This work identifies VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 as phytochrome B–interacting proteins that act redundantly as floral regulators, translocating into the nucleus and mediating the initial steps of phytochrome B signaling to promote flowering.
Abstract
The timing of the transition to flowering in plants is regulated by various environmental factors, including daylength and light quality. Although the red/far-red photoreceptor phytochrome B (phyB) represses flowering by indirectly regulating the expression of a key flowering regulator, FLOWERING LOCUS T (FT), the mechanism of phyB signaling for flowering is largely unknown. Here, we identified two Arabidopsis thaliana genes, VASCULAR PLANT ONE–ZINC FINGER1 (VOZ1) and VOZ2, which are highly conserved throughout land plant evolution, as phyB-interacting factors. voz1 voz2 double mutants, but neither single mutant, showed a late-flowering phenotype under long-day conditions, which indicated that VOZ1 and VOZ2 redundantly promote flowering. voz1 voz2 mutations suppressed the early-flowering phenotype of the phyB mutant, and FT expression was repressed in the voz1 voz2 mutant. Green fluorescent protein–VOZ2 signal was observed in the cytoplasm, and interaction of VOZ proteins with phyB was indicated to occur in the cytoplasm under far-red light. However, VOZ2 protein modified to localize constitutively in the nucleus promoted flowering. In addition, the stability of VOZ2 proteins in the nucleus was modulated by light quality in a phytochrome-dependent manner. We propose that partial translocation of VOZ proteins from the cytoplasm to the nucleus mediates the initial step of the phyB signal transduction pathway that regulates flowering.
INTRODUCTION
Vegetative development in plants is controlled by various environmental cues, such as daylength, light quality, temperature, drought, and nutrition (Fankhauser and Chory, 1997; Bäurle and Dean, 2006). These signals are sensed by a variety of systems, transmitted by different signal transduction pathways, and integrated to control expression of a specific set of downstream genes for whole plant fitness. The molecular mechanisms for such integration have remained largely elusive. Of the various environmental cues, light affects many aspects of plant growth as a source of energy but also provides pivotal information about the environment (Whitelam et al., 1998). To detect the intensity, quality (wavelength), and direction of incident light, plants have evolved a set of photoreceptors: red/far-red light-absorbing phytochromes; blue light receptors, such as cryptochromes and phototropins; and UV-B receptors (Chen et al., 2004; Rizzini et al., 2011). Among these photoreceptors, only the phytochrome family members are sensitive to red and far-red light.
Phytochromes are encoded by a small gene family, and five phytochromes (phytochrome A [phyA] to phyE) have been identified in Arabidopsis thaliana (Sharrock and Quail, 1989; Clack et al., 1994). The photosensory function of the phytochrome resides in its capacity for reversible interconversion between the biologically active Pfr and the inactive Pr (Quail et al., 1995). Of the five phytochromes, phyA and phyB have the most important photosensory functions. phyA is a photolabile photoreceptor accumulated in the dark and triggers very low fluence responses, such as seed germination, and high irradiance responses, such as inhibition of hypocotyl elongation under far-red light. phyB is the dominant phytochrome species in light-grown plants and plays a role in germination, deetiolation, shade avoidance, and flowering repression (Franklin and Quail, 2010). Newly synthesized phyB in the cytoplasm is in the Pr form but is converted into the Pfr form by red light and translocated to the nucleus (Kircher et al., 2002; Nagatani, 2004) and then affects the expression of many downstream genes to induce photomorphogenesis (Devlin et al., 2003; Tepperman et al., 2004).
To determine the components that mediate the phytochrome-dependent signaling cascade, two main experimental approaches have been used: screening for light response mutants and yeast two-hybrid assays to identify phytochrome-interacting proteins (Nagy and Schäfer, 2002). Genetic screenings have identified regulatory networks mediated by phytochromes. For example, the constitutive photomorphogenic/de-etiolated/fusca mutants represent one of the major networks in light signaling and function as negative regulators of photomorphogenesis (Hardtke and Deng, 2000). In darkness, these factors work in concert to target a number of photomorphogenesis-promoting transcription factors for degradation by the proteasome, thus preventing photomorphogenesis (Osterlund et al., 2000; Schwechheimer and Deng, 2000). As an alternative approach, yeast two-hybrid assays have identified several phytochrome-interacting proteins, such as basic helix-loop-helix transcription factors, designated phytochrome-interacting factors (PIFs), whose functions have been characterized in detail (Castillon et al., 2007). PIFs act as negative regulators for various light-regulated responses (Ni et al., 1998; Kim et al., 2003; Leivar and Quail, 2011). PIF proteins are localized constitutively in the nucleus and interact directly with phytochromes in the photoactivated Pfr form (Ni et al., 1999; Shimizu-Sato et al., 2002; Khanna et al., 2004). PIFs that interact with phytochromes are rapidly phosphorylated and then subjected to ubiquitylation and subsequent degradation (Shen et al., 2005; Al-Sady et al., 2006).
Identification of a substantial number of signaling components has enabled an outline of phytochrome signal transduction from light perception to regulation of gene expression to be drafted (Chory, 2010). However, the studies described above have screened mostly responses in early stages of development, such as germination and inhibition of hypocotyl elongation, as indicators of the phytochrome-signaling pathway. For this reason, despite identification of a large number of phytochrome-signaling components, phyB-interacting factors that function in vegetative and reproductive developmental processes such as flowering have not been studied extensively, with the exception of PIF4, which was recently reported to control thermosensory activation of flowering (Kumar et al., 2012).
The timing of the transition from the vegetative to the reproductive phase in flowering plants is tightly controlled by light conditions. In response to daylength, leaves produce a mobile signal that is transported to the shoot apex to induce flowering (Zeevaart, 1976). Recent work on Arabidopsis and several other species identified the FLOWERING LOCUS T (FT) protein as the main component of the long-distance signal florigen (Corbesier et al., 2007; Tamaki et al., 2007; Notaguchi et al., 2008; Navarro et al., 2011). In Arabidopsis, long-day (LD) conditions accelerate flowering, whereas short-day (SD) conditions delay flowering. Spectral light quality also affects flowering time. These processes are regulated by multiple photoreceptors, including phyB (Guo et al., 1998; Mockler et al., 2003). phyB inhibits flowering by repressing FT expression (Cerdán and Chory, 2003); CONSTANS (CO)-dependent and CO-independent mechanisms mediate this repression. CO is a central regulator of photoperiodic flowering, and its expression in an oscillating pattern is controlled by the circadian clock (Suárez-López et al., 2001). In addition, CO is regulated posttranslationally by light quality (Valverde et al., 2004). phyB promotes the degradation of CO early in the day and under red light (Valverde et al., 2004; Jang et al., 2008). The CO-independent mechanism for phyB repression of flowering is not well understood, except for the involvement of PHYTOCHROME AND FLOWERING TIME1 (PFT1), which was identified by genetic screening (Cerdán and Chory, 2003). PFT1 confers a late-flowering phenotype when mutated and regulates FT expression downstream of phyB by both CO-dependent and CO-independent pathways (Cerdán and Chory, 2003; Iñigo et al., 2012). PFT1 encodes the MED25 subunit of the mediator complex (Bäckström et al., 2007). Another flowering regulator is FLOWERING LOCUS C (FLC), a MADS box transcription factor and a major determinant of the response to prolonged cold (vernalization) (Michaels and Amasino, 1999; Sheldon et al., 1999). FLC represses downstream genes that promote flowering, including FT (Helliwell et al., 2006; Searle et al., 2006). During vernalization, the FLC transcript level decreases and plants become competent to flower (Kim et al., 2009). Recently, it was reported that the addition of far-red light to LD conditions promotes flowering by altering the balance between the FLC-mediated repression and the CO-mediated induction of flowering (Wollenberg et al., 2008).
To investigate the mechanism by which phytochrome signaling regulates the growth phase, we performed a yeast two-hybrid screen using phytochrome as the bait against a cDNA library derived from Arabidopsis in the late vegetative phase. We identified the VASCULAR PLANT ONE–ZINC FINGER1 (VOZ1) and VOZ2 genes as phyB-interacting factors. The voz1 voz2 double mutant showed delayed flowering under LD conditions, and genetic analysis demonstrated that VOZ1 and VOZ2 act downstream of phyB. Expression analysis indicated that VOZ1 and VOZ2 upregulate FT expression and downregulate FLC expression. Transgenic studies showed that VOZ2 functions in the nucleus and that VOZ2 abundance in the nucleus is controlled by light quality mediated by phytochromes.
RESULTS
phyB-Interacting Factors VOZ1 and VOZ2 Redundantly Promote Flowering
We performed a yeast two-hybrid screen to identify proteins associated with phyB (see Supplemental Figure 1 online). Among candidates for phyB-interacting factors were VOZ1 and VOZ2, which have a zinc-finger motif and transcriptional activator activities (Mitsuda et al., 2004). VOZ1 and VOZ2 belong to the subgroup VIII-2 of the NAC proteins, which comprise one of the largest transcription factor families in plants (Jensen et al., 2010). In vitro pull-down experiments also confirmed the interaction of phyB with VOZ1 (see Supplemental Figure 1 online).
To evaluate the functions of VOZ1 and VOZ2 in phytochrome signaling in vivo, we examined phenotypes of T-DNA insertional mutants of voz1 (voz1-1 and voz1-2) and voz2 (voz2-1 and voz2-2) (see Supplemental Figure 2 online). No transcripts were detected for the voz1-1, voz1-2, and voz2-2 alleles (see Supplemental Figure 2 online). The voz2-1 mutant allele carries a T-DNA insertion that interrupts the first intron upstream of the start codon. In the voz2-1 mutant, only 5′-truncated transcripts were detected (see Supplemental Figure 2 online). To examine the effect of this T-DNA insertion on VOZ2 protein levels, we performed protein gel blot analysis using anti-VOZ2 antibodies. No VOZ2 protein signal was detected in voz2-1 or voz2-2 mutants, which indicated that both mutants are loss-of-function alleles (see Supplemental Figure 2 online).
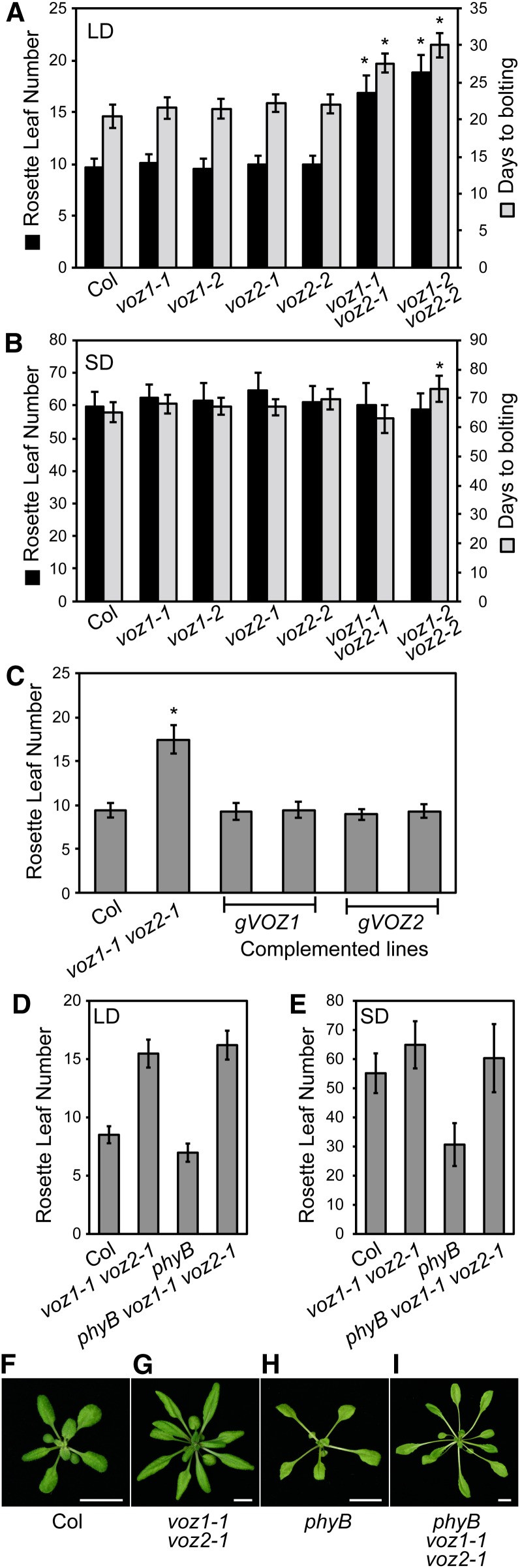
We then examined whether the single and double mutants of voz1 and voz2 showed any visible phenotypes. The double mutants showed an apparent late-flowering phenotype under normal growth conditions. The flowering time of the mutants was measured under different photoperiods (Figures 1A and 1B; see Supplemental Figure 3 online). Under both LD and SD conditions, none of the single mutants showed a significant difference in flowering time. To test the possibility of functional overlap of VOZ1 and VOZ2, we analyzed two lines of double mutants (voz1-1 voz2-1 and voz1-2 voz2-2). Both double mutants showed a late-flowering phenotype under LD conditions. Under SD conditions, the voz1-2 voz2-2, but not voz1-1 voz2-1, mutant displayed a delayed flowering time when measured as number of days to bolting (Figure 1B), which suggested that the voz1-2 voz2-2 mutant exhibited developmental retardation under SD conditions.
Figure 1.
Flowering Phenotype of voz Mutants and Genetic Interaction between phyB and VOZs.
(A) Rosette leaf number and number of days to bolting of the voz1, voz2, and double mutants grown under LD conditions (16 h white light/8 h dark). Data are the mean ± sd (n ≥ 12). Asterisks indicate a significant difference from Col at P < 10−4.
(B) Rosette leaf number and number of days to bolting of the voz1, voz2, and double mutants grown under SD conditions (8 h white light/16 h dark). Data are the mean ± sd (n ≥ 12). Asterisk indicates a significant difference from Col at P < 10−4.
(C) Complementation of voz1-1 voz2-1 mutant by VOZ1 and VOZ2 genomic fragments under LD conditions. Two independent lines were examined for each genomic construct (gVOZ1 and gVOZ2, respectively). Data are the mean ± sd (n ≥ 18). Asterisk indicates significant difference from Col at P < 10−4.
(D) Rosette leaf number at bolting of voz1 voz2, phyB, and the triple mutants grown under LD conditions. Data are the mean ± sd (n ≥ 35).
(E) Rosette leaf number at bolting of voz1 voz2, phyB, and the triple mutants grown under SD conditions. Data are the mean ± sd (n ≥ 10).
(F) to (I) Plants at bolting. Plants were grown under LD conditions. Col (F) and phyB mutant (H) at day 16. voz1-1 voz2-1 mutant (G) and phyB voz1-1 voz2-1 mutant (I) at day 24. Bars = 1 cm.
[See online article for color version of this figure.]
To confirm that the observed flowering phenotypes were caused by the defective VOZ genes, we performed complementation experiments. Either construct that carried a genomic fragment of VOZ1 or VOZ2 rescued the flowering time defect in the voz1-1 voz2-1 double mutant (Figure 1C). This result indicated that VOZ1 and VOZ2 regulate flowering time redundantly.
We did not observe other phytochrome-related phenotypes in the mutants. The hypocotyl length of voz1, voz2, and voz1 voz2 mutants was unaltered compared with that of wild-type seedlings under both red and far-red light conditions (see Supplemental Figure 4 online). The voz1, voz2, and voz1 voz2 mutants also displayed normal plant architecture (see Supplemental Figure 3 online), and the accumulation levels of pigments such as chlorophyll and anthocyanin were largely unaffected (see Supplemental Figure 5 online).
VOZ1 and VOZ2 Function Downstream of phyB in the Flowering Pathway
To examine the genetic relationship between phyB and VOZ1/VOZ2 in the flowering pathway, we measured the flowering time in the phyB voz1-1 voz2-1 triple mutant and the phyB mutant. The voz1-1 voz2-1 double mutation completely suppressed the early-flowering phenotype of the phyB mutant under both LD and SD conditions (Figures 1D and 1E), which strongly suggested that VOZ1 and VOZ2 are essential for phyB regulation of flowering time. The elongated petiole, leaf hyponasty, and reduced leaf area phenotypes of the phyB mutant were unaffected in the phyB voz1-1 voz2-1 mutant (Figures 1F to 1I). This result, together with the normal hypocotyl length phenotype and complete suppression of the phyB early-flowering phenotype, suggested that VOZ1 and VOZ2 function downstream of phyB specifically in the flowering pathway.
VOZ1 and VOZ2 Function in the Vascular Bundle to Regulate Flowering
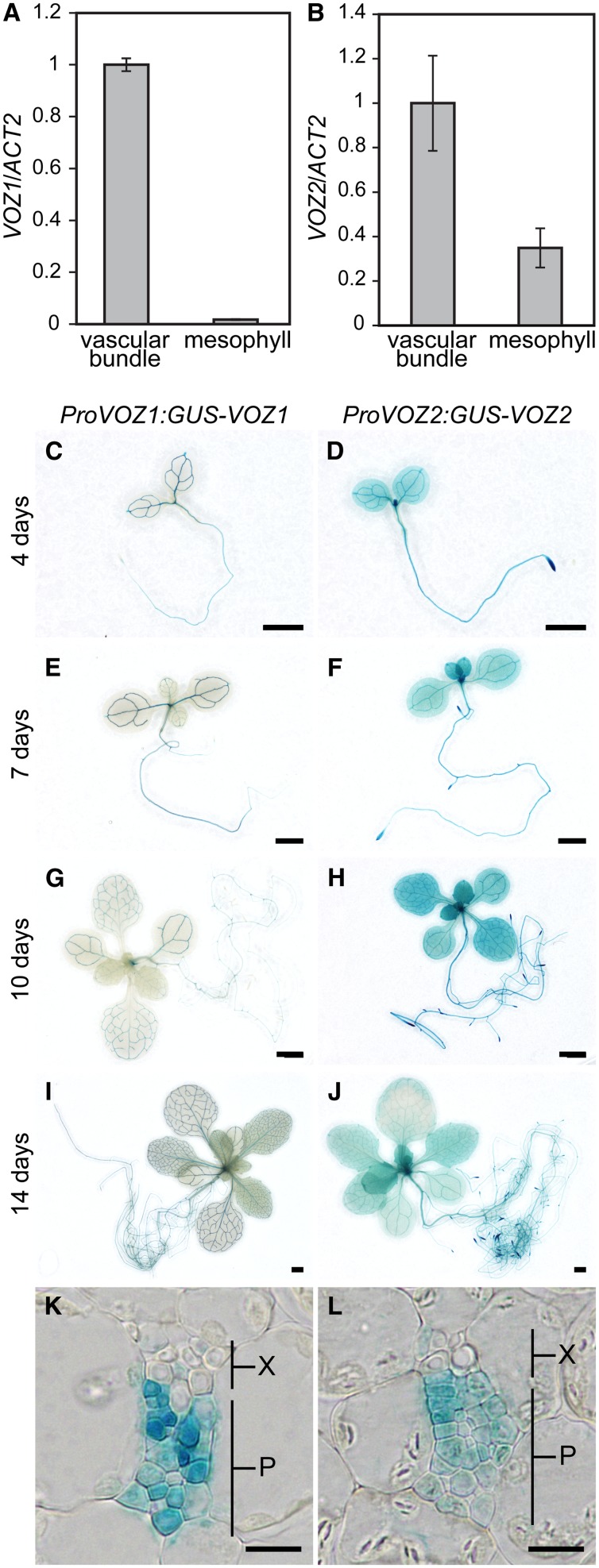
Since VOZ proteins interact with phyB, and phyB represses FT expression in leaves (Cerdán and Chory, 2003; Takada and Goto, 2003), VOZ1 and VOZ2 were predicted to function in leaves. Organ-specific expression of VOZ1 and VOZ2 was not discriminated clearly by RT-PCR analysis (see Supplemental Figure 6 online). We isolated mesophyll protoplasts and vascular bundles from cotyledons to examine tissue-specific expression of VOZ1 and VOZ2 in leaves. We used tissue-specific expression marker genes, ribulose-1, 5-bisphosphate carboxylase/oxygenase small subunit (RBCS) for mesophyll cells and SUCROSE-PROTON SYMPORTER2 (SUC2) for vascular bundles, to verify separation of the tissues (Endo et al., 2005; Endo et al., 2007). RBCS expression in the mesophyll sample was ∼2.7 times higher than that in the vascular bundle sample, whereas SUC2 expression was detected predominantly in the vascular bundle sample (see Supplemental Figure 7 online), which demonstrated separation of the tissues of interest. VOZ1 was readily detectable in vascular bundles but not in mesophyll cells (Figure 2A), whereas VOZ2 was expressed in mesophyll cells as well as vascular bundles (Figure 2B).
Figure 2.
Spatial Patterns of VOZ Expression.
(A) and (B) Mesophyll protoplasts and vascular bundles were isolated from cotyledons and VOZ1 or VOZ2 RNA levels were determined by quantitative RT-PCR and then normalized to ACT2. Seedlings were grown under LD conditions for 10 d. ACT2 was used as a control. RNA extraction was performed three times independently. Data are the mean ± se (n = 3).
(C) to (J) Representative GUS staining of ProVOZ1:GUS-VOZ1 #2 ([C], [E], [G], and [I]) and ProVOZ2:GUS-VOZ2 #1 ([D], [F], [H], and [J]) transgenic lines. LD-grown seedlings were analyzed on day 4 ([C] and [D]), day 7 ([E] and [F]), day 10 ([G] and [H]), and day 14 ([I] and [J]). Bars = 1 mm.
(K) and (L) Transverse sections through cotyledon of ProVOZ1:GUS-VOZ1 #2 (K) and ProVOZ2:GUS-VOZ2 #1 (L) transgenic plants. P, phloem; X, xylem. Bars = 10 μm.
To monitor the temporal and spatial expression patterns of VOZ1 and VOZ2, we generated transgenic lines that expressed translational fusions of the VOZ genomic fragment to the bacterial uidA gene, which encodes β-glucuronidase (GUS), driven by the VOZ promoter in the voz1-1 voz2-1 mutant. The late-flowering phenotype of the voz1-1 voz2-1 mutant was complemented in all of the five independent ProVOZ:GUS-VOZ lines examined (see Supplemental Figure 7 online), and the GUS staining pattern was consistent among lines carrying the construct. Consistent with the results of mRNA expression patterns, GUS-VOZ1 signals were detected exclusively in the phloem, whereas GUS-VOZ2 signals were detected in the leaf as a whole (Figures 2C to 2L; see Supplemental Figure 7 online). The expression patterns of GUS-VOZ proteins remain unchanged during vegetative growth (Figures 2C to 2J). Given that VOZ1 and VOZ2 redundantly regulate flowering (Figure 1C) and that both are expressed in vascular bundles, the functional site of VOZ1 and VOZ2 for flowering was indicated to be the vascular bundles.
VOZ1 and VOZ2 Upregulate FT and Downregulate FLC in Leaves
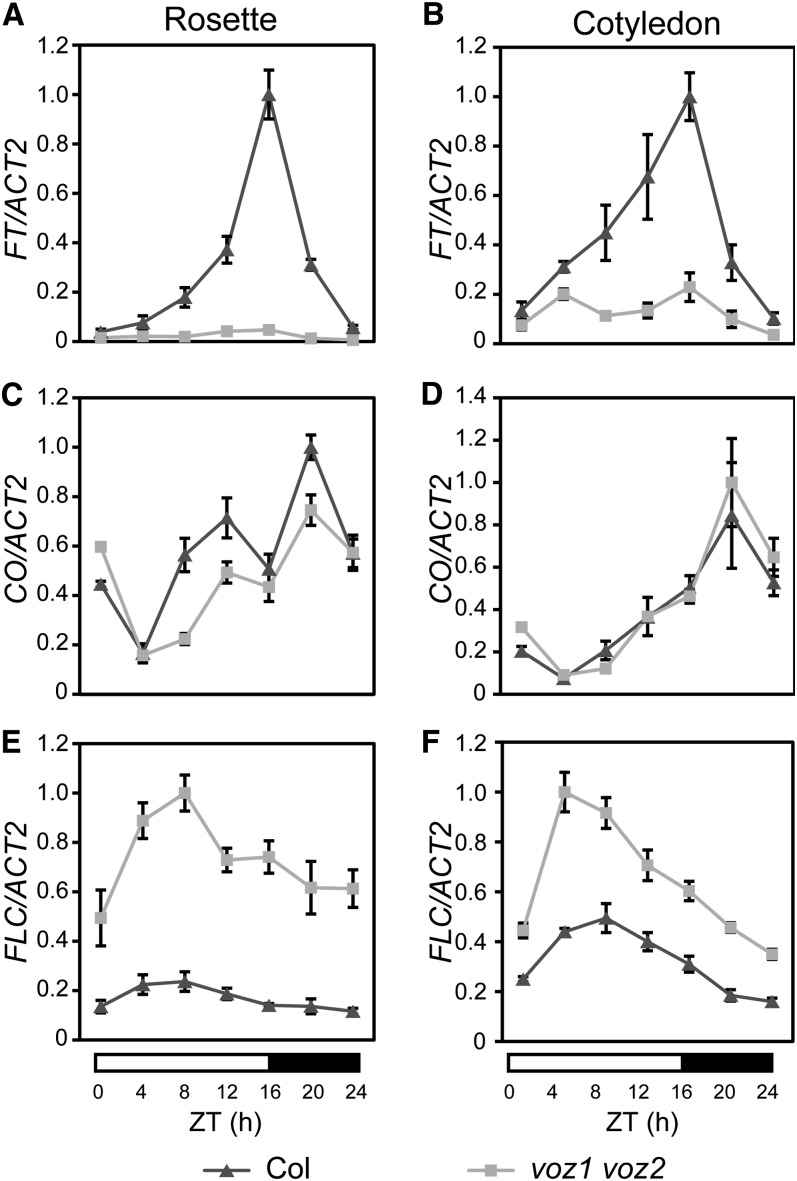
To investigate how VOZ1 and VOZ2 regulate flowering, we examined their role in the regulation of key genes that affect flowering time. FT is an integrator of several flowering pathways and has been proposed to be downregulated by phyB (Cerdán and Chory, 2003). We first analyzed the diurnal expression patterns of FT in cotyledons and rosette leaves using 10-d-old seedlings grown under LD conditions. As reported previously, FT expression in wild-type plants was maximal ∼16 h after dawn under LD conditions (Yanovsky and Kay, 2002) (Figures 3A and 3B). However, FT expression was repressed significantly in the voz1 voz2 mutant, and the peak observed in late daytime in wild-type plants was eliminated. This repression of FT expression was observed in the cotyledons and rosette leaves, although the repression was more prominent in rosette leaves (Figures 3A and 3B).
Figure 3.
Relative Expression Levels of FT, CO, and FLC in the Wild Type and voz1 voz2 Mutant.
Relative expression levels of FT ([A] and [B]), CO ([C] and [D]), and FLC ([E] and [F]) were determined by quantitative RT-PCR in Col (triangles) and voz1 voz2 mutant (squares). Plants were grown for 10 d under LD conditions and harvested at the indicated times. RNA was extracted from rosette leaves ([A], [C], and [E]) or cotyledons ([B], [D], and [F]). ACT2 was used as a control. RNA extraction was performed three times independently. Data are the mean ± se (n = 3). ZT, zeitgeber time.
Next, we examined the expression levels of FT upstream factors. CO is a key factor of the photoperiod pathway and known to be an activator of FT expression. The CO expression pattern was similar in wild-type plants and the voz1 voz2 mutant (Figures 3C and 3D), but the peaks of CO expression in rosette leaves in late daytime and at midnight were less prominent in the voz1 voz2 mutant (Figure 3C). FLC is a floral repressor that functions in the autonomous and vernalization pathways and represses FT expression. Intriguingly, FLC expression levels in the voz1 voz2 mutant were remarkably higher than those of wild-type plants throughout the day (Figures 3E and 3F). The difference in FLC expression between wild-type plants and the voz1 voz2 mutant was greater in rosette leaves than in cotyledons (Figures 3E and 3F). These observations suggested that the late-flowering phenotype of the voz1 voz2 mutant is caused by the increase in FLC expression and subsequent decrease in FT expression in leaves.
VOZ Proteins Mainly Localize in the Cytoplasm but Function in the Nucleus
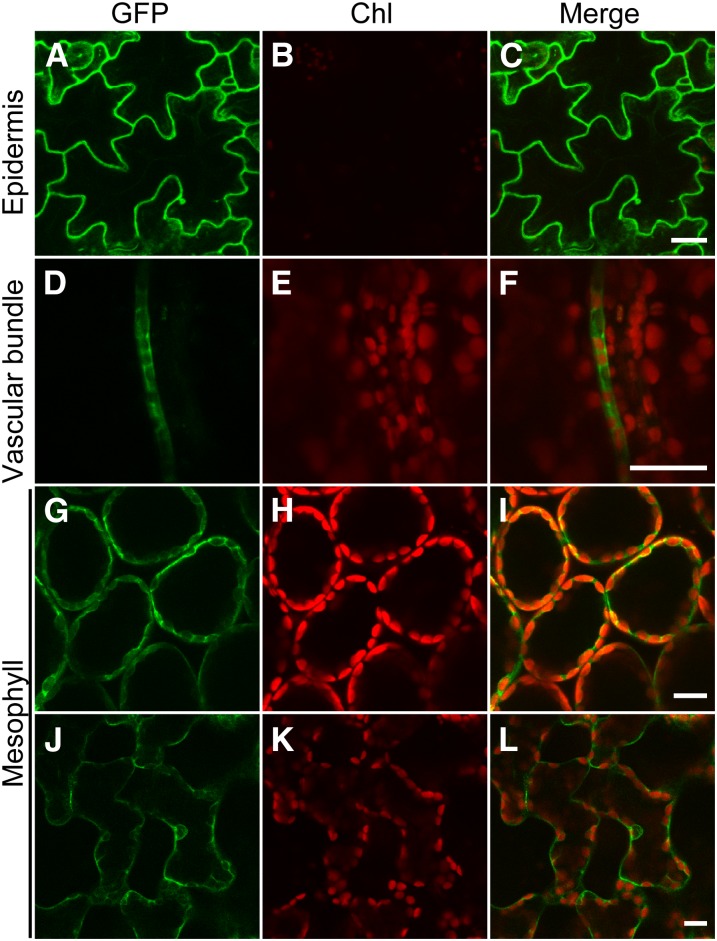
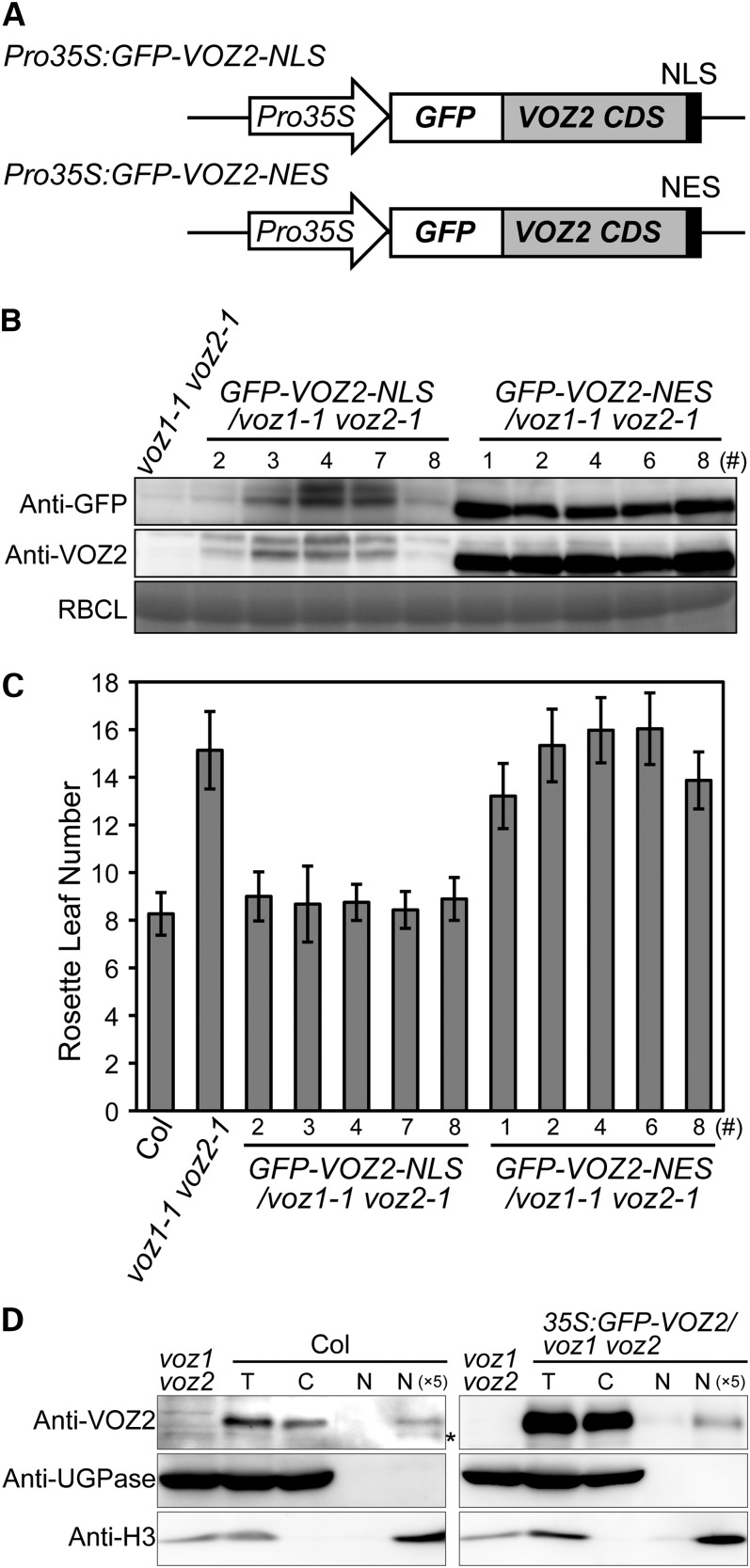
To visualize its subcellular localization, VOZ2 was fused to the C terminus of the green fluorescent protein (GFP). The GFP-VOZ2 coding sequence (CDS) driven by the cauliflower mosaic virus 35S promoter complemented the late-flowering phenotype of the voz1 voz2 mutant (see Supplemental Figure 8 online), which indicated that the fused VOZ2 protein was functional. However, no effects of excessive VOZ2 expression were observed both under LD and SD conditions (see Supplemental Figures 8 and 9 online). We then observed the subcellular localization of GFP-VOZ2 under a laser scanning confocal microscope. In epidermal cells (Figures 4A to 4C), vascular bundle cells (Figures 4D to 4F), and mesophyll cells (Figures 4G to 4L), GFP-VOZ2 signals were detected mainly in the cytoplasm, and their localization was not markedly affected by light quality (red and far-red light) (see Supplemental Figure 8 online).
Figure 4.
Subcellular Localization of GFP-VOZ2 Fusion Protein.
Confocal images of GFP ([A], [D], [G], and [J]), chloroplast autofluorescence (Chl) ([B], [E], [H], and [K]), and merged fluorescence (Merge) ([C], [F], [I], and [L]) from epidermal cells ([A] to [C]), vascular bundle cells ([D] to [F]), palisade mesophyll cells ([G] to [I]), and spongy mesophyll cells ([J] to [L]) in leaves of Pro35S:GFP-VOZ2/voz1 voz2 plants grown under LDs for 7 d. Bars = 20 μm.
Although GFP-VOZ2 signal was detected mainly in the cytoplasm, the presence of a zinc-finger motif and a NAC domain in VOZ proteins strongly suggested that VOZ1 and VOZ2 function in the nucleus. To investigate this enigma, we analyzed the functional intracellular localization of VOZ2. We fused the GFP-VOZ2 protein to a nuclear localization signal (NLS) or a nuclear export signal (NES) and expressed the fusion proteins in the voz1 voz2 mutant (Figure 5A). The GFP-VOZ2 transcripts were detected in all lines examined, and two each representative lines were checked to overexpress VOZ2 mRNA compared with Columbia (Col) (see Supplemental Figure 10 online). The accumulation level of the GFP-VOZ2-NLS protein was significantly lower than that of the GFP-VOZ2-NES protein (Figure 5B), but the late-flowering phenotype of the voz1 voz2 mutant was complemented in the transgenic lines bearing the NLS construct (Figure 5C). By contrast, the GFP-VOZ2-NES protein was readily detected in the transgenic lines but failed to complement the late-flowering phenotype (Figures 5B and 5C). These results indicated that nuclear localization of the VOZ2 protein is essential for its function in flowering. In addition, together with the Pro35S:GFP-VOZ2/voz1 voz2 line, the distinct surplus effect of GFP-VOZ2-NLS overexpression was not observed in the flowering phenotype and flowering gene expression under both LD and SD conditions (Figure 5C; see Supplemental Figure 9 online).
Figure 5.
Subcellular Localization Analysis of Functional VOZ2.
(A) Diagrams of GFP-VOZ2 constructs with NLS or NES.
(B) GFP-VOZ2 protein levels in the seedlings on day 10. Total soluble proteins were subjected to protein immunoblot analysis with anti-GFP and anti-VOZ2 antibodies. Coomassie blue staining of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL) is shown as a loading control.
(C) Rosette leaf number at bolting of NLS and NES lines grown under LD conditions. Data are the mean ± sd (n ≥ 27).
(D) Protein gel blot of cytosolic and nuclear fractions of Col and the Pro35S:GFP-VOZ2/voz1 voz2 line grown under continuous white light for 10 d was probed with anti-VOZ2, anti-UGPase, and anti-histone H3 antibodies. Asterisk represents nonspecific detection. C, cytosolic fraction; N, nuclear fraction; N (×5), fivefold concentrated nuclear fraction; T, total fraction.
To confirm that VOZ proteins function in the nucleus, we performed a cell fractionation experiment to establish the presence of VOZ proteins in the nucleus. We examined the abundance of the VOZ2 and GFP-VOZ2 proteins in the cytosolic and nuclear fraction isolated from 10-d-old seedlings of Col and the Pro35S:GFP-VOZ2/voz1 voz2 line, respectively. Protein blot analysis using antibodies specific to the marker proteins confirmed that each fraction was relatively free from contamination. UDP-Glc pyrophosphorylase (UGPase) was detected only in the cytosolic fraction, and histone H3 was present only in the nuclear fraction (Figure 5D). As indicated by the intensity of the histone H3 signal, a portion of the nuclear fraction was lost during handling (Figure 5D). To detect a clear signal in the nuclear fraction, the fraction was concentrated fivefold. Although VOZ2 and GFP-VOZ2 signals were clearly more abundant in the cytosolic fraction than in the nuclear fraction of Col and the Pro35S:GFP-VOZ2/voz1 voz2 line, respectively, consistent with our GFP-VOZ2 confocal image data (Figure 4), we detected distinct VOZ2 and GFP-VOZ2 signals in the concentrated nuclear fraction (Figure 5D).
VOZ Proteins Interact with phyB in Vivo
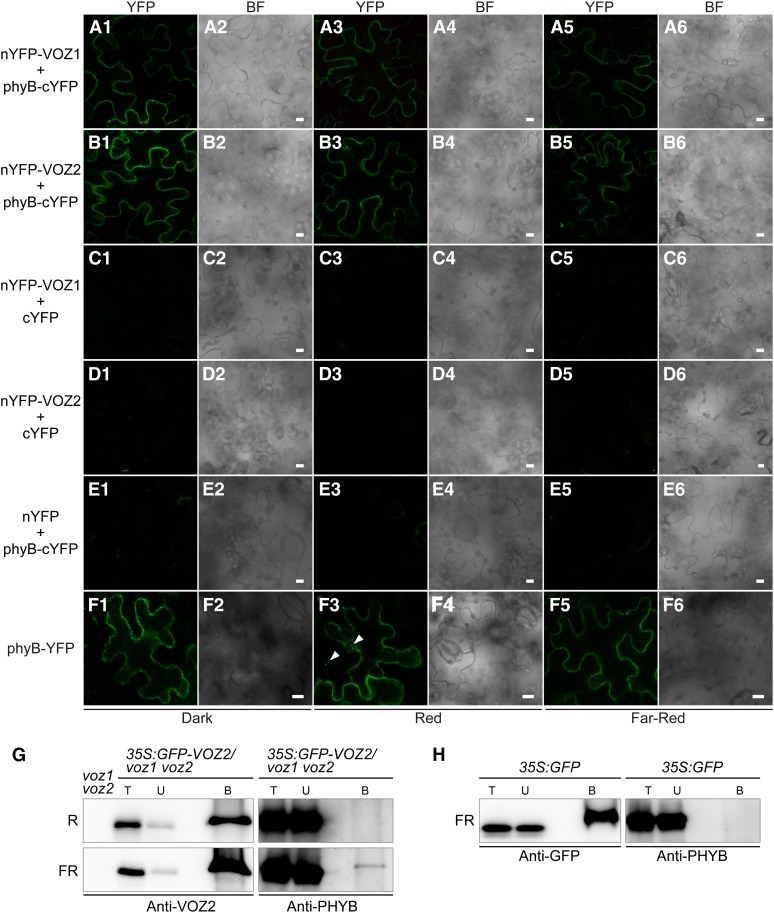
Interactions between phyB and VOZ proteins were detected by bimolecular fluorescence complementation (BiFC) analysis (Figures 6A to 6F), in which plasmids bearing VOZ1 or VOZ2 fused with the N-terminal half of the yellow fluorescent protein (YFP) and PHYB fused with the C-terminal half of YFP were introduced into Nicotiana benthamiana by Agrobacterium tumefaciens infiltration. The subcellular localization of phyB–VOZ interactions was detected only in the cytoplasm and did not change under red, far-red, and dark conditions (Figures 6A1 to 6E6), whereas phyB is reportedly localized in the nucleus under red light (Yamaguchi et al., 1999; Kircher et al., 2002). In our transient expression experiment using N. benthamiana, nuclear translocalization of phyB-YFP under red light was partial (Figure 6F3), possibly because of an excessive amount of the phyB-YFP protein. It should be noted that phyB-YFP signal under red light in Arabidopsis transgenic lines carrying the same construct was observed only in the nucleus (see Supplemental Figure 11 online). These results suggested that the VOZ protein can interact with phyB in the cytoplasm.
Figure 6.
Interaction between phyB and VOZ1/VOZ2 in Vivo.
(A1) to (F6) BiFC analysis of phyB and VOZ1/VOZ2. Confocal images of YFP ([A1] to [F1], [A3] to [F3], and [A5] to [F5]) and bright-field (BF) images ([A2] to [F2], [A4] to [F4], and [A6] to [F6]) from epidermal cells of N. benthamiana infected with Agrobacterium harboring the constructs described below under dark ([A1] to [F1] and [A2] to [F2]), red ([A3] to [F3] and [A4] to [F4]), and far-red ([A5] to [F5] and [A6] to [F6]) conditions. Bars = 10 μm. BiFC analysis of the interaction between VOZ1 and phyB ([A1] to [A6]) and VOZ2 and phyB ([B1] to [B6]). VOZ1 and VOZ2 were fused to nYFP, and phyB was fused to cYFP to generate nYFP-VOZ and phyB-cYFP, respectively. A vector containing only nYFP or cYFP was used as a negative control ([C1] to [C6], [D1] to [D6], and [E1] to [E6]). phyB subcellular localization was observed with the Pro35S:PHYB-YFP construct ([F1] to [F6]). Arrows indicate nuclear speckles.
(G) and (H) Co-IP of GFP-VOZ2 or GFP from Pro35S:GFP-VOZ2 and Pro35S:GFP plant extracts, respectively, using anti-GFP antibody-tagged microbeads. Plants were grown under continuous white light for 9 d and treated with either red (R) or far-red (FR) light for 8 h. B, bound fraction; T, total fraction; U, unbound fraction.
[See online article for color version of this figure.]
To confirm the interactions between phyB and VOZ proteins in vivo, we performed a coimmunoprecipitation (co-IP) experiment using Pro35S:GFP-VOZ2/voz1 voz2 transgenic plants and the anti-PHYB antibody. We obtained evidence of interaction between GFP-VOZ2 and phyB under far-red light irradiation but not under red light irradiation (Figure 6G). When the Pro35S:GFP line was used as a negative control under far-red light conditions, the phyB signal was not detected (Figure 6H). These co-IP data are consistent with the results from BiFC, which showed that phyB and VOZ proteins interacted in the cytoplasm.
Given the abundant cytoplasmic accumulation of GFP-VOZ2 (Figure 4), phyB–VOZ interaction in the cytoplasm (Figure 6), and VOZ function in the nucleus (Figure 5), VOZ might be translocated from the cytoplasm to the nucleus and be subjected to degradation, which could be regulated by light.
VOZ Proteins in the Nucleus Are Degraded under Far-Red and Dark Conditions in a Phytochrome-Dependent Manner
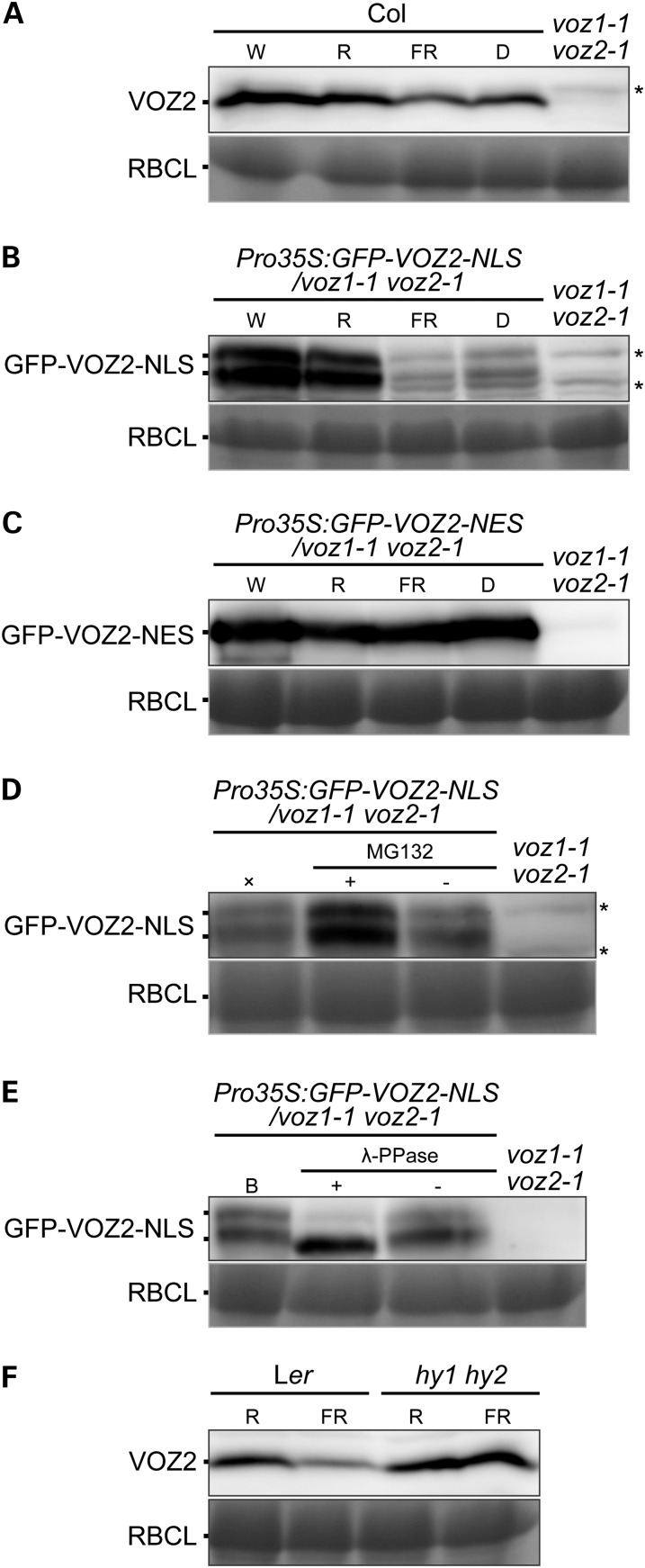
To examine accumulation of the VOZ protein in the nucleus under different light conditions, wild-type and transgenic plants were exposed either to white, red, or far-red light or the dark for 24 h. In wild-type plants, the amount of VOZ2 protein was slightly reduced under far-red and dark conditions (Figure 7A). By contrast, the amount of GFP-VOZ2-NLS protein in the Pro35S:GFP-VOZ2-NLS/voz1 voz2 plant was markedly reduced under far-red and dark conditions (Figure 7B), whereas the amount of GFP-VOZ2-NES protein in the Pro35S:GFP-VOZ2-NES/voz1 voz2 plant was not altered by light conditions (Figure 7C). To evaluate the effect of light on VOZ transcription, we compared the mRNA levels of VOZ1 and VOZ2 in the wild type and GFP-VOZ2 in Pro35S:GFP-VOZ2-NLS/voz1 voz2 plants under different light conditions. In contrast with their protein products, mRNA levels of VOZ genes and GFP-VOZ2 in the wild type and Pro35S:GFP-VOZ2-NLS/voz1 voz2, respectively, were slightly elevated under far-red and dark conditions, except that VOZ1 expression was unaffected in the dark (see Supplemental Figure 12 online). These results suggested that the amount of VOZ proteins is regulated posttranslationally in the nucleus by light. To determine whether any active proteolytic process was involved in the regulation, we treated seedlings of the Pro35S:GFP-VOZ2-NLS/voz1 voz2 lines with the proteasome inhibitor MG132. The MG132 treatment diminished GFP-VOZ2-NLS degradation under far-red light (Figure 7D), which suggested that the degradation of VOZ proteins in the nucleus is mediated by the proteasome system. In the Pro35S:GFP-VOZ2-NLS/voz1 voz2 line, two signals for the GFP-VOZ2-NLS protein were detected (Figures 5B and 7B). To examine the possibility of protein modification by phosphorylation, GFP-VOZ2-NLS was incubated with protein phosphatase. The treatment specifically diminished the larger GFP-VOZ2-NLS form (Figure 7E), thus indicating that a portion of the GFP-VOZ2-NLS protein pool is phosphorylated.
Figure 7.
Degradation and Phosphorylation of VOZ2 Protein.
Protein immunoblotting with anti-VOZ2 antibodies. Coomassie blue staining of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL) is shown as a loading control. Asterisks represent nonspecific detection.
(A) to (C) Protein levels of VOZ2, GFP-VOZ2-NLS, and GFP-VOZ2-NES in Col (A), Pro35S:GFP-VOZ2-NLS/voz1 voz2 line #7 (B), and Pro35S:GFP-VOZ2-NES/voz1 voz2 line #8 (C), respectively. Plants were grown under continuous white light for 10 d and treated with either white (W), red (R), far-red (FR) light, or darkness (D) for 24 h. Each lane contained 60 μg (A), 100 μg (B), or 50 μg (C) of total proteins.
(D) Seedlings grown under continuous white light for 10 d were pretreated with (+) or without (−) 50 μM MG132 for 3 h and transferred to far-red light for 12 h. ×, a control treated with only far-red light. Each lane contained 100 μg of total proteins.
(E) Proteins were extracted from 10-d-old seedlings under continuous white light and incubated with (+) or without (−) λ-PPase. A control sample before λ-PPase treatment is indicated by the letter B. Each lane contained 100 μg of total proteins.
(F) VOZ2 protein levels in Ler and hy1 hy2 mutant. Plants were grown under continuous white light for 10 d and treated with either red or far-red light for 24 h. Each lane contained 65 μg of total proteins.
To investigate the contribution of phytochromes to the far-red- and dark-induced degradation of VOZ2, we examined native VOZ2 levels in phyA, phyB, and hy1 hy2 mutants. phyA is primarily involved in far-red light sensing. HY1 and HY2 encode the enzymes for phytochrome chromophore biosynthesis; thus, the hy1 hy2 mutant has been used as a mutant with no functional phytochrome members (Muramoto et al., 1999; Kohchi et al., 2001; Oka et al., 2011). A lower degree of VOZ2 degradation was observed in the phyA and phyB mutants under far-red light (see Supplemental Figure 13 online). Furthermore, the VOZ2 protein level was largely unaffected in the hy1 hy2 mutant compared with the parental ecotype Landsberg erecta (Ler) under far-red light (Figure 7F). These results suggested that phyA and phyB, as well as the other phytochrome members, are involved in light-dependent VOZ2 degradation.
DISCUSSION
Previous studies demonstrated that Arabidopsis phyB is involved in the control of flowering by repressing FT expression (Cerdán and Chory, 2003). In this study, we identified VOZ1 and VOZ2 as phyB-interacting factors using a yeast two-hybrid screen. Characterization of voz mutants, genetic approaches, and expression analyses indicated that VOZ1 and VOZ2 specifically promote flowering downstream of phyB and regulate the expression of the key flowering-related factors FLC and FT. Histochemical analysis demonstrated that VOZ1 and VOZ2 function in vascular bundles. Moreover, subcellular localization and in vivo interaction analysis suggested that VOZ1 and VOZ2 interact with phyB in the cytoplasm, are translocated to the nucleus at signal transmission, and are subjected to degradation in a phytochrome-dependent manner.
VOZ1 and VOZ2 Promote Flowering Downstream of phyB through FLC and FT
Phytochromes regulate various light responses, including flowering. Molecular and genetic investigations have identified several components involved in the phytochrome-regulated flowering pathway. However, identification of phyB-interacting factors that function in the flowering pathway has proved largely elusive. We have shown that VOZ1 and VOZ2 promote flowering redundantly in Arabidopsis but are not essential for photomorphogenesis (Figure 1; see Supplemental Figures 3 to 5 online). In the phyB mutant background, the voz1 and voz2 mutations resulted in complete suppression of the early-flowering phenotype of phyB mutant (Figures 1D and 1E), and VOZ proteins interacted with phyB in vivo (Figure 6). From these findings, we concluded that VOZ1 and VOZ2 are phyB-interacting factors that regulate flowering time.
Flowering time is influenced by two major environmental factors, namely, photoperiod and temperature (Sung and Amasino, 2004; Imaizumi and Kay, 2006). Plants can also perceive changes in light quality, such as a decrease in the red:far-red ratio of incoming light. CO (Valverde et al., 2004), FT (Halliday et al., 2003), and PFT1 (Cerdán and Chory, 2003) are flowering regulators that act downstream of phyB. CO acts as a critical positive regulator of flowering under LD conditions (Putterill et al., 1995) and promotes expression of FT (Samach et al., 2000). Degradation of CO protein in red light is proposed to be a key regulation in the phyB pathway (Valverde et al., 2004; Jang et al., 2008). PFT1 also regulates FT expression by both CO-dependent and CO-independent mechanisms (Cerdán and Chory, 2003; Iñigo et al., 2012). However, the detailed relationship between CO and PFT1 remains unresolved. In this study, we observed reduced FT expression, almost unchanged CO expression, and elevated FLC expression in the voz1 voz2 mutant under LD conditions (Figure 3). In vitro studies indicate that the potential target sequence of VOZ2, GCGTNx7ACGC, which was identified by in vitro binding studies of the V-PPase promoter (Mitsuda et al., 2004), is not present in the FT and FLC genomic regions. Future experiments are required to reveal whether VOZ proteins directly bind to the FT or FLC promoters in vivo.
Interestingly, FLC expression was reported to be higher in the pft1 mutant (Kidd et al., 2009). Furthermore, flowering of both the voz1 voz2 and pft1 mutants is delayed under LD conditions, the increased petiole length of the phyB mutant is unaffected, and the early-flowering phenotype of the phyB mutant is completely suppressed, although the pft1 mutant has mild effects on hypocotyl elongation (Cerdán and Chory, 2003). PFT1 is the MED25 subunit of the plant mediator complex (Bäckström et al., 2007). Recently, 10 different transcription factors were identified as PFT1-interacting factors, and PFT1 was indicated to cooperate with these transcription factors to function as a hub that integrates environmental cues (Elfving et al., 2011; Ou et al., 2011). VOZ1 and VOZ2 might also cooperate with PFT1 and regulate FT and FLC expression directly or indirectly.
A recent report from field experiments showed that FLC-induced repression of flowering could be overridden other than by vernalization, possibly as a result of natural temperature fluctuations or light conditions (Wilczek et al., 2009). We propose that VOZ1 and VOZ2 promote flowering through a mechanism that involves FLC and integrates light and temperature signals. Future research on the relationship between VOZ proteins and PFT1 and phenotypes of the voz1 voz2 mutant under different temperature and light quality conditions that reflect the natural environment may help to increase our understanding of the signal crosstalk between light and temperature in flowering pathways.
Control of VOZ1 and VOZ2 in the Phloem during Flowering
A classical physiological experiment demonstrated that leaves are the major organs to sense daylength for regulation of flowering (Knott, 1934). VOZ1 and VOZ2 are ubiquitously expressed in all organs, including leaves (see Supplemental Figure 6 online). The transcriptional activation of FT by CO appears to occur specifically in the vascular bundles of leaves, where both CO and FT are expressed (Takada and Goto, 2003; An et al., 2004). phyB acts to regulate flowering through FT, which implies that the phyB-mediated flowering pathway resides in the leaf. In contrast with FT, phyB is expressed in almost all tissues, including the epidermis, mesophyll, and vascular bundles in leaves (Somers and Quail, 1995; Goosey et al., 1997). Recent experiments with enhancer trap lines showed that phyB expressed in mesophyll cells suppresses FT expression in vascular bundles (Endo et al., 2005). The results of tissue-specific RT-PCR and histochemical analyses consistently showed that both VOZ1 and VOZ2 are expressed in vascular bundle cells (Figures 2A to 2J). Given their functional redundancy, it is likely that the site of action for VOZ1 and VOZ2 in flowering is the vascular bundle. GUS-VOZ1 was further shown to be expressed specifically in the phloem (Figure 2K), as is the case for ProFT:GUS (Takada and Goto, 2003). In addition, FLC is expressed widely in the plant, including in vascular bundle cells (Sheldon et al., 2002; Bastow et al., 2004). From these results, we conclude that the site of VOZ1 and VOZ2 function is the phloem, in which FT and FLC are also expressed. It should be noted that phyB expressed in mesophyll cells delayed flowering, whereas phyB expressed in vascular bundles did not (Endo et al., 2005). However, the expression level of phyB in vascular-specific phyB expression lines used in the study was lower than that of wild-type plants (Endo et al., 2005), which could explain the failure to suppress flowering. Also, the expression specificity in the vascular bundles of the vascular-specific phyB expression lines was not revealed (Endo et al., 2005). Therefore, the expression specificity of VOZ1 in vascular tissue (Figure 2K) might differ from that of phyB in the vascular-specific phyB expression lines used by Endo et al. (2005). A role of phyB in vascular bundles in flowering has been suggested (Más et al., 2000; Endo et al., 2007). In Arabidopsis, blue light–sensing cryptochrome2 (cry2) is another major photoreceptor that regulates flowering. It has been reported that cry2 functions in vascular bundles (Endo et al., 2007) and physically interacts with phyB in vivo (Más et al., 2000). The function of VOZ proteins revealed in this work should shed light on the function of phyB in vascular bundles, although this does not exclude the fact that phyB functions also in the mesophyll to suppress flowering.
VOZ Proteins Translocate from the Cytoplasm and Function in the Nucleus in Which Their Stability Is Regulated by Light Quality
Upon light perception, phyB proteins move from the cytoplasm to the nucleus and regulate gene expression (Kircher et al., 1999, 2002; Yamaguchi et al., 1999). Although the nucleus has been considered to be the site of phytochrome signaling, some evidence indicates that the cytosol may also be a site of phytochrome action. Cytoplasmic motility is accelerated by red light within a few seconds (Takagi et al., 2003), and recent work also demonstrated that phytochrome transmits light signals to regulate translation in the cytoplasm (Paik et al., 2012). In addition, the cytoplasmic protein PHYTOCHROME KINASE SUBSTRATE1 (PKS1), which regulates phytochrome signaling, interacts with phytochrome (Fankhauser et al., 1999). However, the molecular mechanism for the PKS1 signal in phytochrome signaling is largely unknown. In this study, the results of yeast two-hybrid, in vitro binding (see Supplemental Figure 1 online), BiFC, and in vivo co-IP (Figure 6) assays supported direct interaction between phyB and VOZ proteins in vivo. The BiFC and co-IP assays further suggested that VOZ1 and VOZ2 interact with phyB in the cytoplasm (Figure 6), and epistasis of VOZ1 and VOZ2 to phyB in flowering function was shown genetically (Figures 1D and 1E).
Although our results demonstrated that VOZ2 functions in the nucleus (Figure 5C), which is consistent with the fact that VOZ proteins are DNA binding NAC domain proteins (Jensen et al., 2010), VOZ2 protein was mainly localized in the cytoplasm (Figures 4 and 5D). Translocalization of rice (Oryza sativa) NAC4, one of the NAC transcription factors, to the nucleus depends on its phosphorylation, which is induced by a pathogen recognition signal (Kaneda et al., 2009). Some other NAC proteins have an α-helical transmembrane motif, which is responsible for plasma membrane or endoplasmic reticulum membrane anchoring, and their nuclear import is regulated by proteolytic cleavage of the anchor, often regulated by a stress signal (Puranik et al., 2012). Data from this fractionation experiment revealed the low proportion of nuclear localization of VOZ2 protein, which suggested that the amount of VOZ proteins in the nucleus was tightly regulated, even in the GFP-VOZ2 overexpression line (Figure 5D). This interpretation is consistent with the observation that the degradation of VOZ2 protein in wild-type plants under far-red and dark conditions was less than that of GFP-VOZ2-NLS proteins in the NLS line (Figures 7A and 7B). In addition, GFP-VOZ2-NES protein accumulation levels in the NES line were not altered by light conditions, which suggested that only the VOZ2 protein localized in the nucleus was affected by light quality.
Genetic evidence indicates that phyB repressed VOZ function (Figures 1D and 1E). What is the genetic relationship between phyB and VOZ in the cell? Although our data showed that VOZ proteins function in the nucleus, the BiFC and co-IP assays revealed that VOZ2 interacts with phyB in the cytoplasm under far-red light conditions (Figure 6). One possibility for this interaction is that phyB tethers VOZ proteins within the cytoplasm and the limited translocation of VOZ proteins under far-red light condition is used to inhibit VOZ arbitrary signaling transmission (see Supplemental Figure 14 online). In addition, phyB might be involved in the modification of VOZ proteins, such as phosphorylation, in the cytoplasm, which could lead to degradation of the VOZ2 proteins after their translocation to the nucleus (see Supplemental Figure 14 online). The significance of the cytoplasmic interaction remains unknown. By contrast, both phyB (Chu et al., 2005) and VOZ (Figures 3 and 5) regulate gene expression in the nucleus. Given the limited translocation of VOZ proteins to the nucleus (Figure 5D) and the instability of VOZ2 protein (Figure 5B), phyB–VOZ interaction in the nucleus might not be detectable by the BiFC and co-IP assays, even if VOZ2 interacts with phyB in the nucleus. Therefore, we cannot exclude the possibility that phyB also directly regulates VOZ function in the nucleus.
Several important light-signaling proteins are involved in light-dependent phosphorylation and degradation of proteins. Phosphorylation of the PIF3 protein, for example, is phytochrome dependent. Phytochromes induce rapid phosphorylation of PIF3 as a result of phy–PIF interaction under red light (Al-Sady et al., 2006), which leads to ubiquitylation and degradation of PIF3. However, neither the protein kinase nor the E3 ubiquitin ligase required for PIF3 degradation have been identified (Leivar and Quail, 2011). COP1 is a major negative regulator of the photomorphogenic response and acts as an E3 ubiquitin ligase that mediates degradation of positive regulators of light signal transduction, for example HY5, in the dark (Osterlund et al., 2000; Yi and Deng, 2005), and the subcellular localization of COP1 is reportedly regulated by photoreceptors, such as phyA and phyB (Osterlund and Deng, 1998). Recent studies reported that COP1 acts as a repressor of flowering by promoting the ubiquitin-mediated proteolysis of CO in the dark, although phyB-mediated turnover of CO early in the morning or in red light does not require COP1 (Jang et al., 2008; Liu et al., 2008). In addition, COP1 preferentially targets phosphorylated phyA for degradation under far-red light (Saijo et al., 2008). Destabilization under far-red and dark conditions and phosphorylation of VOZ proteins (see Supplemental Figure 14 online) seem consistent with this COP1-mediated degradation mechanism. Hence, VOZ proteins might be the targets of degradation by COP1. Alternatively, different ubiquitin ligases might exist to promote VOZ protein degradation. Further experiments are needed to elucidate the precise function of VOZ proteins in protein phosphorylation and degradation.
METHODS
Plant Materials and Growth Conditions
The wild-type plants used were the Col and Ler accessions. Seeds of the voz1-1, voz1-2, voz2-1, and voz2-2 mutants correspond to the T-DNA insertion strains GABI_418B02, WISCDSLOX489-492O10, SALK_021718, and SALK_115813, respectively. The background accessions of the voz1-1, voz1-2, voz2-1, voz2-2, phyB-9, and phyA-211 mutants are Col and that of the double mutant hy1-1 hy2-1 is Ler. The voz1-1 voz2-1, voz1-2 voz2-2, and phyB-9 voz1-1 voz2-1 mutants were produced by crossing. The constructs in binary vectors were introduced into Arabidopsis thaliana using Agrobacterium tumefaciens strain C58 (Clough and Bent, 1998). Plants were grown in soil or on half-strength Murashige and Skoog (MS) agar medium, chilled for 3 to 4 d at 4°C, and then transferred to growth chambers under LDs (16 h light/8 h dark) or SDs (8 h light/16 h dark) with an illumination rate of 90 to 110 μmol m−2 s−1 of white fluorescent light at 22°C.
Light Treatment
For red-light treatment, plants were exposed to 30 to 40 μmol m−2 s−1 of red light supplied by fluorescent tubes (FL 20S-Re66; Toshiba) filtered through a 3-mm-thick red acrylic plate (Shinkolite A102; Mitsubishi Rayon). For far-red light treatment, plants were exposed to 35 to 45 μmol m−2 s−1 of far-red light supplied by fluorescent tubes (FL 20S-FR74; Toshiba) filtered through a 3-mm-thick far-red acrylic plate (Deraglass 102; Asahikasei). The light intensity was measured by an optical power meter (Model 1830 C; New Port).
Plasmid Construction
For the complementation test, a VOZ1 genomic fragment including 3379 bp of its upstream region and a VOZ2 genomic fragment including 1870 bp of its upstream region were subcloned into the pDONR-221 vector (Invitrogen). These constructs were introduced into the pGWB1 plant expression vector by LR reaction of the Gateway system (Invitrogen). To construct ProVOZ1:GUS-VOZ1, a 3417-bp upstream fragment and the CDS plus introns with its 517-bp downstream fragment were amplified from genomic DNA. A GUS-containing fragment was amplified from pGWB3. The three PCR products were assembled by triple-template PCR (Tian et al., 2004). This triple-template PCR product was cloned into the pCAMBIA1300 vector. To construct ProVOZ2:GUS-VOZ2, a 1868-bp upstream fragment and the CDS plus introns with its 455-bp downstream fragment were amplified from genomic DNA. These fragments were assembled as described for the ProVOZ1:GUS-VOZ1 construct. The CDS for Pro35S:GFP-VOZ2 was amplified by PCR with cDNA synthesized from purified total RNA. The amplified fragments were subcloned into pENTR/D-TOPO using the Gateway TOPO cloning kit (Invitrogen). This DNA construct was introduced into the pGWB6 vector by the LR reaction of the Gateway system (Invitrogen). For BiFC analysis, CDSs of PHYB, VOZ1, and VOZ2 were subcloned into pENTR/D-TOPO as described above. The PHYB/pENTR/D-TOPO construct was introduced into pB4CY2 and VOZs/pENTR/D-TOPO into pB5NY0 by the LR reaction. To construct Pro35S:GFP-VOZ2-NLS and Pro35S:GFP-VOZ2-NES, we amplified the VOZ2 sequence with primers fused with the NLS or NES sequence (Matsushita et al., 2003). These fragments were subcloned into pENTR/D-TOPO. This DNA construct was introduced into the pGWB6 vector by the LR reaction. pGWB1, pGWB3, and pGWB6 were donated by T. Nakagawa, Shimane University (Nakagawa et al., 2008). pB4CY2 and pB5NY0 were obtained from S. Mano, National Institute of Basic Biology, Japan.
Isolation of Mesophyll Cells and Vascular Bundles
Mesophyll protoplasts and vascular bundles were isolated from 10-d-old seedlings as described by Endo et al. (2005).
Histological Analysis of GUS Staining
Samples were collected at ZT8 for LD-grown plants. For GUS staining, tissues were incubated for 15 min in 90% (v/v) acetone on ice and infiltrated with staining solution (0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 100 mM sodium phosphate buffer, pH 7.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 0.1% [v/v] Triton X-100) under vacuum for 15 min and incubated at 37°C for ∼15 h in the dark. After staining, samples were cleared in a mixture of ethanol and acetic acid (6:1 [v/v]) for 16 h at room temperature and then cleared in 70% (v/v) ethanol. The GUS histochemical staining was observed with a microscope (SZX16; Olympus). For sectioning, 10-d-old seedlings were incubated for 15 min in 50% (v/v) acetone on ice and then treated with staining solution under vacuum for 15 min and incubated at 37°C for ∼15 h in the dark. After staining, samples were incubated in fixative solution (5% formaldehyde, 5% acetic acid, and 56% ethanol), dehydrated through an ethanol series, embedded in Technovit 7100 (Heraeus Kulzer), sectioned at a thickness of 10 μm with a microtome (Microm), and observed with a microscope (FSX100; Olympus).
RNA Isolation and Expression Analysis
Plants were grown at 22°C for 10 d under LD or continuous white light conditions with fluorescent light and then harvested for RNA isolation at various times. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). Isolated RNA was treated with RNase-free DNase (Qiagen). Reverse transcription was performed on 1 μg total RNA with oligo(dT) primer using the Rever Tra Ace first-strand cDNA synthesis kit (Toyobo). Real-time quantitative PCR was performed with the CFX96 real-time PCR detection system (Bio-Rad) using SYBR Premix Ex Taq (TaKaRa). Expression of ACT2 was used for normalization. The following thermal profile was used for all PCRs: 95°C for 10 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. RT-PCR was performed with Ex Taq (TaKaRa), and PCR products were separated in agarose gel and visualized by staining with ethidium bromide. Primer sets used are listed in Supplemental Table 1 online.
Detection of GFP-VOZ2
A confocal laser scanning microscope (FluoView 1000; Olympus) was used to detect green fluorescence from GFP (observation, 500 to 560 nm; excitation, 488 nm) and red autofluorescence from chlorophyll (observation, 650 to 750 nm; excitation, 458 nm).
BiFC Assay
Agrobacterium cultures carrying plasmids for BiFC were grown overnight at 28°C in 10 mL Luria-Bertani plus selective antibiotics, collected by centrifugation, and adjusted to an OD600 of 1.2 in infiltration medium (10 mM MgCl2, 150 μM acetosyringone, and 10 mM MES, pH 5.6). Cells were kept at room temperature in this infiltration medium for 2 to 3 h and then infiltrated into the abaxial air spaces of 3- to 4-week-old Nicotiana benthamiana plants. After infiltration, plants were kept under continuous white light and then treated with far-red light for 30 min, followed by 6 h of darkness (dark condition). After dark treatment, plants were exposed to red light or far-red light for 6 h. Fluorescence from YFP (observation, 520 to 560 nm; excitation, 515 nm) was observed after the light treatment and ∼30 h after infiltration. Fluorescent signals and bright-field images were captured using a confocal laser scanning microscope (FluoView 1000).
Immunochemical Assay
Total protein was extracted from seedlings by grinding fresh tissue in buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [v/v] Triton X-100, and 1× Complete EDTA-free proteinase inhibitor [Roche]). Proteins were visualized with a standard SDS-PAGE method (6% gel). Protein gel blotting was performed using a standard method (as described in the ECL Plus Reagent protocol; GE Healthcare/Amersham). The VOZ2-specific antibody (prepared using the recombinant GST-fused full-length protein of At-VOZ2 as an antigen) was kindly provided by Masa H. Sato, Kyoto Prefectural University, and was diluted 1:3000. Anti-GFP IgG antibody (Invitrogen) was diluted 1:5000. ECL anti-rabbit IgG antibody (horseradish peroxidase–linked species-specific whole antibody; Amersham) diluted 1:10,000 was used as secondary antibody. Protein blots were visualized using the horseradish peroxidase–based ECL Plus reagent (GE Healthcare/Amersham) with an Image Quant LAS 4010 biomolecular imager (GE Healthcare).
Subcellular Fractionation
The aerial parts of 10-d-old plants (0.3 g) were chopped with a razor blade in a Petri dish on ice in 1 mL chopping buffer (50 mM HEPES-KOH, pH 7.5, 5 mM EDTA, 0.4 M Suc, 1× Complete EDTA-free proteinase inhibitor [Roche], and 50 μM MG132 [Wako Japan]). The homogenate was filtered through a cell strainer (70-μm nylon; BD Biosciences). The filtrate (200 μL) was centrifuged at 1000g for 20 min at 4°C. The supernatant was designated as the cytosolic fraction and diluted to 200 μL volume. The pellet was washed two times in nuclei resuspension buffer (20 mM Tris-HCl, pH 8.0, 25% glycerol, 2.5 mM MgCl2, and 0.5% Triton X-100) (Cho et al., 2006) and resuspended in 200 μL chopping buffer to generate the nuclear fraction or resuspended in 40 μL chopping buffer to generate a concentrated nuclear fraction. Each fraction was subjected to immunoblot analysis. Anti-UGPase (Agrisera) was diluted 1:2000 and anti-H3 (Active motif) was diluted 1:10,000.
Co-IP
Immunoprecipitation was performed with μMACS epitope tag protein isolation kits (Miltenyi Biotec). Pro35S:GFP seeds were kindly provide by Shoji Mano and Mikio Nishimura (Mano et al., 2002). Whole seedlings of Pro35S:GFP-VOZ2/voz1 voz2 or Pro35S:GFP plants (0.5 g fresh weight) were homogenized with liquid nitrogen and solubilized in 1.2 mL of buffer (50 mM Tris-HCl, pH 8.0, 1% [v/v] Triton X-100, 50 mM NaCl, 3× Complete EDTA-free proteinase inhibitor [Roche], and 80 μM MG132 [Wako Japan]). Homogenates were centrifuged at 10,000g for 20 min at 4°C to remove cellular debris. Then, 1 mL of the supernatant was mixed with 50 μL of magnetic beads conjugated to an anti-GFP antibody (Miltenyi Biotec) and then incubated on ice for 30 min. The mixtures were applied to μColumns (Miltenyi Biotec) in a magnetic field. After four washes with the buffer and one rinse with Wash Buffer 2 (Miltenyi Biotec), the microbeads were eluted with 70 μL of 2× SDS sample buffer (60 mM Tris-HCl, pH 6.8, 5% [v/v] SDS, 20% [v/v] glycerol, and 10% [v/v] 2-mercaptoethanol) to obtain ∼90 μL of bound fraction. The immunoprecipitates were subjected to immunoblot analysis. The volumes of each fraction loaded were T:U:B = 5:5:35 (μL). The monoclonal antibody mBA2 against PHYB, provided by Akira Nagatani, was used for detection (Shinomura et al., 1996).
MG132 Treatment
Ten-day-old seedlings grown under continuous white light were transferred to liquid half-strength MS medium supplemented with or without 50 μM MG132 (Wako Japan), incubated under white light for 3 h, and then treated with far-red light for 12 h. Total proteins were extracted and subjected to SDS-PAGE followed by protein gel blotting using anti-VOZ2 antibody.
Phosphatase Treatment
Total proteins were extracted from seedlings grown under continuous white light for 10 d. Protein phosphatase treatment was performed with Lambda protein phosphatase (New England Biolabs) according to the manufacturer’s instructions.
Yeast Two-Hybrid Assay
The Matchmaker two-hybrid system 3 (Clontech) was used for the yeast two-hybrid assay. A cDNA library constructed from mRNA purified from the aerial portions of wild-type plants (Col) at bolting was provided by Miho Takemura, Ishikawa Prefectural University. As bait, full-length Arabidopsis PHYB was subcloned into a modified pGBKT7 vector (the original NdeI site in the multiple cloning site was disrupted and a new NdeI site was created at the start codon of the GAL4 DNA binding domain). Transformants (1.8 × 106) were selected on SD medium lacking His, Trp, and Leu (SD−His/−Trp/−Leu) and supplemented with 1 mM 3-amino 1,2,4-triazol.
In Vitro Binding Assay
The fragments of full-length and N-terminal (1 to 1953 bp) PHYB were cloned into the NdeI site of the pGBKT7 vector. Each encoded protein was synthesized separately in vitro using 35S-Met in the TNT T7/T3 coupled reticulocyte lysate system kit (Promega) in accordance with the manufacturer’s protocol. VOZ1 coding fragments were cloned into the EcoRI-SalI sites of pGEX-6P-1 (GE Healthcare Bio-Sciences) for expression in Escherichia coli strain BL21 (DE3). After 6 h of 1 mM isopropyl-β-d-thiogalactopyranoside induction at 16°C, the fusion proteins were extracted in buffer (50 mM Na-PO4, pH 7.0, 100 mM NaCl, 0.1% [v/v] Triton X-100, 1 mM 2-mercaptoethanol, and 1× Complete proteinase inhibitor [Roche]) and purified on glutathione-Sepharose 4B (GE Healthcare Bio-Sciences). The binding reaction was conducted by mixing phyB with either GST or GST-VOZ1 immobilized on glutathione sepharose beads in PBS buffer with 1× Complete proteinase inhibitor (Roche) and incubating the mixture at room temperature for 2 h. Following nine washes with PBS buffer, the proteins were eluted with 2× sample buffer. Proteins retained on the beads were resolved by SDS-PAGE and visualized by autoradiography.
Hypocotyl Elongation
Seedlings were grown on one-tenth MS agar medium without Suc for 5 d under 30 μmol m−2 s−1 red light or 35 μmol m−2 s−1 far-red light.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: At1g28520 (VOZ1), At2g42400 (VOZ2), At2g18790 (PHYB), At3g18780 (ACT2), At1g65480 (FT), At5g15840 (CO), At5g10140 (FLC), At5g62690 (TUB2), At2g26670 (HY1), At3g09150 (HY2), At5g38420 (RBCS-2B), and At1g22710 (SUC2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Interaction of VOZ Proteins with phyB.
Supplemental Figure 2. T-DNA Insertional Mutants for VOZ Genes.
Supplemental Figure 3. Phenotype of voz Mutants Grown under LD or SD Conditions.
Supplemental Figure 4. Hypocotyl Length Phenotype of voz Mutants.
Supplemental Figure 5. Chlorophyll and Anthocyanin Accumulation Levels.
Supplemental Figure 6. VOZ Gene Expression Patterns.
Supplemental Figure 7. Complementation Test and GUS Staining of ProVOZ:GUS-VOZ/voz1 voz2 Transgenic Plants.
Supplemental Figure 8. Subcellular Localization of GFP-VOZ2 Fusion Protein under Different Light Conditions.
Supplemental Figure 9. Relative Expression Levels of FT, CO, and FLC, and Flowering Time in VOZ2 Overexpression Lines.
Supplemental Figure 10. Expression of GFP-VOZ2 mRNA in NLS and NES Lines.
Supplemental Figure 11. Subcellular Localization of phyB-YFP in Arabidopsis.
Supplemental Figure 12. Expression of VOZ mRNA under Different Light Conditions.
Supplemental Figure 13. VOZ2 Protein Accumulation Levels in phy Mutants under Different Light Conditions.
Supplemental Figure 14. Schematic Illustration of a Model for VOZ Function.
Supplemental Table 1. PCR Primers Used in the Expression Analysis.
Supplementary Material
Acknowledgments
We thank Masa H. Sato, Yusuke Nakai, and Nobutaka Mitsuda for discussion and supplying us with the VOZ2 antibody. We thank Tsuyoshi Nakagawa and Shoji Mano for providing the Gateway binary plasmids, Miho Takemura for the two-hybrid library, Akira Nagatani for the monoclonal antibody mBA2, Mikio Nishimura for Pro35S:GFP seeds, Motomu Endo for technical advice on tissue separation, Makoto Shirakawa for technical advice and discussion, Ryohei Thomas Nakano and Haruko Ueda for technical advice on the in vivo co-IP experiment, and Ayako Yamaguchi, Takashi Yamano, Kimitsune Ishizaki, and Katsuyuki T. Yamato for critical reading of the article. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (21026018 and 23012025 to T.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, for Scientific Research B (18380200 and 23380058 to T.K.) and for the Japan Society for the Promotion of Science Fellows (23-5444 to Y.Y.) from the Japan Society for the Promotion of Science and by Plant Global Education Project of the Nara Institute of Science and Technology.
AUTHOR CONTRIBUTIONS
Y.Y., K.M., and T.K. designed the research. Y.Y., K.M., M.U., A.Y., R.S., and A.N. performed research. Y.Y., K.M., and M.U. analyzed data. Y.Y. and T.K. wrote the article.
Glossary
- PIF
phytochrome-interacting factor
- LD
long-day
- SD
short-day
- GUS
β-glucuronidase
- GFP
green fluorescent protein
- CDS
coding sequence
- NLS
nuclear localization signal
- NES
nuclear export signal
- Col
Columbia
- UGPase
UDP-Glc phyrophosphorylase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- co-IP
coimmunoprecipitation
- Ler
Landsberg erecta
- MS
Murashige and Skoog
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Cerdán P.D., Chory J. (2003). Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cho Y.H., Yoo S.D., Sheen J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Chory J. (2010). Light signal transduction: An infinite spectrum of possibilities. Plant J. 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L.Y., Shao H.B., Li M.Y. (2005). Molecular mechanisms of phytochrome signal transduction in higher plants. Colloids Surf. B Biointerfaces 45: 154–161 [DOI] [PubMed] [Google Scholar]
- Clack T., Mathews S., Sharrock R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Devlin P.F., Yanovsky M.J., Kay S.A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., Nilsson A., Ulfstedt M., Ronne H., Wingsle G., Nilsson O., Björklund S. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Mochizuki N., Suzuki T., Nagatani A. (2007). CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Nakamura S., Araki T., Mochizuki N., Nagatani A. (2005). Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17: 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13: 203–229 [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosey L., Palecanda L., Sharrock R.A. (1997). Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115: 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Halliday K.J., Salter M.G., Thingnaes E., Whitelam G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Deng X.W. (2000). The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 124: 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Kay S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Iñigo S., Alvarez M.J., Strasser B., Califano A., Cerdán P.D. (2012). PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Kaneda T., Taga Y., Takai R., Iwano M., Matsui H., Takayama S., Isogai A., Che F.-S. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott J.E. (1934). Effect of a localized photoperiod on spinach. Proc. Am. Soc. Hortic. Sci. 31: 152–154 [Google Scholar]
- Kohchi T., Mukougawa K., Frankenberg N., Masuda M., Yokota A., Lagarias J.C. (2001). The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43: 331–341 [DOI] [PubMed] [Google Scholar]
- Más P., Devlin P.F., Panda S., Kay S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N., Nagatani A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Hisabori T., Takeyasu K., Sato M.H. (2004). VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Mockler T., Yang H., Yu X., Parikh D., Cheng Y.C., Dolan S., Lin C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100: 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T., Kohchi T., Yokota A., Hwang I., Goodman H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7: 708–711 [DOI] [PubMed] [Google Scholar]
- Nagy F., Schäfer E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53: 329–355 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., Tomita Y., Dohi K., Mori M., Araki T. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49: 1645–1658 [DOI] [PubMed] [Google Scholar]
- Oka Y., Kong S.G., Matsushita T. (2011). A non-covalently attached chromophore can mediate phytochrome B signaling in Arabidopsis. Plant Cell Physiol. 52: 2088–2102 [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Deng X.W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Ou B., Yin K.Q., Liu S.N., Yang Y., Gu T., Wing Hui J.M., Zhang L., Miao J., Kondou Y., Matsui M., Gu H.Y., Qu L.J. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Paik I., Yang S., Choi G. (2012). Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. USA 109: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. (2012). NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Quail P.H., Boylan M.T., Parks B.M., Short T.W., Xu Y., Wagner D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268: 675–680 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Zhu D., Li J., Rubio V., Zhou Z., Shen Y., Hoecker U., Wang H., Deng X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Deng X.W. (2000). The COP/DET/FUS proteins-regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11: 495–503 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R.A., Quail P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Conn A.B., Dennis E.S., Peacock W.J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Hanzawa H., Kubota M., Watanabe M., Furuya M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Quail P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7: 413–427 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2004). Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7: 4–10 [DOI] [PubMed] [Google Scholar]
- Takada S., Goto K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S., Kong S.G., Mineyuki Y., Furuya M. (2003). Regulation of actin-dependent cytoplasmic motility by type II phytochrome occurs within seconds in Vallisneria gigantea epidermal cells. Plant Cell 15: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hudson M.E., Khanna R., Zhu T., Chang S.H., Wang X., Quail P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Whitelam G.C., Patel S., Devlin P.F. (1998). Phytochromes and photomorphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Wollenberg A.C., Strasser B., Cerdán P.D., Amasino R.M. (2008). Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Zeevaart J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27: 321–348 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.