In rice, the two florigens Hd3a and RFT1 are separated by only 11.5 kb in the genome. This study reveals that rice SDG724 specifically affects the histone H3 lysine 36 me2/3 level of RFT1 but not its close paralog Hd3a. Therefore, RFT1 and Hd3a have functionally diverged to control flowering time under long-day and short-day conditions partly via a fine-tuned epigenetic mechanism.
Abstract
Chromatin modifications affect flowering time in the long-day plant Arabidopsis thaliana, but the role of histone methylation in flowering time regulation of rice (Oryza sativa), a short-day plant, remains to be elucidated. We identified a late-flowering long vegetative phase1 (lvp1) mutant in rice and used map-based cloning to reveal that lvp1 affects the SET domain group protein 724 (SDG724). SDG724 functions as a histone methyltransferase in vitro and contributes to a major fraction of global histone H3 lysine 36 (H3K36) methylation in vivo. Expression analyses of flowering time genes in wild-type and lvp1 mutants revealed that Early heading date1, but not Heading date1, are misregulated in lvp1 mutants. In addition, the double mutant of lvp1 with photoperiod sensitivity5 (se5) flowered later than the se5 single mutant, indicating that lvp1 delays flowering time irrespective of photoperiod. Chromatin immunoprecipitation assays showed that lvp1 had reduced levels of H3K36me2/3 at MADS50 and RFT1. This suggests that the divergent functions of paralogs RFT1 and Hd3a, and of MADS50 and MADS51, are in part due to differential H3K36me2/3 deposition, which also correlates with higher expression levels of MADS50 and RFT1 in flowering promotion in rice.
INTRODUCTION
Flowering time is affected by both endogenous genetic factors and exogenous environmental signals. In Arabidopsis thaliana, a complex genetic network controlling the transition to flowering consists of four pathways, the autonomous, photoperiod, vernalization, and gibberellin pathways (Komeda, 2004). In the short-day (SD) plant rice (Oryza sativa), flowering (heading date) is an important agronomic trait for climatic and regional adaption; breeding for this trait allowed rice varieties to be selected for growth at various latitudes, altitudes, and seasons (Izawa, 2007a). Thus, heading date studies are essential for understanding the genetic and biochemical mechanism of flowering and have a significant impact on sustainable agricultural production of rice.
There are at least two independent flowering pathways in rice. The Heading date1 (Hd1) pathway is conserved between rice and Arabidopsis, but the Early heading date1 (Ehd1) pathway is unique to rice (Doi et al., 2004). In rice, homologs of CONSTANS, FLOWERING LOCUS T (FT), and GIGANTEA (GI) are Hd1, Heading date 3a (Hd3a), and Os GI, respectively (Yano et al., 2000; Hayama et al., 2002, 2003; Kojima et al., 2002). Although Os GI regulates Hd1, as in Arabidopsis, Hd1 plays a more enigmatic role in rice. Hd1 represses flowering under long-day (LD) conditions but promotes flowering slightly under SD conditions, by controlling the expression of Hd3a (Hayama et al., 2003; Tamaki et al., 2007; Komiya et al., 2008). Ehd1, which encodes a B-type response regulator, is a unique transcriptional regulator and promotes flowering by controlling FT-like gene expression independent of Hd1 under both SD and LD conditions in rice (Doi et al., 2004; Izawa, 2007a). Recently, mounting evidence indicates that Ehd1 is an important integrator of the floral transition in rice. Several genes belonging to multiple pathways that control the expression level of Ehd1 have been identified (Komiya et al., 2009). PHOTOPERIOD SENSITIVITY5 (SE5), the rice homolog of Arabidopsis HEME OXYGENASE1, encodes a key heme oxygenase involved in phytochrome chromophore biosynthesis (Izawa et al., 2000, 2002). SE5 acts as a floral repressor and delays heading date mainly through suppressing Ehd1 expression (Izawa, 2007b; Andrés et al., 2009). Rice MADS50, a homolog of the Arabidopsis gene SUPPRESSOR OF OVEREXPRESSION OF CO1, is upstream of Ehd1 but works either parallel with or downstream of Os GI (Lee et al., 2004; Ryu et al., 2009). Rice MADS51 is a type I MADS box gene that acts downstream of GI and upstream of Ehd1 and promotes flowering transition under SD conditions (Kim et al., 2007). Ehd2 (also known as ID1/RID1), which encodes a Cys-2/His-2 zinc-finger transcription factor, is a homolog of maize Indeterminate1 (ID1) and promotes flowering under both SD and LD conditions by upregulating expression of Ehd1 (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). Grain number, plant height, and heading date7 (Ghd7) encodes a CCT domain protein, acts upstream of Ehd1, and delays flowering under LD conditions (Xue et al., 2008). Florigen, the mobile signal that moves from an induced leaf to the shoot apex and activates the floral transition at the apical meristem, remained elusive for over 70 years (Knott, 1934) but has now been proposed to be the FT protein in both Arabidopsis and rice (Corbesier et al., 2007; Tamaki et al., 2007). In rice, RICE FLOWERING LOCUS T1 (RFT1) and Hd3a, which are two homologs of FT, were shown to qualify as florigen genes (Komiya et al., 2009; Tsuji et al., 2011). In rice plants with suppressed activity of these two genes, no heading was under either SD or LD conditions (Komiya et al., 2009; Tsuji et al., 2011). Furthermore, many regulators of the RFT1 florigen, such as MADS50, Ehd1, Hd1, Ghd7, and Ehd2/ID1/RID1, form a LD flowering regulatory network in rice (Komiya et al., 2009).
Chromatin structure is important for eukaryotic gene expression, and histone Lys methylation has drawn special attention due to its complex role in this process (Wu et al., 2009; Qiao et al., 2011). Histone Lys methylation in plants functions in biological processes such as flowering transition, floral organ development, carotenoid biosynthesis, shoot and root branching, pollen and macro-trichome development, and the brassinosteroid signaling pathway (Kim et al., 2005; Zhao et al., 2005; Y. Ding et al., 2007; Dong et al., 2008; Xu et al., 2008; Cazzonelli et al., 2009a, 2009b; Grini et al., 2009; Berr et al., 2011; Deal and Henikoff, 2011; Feng and Jacobsen, 2011; Thorstensen et al., 2011; Sui et al., 2012). In Arabidopsis, FLOWERING LOCUS C (FLC) and other flowering time genes are regulated by different kinds of histone Lys methylations (Michaels and Amasino, 1999; He and Amasino, 2005; He, 2009; Berr et al., 2011; Deal and Henikoff, 2011; Feng and Jacobsen, 2011; Thorstensen et al., 2011). SET domain proteins, named after Su(var)3-9, E (z), and Trithorax (TRX), are bona fide histone Lys methyltransferases (HMTase) that modify specific histone Lys residues (Ng et al., 2007; Thorstensen et al., 2011). Histone H3 lysine 36 (H3K36) methylation is likely linked to transcription elongation, and H3K36 HMTase genes are involved in regulating the floral transition of Arabidopsis (Thorstensen et al., 2011). For example, lesions in SET DOMAIN GENE8 (SDG8) cause early flowering due to a decrease in H3K4 and H3K36 methylation levels at the FLC locus (Kim et al., 2005; Zhao et al., 2005; Ko et al., 2010), and lesions in SDG25/ARABIDOPSIS TRITHORAX-RELATED7 also promote flowering due to a suppression of FLC expression (Berr et al., 2009; Tamada et al., 2009).
Currently, there is little evidence that chromatin modification participates in the flowering transition of rice. Previously, we characterized SDG714, a histone H3K9 HMTase coding gene that did not affect flowering time in rice (Y. Ding et al., 2007). Here, we report that a major HMTase-encoding gene, Long vegetative phase 1 (LVP1)/SDG724, is required for H3K36 methylation and promotes heading date in rice. The loss-of-function mutant lvp1 has a late flowering phenotype under both LD and SD conditions, associated with the suppressed expression of MADS50, MADS51, Ehd1, RFT1, and Hd3a. Furthermore, our results suggest a novel mechanism for the epigenetic regulation of flowering in rice, in which SDG724 mediates H3K36me2/3 deposition at the MADS50 and RFT1 loci and promotes flowering through MADS50/MADS51-Ehd1-Hd3a/RFT1 pathways.
RESULTS
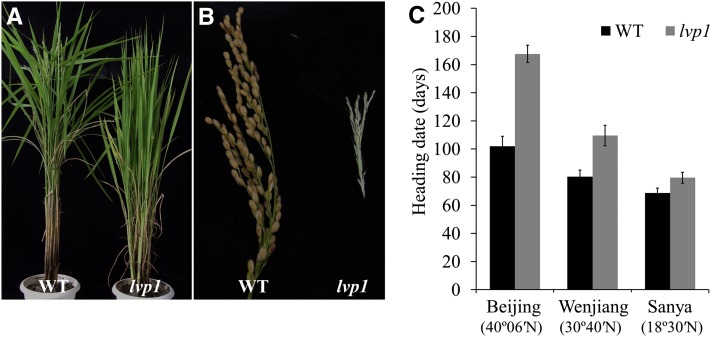
The lvp1 Mutant Has a Daylength-Independent Long Vegetative Phase
We previously generated a large population of T-DNA rice mutants in the Nipponbare cv background (Ma et al., 2009), which we screened under natural LD conditions for flowering time mutants in Beijing, China. One of the late-heading mutants identified was named long vegetative phase1 (lvp1; referred to as lvp1-1). The phenotype of lvp1 did not cosegregate with the T-DNA insertion, and an lvp1 plant without the T-DNA insertion was isolated for further analysis. Under Beijing field conditions, lvp1 plants did not show heading even in November, 160 d after germination, when the weather became too cold for rice growth (Figure 1). Wild-type and lvp1 plants were then grown in three locations with different latitudes, Sanya (18°30′N, 110°01′E, SD), Wenjiang (30°40′N, 103°51′E, LD), and Beijing (40°06′N, 116°24′E, LD). The rice growing season in Sanya coincided with a natural SD photoperiod, the one in Beijing with a natural LD photoperiod, and the one in Wenjiang with an intermediate photoperiod between Sanya and Beijing. We surveyed the heading dates of wild-type and lvp1 plants in all three locations and found that the heading dates of lvp1 were delayed in all three locations compared with the wild type: from 102 to 168 d in Beijing, from 80 to 110 d in Wenjiang, and from 69 to 80 d in Sanya (Figure 1C). We also grew lvp1 mutant and wild-type plants under SD (10 h light/14 h dark) and LD conditions (14 h light/10 h dark) in growth chambers. Under SD conditions, the flowering time of the lvp1 mutant was 76 d, 12 d later than for wild-type plants (64 d; see Supplemental Figure 1 online). Under LD conditions, the lvp1 mutant did not flower after more than 200 d, whereas wild-type plants flowered after 82 d (see Supplemental Figure 1 online). These data suggest that the LVP1 gene plays an important role in the floral transition under both LD and SD conditions in rice. However, the difference in flowering time between wild-type and lvp1 plants was smaller under SD conditions, although the mutation delayed flowering time dramatically under LD conditions.
Figure 1.
Phenotype of the lvp1 Mutant.
(A) Phenotypes of 110-d-old wild-type (WT; left) and lvp1 (right) mutant plants grown in a paddy field at Beijing.
(B) Panicles of 150-d-old wild-type (left) and lvp1 plants (right) plants grown in Beijing. The lvp1 mutant was not heading at this time and only developed immature panicles.
(C) Heading date investigation of wild-type and lvp1 plants grown at three places of different latitudes.
Error bars indicate sd (n = 20).
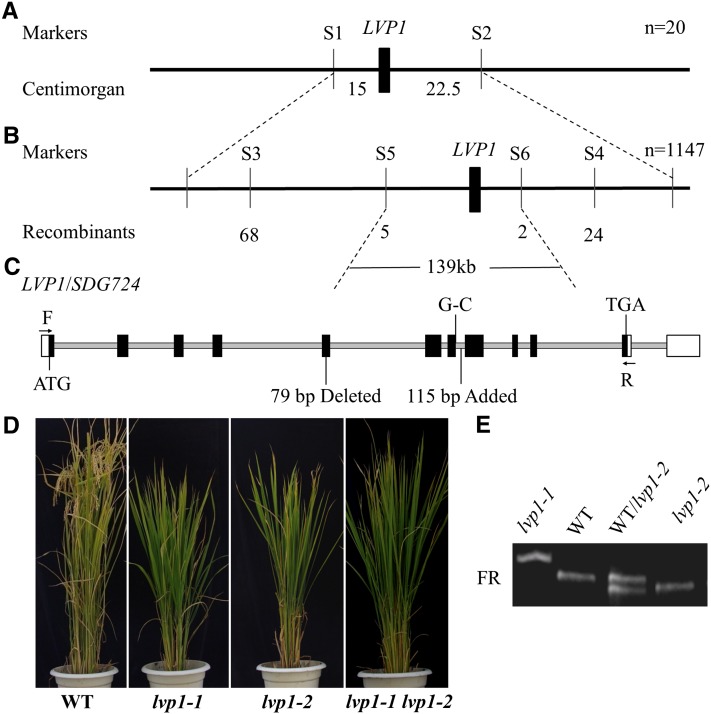
Map-Based Cloning of LVP1
Genetic analysis demonstrated that the late flowering phenotype of lvp1 segregated as a complete monogenic recessive trait. Therefore, we carefully selected 1147 extremely late-heading plants from an F2 population derived from a cross between lvp1 and Minghui 63 and used a map-based cloning strategy to identify the candidate gene. Using simple sequence repeat and sequence-tagged site (STS) markers, we initially mapped the candidate gene to a 6-Mb genomic region between markers S1 and S2 on chromosome 9 (Figure 2A) and then narrowed down the gene to an ∼139-kb genomic region (Figure 2B). According to a rice annotation project database (http://rapdb.dna.affrc.go.jp/), there are 16 putative genes in this region, coding for three hypothetical proteins, 11 retrotransposon proteins, one SET domain–containing protein, and one zinc-finger protein. RT-PCR results showed that the transcribed fragment of the SDG Os09g0307800 (SDG724) was longer in the lvp1-1 mutant than in wild-type plants (Figure 2E). Sequencing results revealed that an AG-to-AC substitution occurred at the splice site between the seventh intron and exon of SDG724 and that the entire 115 bp of the seventh intron was retained in lvp1-1 derived transcripts, leading to a premature stop codon in the transcript of the lvp1-1 allele (Figure 2C). The AG/AC substitution in SDG724 was confirmed in genomic DNA of lvp1 mutant plants.
Figure 2.
Map-Based Cloning of LVP1.
(A) Primary mapping of LVP1 localized the gene to the region between the molecular markers S1 and S2 on chromosome 9.
(B) Fine mapping of LVP1 using 1147 F2 progeny plants further delimited the gene to a 139-kb genomic region on chromosome 9.
(C) Genomic structure of LVP1/SDG724. Black boxes show exons, white boxes show untranslated regions, and brown lines show introns. “G-C” and “115 bp Added” describe the mutation of the lvp1-1 allele; “79 bp Deleted” describes the mutation of the lvp1-2 allele. F and R indicate the primer pair used in (E) and are listed in Supplemental Table 1 online.
(D) Allelism test between lvp1-1 and lvp1-2. WT, wild type.
(E) Genotyping of SDG724 transcripts in wild-type, lvp1-1, and lvp1-2 plants by RT-PCR. The FR primer pairs are indicated in (C) and listed in Supplemental Table 1 online.
Further screening of our mutant rice population yielded a second allele, lvp1-2, in which the fifth exon of the SDG724 transcript was deleted (Figures 2C and 2E). The lvp1-2 allele flowered as late as lvp1-1 plants under both SD and LD conditions. Crosses between lvp1-1 and lvp1-2 mutants produced late flowering in both in F1 and F2 populations, confirming that the two mutants are allelic (Figure 2D).
To confirm that the lesions in SDG724 were responsible for the late-flowering phenotype of lvp1 mutants, a genomic fragment containing the entire 6631-bp SDG724 coding sequence and 1963-bp 5′-upstream region and 1346-bp 3′-downstream noncoding region was obtained from the BAC clone AP006235 and transformed into lvp1-1 mutant plants. We obtained 30 transgenic lines, as confirmed by PCR. Heading date analyses showed that the flowering time defect was rescued in the transgenic plant lines (see Supplemental Figure 2 online). Taken together, these results confirmed that lesions in SDG724 were responsible for the late-flowering phenotype of lvp1 plants.
SDG724 Encodes a SET Domain–Containing Protein
SET domain–containing proteins are well annotated and characterized in Arabidopsis (Springer et al., 2003; He, 2009; Liu et al., 2010; Berr et al., 2011; Deal and Henikoff, 2011; Feng and Jacobsen, 2011; Thorstensen et al., 2011). In the rice genome, at least 35 SET protein coding genes were annotated (Ng et al., 2007), but few have been characterized (Liang et al., 2003; Thakur et al., 2003; B. Ding et al., 2007; Y. Ding et al., 2007; Qin et al., 2010; Sui et al., 2012). The plant SET domain–containing proteins have been divided into seven classes. SDG724 is considered a class II SET domain protein. Class II members possess three conserved domains, an AWS (for Associated with SET) domain, a SET domain, and a Cys-rich post-SET domain (Ng et al., 2007). Rice has four class II SDG genes: SDG708, SDG724, SDG725, and SDG736 (Ng et al., 2007). As a distinguishing feature, SDG724 has a more centrally located SET domain (Ng et al., 2007), which may suggest a unique role for SDG724 in rice. We performed a phylogenetic analysis of class II SDGs using Arabidopsis, rice, and maize (Zea mays) protein sequences (see Supplemental Data Set 1 online). This analysis indicated that SDG110 from maize and SDG7 and SDG24 from Arabidopsis are the closest homologs of SDG724 (see Supplemental Figure 3A online). Therefore, further protein sequence alignment analysis was done among these four SDG members. The result showed that SDG724 shared the highest sequence similarity with SDG110 in maize and differed from the Arabidopsis proteins SDG7 and SDG24 by a 20–amino acid deletion in its C-terminal region, which may suggest functional divergence of the SDGs between monocotyledonous and dicotyledonous plants (see Supplemental Figure 3B online).
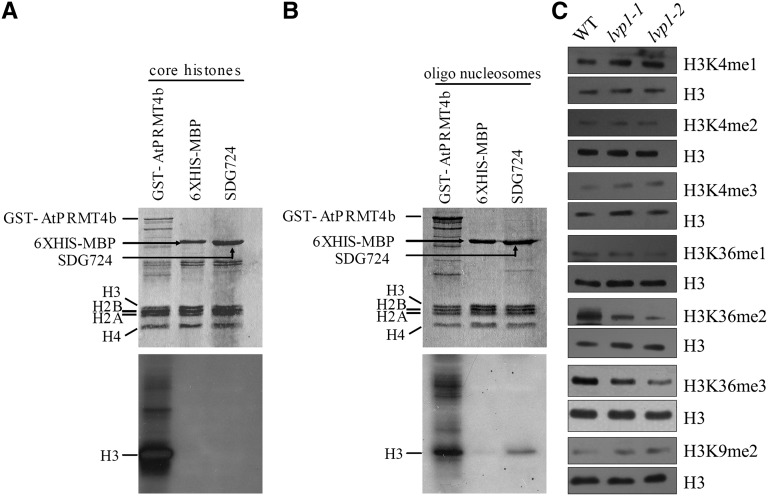
SDG724 Methylates Oligonucleosomes in Vitro
Several plant SDGs have in vitro HMTase activities (Liu et al., 2010; Berr et al., 2011; Deal and Henikoff, 2011; Thorstensen et al., 2011). For instance, SDG714 in rice tends to choose core histones as preferred substrates (Y. Ding et al., 2007), but Arabidopsis SDG8, SDG26, and SDG25 prefer to methylate oligonucleosomes (Xu et al., 2008; Berr et al., 2009). To test in vitro enzymatic activity of SDG724, recombinant protein was expressed in Escherichia coli and purified. After removal of the purification tags, the native SDG724 protein was used in methyltransferase activity assays using tritium-labeled S-[methyl-3H]-adenosyl-l-Met (SAM) as the methyl donor and core histones and oligonucleosomes as substrates. Compared with the positive control, Arabidopsis protein Arg methyltransferase 4b (PRMT4b; Niu et al., 2008), SDG724 showed little enzymatic activity when core histones were used as substrates (Figure 3A). However, when the substrates were changed to oligonucleosomes, a signal was observed at the position corresponding to histone H3 in the SDG724 reaction system (Figure 3B). Thus, SDG724 preferred oligonucleosomes rather than core histones as a substrate and acted as a histone H3 methyltransferase in vitro.
Figure 3.
SDG724 Acts as a SET Domain–Containing Histone Methyltransferase.
(A) and (B) In vitro HMTase assay analyses. Calf thymus core histones (A) and oligonucleosomes (B) were used as substrates, and tritium-labeled SAM was used as methyl donor. Proteins analyzed for enzyme activity are pointed out with arrows. Top panel: Coomassie blue–stained protein gel. Bottom panel: Corresponding autoradiograph.
(C) Analysis of the global levels of H3K36me1/2/3, H3K4me1/2/3, and H3K9me2 markers in wild-type (WT), lvp1-1, and lvp1-2 plants using H3K methylation tag-specific antibodies. Histone-enriched proteins were extracted from leaves of 50-d-old plants grown in Beijing under natural LD conditions. For each immunoblot, the same membrane was stripped and reblotted with an antibody against H3 as a loading control.
SDG724 Is a Major HMTase Responsible for H3K36 Methylation
In Arabidopsis, certain HMTases act as major factors that control specific histone methylation of the entire genome (Liu et al., 2010; Berr et al., 2011; Deal and Henikoff, 2011; Thorstensen et al., 2011). For example, SDG8 is the major in vivo H3K36 HMTase required for global H3K36me2/3 modifications (Zhao et al., 2005; Xu et al., 2008). To investigate whether SDG724 also had a global effect on specific histone H3 sites, immunoblot analyses using a series of antibodies that recognize specific histone Lys methylation modifications were done to identify histone modification differences between lvp1 mutant and wild-type plants. The heading date for wild-type plants grown in Beijing under natural LD conditions is ∼70 d. Thus, to ensure that both wild-type and lvp1 mutant plants used in the assay were at the vegetative stage, crude histone extracts were isolated from the top three leaves of 50-d-old plants. As loading control, the total amount of histone H3 was determined using a specific antibody. The intensities of immunoblot signals were measured by ImageJ software, and the relative abundance of each histone modification was determined (see Supplemental Figure 4 online). Compared with the wild type, the levels of H3K4me1, H3K4me2, H3K4me3, and H3K9me2 showed no obvious differences in the lvp1-1 and lvp1-2 mutants; however, the levels of H3K36me1, H3K36me2, and H3K36me3 were clearly reduced in both mutants (Figure 3C; see Supplemental Figure 4 online). It is interesting to note that the decrease in H3K36me2 levels in lvp1 plants was more pronounced than that of the other two H3K36 methylation types (Figure 3C; see Supplemental Figure 4 online). In summary, lesions in SDG724 lead to a global decrease of H3K36me1, H3K36me3, and especially H3K36me2 levels, indicating that SDG724 is a histone methyltransferase responsible for H3K36 methylation in rice.
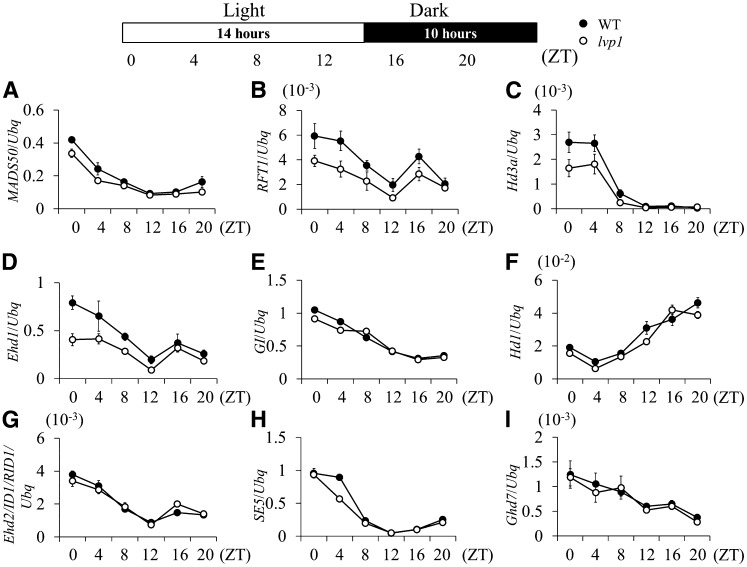
SDG724 Is Required in the MADS50-Ehd1-RFT1 Flowering Pathway under LD Conditions
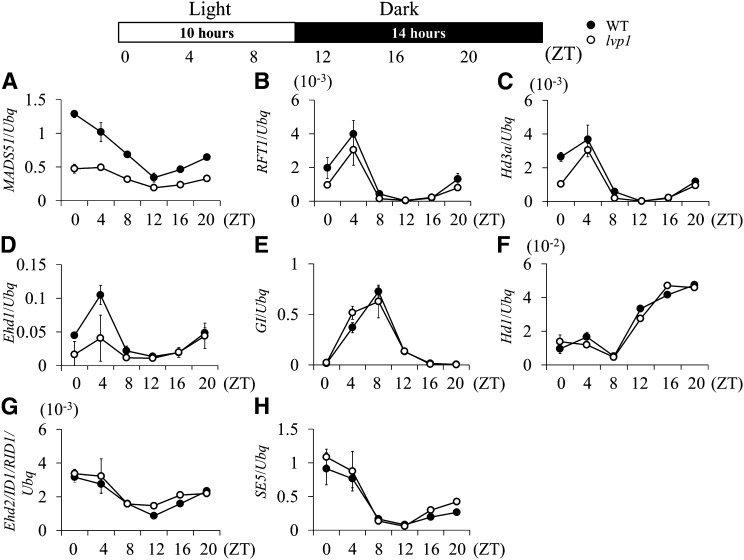
Because the loss-of-function mutant lvp1 had an extremely late-flowering phenotype under LD conditions, we determined whether the diurnal expression of major flowering time genes controlling the floral transition under LD photoperiods was affected. Leaf samples were collected every 4 h from 30-d-old plants grown under LD conditions, and expression of the genes was analyzed. Quantitative RT-PCR results showed that expression levels of GI, Hd1, and SE5 changed little in lvp1 plants; however, expression levels of MADS50 and Ehd1 were decreased in the lvp1 mutant (Figure 4). Due to the decreased level of Ehd1, expression levels of RFT1 and Hd3a were also lower in lvp1 plants. This suggested that SDG724 activity probably affects the Ehd1 but not the Hd1 pathway of flowering. By contrast, Ehd2/ID1/RID1 and Ghd7 levels appeared to be unaffected by the lvp1 mutation, suggesting that these genes are upstream of or parallel to SDG724 in the gene regulatory network of flowering under LD conditions (Figure 4).
Figure 4.
Diurnal Expression Differences of Flowering Time Genes between Wild-type and lvp1 Plants Grown under LD Conditions.
Leaf samples were collected every 4 h from 30-d-old plants grown under LD conditions. Each point represents at least three biological replicates. y axis, relative transcript levels of flowering time genes normalized to those of rice Ubiquitin. Error bars indicate sd (n = 3 or more). WT, wild-type; ZT, Zeitgeber time. Primers are listed in Supplemental Table 1 online.
To provide further evidence that MADS50, Ehd1, Hd3a, and RFT1 were downstream targets of SDG724, we examined the expression levels of those genes at four developmental stages (30, 60, 90, and 120 d after germination [DAG]) in plants grown in Beijing under natural LD conditions. Leaf samples were collected 2 h after dawn, when transcription of the putative downstream genes was at a high level. The results showed that compared with the wild type, transcript levels of the downstream genes were lower in the lvp1 mutant and that transcript levels of the flowering time genes peaked at 60 DAG in wild-type plants (see Supplemental Figure 5 online). These observations further supported the conclusion that under LD conditions, SDG724 activity promoted MADS50 transcription, which leads to upregulation of its downstream targets Ehd1 and FT-like genes. Also notably, MADS50 expression was only marginally changed in the controlled growth chamber under artificial LD conditions at 30 d (Figure 4) but considerably changed under natural LD conditions at 60 DAG (see Supplemental Figure 5 online), when the lvp1 mutant and wild-type plants were all still vegetative.
SDG724 Promotes the MADS51-Ehd1-Hd3a Flowering Pathway under SD Conditions
Because lvp1 mutants also flowered ∼12 d later under SD conditions (see Supplemental Figure 1 online), we examined the diurnal expression levels of genes in the SD flowering pathway. Leaf samples were collected every 4 h from 25-d-old plants grown under SD conditions. Ehd2/ID1/RID1, SE5, MADS51, GI, Ehd1, Hd1, and Hd3a were analyzed as likely members of the SD flowering pathway (Hayama et al., 2003; Kim et al., 2007). Compared with wild-type plants, transcript levels of MADS51 and Ehd1 were decreased, correlating with lower expression levels of Hd3a in lvp1 plants; however, there were no obvious alterations in GI and Hd1 expression levels (Figure 5). These results suggested that SDG724 promoted the flowering transition through MADS51-Ehd1, but not GI-Hd1. Additionally, no significant differences were observed in expression levels of SE5 and Ehd2/ID1/RID1 between the wild-type and lvp1 mutant plants, indicating that these loci are not controlled by SDG724. Taken together, genes involved in the MADS51-Ehd1 pathway may require SDG724 for high expression, which affects expression levels of Hd3a under SD conditions.
Figure 5.
Diurnal Expression Differences of Flowering Time Genes between Wild-Type and lvp1 plants Grown under SD Conditions.
Leaf samples were collected every 4 h from 25-d-old plants grown under SD conditions. Each point represents at least three biological replicates. y axis, relative transcript levels of flowering time genes normalized to those of rice Ubiquitin. Error bars indicate sd (n = 3 or more). WT, wild-type; ZT, Zeitgeber time. Primers are listed in Supplemental Table 1 online.
Mutations in SDG724 Delay Flowering Irrespective of Photoperiod
Lesions in SE5, which encodes a putative heme oxygenase in phytochrome chromophore biosynthesis of rice, produce an extremely early flowering phenotype and a complete lack of photoperiodic response (Izawa et al., 2000, 2002). The early flowering phenotype of se5 mutants under both SD and LD conditions is mainly due to increased expression levels of Ehd1 (Andrés et al., 2009). To investigate the role of SDG724 in a photoperiod-insensitive background, lvp1 se5 double mutants were created using a se5 nonsense mutation in Nipponbare, the same genetic background as for the lvp1 mutant (see Supplemental Figure 6 online). Under both SD and LD conditions, double mutant lvp1 se5 plants flowered earlier than wild-type plants. However, in comparison to se5, the flowering time of lvp1 se5 was delayed by ∼6 d under both SD and LD conditions, suggesting that efficient upregulation of flowering time genes in the se5 mutant background was partially dependent on SDG724 activity. To test this, flowering time genes were analyzed, indicating that compared with the se5 single mutant, expression levels of MADS51, MADS50, Ehd1, Hd3a, and RFT1 were lower in lvp1 se5 double mutant plants under both SD and LD conditions (see Supplemental Figures 7 and 8 online), suggesting that SDG724 was necessary for full activation of Ehd1 expression in the lvp1 se5 double mutant. We speculate that in the lvp1 se5 double mutant, Ehd1 transcript levels are decreased due to reduced expression of MADS50 and MADS51 under both LD and SD conditions, thus causing a late flowering phenotype. By contrast, no obvious differences in Hd1 expression levels between se5 single and lvp1 se5 double mutant plants were observed (see Supplemental Figures 7 and 8 online), further confirming that SDG724 was independent of the Hd1 associated pathway. Taken together, we show that lesions in SDG724 delay flowering via the Ehd1 flowering pathway, even in a genetic background lacking any degree of photoperiodic sensitivity.
Levels of H3K36me2/3 Are Decreased at the MADS50 and RFT1 Loci
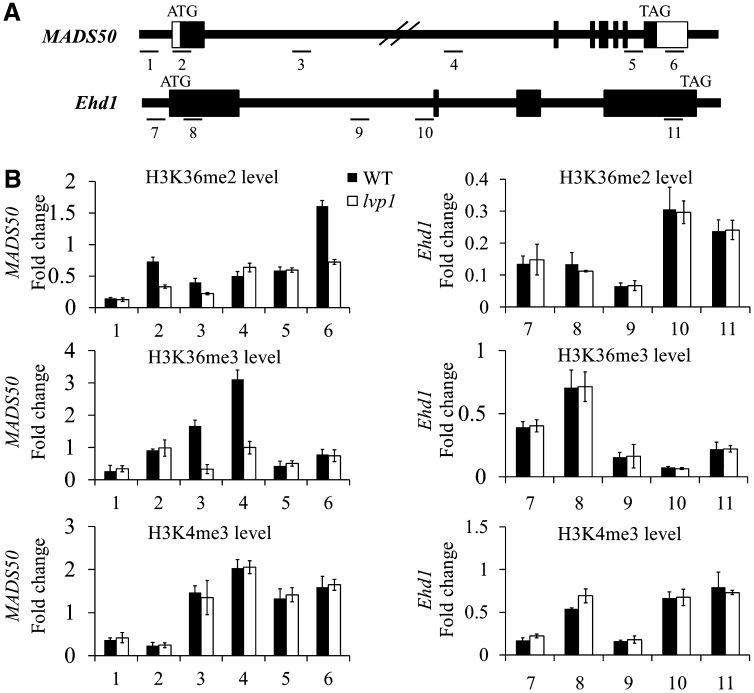
The H3K36 methylation mark is generally associated with transcriptionally active chromatin states (Cazzonelli et al., 2009a; Liu et al., 2010). According to our results, the SDG724 protein functions as an H3K36 methyltransferase and promotes the expression of certain flowering time regulatory genes. This evidence prompted us to determine whether SDG724 was required for depositing H3K36 methylation marks onto the chromatin of the flowering time genes. We collected leaf samples of 50-d-old plants grown under Beijing natural LD conditions and samples of 25-d-old plants grown under artificial SD conditions in a growth chamber. At these two time points, both wild-type and lvp1 plants were at the vegetative growth stage, and the transcripts of SDG724 target genes were all at lower levels in the lvp1 mutant (Figure 4; see Supplemental Figure 5 online). Chromatin immunoprecipitation (ChIP) experiments were done using specific antibodies against H3K36me2 and H3K36me3 because their levels were globally decreased in lvp1 mutants compared with the wild-type plants. As specificity and loading controls, antibodies against H3K4me3 and H3 were used, respectively. Two MADS box genes, MADS50 and MADS51, were shown to be downregulated in the lvp1 mutant. For the ChIP assay, a reduced H3K36me2/3 level was observed within the gene body region of MADS50 in the lvp1 mutant, which is in accordance with its decreased expression level (Figure 6). However, despite the low level of MADS51 transcripts in the lvp1 mutant under SD conditions, no obvious changes in H3K36me2/3 levels were detected between mutants and the wild type at the chromatin regions we examined (see Supplemental Figure 9 online). These results suggested different mechanisms account for the downregulation of the two MADS box genes MADS50 and MADS51 in the lvp1 mutant, and only the expression of MADS50 closely correlated with the chromatin modification levels of H3K36me2/3.
Figure 6.
H3K36me2/3 Levels Are Reduced at the MADS50 Locus in lvp1 Mutant Plants.
ChIP samples were analyzed by quantitative PCR of six regions of MADS50 and five regions of Ehd1. The rice Actin1 gene was used as an internal control. The relative levels of specific histone H3 Lys methylation marks were calculated as the ratio of specific methylation marks over total H3 and normalized as the ratio over total H3 at Actin1; error bars indicate sd (n = 3 or more). Primers are listed in Supplemental Table 1 online.
(A) Genomic structure of the MADS50 and Ehd1 loci. Regions assayed in (B) are indicated by lines and numbered.
(B) ChIP analysis of H3K36me2/3 and H3K4me3 deposited on the chromatin of MADS50 and Ehd1 loci in wild-type (WT) and lvp1 mutant plants.
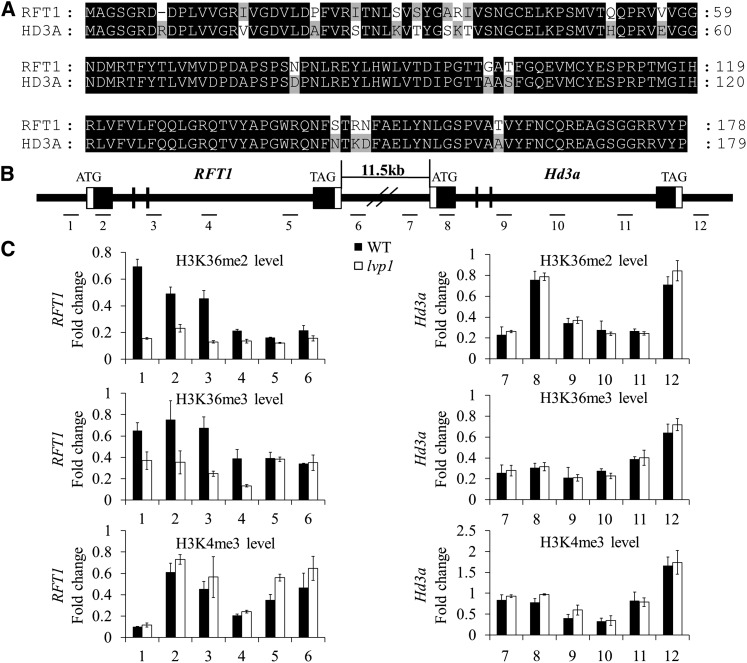
RFT1 and Hd3a encode two rice florigens and are closely linked in the genome, separated by only 11.5 kb. However, RFT1 and Hd3a have functionally diverged to control the LD and SD flowering time pathways, respectively (Komiya et al., 2008). Our ChIP results showed that H3K36me2/3 levels of RFT1 at most chromosomal regions examined were reduced in the lvp1 mutant. However, only subtle changes in H3K36 methylation were detected on the chromatin of Hd3a under either SD or LD conditions (Figure 7; see Supplemental Figure 9 online). Thus, despite of their close chromosomal locations and high degree of similarity, SDG724 specifically affects H3K36me2/3 levels at the RFT1, but not the Hd3a locus. In addition, H3K36me2/3 levels at the Ehd1 locus were also found to be unchanged in lvp1 plants, even though its expression was downregulated in lvp1 compared with wild-type plants (Figure 6). At all the loci examined, H3K4me3 levels were not significantly different between lvp1 mutant and wild-type plants, which demonstrated a specific relationship between SDG724 and H3K36 methylation.
Figure 7.
H3K36me2/3 Levels Are Severely Reduced at the RFT1 Locus in lvp1 Mutant Plants.
ChIP samples were analyzed by quantitative PCR of six different regions of each gene. The rice Actin1 gene was used as an internal control. The relative levels of specific histone H3 Lys methylation marks were calculated as the ratio of specific methylation marks over total H3 and normalized as the ratio over total H3 at Actin1; error bars indicate sd (n = 3 or more). Primers are listed in Supplemental Table 1 online.
(A) Alignment of RFT1 and Hd3a protein sequences using ClustalX and GenDoc. Shades of black indicate conserved amino acids.
(B) Genomic structure of the RFT1 and Hd3a loci on chromosome 6. Black boxes indicate exons, and white boxes indicate untranslated regions. Regions assayed in (C) are indicated by lines and numbered.
(C) ChIP analysis quantifying H3K36me2/3 and H3K4me3 levels in the chromatin of the RFT1 and Hd3a loci in wild-type (WT) and lvp1 mutant plants.
DISCUSSION
In this study, we identified LVP1/SDG724, as a rice histone methyltransferase gene. SDG724 is a class II SET domain protein and is constitutively expressed in various kinds of tissues (see Supplemental Figure 10 online). Members of this class, including SDG4, SDG8, and SDG25 in Arabidopsis and SDG725 in rice, were shown to have H3K36 methylation activities (Zhao et al., 2005; Cartagena et al., 2008; Dong et al., 2008; Xu et al., 2008; Cazzonelli et al., 2009b; Grini et al., 2009; Berr et al., 2010a; Ko et al., 2010; Sui et al., 2012). Our study demonstrated that SDG724 is a major factor for H3K36me3 deposition in rice for the following reasons. First, recombinant SDG724 protein can specifically methylate histone H3 in oligonucleosomes (Figures 3A and 3B), indicating that it has in vitro H3 HMTase activity; second, global levels of H3K36me1, H3K36me2, and H3K36me3 were all reduced in lvp1 plants due to a lesion in SDG724 (Figure 3C); third, lesions in SDG724 specifically decreased the abundance of H3K36me2/3 marks at the MADS50 and RFT1 loci in lvp1 plants (Figures 6 and 7).
In animals and yeast, H3K36 methylation recruits other histone modifiers, such as histone acetyltransferases or deacetylases, thus reflecting the complexity of transcriptionaln mechanisms (Liu et al., 2010). In addition, H3K36me3 also provides a splicing-related mark, which is involved in the histone modification status in Caenorhabditis elegans (Liu et al., 2010). In Arabidopsis, H3K36 methylation may participate in fine-tuning transcription levels at individual gene loci (Guo et al., 2010), but it is not clear whether the regulatory mechanisms of H3K36 methylation are shared by different species (Liu et al., 2010). The identification of the HMTase SDG724 as a major factor in H3K36 methylation in rice will facilitate future studies on how H3K36 methylation may affect chromatin structure and gene transcription in different organisms.
Although the flowering time genes MADS50, MADS51, Ehd1, Hd3a, and RFT1 all displayed significant changes in transcriptional activity due to a lesion in SDG724, H3K36me2/3 levels were only decreased at MADS50 and RFT1 in lvp1 mutant plants. RFT1 and Hd3a are paralogs and members of the phosphatidylethanolamine binding protein gene family (Chardon and Damerval, 2005), and they appear to be the only two major florigen genes in rice (Tamaki et al., 2007; Komiya et al., 2008, 2009). However, several lines of evidence indicate that the functions of the two paralogs have diverged after duplication (Hagiwara et al., 2009). First, it was shown that RFT1 but not Hd3a is the major florigen in the fields of northern Asia under natural LD growth conditions (Komiya et al., 2008). Second, the nucleotide sequences of RFT1 are more highly differentiated in various rice accessions and have thus diverged more rapidly than Hd3a during rice evolution (Hagiwara et al., 2009). Third, it has been observed that the critical photoperiod for RFT1 activation was not as fixed as that for Hd3a (Itoh et al., 2010). Taken together, this evidence indicates that RFT1 and Hd3a are regulated by different genetic mechanisms, and our study provides new evidence for this interpretation. Because lesions in SDG724 specifically lowered H3K36me2/3 levels in chromatin of the RFT1 but not the Hd3a locus (Figure 7), we conclude that different biochemical mechanisms regulate the transcriptional activities of those two florigen genes. Similarly, our ChIP data also suggest that the two MADS box flowering time genes MADS50 and MADS51 may also have a similar epigenetic regulatory mechanism as RFT1 and Hd3a. It will be interesting in future studies to examine whether other histone methylation states are needed to distinguish the roles of the two florigen genes in promoting the flowering transition in rice.
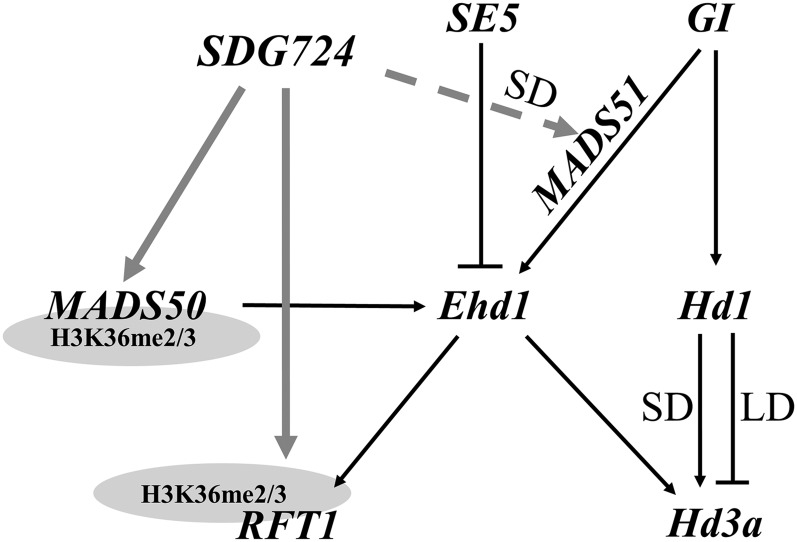
In rice, Hd1 and Ehd1 are two important integrators of the floral transition, and numerous genes controlling their expression have been identified (Komiya et al., 2009; Tsuji et al., 2011). Our results suggest that SDG724 can affect expression levels of Ehd1 via MADS50/MADS51 and independent of Hd1 (Figure 8). Furthermore, the double mutant lvp1 se5 had lower expression levels of Ehd1 than se5 single mutant plants, confirming that SDG724 is required for the Ehd1 associated pathway (see Supplemental Figures 7 and 8 online). However, the japonica rice cultivar Taichung 65 carries loss-of-function alleles of both Hd1 and Ehd1, but it flowers within the range of other accessions and serves as a commercial rice variety (Doi et al., 2004), suggesting that besides the Hd1 and Ehd1 pathways, other not yet identified regulatory mechanisms control the floral transition in rice (Doi et al., 2004; Hagiwara et al., 2009; Saito et al., 2009). Here, we propose that SDG724 may affect the floral transition by activating RFT1, independently of the Hd1 and Ehd1 flowering promotion pathways (Figure 8). SDG724 is responsible for global H3K36me2/3 methylation, which primarily correlates with transcriptionally active chromatin in rice, and lesions in SDG724 reduced RFT1 transcript levels in association with a reduction in the level of H3K36me2/3 marks at the RFT1 locus. Thus, SDG724 might directly control the H3K36me2/3 states of RFT1 chromatin, and the SDG724-RFT1 pathway may be part of a novel Hd1 and Ehd1 independent program for activating the floral transition in rice.
Figure 8.
Proposed Model for the SDG724 Flowering Pathway in Rice.
The black lines show previously identified pathways, and the gray lines indicate pathways added to this model from our study. SDG724 is required for high expression of MADS50, MADS51, Ehd1, RFT1, and Hd3a, but influences H3K36me2/3 levels only in MADS50 and RFT1 chromatin.
In rice, the floral transition is an important agronomic trait for climatic and regional adaptability, and studies on heading date are not only necessary for understanding the genetic mechanisms of flowering, but are also significant for agriculture. Although the northern limit for growing wild rice species is around 28°N, cultivated rice can be grown as far north as around 45°N (Izawa, 2007a). Thus, it is important to identify the floral transition mechanisms in rice under LD conditions, and our work provides evidence for a regulatory mechanism controlled by SDG724 under LD conditions, which could also be useful for commercial rice improvement. Along those lines, a recently launched breeding program for growing rice at low latitudes with almost constant SD conditions aims to develop rice varieties with long basic vegetative growth phases (BVPs). Long BVPs allow rice plants to achieve sufficient vegetative growth by delaying flowering, thus producing high-yielding plants that can be used in double or triple cropping (Saito et al., 2009; Yuan et al., 2009). Ef1 (Ehd1) was the first locus identified in this BVP breeding program (Yuan et al., 2009), which is in agreement with our study showing that SDG724 promotes flowering mainly through Ehd1 under SD conditions. Moreover, because lesions in SDG724 still repress flowering in lvp1 se5 double mutants, which are blind to photoperiod, lvp1 mutant plants could be a useful genetic material for rice breeding to achieve long BVPs in areas with low latitudes.
METHODS
Plant Materials and Growth Conditions
Flowering mutants were screened under natural LD conditions in a paddy field at the experimental station of the Institute of Genetics and Developmental Biology in Beijing, China, in the summer of 2004. An F2 population from a cross between lvp1 and Minghui 63 (Oryza sativa ssp indica) was used to map LVP1 under natural LD conditions in Beijing. Materials used here were also planted in growth chambers in which artificial LD conditions consisted of 14 h light at 30°C followed by 10 h darkness at 28°C, and SD conditions consisted of 10 h light at 30°C followed by 14 h darkness at 28°C.
Map-Based Cloning
We generated a total of 121 simple sequence repeat and STS markers showing polymorphisms between the lvp1 mutant and Minghui 63 for linkage analysis. The novel STS markers were designed based on the difference of DNA sequences between Nipponbare (O. sativa ssp japonica) and 9311 (O. sativa ssp indica) using the Primer Premier 5.0 software (http://www.premierbiosoft.com; Tong et al., 2009). The key mapping markers are listed in Supplemental Table 1 online.
Vector Construction and Plant Transformation
For complementation of the lvp1 mutant, a 1346-bp 3′-downstream fragment was obtained by PCR using primers SDG724-C and inserted into the pCAMBIA1300 vector after digestion with BamHI-PstI. An 8567-bp genomic fragment containing the entire SDG724 transcript was then removed from BAC clone AP006235 by digestion with BamHI-SmaI and introduced into the pCAMBIA1300 vector carrying the 1346-bp 3′-downstream regions. The lvp1 mutant was used as the recipient for transformation by an Agrobacterium tumefaciens–mediated method, as described previously (Liu et al., 2007).
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted and quantitative RT-PCR was performed using the Bio-Rad CFX96 real-time system following a protocol as described previously (Yang et al., 2009). First-strand cDNA was synthesized from 2 μg total RNA. The rice Ubiquitin gene was used as an internal control in quantitative RT-PCR analysis, and all primers for the flowering genes analyzed are listed in Supplemental Table 1 online.
Protein Purification and HMTase Assays
The full-length cDNA of SDG724 was amplified by PCR using primers SDG724-His-MBP (see Supplemental Table 1 online) and cloned into the pMCSG7 vector containing His-MBP tags. The SDG724 fusion gene was expressed in Escherichia coli (BL21 [RIL]; Invitrogen), and recombinant protein was purified using a His-Bind purification kit (EMD Chemicals). Recombinant protein was then cleaved using the tobacco etch virus protease, and the mature SDG724 protein without His-MBP tags was used for HMTase assays. HMTase assays were performed using tritium-labeled SAM (TRK865; Amersham Biosciences) to monitor the incorporation of tritium-labeled methyl groups into different histone substrates. After termination of the HMTase assay, the reaction mix was separated by SDS-PAGE gel electrophoresis, dried, and exposed on x-ray film (Y. Ding et al., 2007; Jacob et al., 2009).
Histone Extraction and Immunoblot Analysis
Crude histone extracts were made from 50-d-old leaves of wild-type and lvp1 plants grown under natural LD conditions in Beijing, and immunoblot analyses were performed as described (Guo et al., 2010). Detection of histone H3 protein was used as loading control. For each immunoblot analyzing a specific modification, the same membrane was stripped and blotted with an antibody against H3. Specific antibodies used were as follows: ab1791 (anti-H3; Abcam; 1:10,000 dilution), 07-436 (anti-H3K4me1; Upstate; 1:10,000 dilution), 07-030 (anti-H3K4me2; Upstate; 1:10,000 dilution), 07-473 (anti-H3K4me3; Upstate; 1:5000 dilution), 07-548 (anti-H3K36me1; Upstate; 1:3000 dilution), 07-369 (anti-H3K36me2; Upstate; 1:3000 dilution), and ab9050 (anti-H3K36me3; Abcam; 1:1000 dilution).
ChIP
Considering their different expression patterns, MADS50, MADS51, Ehd1, Hd3a, and RFT1 were all at a high level in the lvp1 mutant and wild type at Zeitgeber time 2, leaves of 50-d-old plants grown under Beijing natural LD conditions and 25-d-old plants grown under artificial SD conditions in a growth chamber were collected 2 h after dawn for ChIP experiments as previously described (Berr et al., 2010b). The antibodies 07-473 (anti-H3K4me3; Upstate), 07-369 (anti-H3K36me2; Upstate), ab9050 (anti-H3K36me3; Abcam), and ab1791 (anti-H3; Abcam) were used to pull down chromatin containing the specific methylation marks. Detection of MADS50, MADS50, Ehd1, Hd3a, and RFT1 chromatin regions using quantitative PCR as described previously (Guo et al., 2010). The rice Actin1 gene was used as an internal control for the quantitative PCR analysis, and all the primers used are listed in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found at http://www.chromdb.org/ under the following accession numbers: Os09g13740 (SDG724, rice), Os04g34980 (SDG708, rice), Os02g34850 (SDG725, rice), Os02g39800 (SDG736, rice), At2g44150 (SDG7, Arabidopsis), At3g59960 (SDG24, Arabidopsis), At4g30860 (SDG4, Arabidopsis), At1g77300 (SDG8, Arabidopsis), At1g76710 (SDG26, Arabidopsis), AF545814 (SDG110, maize), and AY122273 (SDG102, maize). Other accession numbers, which can be found in National Center for Biotechnology Information databases, are listed in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heading Date Investigation of Wild-Type and lvp1 Plants under SD and LD Conditions.
Supplemental Figure 2. Rescue of lvp1 Phenotype by Transformation with SDG724.
Supplemental Figure 3. Phylogenetic Analysis of SDG724 Homologs in Plants.
Supplemental Figure 4. Quantification of Immunoblot Signals.
Supplemental Figure 5. Expression Levels of MADS50, Ehd1, Hd3a, and RFT1 at Different Developmental Stages of LD Grown Plants.
Supplemental Figure 6. Analysis of Double Mutant lvp1 se5 Plants.
Supplemental Figure 7. Expression Analysis of Flowering Genes in se5 Single and lvp1 se5 Double Mutant Plants Grown under SD Conditions.
Supplemental Figure 8. Expression Analysis of Flowering Genes in se5 Single and lvp1 se5 Double Mutant Plants Grown under LD Conditions.
Supplemental Figure 9. H3K36me2/3 Levels Are Not Affected at the MADS51 and Hd3a Loci in lvp1 Mutant Plants Grown under SD Conditions.
Supplemental Figure 10. Expression Pattern Analysis of SDG724 in Different Tissues by Quantitative RT-PCR.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Michael Schläppi from Marquette University for his critical reading of this article and extensive English editing. We thank Shouyun Cao for her technical assistance on all rice transformations and Gupo Li for his greenhouse and paddy field management. This research was supported by grants from Ministry of Science and Technology (2009CB118506), the Ministry of Agriculture (2011ZX08001-004), and the National Natural Science Foundation of China (30771314, 30825029, and 30921061).
AUTHOR CONTRIBUTIONS
C.S., J.F., and C.C. designed the experiments, analyzed the data, and wrote the article. J.F., J.T., and Q.Q. identified the mutant materials. C.S., J.F., and X.D. constructed and identified the double mutant lvp1 se5. C.S., J.F., and F.C. performed map-based cloning, vector construction, and plant transformation. C.S., F.Z., and L.L. performed RNA extraction and quantitative RT-PCR. C.S., T.Z., and G.Z. performed protein purification and HMTase assays, histone extraction, and immunoblot analysis. C.S., T.Z., B.X., and X.C. performed ChIP.
Glossary
- SD
short-day
- LD
long-day
- HMTase
histone lysine methyltransferase
- H3K36
histone H3 Lys 36
- STS
sequence-tagged site
- SAM
S-[methyl-3H]-adenosyl-l-Met
- DAG
d after germination
- ChIP
chromatin immunoprecipitation
- BVP
basic vegetative growth phases
References
- Andrés F., Galbraith D.W., Talón M., Domingo C. (2009). Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice. Plant Physiol. 151: 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., McCallum E.J., Alioua A., Heintz D., Heitz T., Shen W.H. (2010a). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., McCallum E.J., Ménard R., Meyer D., Fuchs J., Dong A., Shen W.H. (2010b). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22: 3232–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Shafiq S., Shen W.H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809: 567–576 [DOI] [PubMed] [Google Scholar]
- Berr A., Xu L., Gao J., Cognat V., Steinmetz A., Dong A., Shen W.H. (2009). SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 151: 1476–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartagena J.A., Matsunaga S., Seki M., Kurihara D., Yokoyama M., Shinozaki K., Fujimoto S., Azumi Y., Uchiyama S., Fukui K. (2008). The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev. Biol. 315: 355–368 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C.I., Cuttriss A.J., Cossetto S.B., Pye W., Crisp P., Whelan J., Finnegan E.J., Turnbull C., Pogson B.J. (2009b). Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C.I., Millar T., Finnegan E.J., Pogson B.J. (2009a). Promoting gene expression in plants by permissive histone lysine methylation. Plant Signal. Behav. 4: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F., Damerval C. (2005). Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Zhu Y., Gao J., Yu Y., Cao K.M., Shen W.H., Dong A.W. (2007). Molecular characterization of three rice SET-domain proteins. Plant Sci. 172: 1072–1078 [Google Scholar]
- Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., Liu C., Bi Y., Qian Q., Cheng Z., Chu C., Cao X. (2007). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G., Ma D.P., Li J. (2008). The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 373: 659–664 [DOI] [PubMed] [Google Scholar]
- Feng S., Jacobsen S.E. (2011). Epigenetic modifications in plants: An evolutionary perspective. Curr. Opin. Plant Biol. 14: 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini P.E., Thorstensen T., Alm V., Vizcay-Barrena G., Windju S.S., Jørstad T.S., Wilson Z.A., Aalen R.B. (2009). The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 4: e7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu Y., Law J.A., Zhang X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine [corrected] 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 18557–18562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara W.E., Uwatoko N., Sasaki A., Matsubara K., Nagano H., Onishi K., Sano Y. (2009). Diversification in flowering time due to tandem FT-like gene duplication, generating novel Mendelian factors in wild and cultivated rice. Mol. Ecol. 18: 1537–1549 [DOI] [PubMed] [Google Scholar]
- Hayama R., Izawa T., Shimamoto K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43: 494–504 [DOI] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- He Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2: 554–564 [DOI] [PubMed] [Google Scholar]
- He Y., Amasino R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10: 30–35 [DOI] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Izawa T. (2007a). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58: 3091–3097 [DOI] [PubMed] [Google Scholar]
- Izawa T. (2007b). Daylength measurements by rice plants in photoperiodic short-day flowering. Int. Rev. Cytol. 256: 191–222 [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Tokutomi S., Okuno K., Shimamoto K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Jacob Y., Feng S., LeBlanc C.A., Bernatavichute Y.V., Stroud H., Cokus S., Johnson L.M., Pellegrini M., Jacobsen S.E., Michaels S.D. (2009). ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.L., Lee S., Kim H.J., Nam H.G., An G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., He Y., Jacob Y., Noh Y.S., Michaels S., Amasino R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott J.E. (1934). Effect of a localized photoperiod on spinach. Proc. Soc. Hort. Sci. 31: 152–154 [Google Scholar]
- Ko J.H., Mitina I., Tamada Y., Hyun Y., Choi Y., Amasino R.M., Noh B., Noh Y.S. (2010). Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komeda Y. (2004). Genetic regulation of time to flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim J., Han J.J., Han M.J., An G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Liang Y.K., Wang Y., Zhang Y., Li S.G., Lu X.C., Li H., Zou C., Xu Z.H., Bai S.N. (2003). OsSET1, a novel SET-domain-containing gene from rice. J. Exp. Bot. 54: 1995–1996 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Liu X.Q., Bai X.Q., Wang X.J., Chu C.C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Ma Y.M., et al. (2009). Molecular analysis of rice plants harboring a multi-functional T-DNA tagging system. J. Genet. Genomics 36: 267–276 [DOI] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Wang Z.X., Minobe Y., Izawa T., Yano M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 148: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D.W., Wang T., Chandrasekharan M.B., Aramayo R., Kertbundit S., Hall T.C. (2007). Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Zhang Y., Pei Y., Liu C., Cao X. (2008). Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 148: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., et al. (2008). Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56: 1018–1029 [DOI] [PubMed] [Google Scholar]
- Qiao Q., Li Y., Chen Z., Wang M., Reinberg D., Xu R.M. (2011). The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J. Biol. Chem. 286: 8361–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F.J., Sun Q.W., Huang L.M., Chen X.S., Zhou D.X. (2010). Rice SUVH histone methyltransferase genes display specific functions in chromatin modification and retrotransposon repression. Mol. Plant 3: 773–782 [DOI] [PubMed] [Google Scholar]
- Ryu C.H., Lee S., Cho L.H., Kim S.L., Lee Y.S., Choi S.C., Jeong H.J., Yi J., Park S.J., Han C.D., An G. (2009). OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 32: 1412–1427 [DOI] [PubMed] [Google Scholar]
- Saito H., Yuan Q., Okumoto Y., Doi K., Yoshimura A., Inoue H., Teraishi M., Tsukiyama T., Tanisaka T. (2009). Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 119: 315–323 [DOI] [PubMed] [Google Scholar]
- Springer N.M., Napoli C.A., Selinger D.A., Pandey R., Cone K.C., Chandler V.L., Kaeppler H.F., Kaeppler S.M. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132: 907–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui P., Jin J., Ye S., Mu C., Gao J., Feng H., Shen W.H., Yu Y., Dong A. (2012). H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 70: 340–347 [DOI] [PubMed] [Google Scholar]
- Tamada Y., Yun J.Y., Woo S.C., Amasino R.M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Thakur J.K., Malik M.R., Bhatt V., Reddy M.K., Sopory S.K., Tyagi A.K., Khurana J.P. (2003). A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear-localized protein expressed preferentially in young seedlings and during reproductive development. Gene 314: 1–13 [DOI] [PubMed] [Google Scholar]
- Thorstensen T., Grini P.E., Aalen R.B. (2011). SET domain proteins in plant development. Biochim. Biophys. Acta 1809: 407–420 [DOI] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Tsuji H., Taoka K., Shimamoto K. (2011). Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14: 45–52 [DOI] [PubMed] [Google Scholar]
- Wu C., You C., Li C., Long T., Chen G., Byrne M.E., Zhang Q. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA 105: 12915–12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.I., Lessard J., Crabtree G.R. (2009). Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J.P., Steinmetz A., Shen W.H. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wu Y., Li Y., Ling H.Q., Chu C. (2009). OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol. Biol. 70: 219–229 [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Saito H., Okumoto Y., Inoue H., Nishida H., Tsukiyama T., Teraishi M., Tanisaka T. (2009). Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor. Appl. Genet. 119: 675–684 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Yu Y., Meyer D., Wu C., Shen W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7: 1256–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.