Abstract
To sustain plants’ postembryonic growth and development in a structure of cells fixed in cell walls, a tightly controlled short distance cell–cell communication is required. The focus on phytohormones, such as auxin, has historically overshadowed the importance of small peptide signals, but it is becoming clear that secreted peptide signals are important in cell–cell communication to coordinate and integrate cellular functions. However, of the more than 1000 potential secreted peptides, so far only very few have been functionally characterized or matched to a receptor. Here, we will describe our current knowledge on how small peptide signals can be identified, how they are modified and processed, which roles they play in Arabidopsis thaliana development, and through which receptors they act.
INTRODUCTION
Cells within an organism must communicate over both short and long physiological ranges to ensure proper patterning and functional connections. There are several ways to achieve this in plants, including phytohormones, mobile transcription factors, noncoding RNAs, and small signaling peptides (Busch and Benfey, 2010; Van Norman et al., 2011). Most prominently, directional transport of the phytohormone auxin from one cell to the other provides cues for patterning and development (Vanneste and Friml, 2009; Grunewald and Friml, 2010). However, signaling through direct symplastic transport of transcription factors between cells, via plasmodesmata, has also been extensively explored (Busch and Benfey, 2010; Wu and Gallagher, 2011). Major examples of transcription factors that are mobile between cells are TARGET OF MONOPTEROS7 (involved in specification of the hypophysis during embryogenesis) (Schlereth et al., 2010) and SHORT ROOT (involved in the production of cortex and endodermis tissues) (Helariutta et al., 2000; Cui et al., 2007). In addition, microRNAs can also be considered to be intercellular signaling molecules; for example, a gradient of miRNA165/166 is required for specification of root xylem cell fate (Carlsbecker et al., 2010).
While the importance of signaling peptides in cell–cell communication has been recognized in animals for many years, in plants, the focus on nonpeptide, lipophillic phytohormones, such as auxin, has historically overshadowed the importance of signaling peptides (which should not be confused with signal peptide, a presequence directing a protein to the secretory pathway) (Vanneste and Friml, 2009). Nevertheless, in just over a decade, several secreted peptides have been recognized as important in cell–cell communication in plants, coordinating and integrating cellular functions in complex developmental processes (Matsubayashi, 2011a, 2011b). The identification of receptors, such as leucine-rich repeat–receptor-like kinases (LRR-RLKs), further underlines the importance of signaling peptides in plant development (De Smet et al., 2009).
Here, we will describe our current knowledge on the role signaling peptides play in Arabidopsis thaliana development and through which receptors they (likely) act. In addition, we will describe how small signaling peptides can be identified and how they are modified and processed.
LOOKING FOR SOMETHING SMALL
Peptide signals are small in size (usually <20 amino acids in the mature form and rarely more than ∼120 amino acids as a full-length precursor) and often present in very low (nanomolar range) physiological concentrations, so finding them presents a challenge. Furthermore, microarrays and other tools designed for identification of differentially expressed genes have not been a particularly useful tool in signaling peptide characterization: Small genes are often overlooked or not adequately represented on arrays and are poorly predicted using gene prediction algorithms as they are difficult to distinguish from short, random open reading frames (Olsen et al., 2002; Lease and Walker, 2006). The problem is further compounded due to the low expression levels of signaling peptides. In addition, microarrays are blind to how posttranslational modifications affect protein activity, a drawback that is particularly pertinent due to the crucial role of posttranslational modification processes in the activation of many signaling peptides. By measuring gene expression levels, it cannot be distinguished whether a modified, active form of a signaling peptide or the unmodified, inactive form is prevalent in the relevant sample (Kodadek, 2001).
The secreted signaling peptides discovered in plants thus far can be considered to fall into two broad groups: small posttranslationally modified peptides and Cys-rich peptides (Matsubayashi, 2012) (Figure 1A). The small posttranslationally modified peptides are a group of secreted signaling peptides characterized by small (<20 amino acid) mature peptides, which are cleaved from a longer translated precursor peptide, with a general paucity of Cys residues, and which are subject to one or more posttranslational modifications. Several small posttranslationally modified peptides have been identified, including CLAVATA3 (CLV3), CLV3/EMBRYO SURROUNDING REGION-RELATED (CLE), TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF), PHYTOSULFOKINE (PSK), PLANT PEPTIDE CONTAINING SULFATED TYROSINE1 (PSY1), C-TERMINALLY ENCODED PEPTIDE1 (CEP1), INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), and ROOT GROWTH FACTOR (RGF)/GOLVEN (GLV)/CLE-LIKE (CLEL). The Cys-rich peptides have several uniting features despite a generally high amino acid sequence divergence between individual peptides: Though often larger than small posttranslationally modified peptides, they are still small (<160 amino acids), they are cationic, and they have a conserved N-terminal signal peptide. In contrast with small posttranslationally modified peptides, their C-terminal domain is Cys rich, typically containing four to 16 Cys residues. The presence of Cys residues is thought to be required for the formation of disulphide bridges to hold the mature peptide in an active conformation (Pearce et al., 2001a). The Cys-rich peptide family of signaling peptides includes the RAPID ALKALINISATION FACTOR (RALF)/RALF-LIKE (RALFL), EPIDERMAL PATTERNING FACTOR1 (EPF1), EPF2, TAPETUM DETERMINANT1 (TPD1), STOMAGEN/EPF-LIKE9 (EPFL9), and EARLY NODULIN peptides. The discoveries of the currently known plant signaling peptides encompass various and evolving approaches, from the earliest biochemical assay approaches, through genetic screens, to in silico approaches. We discuss these methods below, using appropriate examples.
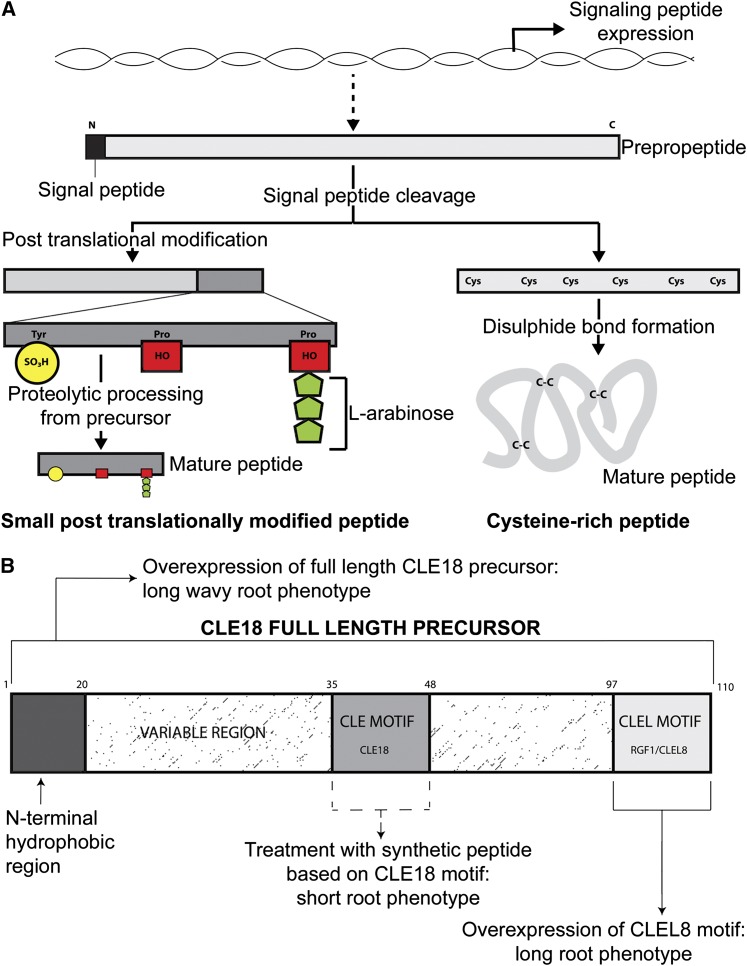
Figure 1.
Posttranslational Modifications and Processing of Small Signaling Peptides.
(A) Following entry of the full-length mRNA-encoded prepropeptide to the secretory pathway, the N-terminal hydrophobic signal peptide is cleaved by a signal peptidase. Small posttranslationally modified peptides and Cys-rich peptides then follow different pathways to produce an active peptide. All small posttranslationally modified peptides discovered in plants thus far undergo one or more of three types of post translational modification: Tyr sulfation (yellow), Pro hydroxylation (red), and/or Hyp arabinosylation (green). Proteolytic cleavage of the modified peptide from the precursor sequence completes activation. Cys-rich peptides often do not require proteolytic processing from a precursor, though there are exceptions. Activation of Cys-rich peptides is completed upon formation of disulphide bonds between the conserved Cys residues of the peptide, thus bringing the peptide to an active conformation. Schematic is based on Matsubayashi et al. (2012) with permission.
(B) The CLE18 precursor contains two motifs with sequence similarity to the canonical CLE motif. CLE18 is found within the variable region of the peptide at amino acid residues 35 to 48, rather than at the extreme C terminus, like other currently known CLE motifs. At the C terminus, there is a sequence of 13 amino acids with a CLEL sequence. At the N terminus is a region of hydrophobic amino acids, thought to act as a secretion signal (Meng et al., 2012). Overexpression of the full-length precursor gives a long root phenotype (Strabala et al., 2006); conversely, treatment with a synthetic peptide based on the CLE18 motif causes a short root phenotype (Ito et al., 2006). Overexpression of the CLEL8/RGF1 motif (derived from the CLE18 precursor) and related CLELs (RGF6/CLEL6, RGF5/CLEL7, and RGF9/GLV2/CLEL19), derived from other precursors, also results in a long root phenotype, suggesting that the extreme C-terminal motif represents the sequence of the active peptide (Matsuzaki et al., 2010; Meng et al., 2012; Whitford et al., 2012).
Biochemical Assays
The first plant signaling peptide, tomato systemin (TomSys), was discovered through observations that extracts from wounded tomato (Solanum lycopersicum) leaves, when added in water to young tomato plants, induced production of proteinase inhibitors I and II (Ryan, 1974). Interestingly, the effect could not be generated through substitution of the unknown active ingredient with a variety of plant hormones, including auxin, gibberellic acid, abscisic acid, and ethylene, which were (at the time) the main known candidate factors that could initiate a signaling pathway. Purification of a peptide from tomato leaf extract by HPLC and testing by application of both eluted fractions and a later-generated synthetic peptide to young plants, revealed the identity of the active ingredient as an 18–amino acid polypeptide factor nearly 20 years after its initial discovery (Ryan, 1974; Pearce et al., 1991).
Two signaling peptides with no sequence similarity to TomSys, but with similar functionality, named tobacco systemin I (TobHypSysI) and TobHypSysII (“Hyp” referring to hydroxyprolination of these peptides), were later discovered in wounded tobacco (Nicotiana tabacum) leaves using similar methods. However, the method used to identify TobHypSys also incorporated a new assay: the alkalinization assay. This assay was developed following the observation that TomSys causes an alkalinization of suspension culture medium, and it was hypothesized that other active peptides might have a similar effect (Meindl et al., 1998; Schaller and Oecking, 1999; Pearce et al., 2001b). Using this alkalinization assay, crude tobacco leaf extracts were run on HPLC columns, the eluted fractions were added to aliquots of tobacco suspension culture medium, and the pH of the medium measured. Increases in the pH of the media identified two major peaks corresponding to TomHypSysI and TomHypSysII.
This alkalinization assay was also used to identify RALF from tobacco, which alkalinized suspension culture media and induced mitogen-activated protein kinase activity. But, unlike the systemins, RALF was not involved in wound response (Pearce et al., 2001b). RALF-mediated alkalinization of the media occurs due to a receptor-mediated response leading to the inhibition of a membrane-bound ATPase (Meindl et al., 1998; Stratmann et al., 2000).
In a process that echoed the identification of TomSys as the active ingredient from a crude plant extract, PSK was discovered during attempts to identify conditioned media factors. Conditioned media containing extracts prepared from rapidly growing cells in culture had been shown to be necessary for the proliferation of cells in low-density plant cell culture, but it was unclear which unknown substance(s) within the extracts was active in promoting the proliferation. A bioassay based on the mitotic activity of asparagus (Asparagus officinalis) mesophyll cells in liquid culture identified an active, growth-promoting compound, purified through chromatography, PSK-α (Matsubayashi and Sakagami, 1996). Following its discovery in asparagus, PSK has been found to be widely conserved in many other plant species, including rice (Oryza sativa) and Arabidopsis, in which five paralogous PSK genes have been identified, designated PSK1–5, that are expressed in various tissues (Matsubayashi et al., 1997, 2006).
Likewise, TDIF was discovered during attempts to isolate xylogen, an arabinogalactan protein that mediates cell interaction in vascular development, from Zinnia elegans mesophyll cell culture. Tracheary element differentiation in these cells was being inhibited by an unknown extracellular factor, which was subsequently isolated and purified from the conditioned cell medium as a 12–amino acid peptide (Ito et al., 2006). Homology searches revealed that TDIF was identical to the C-terminal 12 amino acids of Arabidopsis CLE41 and CLE44 and highly homologous to CLE42 and CLE46. Indeed, dodecapeptides based on the CLE motifs of CLE41 and CLE44 showed strong TDIF activity, in terms of promoting tracheary element differentiation of Z. elegans mesophyll cells in culture and in Arabidopsis (Ito et al., 2006; Hirakawa et al., 2008; Whitford et al., 2008; Etchells and Turner, 2010).
However, the typically low physiological concentrations of signaling peptides in tissues may evade detection through bioassay methods. Using a different, peptidomics-based approach, taking into account the importance of posttranslational peptide modifications (see below), it is possible to enrich sulfated peptides, present at low concentrations in a complex peptide mixture, using anion exchangers (Amano et al., 2007). This approach provided a pool of likely candidates for potential signaling peptides, including PSY1, which was found to promote cellular proliferation and expansion in various tissues (Amano et al., 2007).
Genetic Approaches
Forward and reverse genetics approaches have allowed signaling peptide biology to progress further. For example, CLV3 was identified through genetic screening approaches as a putative signaling peptide (Clark et al., 1995; Fletcher et al., 1999). The ethyl methanesulfonate–induced clv3 mutant has an enlarged shoot apical meristem (SAM) and superfluous floral meristems, suggesting a role in regulating meristematic growth (Clark et al., 1996).
Still, one major issue with peptides is the limited number of (characterized) loss-of-function mutants, since most have no useful, available T-DNA insertions. Partly because small genes, such as potential secreted signaling peptides, are less likely to be directly hit by a T-DNA. To complicate matters further, the functional redundancy of some signaling peptides can mask phenotypes when only one family member is successfully disrupted. Nevertheless, two signaling peptides have been discovered through screening T-DNA insertion mutants, IDA and TPD1. A T-DNA insertion in the promoter of a gene at locus At1G68765 gave rise to a phenotype that failed to undergo floral organ abscission following pollination, which were loss-of-function ida mutants (Butenko et al., 2003). Five paralogous genes were also found in Arabidopsis, called IDA-LIKE (IDL). The structure of IDA has not been fully elucidated; however, it is known that the IDA gene codes for a 77–amino acid polypeptide (a possible precursor) and, in common with other small signaling peptides, contains a conserved Pro-rich domain at the C terminus. TPD1 was discovered through screens of gene and enhancer trap lines, where one line produced no pollen (Yang et al., 2003).
Based on observations that previously discovered signaling peptides, such as PSK and PSY1, often require Tyr sulfation by a TYROSYLPROTEIN SULFOTRANSFERASE (TPST), it is logical to assume that a tpst loss-of-function mutant might display a phenotype arising from a deficiency in active signaling peptides, and this could help identify signaling peptide candidates. The tpst loss-of function mutant displays many pleiotropic phenotypes, including severely shortened roots and reduced root meristematic activity with failure of root stem cell maintenance (Komori et al., 2009; Zhou et al., 2010). The tspt mutant and a reverse-genetics screen were used to characterize RGF/GLV/CLEL peptides (Matsuzaki et al., 2010).
Bioinformatics Digs Deeper
The identification of several signaling peptide encoding genes shows that in silico methods using bioinformatics tools can be useful for initial identification of small signaling peptides. For example, SignalP is an algorithm that can predict the presence and location of signal peptides and cleavage sites in an amino acid sequence. It has been used as a tool during the discovery and characterization of several secreted peptides, including IDA and CEP1 (Butenko et al., 2003; Ohyama et al., 2009), and was the starting point for the discovery of the RALFL family (Olsen et al., 2002). Some of the results generated from an initial SignalP search of an Arabidopsis genome database were compared for homology to known peptides, using various types of BLAST search and ClustalW alignments, leading to discovery of the uncharacterized RALFL gene family in Arabidopsis (Olsen et al., 2002). Similarly, computer programs (PHDsec and SOPMA) were used to compare the secondary structure of the identified peptides. Some members of this family had high sequence similarity to RALF peptides, leading to them being designated the RALFL family (Olsen et al., 2002; Haruta and Constabel, 2003). Arabidopsis has been shown to have around 40 RALFL genes, which are expressed throughout the plant and impact various developmental processes, including seedling development, stem development, and root development (Matos et al., 2008; Srivastava et al., 2009; Bedinger et al., 2010; Mingossi et al., 2010).
CEP1 was identified through an in silico approach by analysis of an Arabidopsis genome database, by searching for paralogs to the peptide CLE44 (Ohyama et al., 2009). The reasoning behind this approach was that signaling peptides, such as PSK, PSY1, and CLE, are encoded by several paralogous genes and have many specific features that can be programmed in to computational searches to maximize the quality of the final candidate pool, such as size limitations, presence of an N-terminal signal sequence, and Cys paucity. Tellingly, the candidate pool identified by this search consisted of ∼40% of peptides with known or predicted function, such as PSK, PSY precursors, IDA, IDL, and several CLE family peptides. Identification of one uncharacterized gene family led to identification of a 15–amino acid hydroxyprolinated peptide, namely, CEP1 (Ohyama et al., 2009).
This in silico approach was then adopted using similar search parameters as those used earlier to identify CEP1, but including additional search criteria to identify peptides with an Asp-Tyr sulfation motif. From this search, the RGF/GLV/CLEL polypeptide family was found with features matching all the search criteria. Mass spectrometry of peptides derived from plants overexpressing one member identified a 13–amino acid Tyr-sulfated peptide, derived from the C-terminal domain of the full-length cDNA-encoded protein. In parallel, homology searches using the C-terminal sequence of CLE18 also revealed 10 members of the same RGF/GLV/CLEL family. It has been suggested that the active peptide sequence of RGF1 is derived from the CLE motif of its precursor. Interestingly, the RGF1/CLE18 precursor in fact contains not one, but two functional motifs within its sequence: the RGF1/CLE18 13–amino acid CLE motif, spanning residues 35 to 48 (not the extreme C terminus, as would be expected for a CLE motif), and a second, previously undiscovered motif spanning residues 75 to 87, designated the CLEL motif because of the similarity to the classical CLE motif (Matsuzaki et al., 2010; Meng et al., 2012) (Figure 1B). Homology searches revealed that similar motifs were found in nine other proteins besides the RGF1/CLE18 precursor.
Many of the Cys-rich peptides have been identified through computational analysis of several model plant genomes (Graham et al., 2004; Silverstein et al., 2005, 2007). For example, the Cys-rich peptide EPF1 was discovered through systematic individual 35S promoter–driven overexpression of 153 genes that were predicted to encode small secreted peptides in Arabidopsis followed by phenotypic analysis, and one of the overexpressing lines had reduced stomatal density (Hara et al., 2007). Homology searches for EPF1 identified 10 homologs in Arabidopsis: EPF2 and EPFL1 to 9 (Hara et al., 2009; Hunt and Gray, 2009). STOMAGEN/EPFL9 was identified in the same large-scale screen used to identify EPF1 and in an in silico screen to identify genes that are coexpressed with stomatal regulators (Kondo et al., 2010; Sugano et al., 2010).
FROM CODING SEQUENCE TO MATURE, BIOACTIVE SECRETED PEPTIDE
Many signaling peptide genes encode a protein that is longer than the mature, active form of the peptide. Several of them are expressed as ∼100–amino acid prepropeptides, which require proteolytic processing by endopeptidases, and cleavage of a signal peptide by signal peptidases, to produce a functional peptide. The small posttranslationally modified peptides and the Cys-rich peptide groups require different types of processing: Both usually undergo removal of signal sequences (by signal peptidases) and removal of extra mRNA-encoded precursor sequence (some Cys-rich peptides do not require this step). Before processing, the small posttranslationally modified peptides undergo enzyme-catalyzed modifications to selected residues of the mature-length peptide. Following processing, Cys-rich peptides are predicted to form disulphide bonds between the conserved Cys residues in their sequence to give the peptides their final active conformation.
Posttranslational Modifications
So far, three types of posttranslational modification, which possibly act to enhance receptor binding, have been observed to occur to signaling peptides in plants: Tyr sulfation, Pro hydroxylation, and Hyp arabinosylation (Matsubayashi, 2011b) (Figure 1A, Table 1). Tyr sulfation is thought to be catalyzed by the plant-specific TPST, a 62-kD type I transmembrane protein localized in the Golgi (Komori et al., 2009). The peptide substrate is known to require, as a minimum, an Asp residue N-terminally adjacent to a Tyr residue (an Asp-Tyr motif), and TPST requires the cosubstrate 3′-phosphoadenosine 5′-phosphosulfate as a sulfate donor (Matsubayashi, 2012). Pro hydroxylation is catalyzed by PROLYL-4-HYDROXYLASE (P4H), a type II Golgi and ER membrane protein of the 2-OXOGLUTARATE-DEPENDANT DIOXYGENASE family (Myllyharju, 2003). Thirteen genes for P4H have been identified in Arabidopsis, but no substrate consensus sequence for Pro hydroxylation has been established yet (Matsubayashi, 2012). Following the conversion of Pro to Hyp, a subset of peptides undergoes a further posttranslational modification, Hyp arabinosylation. In a posttranslational modification unique to plants, an O-linked l-arabinose chain (triarabinoside) is added to certain Hyp residues within the peptide. The linkage of the triarabinoside chain to the Hyp residue and the linkage of the sugar moieties within the chain to each other are separate reactions and hypothesized to occur in two steps mediated by different enzymes. A Hyp arabinosyltransferase catalyzes a β-linkage between the hydroxyl group of Hyp to an arabinose, and an arabinosyltransferase mediates the β1,2-linkage between the arabinose residues themselves (Matsubayashi, 2012). However, the enzymes responsible for this process in plants are yet to be identified.
Table 1. Posttranslational Modifications and Proteolytic Processing of Small Signaling Peptides.
| Peptide | Precursor Length (Amino Acids) | Mature Length (Amino Acids) | Posttranslational Modifications | Proteolytic Processing Enzyme(s) | Posttranslational Modification Enzyme(s) |
|---|---|---|---|---|---|
| CEP1 | 91 | 15 | Pro hydroxylation (2 residues) | ? | Prolyl-4-hydroxylase (P4H)* |
| CLV3 | 96 | 13 | Pro hydroxylation (2 residues) | ? | Prolyl-4-hydroxylase (P4H)* |
| Hyp arabinosylation (1 residue) | |||||
| PSK | ∼80 | 5 | Tyr sulfation (2 residues) | Subtilisin-like Ser protease SBT1.1 | Tyrosylprotein sulfotransferase |
| (77–87 for PSK1-5) | |||||
| PSY1 | 75 | 18 | Tyr sulfation (1 residue) | ? | Tyrosylprotein sulfotransferase |
| Pro hydroxylation (two residues) | Prolyl-4-hydroxylase (P4H)* | ||||
| Hyp arabinosylation (one residue) | |||||
| RGF1 | 116 | 13 | Tyr sulfation (one residue) | ? | Tyrosylprotein sulfotransferase |
| (79–141 for RGF2-9) | Pro hydroxylation (one residue) | Prolyl-4-hydroxylase (P4H)* | |||
| TDIF (CLE41, CLE42, CLE44) | 88–112 | 12 | Pro hydroxylation (two residues) | ? | Prolyl-4-hydroxylase (P4H)* |
| ? | ? | ? | ? | SDD1 | ? |
?, not yet identified; *, likely/predicted.
CLE family proteins are united by their possession of a conserved 12– to 14–amino acid CLE motif, usually positioned at the extreme C terminus of the full-length translated precursor protein, and a hydrophobic N-terminal signal sequence (Cock and McCormick, 2001; DeYoung and Clark, 2001). Cleavage of the CLE motif from the precursor is necessary for production of the mature 12– to 14–amino acid peptide, which may then undergo further posttranslational modification (Fiers et al., 2005, 2006; Ito et al., 2006; Ni and Clark, 2006; Ohyama et al., 2009; Katsir et al., 2011). The CLE motif contains four conserved Pro residues at positions 4, 7, and 9: Pro-4 and Pro-7 appear to have particular significance as the targets for posttranslational modification (hydroxylation: Pro-4/Pro-7; arabinosylation: Pro-7). The CLV3 gene is expressed as a protein of 96 amino acids, including an 18 hydrophobic N-terminal amino acid signal peptide to direct the peptide for secretion, a typical feature of CLE peptides. Cleavage occurs at the Arg at the beginning of the CLE motif (Arg-70), and this residue is conserved absolutely through all CLE proteins, including CLV3 (Ni and Clark, 2006; Oelkers et al., 2008). Recent evidence suggests that the cleavage of the CLE motif from the precursor is likely to be due to the action of an as yet unknown Ser protease, based on the ability of the Ser protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride and bestatin to block the proteolytic processing of the CLE domain (Ni et al., 2011). Following processing, CLV3 undergoes further posttranslational modification. The amino acid sequence of CLV3 contains three Pro residues, of which the first two N-terminal Pro residues are hydroxylated and one Hyp (Hyp-7) then undergoes Hyp arabinosylation. The arabinosylation of Hyp-7 has been shown to enhance binding of CLV3 to the receptor CLV1 (Kondo et al., 2006). TDIF (CLE41/CLE42) is cleaved from a 99– to 112–amino acid protein precursor before proteolytic processing to separate the 12–amino acid CLE domain from the precursor (Ito et al., 2006).
Similar mechanisms may also apply to the RGF/GLV/CLEL family peptides with regards to the processing of the CLE motif from the precursor. However, though it seems likely, it is unknown if CLEL motifs are extracted from the full-length precursor in the same way as the CLE motifs. In addition, it is unclear whether CLEL motif peptides require any posttranslational modification, such as Tyr sulfation, for optimal activity, as both the sulfated and unsulfated forms of the RGF1/CLEL8 peptide have been proposed as the active form in planta (Matsuzaki et al., 2010; Meng et al., 2012). Nevertheless, in agreement with the findings for the RGF1 peptide, Tyr sulfation of synthetic peptides representing the proposed active form of RGF6/GLV1 and RGF4/GLV3 increased their bioactivity by two to three orders of magnitude. This opens the possibility that other precursors might also give rise to multiple functional peptides which are subject to distinct proteolytic processing and posttranslational modifications (Figure 1B).
Amino acid sequencing of purified PSK-α suggested that PSK was subject to posttranslational Tyr O-sulfation. The sulfation of two Tyr residues, namely, Tyr-1 and Tyr-3, of the PSK-α peptide was later confirmed through synthesis and subsequent sulfation of the peptide using arylsulfotransferase. Solid-phase peptide synthesis and sulfation of PSK-α and 12 analogs with an adimethylformamide-sulfurtrioxide complex confirmed the active peptide core of PSK as Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln. The second peptide identified, PSK-β, was found to be a C-terminal truncated tetrapeptide of PSK-α, which also underwent Tyr sulfation: Tyr(SO3H)-Ile-Tyr(SO3H)-Thr (Matsubayashi et al., 1996; Matsubayashi and Sakagami, 1996). Acidic residues surrounding the Tyr residues appear essential for optimal sulfation activity, probably by a class of tyrosylprotein sulfotransferases unique to plants (Hanai et al., 2000). Studies on PSK1 in rice show that the mature pentapeptide of PSK-α is proteolytically processed from an ∼80–amino acid precursor, preprophytosulfokine-α (the exact precursor length varies according to species and PSK paralog). Preprophytosulfokine-α includes a hydrophobic 22–amino acid signal peptide at the N terminus, targeting it for inclusion in secretory pathways (Yang et al., 1999, 2000).
PSY1, which was found to promote cellular proliferation and expansion in various tissues, is expressed as a precursor of 75 amino acids and is proteolytically processed to a mature form of 18 amino acids: In addition to Tyr sulfation, PSY1 also undergoes Pro hydroxylation and subsequent Hyp arabinosylation (Amano et al., 2007; Matsubayashi, 2011a).
Tobacco RALF cDNA encodes a 115–amino acid polypeptide, which includes an N-terminal signal sequence (it is predicted by TargetP that all RALF prepropetides are targeted to the endomembrane system [Bedinger et al., 2010]), and a 49–amino acid C-terminal sequence representing the mature RALF peptide. PreRALF has a conserved dibasic site ∼50 amino acids upstream from the C terminus. In animals, precursors to signaling peptides typically require such dibasic motifs to mark the site of mature peptide excision. These are recognition sites for a family of subtilisin/kexin-like preprotein convertases required for processing. However, most plant signaling peptide precursors discovered so far do not appear to require dibasic motifs for proteolytic processing. Thus, the conserved dibasic site found in preRALF is quite unusual. Within the conserved C-terminal sequence are four conserved Cys residues involved in the formation of two disulphide bridges (Pearce et al., 2001b). These disulphide bridges are thought to be essential for the active conformation of the protein and are absolutely required for function: Reduction of RALF with iodoacetamide renders the peptide unable to alkalize the cell culture medium in alkalization assays (Pearce et al., 2001a).
EPF1, EPF2, and EPFL1 to 9 all have a predicted secretory signal sequence and six C-terminal conserved Cys residues (Hara et al., 2009). The STOMAGEN/EPFL9 gene encodes a 102–amino acid peptide. The N-terminal signal peptide is followed by a predicted cleavage site between residues 31 and 32. Members of the EPF family all contain a long loop between the 4th and 5th conserved Cys: The loop itself has very high sequence diversity between the members of the EPF family, which is thought to explain the functional diversity of the peptides (Kondo et al., 2010). Purification and mass spectrometry (matrix-assisted laser desorption/ionization-time of flight and liquid chromatography–mass spectrometry, respectively) of endogenously derived trypsin and chemotrypsin/lysyl endopeptidase-digested STOMAGEN protein revealed that the active STOMAGEN peptide is 45 amino acids in length, is processed from a 120–amino acid precursor, and forms three disulphide bonds (Kondo et al., 2010; Sugano et al., 2010). The disulphide bonds are crucial to STOMAGEN function. While STOMAGEN is active at nanomolar (∼10 nM) concentrations (a typical active concentration range for signaling peptides), both unfolded STOMAGEN and STOMAGEN where Cys residues were replaced with Ser residues were unable to increase stomatal density even at very high (10 µM) concentrations (Ohki et al., 2011).
Proteolytic Processing
The genes of the small posttranslationally modified peptides and many of the Cys-rich peptides encode a full-length protein that is longer than the length of the active peptide (Figure 1A, Table 1). The active peptide is freed from additional sequence by proteolytic processing to produce the mature-length peptide. The enzymes responsible for processing individual signaling peptide precursors remain largely unknown but are likely to be members of the subtilase group of Ser proteases. Subtilases are found in archaea, bacteria, yeast, and higher eukaryotes, but plants have many more subtilisin-like enzymes than animals: 56 in Arabidopsis compared with nine in humans, but very few loss-of-function mutants show obvious visible phenotypes (Rautengarten et al., 2005; Ottmann et al., 2009).
A subtilisin-like Ser protease, SBT1.1, is required for processing of the PSK4 precursor (Srivastava et al., 2008). Overexpression of SBT5.4 gives a clv-like phenotype, though it was not demonstrated to cleave CLV3 in vitro (Liu et al., 2009). Two other members of this family of subtilisin-like enzymes, SITE 1 PROTEASE (S1P) and STOMATAL DENSITY AND DISTRIBUTION (SDD1), have been suggested to be involved in proteolytic processing of peptide signals. In the Cys-rich peptide family, RALF23 has been shown to require S1P for processing. S1P is a Golgi and endoplasmic reticulum–localized Ser protease that requires a dibasic RR motif (typical for animal subtilase substrates) for substrate recognition (Srivastava et al., 2009). SDD1, an extracellular protease, has been suggested to play a role in processing of EPF1 (Berger and Altmann, 2000), though conversely the phenotypes of mutants for epf1 and epf2 are more severe when crossed with sdd1 to create a double mutant. The additive phenotype suggests EPF1 and EPF2 act independently from SDD1 (Hara et al., 2007, 2009; Hunt and Gray, 2009); thus, another enzyme is likely responsible for processing EPF family peptides (Table 1).
SIGNALING PEPTIDES IN ARABIDOPSIS DEVELOPMENT
At present, much of our knowledge relies on a few family members that have been investigated in depth, not in the least the CLE family, which has been shown to be associated with development in multiple Arabidopsis organs. Already at the first stages of plant life, cell–cell communication mediated by small signaling peptides and receptor kinases plays a key role (Escobar-Restrepo et al., 2007; Okuda et al., 2009; Kessler et al., 2010). Below, we will illustrate the essential involvement of various peptide families in plant organ and tissue development.
Anther and Pollen Development
The anther is a four-lobed structure that develops from three distinct cell layers, known as L1, L2, and L3. The L1 layer generates the epidermis, the L2 layer gives rise to endothecium, middle layer, tapetum, and the microsporocytes, and the L3 layer generates the connective tissues and vasculature within the center of the anther. Microsporocytes, or pollen mother cells, meiotically divide into four daughter cells known as microspores, which will develop into mature pollen granules. During L2 layer formation, the cells divide periclinally to form primary parietal cells. The parietal cells will further divide into secondary parietal cells, which, depending on location, can further divide periclinally into endothecium, middle layer, or tapetum. The tapetal layer is important to pollen development as it provides essential lipids and small molecules that aid in pollen formation and pollen wall generation (Goldberg et al., 1993; Zhao, 2009; Ge et al., 2010).
The tpd1 mutant shows a complete lack of tapetal layer, zero pollen granules, and extra microsporocyte cells within the anther, since tapetal cell precursors develop into microsporocytes instead of tapetal cells (Yang et al., 2003). This suggests that the microsporocyte stage of development occurs before the tapetal development stage and that the TPD1 peptide may act somewhere between the two, initiating tapetal cell differentiation (Ge et al., 2010).
Recently, a number of CLE peptides have also been shown to be expressed during anther development. CLE1 has been located in the tapetum of the anther, as well as within the pollen granules. CLE11, CLE12, and CLE13 are all found within the pollen granules, during various stages of pollen development, while CLE25 is expressed only during early anther development (Jun et al., 2010). However, their role in anther and pollen development remains to be demonstrated.
Embryo
The female gametophyte receives two sperm cells from a pollen tube: One sperm cell fertilizes the egg, which will develop into the embryo, and the other leads to endosperm formation, which are cells that regulate and nourish the embryo during development. The embryo, encased in the protective seed coat, will undergo a series of stereotypical divisions to form a miniature seedling (De Smet et al., 2010; Lau et al., 2012).
Several CLE peptides recently have been shown to be expressed during embryogenesis; specifically, CLE8 has been directly linked to embryo and endosperm proliferation and differentiation (Fiers et al., 2004; Jun et al., 2010; Fiume et al., 2011; Fiume and Fletcher, 2012). The cle8 knockout mutants and CLE8 artificial microRNA lines do not display any obvious vegetative aberrant phenotypes, but small, defective, underdeveloped, and misshaped seeds often occur within mature siliques. This correlated with deviations in early embryo cell division patterns and in endosperm proliferation and differentiation. Transgenic lines overexpressing CLE8 show a slight increase in embryo length and overall size, suggesting a positive correlation between the CLE8 peptide and seed size. CLE8 has been shown to act upstream of WUSCHEL-LIKE HOMEOBOX8 (WOX8), evidenced by severely reduced WOX8 expression in the cle8 mutant. Conversely, overexpression of CLE8 shows a marked increase in WOX8 expression, suggesting that these two proteins act synergistically in suspensor and endosperm development (Fiume and Fletcher, 2012).
SAM
The leaves, stems, and floral parts of plants develop from the SAM, a conserved stem cell niche that resides at the top of the apical-basil axis of the plant. It is a tightly organized and regulated structure separated in three zones: the peripheral zone (PZ), the central zone, and the rib zone. The central zone consists of a highly conserved stem cell population, residing directly above the organizing center (OC), whose daughter cells are displaced into the PZ. It is the cells located in the PZ that generate the aerial tissues and organs of the plant (Fletcher and Meyerowitz, 2000; Stahl and Simon, 2005) (Figure 2A).
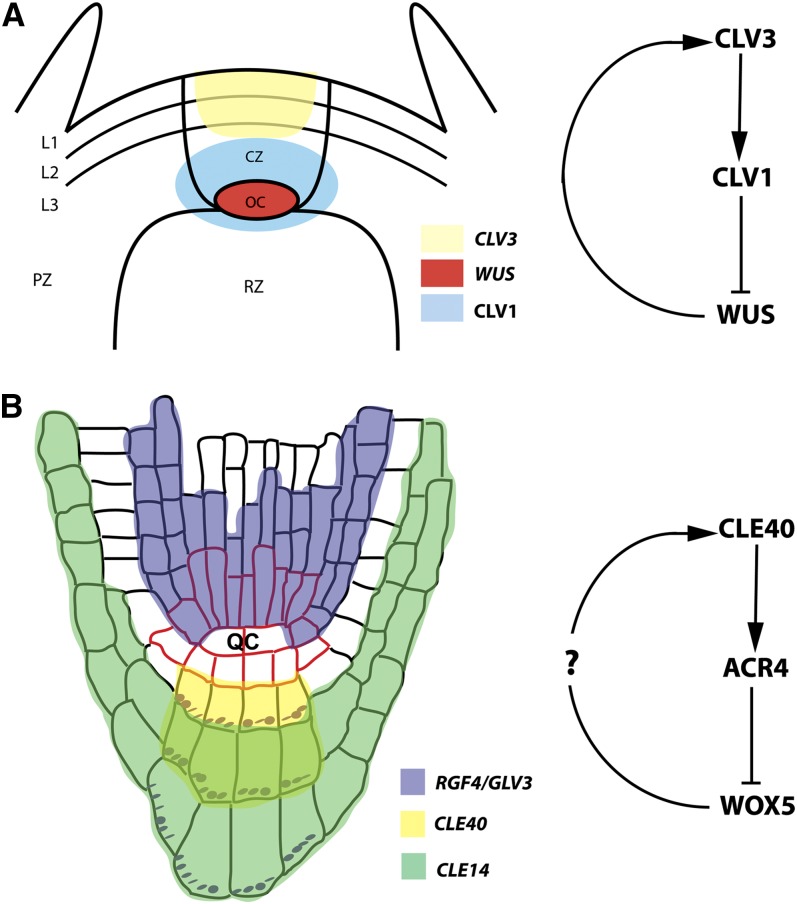
Figure 2.
SAM and RAM.
(A) CLV3 (yellow) is expressed in the central zone (CZ) of the SAM and inhibits expression of WUS (red) in the OC. WUS itself promotes CLV3 expression. The CLV1 receptor is expressed below layers one and two of the SAM (L1 and L2). On the right is the proposed CLV3-CLV1-WUS signaling module. PZ, peripheral zone; RZ, rib zone.
(B) Several peptides, such as CLE14, CLE40, and RGF4/GLV3, are expressed in the RAM in overlapping domains to control cell identity and meristematic activity. On the right is the proposed CLE40-ACR4-WOX5 signaling module. At present, there is no evidence for WOX5 controlling CLE40 expression (?). Red outline, stem cells and QC; purple, starch granules.
In shoot development, the peptide ligand CLV3 is a major regulator of the stem cell niche (Fletcher et al., 1999; Kondo et al., 2006) (Figure 2A). The clv3 mutant results in overproliferation of the SAM stem cell population, to sizes often 1000 times larger than that of wild-type SAMs, resulting in a club-like shaped stem (clava meaning club) (Koornneef et al., 1983; Clark et al., 1995). By contrast, overexpression of CLV3 eliminates the stem cell niche completely, terminating the SAM. These CLV3 overexpression plants cease to initiate organs after the emergence of the first leaves, and, in a subset of the lines, flowers formed without the ability to generate inner whorls with stamens or carpels (Brand et al., 2000). CLV3 is shown to be involved in a negative feedback signaling pathway with the stem cell promoting homeodomain transcription factor protein WUSCHEL (WUS) (Mayer et al., 1998; Fletcher et al., 1999; Schoof et al., 2000; Nimchuk et al., 2011). CLV3 is expressed in stem cells at the meristem apex, and the CLV3 peptide is thought to diffuse down toward the OC to restrict the expression of WUS, thereby regulating and inhibiting the expansion of stem cells in the SAM (Kinoshita et al., 2010; Nimchuk et al., 2011; Yadav et al., 2011). The expression of WUS defines the boundaries of the OC and surrounding stem cell niche and actively promotes the proliferation and division of stem cells in the apical cell layers (Buechel et al., 2010). Furthermore, WUS migrates between cells, toward the CLV3 expressing cell layers, where WUS directly binds to genomic regions of CLV3, activating its transcription in the central zone at the meristem apex (Yadav et al., 2011). This negative feedback loop between WUS and CLV3 is well established (Figure 2A): clv3 mutants overproduce stem cells, resulting in an enlarged SAM, whereas wus mutants eliminate stem cell populations and result in SAM termination (Laux et al., 1996; Nimchuk et al., 2011). By contrast, CLV3 overexpression results in a wus-like phenotype and WUS overexpression results in a clv3-like phenotype (Kinoshita et al., 2010; Nimchuk et al., 2011). Thus, the homeostatic model shows that as stem cells decrease in number, CLV3 expression is lowered, which raises WUS expression, inducing stem cell divisions. If stem cells become too abundant, CLV3 expression increases, lowering WUS levels and thus lowering the rate of stem cell divisions (Brand et al., 2000; Schoof et al., 2000; Müller et al., 2006; Yadav et al., 2011).
In addition to CLV3, there are over 30 CLE-related peptides containing the CLE motif, of which, CLE16, CLE17, and CLE27 have also been identified in and around the SAM (Lenhard and Laux, 2003; Sharma et al., 2003; Fiers et al., 2005; Jun et al., 2010). With the known redundancy of CLE peptides, there is a strong possibility that these CLE peptides diffuse from the SAM periphery and have significant functional roles within SAM development (Brand et al., 2000; Schoof et al., 2000; Jun et al., 2010). For example, it has been shown that many CLE motif–containing peptides can replace CLV3 function in the SAM (Ni and Clark, 2006). Overexpression of CLE2, 3, 4, 5, 6, 7, 9, 10, 11, and 13 all results in phenotypes similar to overexpression of CLV3 or wus lines (Brand et al., 2000; Hobe et al., 2003; Strabala et al., 2006). With the majority of CLV3, CLE9, CLE10, CLE11, and CLE13 overexpression lines dying significantly earlier than the wild type, it appears that these, possibly redundant, CLE peptides are integral to normal SAM development (Strabala et al., 2006). Possibly though, the CLE peptides may in fact be activating the endogenous CLV3 receptors, mimicking CLV3 overexpression.
Also, during axillary bud formation, CLE41, CLE44, and CLE42 play an important role through regulating meristematic activity as demonstrated through application of synthetic peptides and overexpression studies (Yaginuma et al., 2011).
Lastly, EPFL peptides have been shown to play a role in inflorescence growth. While the single knockout mutants epfl4/challah-like2 (cll2), epfl5/cll1, and epfl6/chal show no distinguishable phenotype from the wild type, epfl4 epfl6 and chal cll1 cll2 mutants show a strong compacted inflorescence with clustered flowers (Abrash et al., 2011; Uchida et al., 2012).
Primary Root Development
The underground parts of the plant, the main root, lateral roots, and root hairs, all of which play a role in water and nutrient uptake (De Smet et al., 2012; Smith and De Smet, 2012), are derived from cells originating in the root apical meristem (RAM). The RAM consists of stem cells surrounding the quiescent center (QC), which transmits developmental cues to the neighboring stem cells. Stem cells can generate further stem cells, in a nondifferentiated state, as well as daughter cells that differentiate into the respective cell layers. The formation of the columella represents an elegant example, where columella stem cells give rise to two daughter cells of which the lower one differentiates in the columella (containing starch granules) (van den Berg et al., 1998) (Figure 2B).
Several signaling peptides are expressed in the root tip, and several of these have been assigned a role in primary root development. Among the peptides involved in developmental processes in the RAM, the CLE and the RGF/GLV/CLEL families of peptides are the most prominent (Casamitjana-Martínez et al., 2003; Hobe et al., 2003; Stahl et al., 2009; Matsuzaki et al., 2010; Meng et al., 2012; Whitford et al., 2012) (Figure 2B).
Communication between QC and distal columella (stem) cells in the RAM has been elucidated as a signal transduction pathway between WOX5 and CLE40 (Figure 2B). CLE40 is expressed in the inflorescence apices, leaves, and roots of the plant but has only been observed at functionally significant levels within the root tip. Overexpression of CLE40 results in stunted primary root growth shortly after germination and these lack stem cells surrounding the QC but instead consists of large, differentiated cells. The cle40 mutant shows a significant enlarged root phenotype due to rapid and aberrant division of columella initial cells. This overproliferation of initial cells is connected to an expansion of the WOX5 domain, suggesting that CLE40 has a role in negatively regulating WOX5, supporting its proposed role in controlling or inhibiting the signals transmitted from the QC to the surrounding stem cells (Hobe et al., 2003; Stahl et al., 2009).
Various other CLE family peptides have been shown to be expressed in the RAM: CLE12, CLE14, CLE16, CLE17, CLE22, and CLE27 (Jun et al., 2010; Meng et al., 2010). For example, CLE14 (root cap and root hairs), CLE16 (root cap and root elongation zone), and CLE17 (root meristematic zone) are expressed in specific tissues of the root, suggesting specific roles. While cle19 mutants give no phenotype, when overexpressed or applied exogenously, CLE14, CLE19, and CLE20 can irreversibly induce differentiation of stem cells rather than division and elongation, resulting in early consumption of the RAM and causing a strong short root phenotype (Casamitjana-Martínez et al., 2003; Fiers et al., 2004, 2005; Jun et al., 2010; Meng et al., 2010). Overexpression of CLE9, CLE10, CLE11, or CLE13 results in severe inhibition of primary root elongation, while CLE18, CLE25, and CLE26 overexpression leads to enhanced root elongation (Strabala et al., 2006). How CLE family peptides, with highly similar homologies, can have antagonistic roles in root elongation has yet to be elucidated, but similar antagonistic roles of related signaling peptides have also been observed during stomatal development (see below) (Kondo et al., 2008). Interestingly, the phytohormone cytokinin was shown to be involved in processing or transmitting CLE peptide signals, thus regulating the effects of CLE14 and CLE20 in planta. While exogenous application of CLE14 and CLE20 results in a strong short root phenotype, the combination with cytokinin largely rescued the reduction in primary root length (Meng and Feldman, 2010).
Two other signaling peptides expressed in roots, PSK and PSY1, are also involved in root elongation (Amano et al., 2007; Matsuzaki et al., 2010). These peptides can restore cell elongation activity in the elongation differentiation zone of roots in tpst but cannot restore meristem activity to that of the wild type, as evidenced by additional QC cells and aberrant columella cells (Matsuzaki et al., 2010). To restore the meristem size of tpst to a comparable level to that of the wild type, application of RGF1/GLV11/CLEL8 is required, suppressing formation of additional QC cells and restoring columella stem cell numbers. RGF1/GLV11/CLEL8 is expressed in the QC and columella stem cells (with RGF2 and RGF3 expression in the innermost layers of the columella) and the protein diffuses into surrounding cell types forming a concentration gradient (Matsuzaki et al., 2010). While a single rgf1 mutant did not show a significant phenotype, the rgf1 rgf2 rgf3 triple mutant exhibits a similar phenotype as that of tpst, being a short root phenotype with a reduction in meristematic cells. Exogenous application of RGF1/GLV11/CLEL8 to rgf1 rgf2 rgf3 rescued cell numbers to that of the wild type, suggesting a strong functional redundancy between these peptides. The overexpression of RGF1/GLV11/CLEL8 shows the opposite phenotype, with seedlings possessing enlarged meristems, due to additional columella cells (Matsuzaki et al., 2010). Thus, the redundant, sulfated RGF/GLV/CLEL family of peptides is clearly inherent to the maintenance of the stem cell population within the root and appears to act on PLETHORA (PLT) transcription factors (Matsuzaki et al., 2010). PLTs are expressed in response to auxin and, in concert with PIN FORMED (PIN) proteins, act on auxin signaling required for root stem cell specification (Aida et al., 2004; Galinha et al., 2007). Interestingly, RGF/GLV/CLEL peptides also have an effect on auxin-dependent gravitropic response within roots, resulting in a waving phenotype, when overexpressed or exogenously applied. Specifically, RGF1/GLV11/CLEL8 interacts with the PIN2 protein, namely, accumulating the PIN2 proteins at the plasma membrane, through an unknown process, thereby affecting auxin transport (Meng et al., 2012; Whitford et al., 2012).
The small 15–amino acid hydroxyprolinated peptide CEP1 affects root growth but it is as yet unknown through what pathway. Both overexpression of CEP1 and exogenous application of synthetic CEP1 results in reduced primary root growth. Detailed analyses revealed that there are fewer cells in the RAM when CEP1 is overexpressed, with a reduced cell size in the mature region of the root tip. No reduction in QC cells was observed, however, suggesting a role in cell division potential and efficacy, without interfering with QC stem cell capabilities. Strikingly, no CEP1 expression was observed in the RAM, suggesting a nonspecific effect of generally increasing CEP1 levels (Ohyama et al., 2008).
The RALF peptide also reversibly inhibits Arabidopsis root growth and root hair initiation when seedlings are cultured in synthetic RALF-containing media (Pearce et al., 2001b; Bedinger et al., 2010). Overexpression of RALF23 produces slow-growing seedlings, with roots that have a reduced capacity to acidify the rhizosphere, which may have developmental implications, resulting in a semidwarf phenotype (Srivastava et al., 2009). Exogenous application of RALF1 also causes a significant rise in cytoplasmic Ca2+ concentration, suggesting a functional role in Ca2+ modulation and signal transduction (Haruta et al., 2008).
Finally, members of the IDA family are also expressed in the RAM, such as columella cells; however, a functional role for the peptide in RAM management has yet to be elucidated (Stenvik et al., 2006).
Lateral Root Development
Lateral roots develop to aid in nutrient mining and water uptake and develop from a subset of pericycle cells at the xylem poles in the root stele. These pericycle cells and their daughter cells undergo a series of anticlinal and periclinal cell divisions generating a lateral root primordium (De Smet et al., 2008; De Smet and Beeckman, 2011; De Smet, 2012).
While the role of phytohormones during lateral root initiation has been highlighted extensively, only recently has the RGF/GLV/CLEL family of peptides been shown to significantly delay lateral root development when overexpressed (Péret et al., 2009; De Smet, 2012; Meng et al., 2012). Specifically, overexpression of CLEL6 and CLEL7 results in inhibition of pericycle division in the second round of pericycle cell divisions, arresting the initiation of lateral roots. Interestingly, analyses on CLEL-overexpressing plants in conjunction with auxin-containing media have shown that the CLEL peptides actually act in an auxin-independent manner (Meng et al., 2012). In addition, the CLE20 peptide is also expressed in lateral roots, suggesting a possible dual role of the peptide in RAM maintenance and lateral root development (Meng and Feldman, 2010). Finally, CEP1 is expressed in lateral root primordia, with expression of CEP1 peaking during lateral root emergence and decreasing significantly after the lateral roots have elongated, suggesting that CEP1 might be involved in lateral root elongation, similar to its effect on the primary root (Ohyama et al., 2008).
Vascular Development
Within the roots and shoot of the plant lies the vasculature, the cells that are responsible for transporting water, minerals, nutrients, hormones, and even small signaling peptides throughout the organism. The vascular system in plants consists fundamentally of two functional cell types with vastly different functions: xylem and phloem (Sieburth and Deyholos, 2006) (Figure 3). Among the many hormone, transcription factor, and microRNA signals that are required for plant vasculature development, signaling peptides, especially the CLE family, have also become a prominent player (Figure 3) (Fiers et al., 2007; Elo et al., 2009; Hirakawa et al., 2011; Kondo et al., 2011). TDIF (CLE41/44) is shown to promote the proliferation of procambial cells in leaf and hypocotyl vasculature development, while it also inhibits the differentiation of procambial cells into tracheary elements (Ito et al., 2006; Whitford et al., 2008). Overexpression of CLE41 results in a strong stunted and bushy phenotype, with disrupted secondary and mature xylem bundles within the hypocotyls (Whitford et al., 2008), while cle41 and cle44 mutants show a reduction in the number of procambial cells (Hirakawa et al., 2010). Specifically, overexpression of CLE41 results in a wide range of cellular orientations and divisions in contrast with the highly orientated cell divisions, disrupting the localized pattern between procambium and cambium, resulting in overproliferation of vascular bundles and increased vascular root tissue (Hirakawa et al., 2008; Etchells and Turner, 2010). CLE10 plays a role in xylem formation as CLE10 overexpression results in significant inhibition of the early stages of protoxylem vessel formation, which, in roots, appears to occur through activation of cytokinin signaling (Kondo et al., 2011). CLE10 has the ability to inhibit protoxylem differentiation in the root, by the repression of ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, ARR5 and ARR6, which are known to negatively regulate cytokinin signaling, allowing suppression of procambium differentiation into protoxylem vessels (Hirakawa et al., 2011). Finally, CLE19 overexpression results in the xylem network to fail at making viable connections between elements (Fiers et al., 2007).
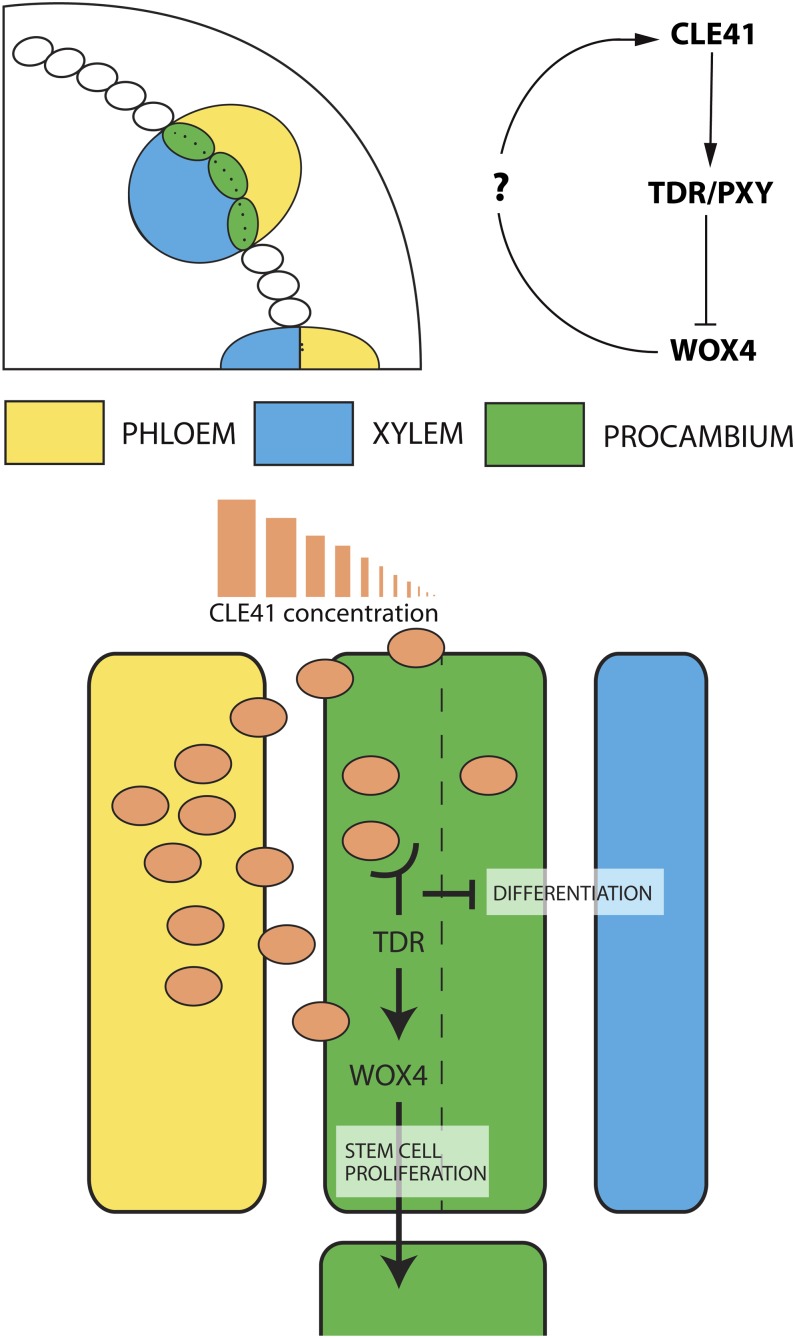
Figure 3.
Vascular Differentiation.
Schematic of a stem section highlighting the vascular bundles (top left). Phloem cells (yellow) generate and secrete CLE41 peptides (peach) into procambial cells (green) where they are bound by membrane-localized TDR/PXY. TDR/PXY inhibits differentiation of procambium into xylem cells (blue) and upregulates the expression of the transcription factor WOX4, initiating stem cell proliferation (green). As CLE41 concentration lowers (toward plane of division: dashed line), procambial cells (green) can differentiate into xylem cells (blue) without inhibition from TDR. On the top right is the proposed CLE41-TDR/PXY-WOX4 signaling module. At present, there is no evidence for WOX4 controlling CLE41 expression (?).
Stomatal Development
Stomata, which are composed of two guard cells, are the organs responsible for the vital gas exchange required to perform respiration. Stomata in Arabidopsis are usually separated by at least one nonstomatal cell to optimize spatial arrangement between the photosynthetic epidermal surface area and the gas exchange organs. The developmental processes behind stomatal differentiation rely heavily on asymmetric cell division derived from positional cues from one leaf cell to the next, but also environmental cues play a role in leaf stomatal density. The guard cells are derived from multipotent epidermal stem cells, the meristemoid mother cells (MMCs), which divide asymmetrically to provide two daughter cells of different sizes. The smaller triangular shaped cells (meristemoids) have stem cell–like capabilities and generally engage in further asymmetric division. The larger daughter cells, while capable of further asymmetric divisions, generally terminally differentiate into guard mother cells (GMCs). The GMCs divide symmetrically to yield the two guard cells, surrounding a pore, to become the stomata (Nadeau, 2009; Serna, 2009; Dong and Bergmann, 2010; Rowe and Bergmann, 2010) (Figure 4).
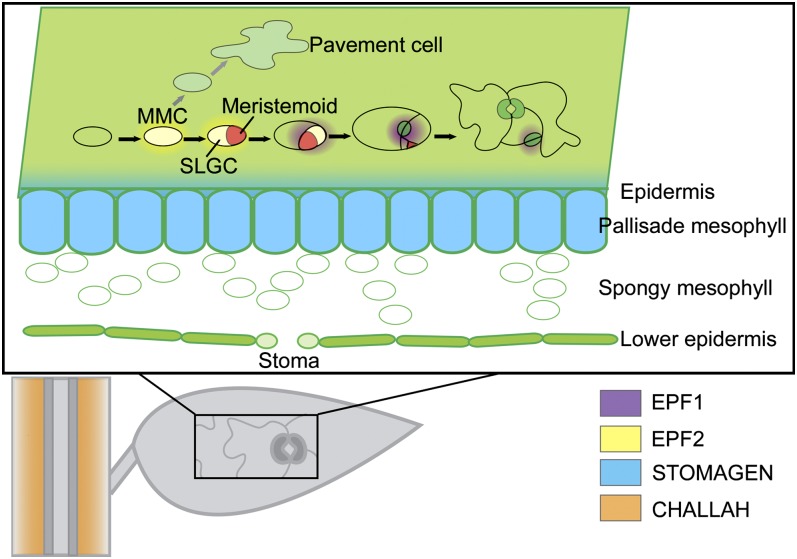
Figure 4.
Stomatal Development.
Stomatal development begins when protodermal cells differentiate into MMCs. Protodermal cells that do not differentiate into MMCs instead differentiate into epidermal pavement cells. The MMC then divides asymmetrically to form a small triangular meristemoid cell (red) and a larger type of daughter cell, a stomatal lineage ground cell (SLGC). At these early stages of stomatal development, cells secrete EPF2 (yellow); after these stages, there is a switch to EPF1 expression (purple). Meristemoids often further divide to amplify the number of cells in the stomatal lineage, before finally differentiating into GMCs. GMCs then divide symmetrically to form two guard cells surrounding a pore, completing stomatal development. Once guard cell maturation is complete, EPF1 secretion is terminated. STOMAGEN (light blue) is expressed in the mesophyll and diffuses to the epidermis, promoting stomatal development. CHAL (orange) is secreted by cells surrounding the vascular bundle and diffuses to the epidermis, restricting stomatal development.
Various members of the EPF family of Cys-rich small peptides, specifically, EPF1, EPF2, EPF-LIKE6 (EPFL6)/CHALLAH (CHAL), and EPFL9/STOMAGEN, are involved in regulating stomatal density and positioning (Figure 4) (Hara et al., 2007; Abrash and Bergmann, 2010; Kondo et al., 2010; Rowe and Bergmann, 2010; Sugano et al., 2010; Shimada et al., 2011). The overexpression of EPF1 or EPF2 results in reduced stomatal density, but loss-of-function mutants indicated they have differing actions (Hunt and Gray, 2009; Kondo et al., 2010). In epf2 mutants, asymmetrical cell divisions occur more frequently than the wild type and stomatal densities significantly increase, while epf1 mutants form random stomatal division patterns, which often leads to stomata in contact with each other (Hara et al., 2007). Promoter swapping experiments between the related EPF1 and EPF2 suggest that EPF2 is expressed earlier in development than EPF1, as EPF2 has the ability to partially rescue stomatal clustering in epf1 with expression of pEPF1:EPF2 but not pEPF2:EPF1 (Hunt and Gray, 2009; Rowe and Bergmann, 2010). This shows that EPF1 efficacy in stomatal patterning is somehow related to EPF2 expression (Rowe and Bergmann, 2010). Indeed, transcriptomics analysis has shown higher levels of EPF2 expression in juvenile leaves, prior to EPF1 expression (Hunt and Gray, 2009). EPF2 may act early in development to control differentiation in meristemoids, controlling asymmetric cell division and proliferation rates, while EPF1 may affect the directionality and spacing of the developing stomata (Hunt and Gray, 2009; Rowe and Bergmann, 2010; Shimada et al., 2011; J.S. Lee et al., 2012). This theory is strengthened by the expression patterns, with EPF2 expressed in mother meristemoid cells and early meristemoids, while EPF1 is expressed in GMCs and young guard cells, showing a clear temporal-spatial division between the two peptides (Shimada et al., 2011) (Figure 4).
EPFL9/STOMAGEN is produced in the mesophyll cells of the plant and positively regulates stomatal development in the epidermis (Hunt et al., 2010; Sugano et al., 2010) (Figure 4). Overexpression of STOMAGEN greatly increases stomatal density to a degree where guard cells are often surrounded by other stomatal precursor cells (Hunt et al., 2010; Sugano et al., 2010). Conversely, knockout mutants show a significant decrease in stomatal density (Hunt et al., 2010; Sugano et al., 2010; Shimada et al., 2011). Lastly, EPFL6/CHAL negatively regulates stomatal development when overexpressed (Shimada et al., 2011; Uchida et al., 2012). Paralagous to EPFL6/CHAL are EPFL4 (CHAL-LIKE2) and EPFL5 (CHAL-LIKE1), together known as CHAL family (CHALf) ligands (Abrash et al., 2011). Nevertheless, single epfl6, epfl4, and epfl5 mutants as well as epfl4 epfl5 epfl6 triple mutants show no difference in stomatal density nor positioning compared with the wild type (Abrash et al., 2011; Uchida et al., 2012). EPFL6/CHAL expression is seen in the hypocotyl and stem of the plant, unlike EPF1 and EPF2, which are expressed in the leaf, showing that there is an organ-specific role of these peptides (Shimada et al., 2011; Uchida et al., 2012).
Abscission
Abscission, whereby entire organs are shed from the plant, involves the organized and programmed cell separation of the middle lamella in plant cells. The middle lamella is the area of the cell wall that is shared between two neighboring cells, solidifying and holding the cells together. Abscission during development is essential for optimal plant growth when specific cells and organs are no longer necessary or functional (Stenvik et al., 2006, 2008).
The majority of signaling molecules known to play a role in abscission are plant hormones (Jinn et al., 2000). However, the small signaling peptide IDA has been shown to stimulate abscission through promotion of cell separation or through inhibiting cellular repair processes (Stenvik et al., 2006, 2008). IDA, together with IDL genes, belongs to a family of six genes and IDA is predominantly expressed during floral abscission. The ida knockout mutants lack the ability to abscise floral organs, with plants fully retaining the petals, sepals, and filaments indefinitely. Furthermore, overexpression of IDA and IDL shows premature floral abscission, with overproliferation of abscission zone cells; however, it varies in intensity between the members of the family. Using IDA upstream and downstream cis-regulatory elements to drive IDL expression in ida knockout mutants, it was shown that only IDL1 had the capability to fully rescue and replace native IDA activity, whereas, IDL2 to IDL5 had very little effect. IDA and IDL proteins have an extended PIP motif (EPIP), which has been shown as the active part of the peptide used in abscission. Arabidopsis plants subjected to synthetic IDA EPIP peptides showed phenotypes similar to IDA overexpression lines, with abscission occurring prematurely, whereas ida knockout lines subjected to IDA EPIP showed a partial rescue of phenotype. Due to the activity of the EPIP motif, it is further hypothesized that IDA and IDL peptides are cleaved, through a similar process as CLV3, where the active EPIP site becomes available to downstream effectors, thereby initiating floral abscission (Stenvik et al., 2008).
SIGNALING PEPTIDES: IDENTIFYING THE CORRESPONDING RECEPTOR
While plant genomes encode >600 putative RLKs and >1000 potential secreted peptides, so far only very few could be shown to form pairs (Torii, 2004; Butenko et al., 2009). Nevertheless, unequivocal identification of the ligand is crucial to fully understand receptor kinase–mediated signaling pathways, as was recently demonstrated for a number of ligand-receptor pairs (Hara et al., 2007; Hirakawa et al., 2008; Jia et al., 2008; Ogawa et al., 2008; Etchells and Turner, 2010; Kinoshita et al., 2010; J.S. Lee et al., 2012; Shinohara et al., 2012; Uchida et al., 2012). Unfortunately, identification of ligand-receptor pairs is technically very challenging and requires strategies that combine transcriptional analyses at cellular resolution, microscopy characterization of loss- and gain-of-function plants, easily to monitor readouts (as, for example, monitoring luciferase activity in protoplasts transiently expressing signaling components [Albert et al., 2010; Mueller et al., 2012a]), and genetic interaction studies to identify candidate pairs, followed by biochemical assays to demonstrate direct physical interactions. In this section, we will describe some of the known peptide ligand-receptor kinase pairs and comment on some likely candidates to others.
Pollen and Anthers
EXCESS MICROSPOROCYTES1 (EMS1)/EXTRA SPOROGENOUS CELLS encodes an LRR-RLK that is localized to the cell surface and is needed for tapetal cell development (Ge et al., 2010). The ems1 mutant displays a lack of tapetal cell development and an excess production of microsporocytes (Albrecht et al., 2005). The nearly identical phenotypic similarities between tpd1, ems1, and tpd1 ems1 mutants suggest a strong link between receptor and putative ligand during tapetal cell and pollen development (Yang et al., 2003; Albrecht et al., 2005). Through yeast two-hybrid and in planta coimmunoprecipitation assays, it has been shown that TPD1 directly interacts with EMS1 in vitro and in vivo and that TPD1 activates phosphorylation of the EMS1 kinase domain (Jia et al., 2008). As TPD1 signaling depends on functional EMS1, these data clearly establish a peptide ligand-receptor kinase pair (Jia et al., 2008). SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) and SERK2 are also proposed receptors for the TPD1 ligand, as serk1 serk2 also results in a lack of tapetal layer, with extra meiotic cells, but biochemical evidence is lacking at present (Albrecht et al., 2005; Ge et al., 2010).
The BARELY ANY MERISTEM1 (BAM1) and BAM2 receptors have also been shown to play a role in anther and pollen development (Hord et al., 2006; Deyoung and Clark, 2008). Since the BAM receptors have been shown to directly bind with CLE family peptides, it is possible that the BAM1 and BAM2 perceive CLE1, CLE11, CLE12, CLE13, and/or CLE25 anther and pollen expressed peptides (Hord et al., 2006; Jun et al., 2010; Shinohara et al., 2012).
Embryo
Little is known about the targets of the CLE8 signaling peptide involved in embryonic development. Several RLKs have already been identified as integral to normal embryo and seed development (Luo et al., 2005; Nodine et al., 2011; Fiume and Fletcher, 2012; Shinohara et al., 2012). As there is often a strong similarity between binding affinities within CLE family peptides, it is likely that the target for CLE8 signaling is an LRR-RLK.
SAM
CLV3 is proposed to interact with four major membrane-associated protein receptors: CLV1, CLV2-CORYNE (CRN), TOAD2/RPK2 (Clark et al., 1997; Jeong et al., 1999; Müller et al., 2008), and, more recently, FLS2 (Lee et al., 2011). The CLV3 and the LRR-RLK CLV1 expression domains significantly overlap, and through competitive binding assays, CLV3 has been shown to directly bind to the CLV1 protein receptor (Fletcher and Meyerowitz, 2000; Ogawa et al., 2008; Bleckmann et al., 2010; Nimchuk et al., 2011). CLV2 has been shown to associate with the CRN protein to form a receptor-like protein complex, and CRN requires CLV2 to localize to the plasma membrane (Bleckmann et al., 2010). The CLV2-CRN complex is thought to associate with or be involved in the CLV3 signaling pathway. However, as no direct interaction between CLV3 and the CLV2 extracellular domain has been shown, the CLV2-CRN complex is seen as a secondary pathway through which apical meristem development is regulated (Nimchuk et al., 2011). The interaction with RPK2 is not well characterized: single rpk2 and clv1 clv2 rpk2 triple mutant lines display phenotypes highly similar to clv3 mutants, suggesting a functional role for RPK2 in CLV3 perception and repressing WUS expression in the meristem (Kinoshita et al., 2010). Finally, CLV3 was reported to interact with the FLS2 receptor (Lee et al., 2011; H. Lee et al., 2012), which is known to recognize the unrelated bacterial flagellin-derived peptide flg22 and activate plant innate immune responses (Gómez-Gómez and Boller, 2000). The CLV3 interaction with FLS2 in the meristem might play a role in protection of the meristem from bacterial pathogen infection (Lee et al., 2011). However, as other researchers have been unable to confirm binding of CLV3 to FLS2 under a variety of conditions (Mueller et al., 2012a, 2012b; Segonzac et al., 2012), it appears that more definitive results are necessary to support this interaction and its importance in vivo. Therefore, at present, the binding of CLV3 by FLS2 and a potential role in immune responses in the meristem remains an open question.
Another well-established receptor in floral stem cell maintenance is the BAM family of receptors: BAM1 to BAM3 genes encode a RLK with homology to CLV1 (DeYoung et al., 2006). Their homology and location in the SAM would suggest a functional similarity; however, this appears not to be the case. Single bam1, bam2, and bam3 mutants do not produce obvious phenotypes, but double and triple mutants produce striking arrested development of meristematic tissues, the opposite phenotype to clv1. The lack of phenotype with bam single mutants suggests a functional overlap between BAM1, BAM2, and BAM3 in the SAM. Interestingly, CLV1 was sufficient to replace BAM1 and BAM2 functionality in bam1 bam2, fully rescuing the phenotype to the wild type. Conversely, BAM1 and BAM2 can partially rescue the clv1 phenotype when expressed in the meristem (DeYoung et al., 2006). It is currently unknown what role BAMs play in the CLV3-CLV1 signaling pathway. BAM1 has been shown to bind with a high degree of affinity to numerous CLE peptides (CLE8 to CLE14), which are expressed during organ-specific developmental processes (Jun et al., 2010; Shinohara et al., 2012). Since CLV1 and BAM expression profiles do not overlap, it has been suggested that BAMs, located in the periphery of the meristem, may sequester CLE (or similar) peptides from entering the meristem, thereby influencing WUS expression (Deyoung and Clark, 2008; Butenko et al., 2009; Wang and Fiers, 2010).
In addition to the above receptors, TDIF RECEPTOR (TDR) appears to be a major receptor for CLE42 during axillary bud formation, but other receptors likely play a role as the tdr mutation did not completely suppress the CLE42 effect (Yaginuma et al., 2011).
Finally, the RLK ERECTA (ER) is generally associated with stomatal development but has been recently shown to be involved in inflorescence development (Uchida et al., 2012). Single er mutants show a strong compacted inflorescence phenotype, very similar to epfl4 epfl6 double mutants. Knockout experiments have shown a negligible role of ERECTA-LIKE1 (ERL1) and ERL2 during inflorescence development. Transcriptional analysis has shown that EPFL4 and EPFL6 are predominantly expressed in endodermal tissue and, through coimmunoprecipitation assays, have been shown to interact directly with ER in neighboring phloem tissue following their diffusion through cell layers (Uchida et al., 2012). Interestingly, both auxin-regulated genes, such as ARGOS that is required for meristematic competence of aboveground organs, and gibberellin metabolic genes were differentially regulated in er and epfl4 epfl6 (Uchida et al., 2012). This suggests that potential interactions between phytohormones and signaling peptides participate in non-cell-autonomous control of inflorescence development.
RAM
For the RAM, it has been suggested that CLE40 acts as a secreted peptide ligand to the RLK ARABIDOPSIS CRINKLY4 (ACR4) (Stahl et al., 2009). ACR4 is expressed in the columella stem cells, in close proximity of CLE40 (De Smet et al., 2008; Stahl et al., 2009), and cle40 and acr4 mutants have similar phenotypes, namely, an expansion in the WOX5 domain and additional layers of columella stem cells (De Smet et al., 2008; Stahl et al., 2009). As CLE40 is expressed in differentiated columella cells, the expression overlaps with an area of weak ACR4 expression (Stahl and Simon, 2009). This area of weak ACR4 expression has been shown to be upregulated by CLE40 treatment. ACR4 is thought to act as a dynamic buffer when CLE40 levels are too high. Increased ACR4 levels will sequester CLE40, allowing the expression of WOX5. WOX5 in turn expands and protects the quiescent center, counteracting the effects of CLE40 (Stahl and Simon, 2009). Alternatively, CLE40 could activate ACR4 signaling, but this remains to be demonstrated biochemically. Intriguingly, this CLE40-ACR4-WOX5 signaling pathway in the RAM is highly similar to the CLV3-CLV1-WUS signaling pathway in the SAM (Figure 2).
The peptides CLE14, CLE19, and CLE20 also appear to act along the CLV2-CRN receptor pathway in the root tip, a pathway that is prevalent in the development of the SAM. The cle14, cle19, and cle20 knockout mutants show a strong short root phenotype that is highly similar to the short root phenotype observed in clv2 and crn mutants. Thus, the CLV2-CRN receptor complex is not only vital to SAM stem cell maintenance through CLV3 signaling, but is also inherent in regulating the RAM through CLE14, CLE19, and CLE20 signaling (Meng and Feldman, 2010).
Lateral Root Initiation
Auxin plays an important role at various stages of lateral root development (De Smet, 2012). However, ACR4 was also shown to be key in the initiation and development of lateral roots (De Smet et al., 2008). ACR4 has been shown to be required for specifying the fate of neighboring cells during pericycle cell division in lateral root initiation and restricting the division of neighboring pericycle cells, thereby controlling the pattern of cell division (De Smet et al., 2008). ACR4 likely acts as signal receiver for one, or many, small signaling peptides generated in or surrounding the lateral root initiation site, such as RGF/GLV/CLEL peptides acting during lateral root initiation (Meng et al., 2012). However, the majority of known receptors for CLE peptides are LRR-RLKs (e.g., CLV1, TDR/PHLOEM INTERCALATED WITH XYLEM [PXY], and BAM), and the active domain of ACR4 has been shown to consist of extracellular crinkly repeats, resulting in a critical difference in receptor structure compared with LRR-RLKs (Gifford et al., 2005), suggesting that other, yet unidentified, signaling peptides and/or receptor kinases could be involved.
Vasculature
The receptor for TDIF has been identified as the LRR-RLK TDR/PXY (Fisher and Turner, 2007; Hirakawa et al., 2008; Etchells and Turner, 2010). TDR/PXY was identified through selecting 56 LRR RLK genes predicted to be expressed in procambium cells (Hirakawa et al., 2008). From this gene set, three T-DNA insertion lines in the same gene were found to be insensitive to TDIF, and this gene, previously described as PXY, was designated as TDR (Fisher and Turner, 2007; Hirakawa et al., 2008). TDR/PXY is primarily expressed in procambium cells, while TDIF is expressed in adjacent phloem cells, in nonoverlapping expression domains. CLE41 must then be secreted toward the xylem where it is received by TDR/PXY, to transmit the correct positional information (Etchells and Turner, 2010; Hirakawa et al., 2011). The TDIF-TDR/PXY pathway controls the WUS-related HOMEOBOX gene, WOX4, which is required for controlling the fates of the vasculature cell fates (Suer et al., 2011) (Figure 3). WOX4 is essential for procambial/cambial cell proliferation, as wox4 mutants show restricted proliferation; however, it is not essential for the inhibition of cambial cells into xylem cells. Thus, two pathways can be distinguished, namely, a TDIF-TDR/PXY-WOX4 pathway, which promotes procambial/cambial stem cell proliferation, and TDIF-unknown factors, which inhibits the differentiation of cambial cells into xylem cells (Hirakawa et al., 2010; Suer et al., 2011). Both TDIF pathways, however, show crosstalk between xylem and phloem cells, as the TDIF derived from phloem is then responsible for the maintenance of the procambial/cambial cells via their differentiation and proliferation capabilities (Hirakawa et al., 2011).
Finally, the signaling peptide CLE10 is thought to act through the CLV2 receptor. CLE10 expression normally inhibits protoxylem formation in roots; however, in clv2 knockout lines, protoxylem formation is no longer inhibited, suggesting a link between the CLE10 signaling peptide and the CLV2 receptor (Kondo et al., 2011).
Stomata
Several receptor-like proteins and RLKs, such as TOO MANY MOUTHS (TMM), ER, and ERL are involved in stomatal development (Yang and Sack, 1995; Shpak et al., 2005). Through coimmunoprecipitation assays of tagged proteins in Nicotiana benthamiana, it has been shown that EPF1 and EPF2 strongly bind to ER and ERL1 receptors and that EPF2 also binds to TMM (J.S. Lee et al., 2012). Since TMM lacks an intracellular domain, this suggests that signal transduction occurring through TMM requires the interaction with a protein with a functional intracellular domain (Shpak et al., 2005). Indeed, TMM has been shown to interact directly with the ER receptor, thereby influencing EPF signaling through signal modulation in a putative ER-TMM complex (J.S. Lee et al., 2012). STOMAGEN/EPFL9, the EPFL peptide with opposite function to EPF2, has also been shown to require TMM to function (Shimada et al., 2011). These two signals acting antagonistically on the TMM receptor may in fact be a novel regulatory system, as this is the first time two antagonistic peptides have been shown to work through the same receptor in a plant system (Shimada et al., 2011). CHALf peptides have also been shown to act through the TMM receptor (Abrash et al., 2011). In a tmm background, CHALf overexpression leads to a striking reduction in stomatal density, suggesting that TMM binds CHALf to buffer signaling and block transmission into stomatal lineage cells. CHALf overexpression phenotypes are also mediated by ER family mutations, suggesting an additional pathway through which CHALf signaling is moderated (Abrash et al., 2011).
Finally, BAM receptors have also been identified in aerial organs during stomatal development and bind with high affinity to peptides like CLE9. As CLE9 is expressed during stomatal development, it is likely that BAMs are functionally involved in stomatal development (Jun et al., 2010; Shinohara et al., 2012).
Abscission
IDA and IDL peptides are thought to act as the peptide ligand to the LRR-RLK HAESA (HAE) and to a family of HAESA-LIKE (HSL) receptor kinases, which have been shown to be expressed in floral abscission zones. While the hae and hsl2 mutants were indistinguishable from wild-type plants, the hae hsl2 double mutants completely lacked floral abscission. The highly similar expression patterns of HAE and HSL2, along with the double mutant results, suggest a functional redundancy within this family of receptors. Interestingly, hae hsl2 plants overexpressing IDA showed all the characteristics of the double mutant phenotype and none of the IDA overexpression phenotype. In addition, exposure of hae hsl2 to the synthetic IDA EPIP-C, IDA EPIP, and IDL1 EPIP peptides did not induce floral abscission. Taken together, this suggests that IDA and HAE-HSL2 may act along the same pathway or may in fact be ligand-receptor pairs (Stenvik et al., 2008). IDA possibly regulates floral organ abscission by acting as a ligand in HAE-HSL2–mediated suppression of BP/KNAT1, which are repressors of the abscission-promoting transcription factors KNAT2 and KNAT6 (Shi et al., 2011).
CONCLUSION AND FUTURE PERSPECTIVES
In the past decade, our knowledge of small peptide signals has increased enormously, not only with respect to their identification through biochemical assays or in silico predictions but, more importantly, also at the level of their functional characterization. However, both genetic and bioassay approaches have drawbacks since many signaling peptides are subject to functional redundancy, hindering the genetic approach, and the typically low physiological concentrations of peptide hormones in tissues may evade detection through bioassay methods. Nonetheless, an increased focus on obtaining knockout mutants, for example, through TILLING, will be essential to elucidate the specific role(s) of peptide signals in developmental processes. Loss-of-function data are especially important since exogenous application of peptides and broad overexpression often results in nonspecific or unrelated phenotypes.
Many of the enzymes involved in posttranslational modification of peptide signals, and virtually all of the enzymes involved in proteolytic processing, remain unknown (Table 1). Given their importance for correct activation of many known small peptide signals, the identification of the genes and/or gene families that encode these enzymes could prove important not only for a more complete understanding of known peptide signals but also give a useful starting point for discovering new peptide signals through genetic methods.
Furthermore, a major gap in our knowledge is that of the enormous number of possible peptide ligand-receptor (kinase) pairs, so far only very few have been shown to form pairs (Butenko et al., 2009). While peptide signals can be matched to their corresponding receptor through a variety of methods, eventually a biochemical approach is required to demonstrate physical interaction. To achieve this, methods need to be applied to investigate these interactions on a larger scale and tools to study the in vivo dynamics of peptide ligand−receptor and receptor−receptor interactions need to be developed. It has been shown on numerous occasions that one RLK can bind several signaling molecules within a peptide family, showing a strong functional redundancy between peptide signals. Conversely, it has been shown that some receptors may functionally replace one another, when expressed in non-native expression profiles. Understanding why and how these proteins can act interchangeably will help to elucidate further peptide signaling pathways whose ligand or receptor pairs are currently unknown. Such an approach will likely further support the observations that there is not a single peptide-receptor pair but that multiple possible interactions exist. Identifying peptide ligand-receptor (kinase) pairs will furthermore provide insight in the (distinct) roles of the numerous peptides in overlapping expression domains. However, the debate on the CLV3-FLS2 pair (Lee et al., 2011; H. Lee et al., 2012; Mueller et al., 2012a, 2012b; Segonzac et al., 2012) illustrates the complexities of peptide signaling and the difficulties of working with these small peptides. In general, conclusions need to take into account experimental setup, differential effects of posttranslational modifications, sensitivity of the receptor for the ligand, and even inadvertent contamination of synthetic peptides.
As mentioned above, knowledge of crosstalk between phytohormones and signaling peptides is only just emerging, with a focus on auxin and cytokinin. For example, RGF/GLV/CLEL peptides influence auxin transport during root and hypocotyl gravitropism by modulating PIN2 trafficking dynamics, ensuring that the asymmetrical auxin gradient required for the gravitropic response is maintained. Additionally, GLV1 and GLV2 expression is rapidly induced by auxin, providing an example of an auxin signaling input modulating an auxin transport output via a signaling peptide, thus controlling the initial trigger (Whitford et al., 2012). In addition, CLE peptides are regulated through cytokinin signaling during root and vasculature development (Meng and Feldman, 2010). The exact nature of this crosstalk remains to be elucidated.
In conclusion, small signaling peptides play an essential role in plant growth and development, but we have only revealed the tip of the iceberg with respect to their signaling potential, including the interaction with receptors and downstream changes.
Acknowledgments
This work was supported by a Biotechnology and Biological Science Research Council David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration grant (PERG06-GA-2009-256354) to I.D.S. We thank the School of Biosciences and Malcolm. J. Bennett for studentship funding and acknowledge the University of Nottingham research committee. S.S. received a Biotechnology and Biological Science Research Council Doctoral Training Grant studentship.
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
References
- Abrash E.B., Bergmann D.C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Abrash E.B., Davies K.A., Bergmann D.C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Albert M., Jehle A.K., Mueller K., Eisele C., Lipschis M., Felix G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285: 19035–19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C., Russinova E., Hecht V., Baaijens E., de Vries S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P.A., Pearce G., Covey P.A. (2010). RALFs: Peptide regulators of plant growth. Plant Signal. Behav. 5: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Buechel S., Leibfried A., To J.P., Zhao Z., Andersen S.U., Kieber J.J., Lohmann J.U. (2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89: 279–284 [DOI] [PubMed] [Google Scholar]
- Busch W., Benfey P.N. (2010). Information processing without brains—The power of intercellular regulators in plants. Development 137: 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M. (2009). Plant peptides in signalling: Looking for new partners. Trends Plant Sci. 14: 255–263 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1995). Clavata3 is a specific regulator of shoot and floral meristem development affecting the same processes as Clavata1. Development 121: 2057–2067 [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cock J.M., McCormick S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H.C., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- De Smet I. (2012). Lateral root initiation: One step at a time. New Phytol. 193: 867–873 [DOI] [PubMed] [Google Scholar]
- De Smet I., Beeckman T. (2011). Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12: 177–188 [DOI] [PubMed] [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis - The humble beginnings of plant life. Plant J. 61: 959–970 [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voβ U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2012). Analyzing lateral root development: How to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B.J., Bickle K.L., Schrage K.J., Muskett P., Patel K., Clark S.E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45: 1–16 [DOI] [PubMed] [Google Scholar]
- DeYoung B.J., Clark S.E. (2001). Signaling through the CLAVATA1 receptor complex. Plant Mol. Biol. 46: 505–513 [DOI] [PubMed] [Google Scholar]
- Deyoung B.J., Clark S.E. (2008). BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Bergmann D.C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91: 267–297 [DOI] [PubMed] [Google Scholar]
- Elo A., Immanen J., Nieminen K., Helariutta Y. (2009). Stem cell function during plant vascular development. Semin. Cell Dev. Biol. 20: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Etchells J.P., Turner S.R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., van der Schors R., van der Geest L., Li K.W., Stiekema W.J., Liu C.M. (2006). The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 141: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C.M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., van der Geest L., van Lookeren Campagne M., Liu C.M. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fiers M., Ku K.L., Liu C.M. (2007). CLE peptide ligands and their roles in establishing meristems. Curr. Opin. Plant Biol. 10: 39–43 [DOI] [PubMed] [Google Scholar]
- Fisher K., Turner S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fiume E., Fletcher J.C. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24: 1000–1012
- Fiume E., Monfared M., Jun J., Fletcher J.C. (2011). CLE polypeptide signaling gene expression in Arabidopsis embryos. Plant Signal. Behav. 6: 443–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Meyerowitz E.M. (2000). Cell signaling within the shoot meristem. Curr. Opin. Plant Biol. 3: 23–30 [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Ge X., Chang F., Ma H. (2010). Signaling and transcriptional control of reproductive development in Arabidopsis. Curr. Biol. 20: R988–R997 [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Robertson F.C., Soares D.C., Ingram G.C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R.B., Beals T.P., Sanders P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Graham M.A., Silverstein K.A., Cannon S.B., VandenBosch K.A. (2004). Computational identification and characterization of novel genes from legumes. Plant Physiol. 135: 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai H., Nakayama D., Yang H., Matsubayashi Y., Hirota Y., Sakagami Y. (2000). Existence of a plant tyrosylprotein sulfotransferase: Novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine variants in vitro. FEBS Lett. 470: 97–101 [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Haruta M., Constabel C.P. (2003). Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol. 131: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Monshausen G., Gilroy S., Sussman M.R. (2008). A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: Identification of AtRALF1 peptide. Biochemistry 47: 6311–6321 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2011). Establishment and maintenance of vascular cell communities through local signaling. Curr. Opin. Plant Biol. 14: 17–23 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe M., Müller R., Grünewald M., Brand U., Simon R. (2003). Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213: 371–381 [DOI] [PubMed] [Google Scholar]
- Hord C.L., Chen C., Deyoung B.J., Clark S.E., Ma H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Bailey K.J., Gray J.E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186: 609–614 [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H.A., Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 105: 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn T.L., Stone J.M., Walker J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A.H., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Davies K.A., Bergmann D.C., Laux T. (2011). Peptide signaling in plant development. Curr. Biol. 21: R356–R364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.A., Shimosato-Asano H., Keinath N.F., Wuest S.E., Ingram G., Panstruga R., Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kodadek T. (2001). Protein microarrays: Prospects and problems. Chem. Biol. 8: 105–115 [DOI] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. (2009). Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 15067–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., Masuda Y., Irie K., Tanaka Y., Takada S., Kakimoto T., Sakagami Y. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51: 1–8 [DOI] [PubMed] [Google Scholar]
- Kondo T., Nakamura T., Yokomine K., Sakagami Y. (2008). Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem. Biophys. Res. Commun. 377: 312–316 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Hirakawa Y., Kieber J.J., Fukuda H. (2011). CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Vaneden J., Hanhart C.J., Stam P., Braaksma F.J., Feenstra W.J. (1983). Linkage map of Arabidopsis thaliana. J. Hered. 74: 265–272 [Google Scholar]
- Lau S., Slane D., Herud O., Kong J., Jürgens G. (2012). Early embryogenesis in flowering plants: Setting up the basic body pattern. Annu. Rev. Plant Biol. 63: 483–506 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chah O.K., Sheen J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Khatri A., Plotnikov J.M., Zhang X.-Z., Sheen J. (2012). Complexity in differential peptide-receptor signaling: Response to Segonzac et al. and Mueller et al. commentaries. Plant Cell 24: 3177–3185
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M., Laux T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130: 3163–3173 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Howell S. (2009). Overexpression of an Arabidopsis gene encoding a subtilase (AtSBT5.4) produces a clavata-like phenotype. Planta 230: 687–697 [DOI] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J.L., Fiori C.S., Silva-Filho M.C., Moura D.S. (2008). A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 582: 3343–3347 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011a). Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011b). Small post-translationally modified peptide signals in Arabidopsis. The Arabidopsis Book 9: e0150, doi/10.1199/tab0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y. (2012). MBSJ MCC Young Scientist Award 2010. Recent progress in research on small post-translationally modified peptide signals in plants. Genes Cells 17: 1–10 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Hanai H., Hara O., Sakagami Y. (1996). Active fragments and analogs of the plant growth factor, phytosulfokine: Structure-activity relationships. Biochem. Biophys. Res. Commun. 225: 209–214 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Kihara H., Niwa M., Sakagami Y. (2006). Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Takagi L., Sakagami Y. (1997). Phytosulfokine-alpha, a sulfated pentapeptide, stimulates the proliferation of rice cells by means of specific high- and low-affinity binding sites. Proc. Natl. Acad. Sci. USA 94: 13357–13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meindl T., Boller T., Felix G. (1998). The plant wound hormone systemin binds with the N-terminal part to its receptor but needs the C-terminal part to activate it. Plant Cell 10: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Feldman L.J. (2010). CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Buchanan B.B., Feldman L.J., Luan S. (2012). CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Ruth K.C., Fletcher J.C., Feldman L. (2010). The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol. Plant 3: 760–772 [DOI] [PubMed] [Google Scholar]
- Mingossi F.B., Matos J.L., Rizzato A.P., Medeiros A.H., Falco M.C., Silva-Filho M.C., Moura D.S. (2010). SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol. Biol. 73: 271–281 [DOI] [PubMed] [Google Scholar]
- Mueller K., Bittel P., Chinchilla D., Jehle A.K., Albert M., Boller T., Felix G. (2012a). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Chinchilla D., Albert M., Jehle A.K., Kalbacher H., Boller T., Felix G. (2012b). Inadvertent cross-contamination as a risk for work with synthetic peptides: flg22 as an example of a pirate peptide in commercial peptide preparations. Plant Cell 24: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Borghi L., Kwiatkowska D., Laufs P., Simon R. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J. (2003). Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 22: 15–24 [DOI] [PubMed] [Google Scholar]
- Nadeau J.A. (2009). Stomatal development: New signals and fate determinants. Curr. Opin. Plant Biol. 12: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Clark S.E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Guo Y., Jin H., Hartsell J., Clark S.E. (2011). Characterization of a CLE processing activity. Plant Mol. Biol. 75: 67–75 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z.L., Tarr P.T., Ohno C., Qu X.A., Meyerowitz E.M. (2011). Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Bryan A.C., Racolta A., Jerosky K.V., Tax F.E. (2011). A few standing for many: Embryo receptor-like kinases. Trends Plant Sci. 16: 211–217 [DOI] [PubMed] [Google Scholar]
- Oelkers K., Goffard N., Weiller G.F., Gresshoff P.M., Mathesius U., Frickey T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohki S., Takeuchi M., Mori M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2: 512. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. (2008). Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Mundy J., Skriver K. (2002). Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. In Silico Biol. (Gedrukt) 2: 441–451 [PubMed] [Google Scholar]
- Ottmann C., Rose R., Huttenlocher F., Cedzich A., Hauske P., Kaiser M., Huber R., Schaller A. (2009). Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc. Natl. Acad. Sci. USA 106: 17223–17228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. (2001a). Production of multiple plant hormones from a single polyprotein precursor. Nature 411: 817–820 [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A., Jr (2001b). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897 [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Rautengarten C., Steinhauser D., Büssis D., Stintzi A., Schaller A., Kopka J., Altmann T. (2005). Inferring hypotheses on functional relationships of genes: Analysis of the Arabidopsis thaliana subtilase gene family. PLOS Comput. Biol. 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M.H., Bergmann D.C. (2010). Complex signals for simple cells: The expanding ranks of signals and receptors guiding stomatal development. Curr. Opin. Plant Biol. 13: 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A. (1974). Assay and biochemical properties of the proteinase inhibitor-inducing factor, a wound hormone. Plant Physiol. 54: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A., Oecking C. (1999). Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W.L., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Segonzac C., Nimchuk Z.L., Beck M., Tarr P.T., Robatzek S., Meyerowitz E.M., Zipfel C. (2012). The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell 24, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L. (2009). Cell fate transitions during stomatal development. Bioessays 31: 865–873 [DOI] [PubMed] [Google Scholar]
- Sharma V.K., Ramirez J., Fletcher J.C. (2003). The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Shi C.L., Stenvik G.E., Vie A.K., Bones A.M., Pautot V., Proveniers M., Aalen R.B., Butenko M.A. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Sugano S.S., Hara-Nishimura I. (2011). Positive and negative peptide signals control stomatal density. Cell. Mol. Life Sci. 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Moriyama Y., Ohyama K., Matsubayashi Y. (2012). Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70: 845–854 [DOI] [PubMed] [Google Scholar]
- Shpak E.D., McAbee J.M., Pillitteri L.J., Torii K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Sieburth L.E., Deyholos M.K. (2006). Vascular development: The long and winding road. Curr. Opin. Plant Biol. 9: 48–54 [DOI] [PubMed] [Google Scholar]
- Silverstein K.A., Graham M.A., Paape T.D., VandenBosch K.A. (2005). Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 138: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A., Jr, Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Smith S., De Smet I. (2012). Root system architecture: Insights from Arabidopsis and cereal crops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Guo H., Yin Y., Howell S.H. (2009). Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Howell S.H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56: 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. (2005). Plant stem cell niches. Int. J. Dev. Biol. 49: 479–489 [DOI] [PubMed] [Google Scholar]
- Stahl Y., Simon R. (2009). Is the Arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signal. Behav. 4: 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stenvik G.E., Butenko M.A., Urbanowicz B.R., Rose J.K., Aalen R.B. (2006). Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.E., Tandstad N.M., Guo Y., Shi C.L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strabala T.J., O’donnell P.J., Smit A.M., Ampomah-Dwamena C., Martin E.J., Netzler N., Nieuwenhuizen N.J., Quinn B.D., Foote H.C., Hudson K.R. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann J., Scheer J., Ryan C.A. (2000). Suramin inhibits initiation of defense signaling by systemin, chitosan, and a beta-glucan elicitor in suspension-cultured Lycopersicon peruvianum cells. Proc. Natl. Acad. Sci. USA 97: 8862–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suer S., Agusti J., Sanchez P., Schwarz M., Greb T. (2011). WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23: 3247–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Torii K.U. (2004). Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int. Rev. Cytol. 234: 1–46 [DOI] [PubMed] [Google Scholar]
- Uchida N., Lee J.S., Horst R.J., Lai H.H., Kajita R., Kakimoto T., Tasaka M., Torii K.U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109: 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Weisbeek P., Scheres B. (1998). Cell fate and cell differentiation status in the Arabidopsis root. Planta 205: 483–491 [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Van Norman J.M., Breakfield N.W., Benfey P.N. (2011). Intercellular communication during plant development. Plant Cell 23: 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Fiers M. (2010). CLE peptide signaling during plant development. Protoplasma 240: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R., et al. (2012). GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev. Cell 22: 678–685 [DOI] [PubMed] [Google Scholar]
- Wu S., Gallagher K.L. (2011). Mobile protein signals in plant development. Curr. Opin. Plant Biol. 14: 563–570 [DOI] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma H., Hirakawa Y., Kondo Y., Ohashi-Ito K., Fukuda H. (2011). A novel function of TDIF-related peptides: Promotion of axillary bud formation. Plant Cell Physiol. 52: 1354–1364 [DOI] [PubMed] [Google Scholar]
- Yang H., Matsubayashi Y., Hanai H., Sakagami Y. (2000). Phytosulfokine-alpha, a peptide growth factor found in higher plants: Its structure, functions, precursor and receptors. Plant Cell Physiol. 41: 825–830 [DOI] [PubMed] [Google Scholar]
- Yang H., Matsubayashi Y., Nakamura K., Sakagami Y. (1999). Oryza sativa PSK gene encodes a precursor of phytosulfokine-alpha, a sulfated peptide growth factor found in plants. Proc. Natl. Acad. Sci. USA 96: 13560–13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Sack F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.L., Xie L.F., Mao H.Z., Puah C.S., Yang W.C., Jiang L., Sundaresan V., Ye D. (2003). Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. (2009). Control of anther cell differentiation: A teamwork of receptor-like kinases. Sex. Plant Reprod. 22: 221–228 [DOI] [PubMed] [Google Scholar]
- Zhou W., Wei L., Xu J., Zhai Q., Jiang H., Chen R., Chen Q., Sun J., Chu J., Zhu L., Liu C.M., Li C. (2010). Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]