-d-old clv3-2 seedlings using the squash method as described by Lee et al. (2011). Only developing leaf primordia are visible. Side view squash (G); top view squash (H).
Bars = 5 μm in (A) to (E) and 10 μm in (G) and (H).
In conclusion, the inability to detect immune responses in Arabidopsis mesophyll protoplasts and seedlings by active exogenous or endogenous CLV3 peptides in conditions where FLS2 is biologically active (this study and Mueller et al., 2012) challenge the biological significance of the previously reported CLV3p binding to FLS2 and the associated activation of FLS2-dependent immune responses (Lee et al., 2011). In addition, our results clearly show that SAMs cannot be infected by the bacterium Pto DC3000 under the conditions used in both studies. Therefore, our results obtained from whole Arabidopsis seedlings cast doubt about the overall validity and biological relevance of the previous claims that the immune receptor FLS2 perceives the meristematic regulatory peptide CLV3p in mesophyll, seedlings, and SAM cells and that CLV3p contributes to SAM immunity against bacterial infection.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotypes/genotypes used in this studies are Ler-0, clv2-1 (Jeong et al., 1999), clv3-2 (Fletcher et al., 1999), clv3-2/Dex:CLV3 transgenic line (Nimchuk et al., 2011), Columbia-0 (Col-0), bak1-4 (SALK_116202; Chinchilla et al., 2007), and fls2 (SALK_093905; Heese et al., 2007). The Ler-0 and clv3-2 pUBQ10:PM-dtTOMATO were generated as described for marker lines by Nimchuk et al. (2011). Seedlings were grown for 14 d in 24-well plates containing liquid Murashige and Skoog medium supplemented with 1% Suc under 16 h light at 22°C. Plates were refilled with fresh medium 1 d before the experiments.
Peptides and Chemicals
Flg22 and elf18 were synthesized by Peptron and resuspended in sterile water. The unmodified 13–amino acid CLV3p (Ohyama et al., 2009) was synthesized by Biomatik. Dexamethasone is from Sigma-Aldrich.
MAPK Activation
MAPK activation assays were performed as described by Boutrot et al. (2010). Briefly, seedlings were treated with water, 1 μM flg22, 1 μM elf18, or 10 μM CLV3p for 10 or 30 min. Tissue was immediately frozen and ground in liquid nitrogen. Proteins were extracted in Lacus buffer and quantified by Bradford. MAPK activation was monitored by protein immunoblots with antibodies that recognize the dual phosphorylation of the activation loop of MAPK (pTEpY). Phospho-p44/42 MAPK (Erk1/2) (Thr-202/Tyr-204) rabbit monoclonal antibodies from Cell Signaling were used according to the manufacturer’s instructions. Blots were stained with Coomassie Brilliant Blue to verify equal loading.
RNA Isolation and Quantitative RT-PCR
RNA isolation was performed as described by Boutrot et al. (2010). Briefly, seedlings were treated with water, 100 nM flg22, 100 nM elf18, or 10 μM CLV3p for 1 or 3 h. Tissue was immediately frozen and ground in liquid nitrogen. Total RNAs were isolated with TRI reagent (Invitrogen) following the manufacturer’s instructions. RNA samples were treated with Turbo DNA-free DNase (Ambion) and quantified with a Nanodrop spectrophotometer (Thermo Scientific). First-strand cDNA was synthesized from 5 μg RNA using SuperScript RNA H-Reverse Transcriptase (Invitrogen) and an oligo(dT) primer, according to the manufacturer’s instructions. cDNA was amplified in triplicate by quantitative PCR using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) and the CFX96 real-time system (Bio-Rad). The relative expression values were determined using U-box as reference gene and the comparative cycle threshold method (2-ΔΔCt). Primers used for quantitative PCR are as follows: SIRK/FRK1 (At2g19190) forward 5′-ATCTTCGCTTGGAGCTTCTC-3′ and reverse 5′-TGCAGCGCAAGGACTAGAG-3′; At2g17740 gene forward 5′-TGCTCCATCTCTCTTTGTGC-3′ and reverse 5′-ATGCGTTGCTGAAGAAGAGG-3′; U-box (At5g15400) forward 5′-TGCGCTGCCAGATAATACACTATT-3′ and reverse 5′-TGCTGCCCAACATCAGGTT-3′; CLV3 (At2g27250) forward 5′-ACGTTCAAGGACTTTCCAACCGCA-3′ and reverse 5′-GTGCAACGGGTCAGG-TCCCG-3′.
Protein Immunoprecipitation
Protein extraction and immunoprecipitation were done as described by Schwessinger et al. (2011). Seedlings (2 g fresh weight) were treated with water, 100 nM flg22, or 10 μM CLV3p for 10 min, ground in liquid nitrogen, and then extracted with buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 5 mM DTT, 1 mM PMSF, 10% glycerol, 1% Nonidet P-40, and 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich]) added at 1 mL/g powder. Samples were centrifuged 15 min at 4°C and 10,000g. Supernatants were adjusted to 2 mg/mL protein and incubated 3 h at 4°C with 20 μL Rabbit IgG TrueBlot IP beads (eBioscience) and 15 μL anti-BAK1 antibodies with gentle agitation. Following incubation, the beads were collected and washed four times with the extraction buffer. SDS loading buffer was then added to the beads that were boiled for 10 min. Proteins were separated by 8% SDS-PAGE and further analyzed by protein gel blot using rabbit polyclonal anti-FLS2 antibodies (Gimenez-Ibanez et al., 2009) or rabbit polyclonal anti-BAK1 antibodies (Schulze et al., 2010).
SAM Infection Assays
SAM infection assays were performed as described by Lee et al. (2011). Seedlings were either squashed using the method described by Lee et al. (2011), or a single cotyledon was removed using sharpened lab forceps. The experiment was repeated three times, and in each case 15 to 25 seedlings were analyzed per genotype.
Microscopy
Confocal imaging for FLS2 endocytosis was performed as described by Salomon et al. (2010) with the Opera microscope (Perkin-Elmer Cellular Technologies Germany) equipped with three 1.3-megapixel charge-coupled device cameras and using a Nipkow spinning disc. Excitation of the samples was performed at 488 nm for GFP; the emission spectrum was taken from 502 to 577 nm. Leaves were prepared in 96-well plates with optical glass bottom (Greiner Bio-One). Detached cotyledons of 2-week-old Arabidopsis plants (Col-0/FLS2-GFP; Chinchilla et al., 2007) were used and incubated with water, CLV3p (1 μM), or flg22 (1 μM) for 30 min. Images of a consecutive series of 21 planes with a distance of 1 μm were taken and displayed as a maximum projection using Acapella Software (Perkin-Elmer Cellular Technologies Germany).
For SAM imaging, a Zeiss 510 confocal microscope was used as by Nimchuk et al. (2011). Image stacks were reconstructed in Amira using a consecutive series.
Acknowledgments
This work was funded by the Gatsby Charitable Foundation (C.Z. and S.R.), by UK Biotechnology and Biological Sciences Research Council Grant BB/G024944/1 (ERA-NET Plant Genomics Pathonet) (C.Z.), and by U.S. National Institutes of Health Grant 1R01GM086639 (E.M.M.). We thank Jeffery Dangl and Detlef Weigel for useful discussions.
AUTHOR CONTRIBUTIONS
E.M.M., Z.L.N., C.S., and C.Z. designed the research; M.B., Z.L.N., C.S., and P.T.T. performed research and analyzed data; and E.M.M., Z.L.N., S.R., and C.Z. wrote the article.
References
- Alfano J.R., Collmer A. (1996). Bacterial pathogens in plants: Life up against the wall. Plant Cell 8: 1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D., Jonak C., Belcram K., Beemster G.T.S., Kronenberger J., Grandjean O., Inzé D., Traas J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M. (2009). Plant peptides in signalling: Looking for new partners. Trends Plant Sci. 14: 255–263 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton N., Caño-Delgado A., Harrison K., Montoya T., Chory J., Bishop G.J. (2007). Tomato BRASSINOSTEROID INSENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell 19: 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton N., Harrison K., Yokota T., Bishop G.J. (2008). Tomato BRI1 and systemin wound signalling. Plant Signal. Behav. 3: 54–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A.H.K., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Davies K.A., Bergmann D.C., Laux T. (2011). Peptide signaling in plant development. Curr. Biol. 21: R356–R364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Lanfermeijer F.C., Staal M., Malinowski R., Stratmann J.W., Elzenga J.T.M. (2008). Micro-electrode flux estimation confirms that the Solanum pimpinellifolium cu3 mutant still responds to systemin. Plant Physiol. 146: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chah O.-K., Sheen J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. (2010). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13: 509–514 [DOI] [PubMed] [Google Scholar]
- Malinowski R., Higgins R., Luo Y., Piper L., Nazir A., Bajwa V.S., Clouse S.D., Thompson P.R., Stratmann J.W. (2009). The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Mol. Biol. 70: 603–616 [DOI] [PubMed] [Google Scholar]
- Montoya T., Nomura T., Farrar K., Kaneta T., Yokota T., Bishop G.J. (2002). Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14: 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Chinchilla D., Albert M., Jehle A.K., Kalbacher H., Boller T., Felix G. (August 24, 2012). Contamination risks in work with synthetic peptides: flg22 as an example of a pirate in commercial peptide preparations. Plant Cell 24: 3193–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Clark S.E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z.L., Tarr P.T., Ohno C., Qu X., Meyerowitz E.M. (2011). Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S., Grunewald D., Stüber K., Schaaf S., MacLean D., Schulze-Lefert P., Robatzek S. (2010). High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol. 154: 1096–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Pearce G., Ryan C.A. (2003). Generation of systemin signaling in tobacco by transformation with the tomato systemin receptor kinase gene. Proc. Natl. Acad. Sci. USA 100: 10114–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Ryan C.A., Jr (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99: 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Zipfel C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14: 54–61 [DOI] [PubMed] [Google Scholar]
- Song X.-F., Yu D.L., Xu T.-T., Ren S.-C., Guo P., Liu C.-M. (2012). Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol. Plant 5: 515–523
- Wang G., Fiers M. (2010). CLE peptide signaling during plant development. Protoplasma 240: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]