In flowering plants, two female gametes, the egg and the central cell, interact with two sperm cells during double fertilization to give rise to an embryo and the endosperm. This work shows that the two events are uncoupled in the glauce mutant where no endosperm evolves and that the central cell is actively involved in sperm reception, by expressing a member of the BAHD superfamily.
Abstract
Double fertilization of the egg cell and the central cell by two sperm cells, resulting in the formation of the embryo and the endosperm, respectively, is a defining characteristic of flowering plants. The Arabidopsis thaliana female gametophytic mutant glauce (glc) can exhibit embryo development without any endosperm. Here, we show that in glc mutant embryo sacs one sperm cell successfully fuses with the egg cell but the second sperm cell fails to fuse with the central cell, resulting in single fertilization. Complementation studies using genes from the glc deletion interval identified an unusual genomic locus having homology to BAHD (for BEAT, AHCT, HCBT, and DAT) acyl-transferases with dual transcription units and alternative splicing that could rescue the sterility defect of glc. Expression of these transcripts appears restricted to the central cell, and expression within the central cell is sufficient to restore fertility. We conclude that the central cell actively promotes its own fertilization by the sperm cell through a signaling mechanism involving products of At1g65450. Successful fertilization of the egg cell is not blocked in the glc mutant, suggesting that evolution of double fertilization in flowering plants involved acquisition of specific functions by the central cell to enable its role as a second female gamete.
INTRODUCTION
Reproduction in the flowering plants involves double fertilization, in which two male gametes, the sperm cells, fuse with two female gametes, the egg cell (EC) and the central cell (CC). The pollen tube transports the sperm cells to the ovule, which is the reproductive organ that contains the embryo sac (female gametophyte), where the female gametes reside. According to current models of double fertilization, the pollen tube enters and discharges the two sperm cells into one of the two synergid cells (SCs) within the embryo sac, following which the sperm cells are transported to their final destination where they fuse with the EC or CC (plasmogamy). The subsequent fusion of two pairs of gametic nuclei (karyogamy) concludes the process of double fertilization. The fertilized EC forms a diploid embryo, whereas the CC fertilization product gives rise to the triploid endosperm, a tissue that nourishes the growing embryo (Berger et al., 2008). Development of new molecular tools, such as female gametophyte-specific markers (Chen et al., 2007; Gross-Hardt et al., 2007; Steffen et al., 2007), has allowed a more precise characterization of the processes involved in double fertilization. These in conjunction with a sperm cell–specific marker (Ingouff et al., 2007) enabled live imaging studies that contributed significantly to our basic understanding of the double fertilization process in Arabidopsis thaliana (Hamamura et al., 2011).
In the female gametophytic mutant glauce (glc), a sexual globular embryo is produced upon fertilization in the absence of endosperm (Ngo et al., 2007). In addition, the glc mutation suppresses autonomous endosperm development in fertilization-independent seed class mutants (Ngo et al., 2007), a phenotype that is not addressed in this study. A segment of 215 kb was found to be deleted from chromosome 1 of glc mutants, and another 135-kb segment is duplicated elsewhere in the genome. However, the molecular identity of GLC has remained unknown up to now. Other features of the glc phenotype, such as which specific steps are disrupted in the process of double fertilization leading to the absence of endosperm formation, also remained to be determined.
Here, we report that a failure of sperm cell fusion with the CC occurs in glc embryo sacs. Moreover, we found that a gene encoding a BAHD (for BEAT, AHCT, HCBT, and DAT) transferase and its isoforms, which are encoded within the 215-kb deletion, were able to rescue the fertilization defect of glc mutants. The BAHD acyl-transferases form a superfamily of plant acyl-CoA using enzymes, which are thought to be involved in secondary metabolism (D’Auria, 2006). Our findings indicate that the two gametic fusion events are distinct and that fusion of the CC with the sperm cell for double fertilization requires the function of this BAHD acyl-transferase.
RESULTS
The Double Fertilization Process in glc Embryo Sacs
Upon fertilization, glc ovules were found to exhibit a remarkable phenotype in which the embryo develops without any endosperm (Ngo et al., 2007). To further characterize the glc phenotype and determine why no endosperm developed, the process of the double fertilization was further studied in glc mutant embryo sacs.
Effect on Pollen Tube Guidance in glc Ovules
First we checked for any possible effects of the glc mutation on pollen tube guidance by pollinating emasculated wild-type and glc/GLC flowers with pollen of the LAT52:GUS (for β-glucuronidase) pollen-specific marker. Pistils were sampled 8 and 14 h later, and GUS presence was detected at the micropylar end of the embryo sac. At 8 h after pollination (HAP), a significant difference between the wild type and glc was observed, where pollen tubes reached over 90% of wild-type ovules (144/158) and burst into their SC, while in glc/GLC pistils pollen tubes reached only 64% (148/230) of the ovules (see Supplemental Figure 1 online).
Considering the heterozygous genetic background of glc/GLC in which half of its embryo sacs are normal, most of the pollen tube arrivals detected at this time point (∼45%; since wild-type ovules are 50% of the total and have a 91% arrival rate) were likely to be at wild-type ovules. This implies that ∼62.5% of the glc mutant ovules did not yet receive any pollen tubes (see Supplemental Figure 1 online). By 14 HAP, however, 90% of the ovules in glc/GLC pistils received a pollen tube, a level that does not differ statistically from wild-type plants (see Supplemental Figure 1 online). Therefore, we conclude that the glc mutation results in an initial delay in pollen tube arrival, possibly as a consequence of weaker pollen tube attraction.
Nonpollinated mature glc/GLC pistils usually contain ∼50 to 60 normal-looking ovules, indistinguishable from one another. However, ∼24 HAP, the ovule population segregates into approximately two even groups: big, normal-looking ovules and small retarded ones (see Supplemental Figure 2 online and Figure 1C). This delineation persists into the mature silique, in which the normal ovules go through successful seed formation, whereas the small ovules abort, resulting in the semisterile phenotype of glc/GLC plants (see below). This observation also confirms that the small ovules indeed harbor nonfunctional female gametophytes.
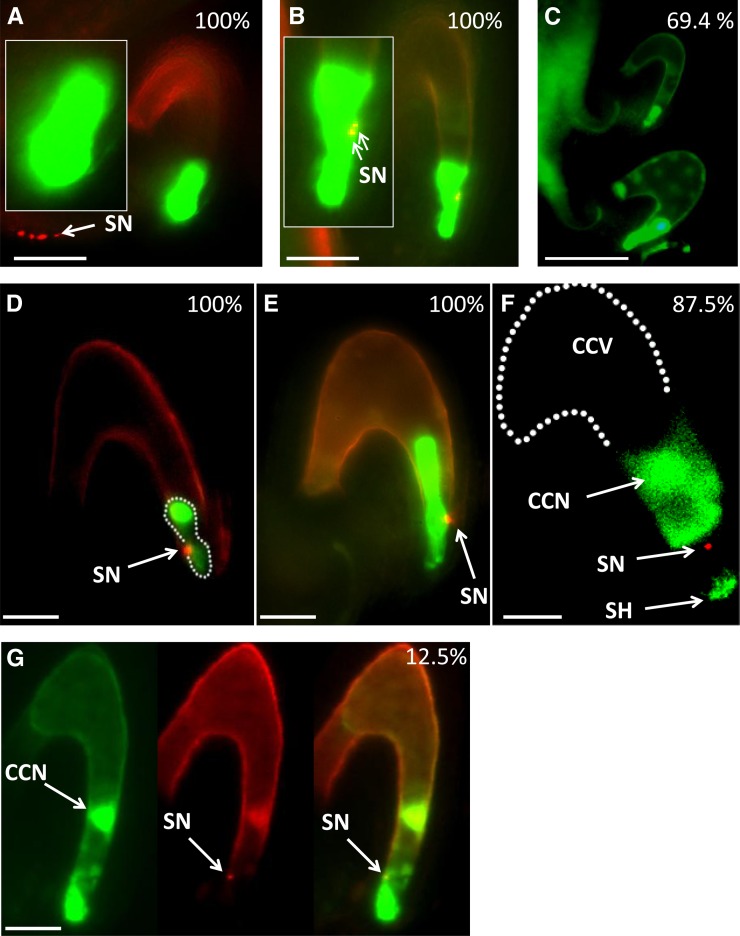
Figure 1.
The Double Fertilization Process in glc Mutant Embryo Sacs.
Representative fluorescence microscopy micrographs of glc embryo sacs expressing GFP markers in the SCs (DD31) ([A] and [B]), EC (DD45) ([C] to [E]), and CC (DD65) ([F] and [G]). In all micrographs except (C), the green and red fluorescent channels were merged and basal autofluorescence was captured to ensure visualization of the embryo sac. Image (F) is part of a single confocal section (captured by a MRC-1024 confocal microscope). The complete Z-stack images can be found in Supplemental Figure 3 online. Emasculated glc/GLC flowers were pollinated with pollen carrying the HTR10 sperm-specific RFP marker. Images were captured at least 24 HAP. White dashed lines indicate the late zygote stage in (D) and the central cell vacuole (CCV) in (F). White arrows throughout the figure point to sperm nuclei (SN; note that in [A], the white arrow points to four sperm nuclei that belong to two pollen tubes arriving at the funiculus); in (F), white arrows points to the probable location of the central cell nucleus (CCN) and to a central cell cytoplasmic protrusion at the micropylar end of the embryo sac called the synergid hook (SH). An embryo is shown in (F). n = 40 ovules for DD31 in (A) and (B) each, 70 for DD45 in (C), 30 for (D) and (E) each, and 40 for DD65. Occurrence percentages are presented in white, at the top right-hand side of the images. Bars = 100 μm in (C), 50 μm in (A) and (B), 25 μm in (D), (E), and (G), and 20 μm in (F).
Sperm Cells Persist Longer in the Micropylar End of glc Ovules
To elucidate the fertilization process in glc embryo sacs, we pollinated glc/GLC heterozygotes with pollen carrying the sperm cell–specific histone HTR10 fused to the red fluorescent protein (RFP) as a marker for the sperm cell (Ingouff et al., 2007). Knowing that by 14 HAP the pollen tube burst occurred in most of the glc ovules, we determined the fate of the discharged sperm cells in these ovules at 24 HAP or later. At that time, the presence of sperm cells was detected in only 15 wild-type ovules out of the 308 examined (4.9%), whereas in the glc/GLC mutant, sperm cells were detected in 132 ovules out of 451 (29.3%) (see Supplemental Table 1 online). Bearing in mind the heterozygous genetic background of the glc/GLC mutant, the actual percentage of the sperm containing glc ovules was ∼55%, indicating disturbance of the fertilization process in the mutated ovules. Moreover, 53.7% (71/132) of the sperm-containing ovules observed in glc/GLC pistils contained only single sperm cells, which we never observed in wild-type pistils (see Supplemental Figure 2 online).
The Progress of the Double Fertilization Process in glc Ovules
To further characterize the persistent single sperm cells observed in the embryo sacs of glc/GLC pistils, we introduced the following embryo sac cell–specific green fluorescent protein (GFP) marker lines (Steffen et al., 2007) into the glc/GLC mutant background: DD31 (SCs), DD45 (EC), and DD65 (CC). Emasculated flowers of F1 progenies from these crosses were pollinated with HTR10:RFP pollen, and 24 HAP or later glc/GLC pistils were sampled. Based on their small size, glc ovules were dissected out and the location of the sperm nuclei in their embryo sacs at the time of dissection was observed using the fluorescent markers.
Prior to sperm discharge, the SCs were round shaped and turgid in all the observed cases (n = 40/40) (Figure 1A). SCs observed after sperm discharge exhibited shape changes (Figure 1B, compare with SC shape in Figure 1A), suggesting that at this stage the glc SC might be undergoing degeneration (Sandaklie-Nikolova et al., 2007).
The delay in pollen tube arrival and burst into glc ovules followed by longer persistence of the discharged sperm cells in the glc embryo sac micropylar end even at 24 HAP or later led to delayed EC fertilization in glc ovules. As reported by Steffen et al. (2007), the DD45:GFP reporter, which marks the EC, persists further into postfertilization stages, from the zygote to early stages of embryo development. The two ovules shown in Figure 1C both expressed the DD45 marker. The right ovule had a well-developed embryo and several endosperm nuclei indicated by traces of autofluorescence, whereas in the smaller ovule, the EC had not yet been fertilized, as indicated by the fact that it had not yet adopted the typical compacted structure of the zygote. The percentage of unfertilized glc ovules at 24 HAP was 69.4% (n = 50/70), while in wild-type ovules, fertilization had already occurred in 94% (n = 62/66) of the ECs, based on using the DD45 marker to monitor their change into the characteristic shape of the zygote.
Since fertilization of the EC in glc ovules was delayed, a lag phase in development of glc embryos as compared with wild-type embryos was expected and indeed observed previously (Ngo et al., 2007). The lack of support of the nourishing endosperm in glc ovules probably enhanced the retarded development of glc embryos when reaching the preglobular stage, and these embryos eventually abort (Ngo et al., 2007).
CC Fertilization Is Prevented in glc Embryo Sacs as Demonstrated by the Failure of the Second Sperm to Enter the CC
After sperm cell release, embryo formation could be detected in the embryo sac of the small defective ovules, but the HTR10:RFP signal corresponding to a single sperm nucleus persisted in the vicinity of the egg apparatus (see Supplemental Figure 2 online). Once the sperm nucleus fused with the EC nucleus, the HTR10:RFP was rapidly removed. This removal is thought to play a role in remodeling of the zygotic chromatin (Ingouff et al., 2007). In glc embryo sacs, EC fertilization is performed successfully; however, glc embryos were observed to exhibit some inhibition of paternal gene expression (Ngo et al., 2007). These observations raised the possibility that the normal rapid removal of HTR10 might be disrupted in the glc zygote. In that case, the persisting HTR10:RFP signal would colocalize with the EC nucleus in the postfertilization stages rather than disappear as in the wild type. Observations of glc late zygotes (Figure 1D) and embryos (Figure 1E) clearly showed that the HTR10:RFP and the DD45 GFP signals did not overlap (the yellow portion of the red spot in Figure 1E is due to partial overlay of red and green signals coming from multiple layers), indicating exclusion of this sperm cell signal from the EC. Therefore, the persisting HTR10:RFP signal after EC fertilization must arise from the second sperm cell, which would normally have fertilized the CC, and is not due to defective removal of the HTR10 from the sperm nucleus fertilizing the EC nucleus. In 100% of the observation samples (30/30), the second sperm nucleus was associated with the micropylar end of the late zygote/embryo.
We then examined the cause underlying the persistence of this second sperm cell nucleus. This could arise from either a failure of fusion of the sperm cell with the CC (plasmogamy) or a failure of the migration of the sperm nucleus after entry into the CC cytoplasm and the consequent failure of karyogamy (i.e., fusion with the CC nucleus). Therefore, glc/GLC plants carrying the marker DD65:GFP, in which GFP was expressed specifically in the CC, were pollinated with HTR10:RFP pollen and observed. Observations of fertilized glc embryo sacs expressing the DD65 CC marker showed in 87.5% of the cases (35/40) that the localization of the persisting RFP signal was clearly external to the CC (Figure 1F; see full Z-stack images in Supplemental Figure 3 online). We conclude that in the glc mutant, failure of the second fertilization event is due to failure of fusion of the sperm cell and the CC, leading to the absence of endosperm formation from the glc CC in these ovules.
However, in 12.5% of the observations (5/40), we noted that the sperm RFP and the CC GFP signals did overlap (Figure 1G). The latter observations were performed at 30 HAP or later, when the SCs had already degenerated and their space was occupied by the cytoplasmic protrusions of the CC at the micropylar end (called synergid hooks; Russell, 1993), which normally develops into the micropylar endosperm after fertilization. The expansion of the CC into this zone might lead to the CC surrounding the remaining sperm cell, resulting in the observed overlap of signals even in the absence of plasmogamy. However, we also cannot rule out that in some cases, plasmogamy was successful but that the subsequent migration of the sperm nucleus to the CC nucleus was disrupted. The latter interpretation is partially supported by previous observations that a low rate (5%) of two to eight endosperm nuclei was observed in glc ovules (Ngo et al., 2007). It is possible that in these cases there was successful cell fusion but no karyogamy, which could trigger autonomous endosperm formation. In a study of fertilization by pollen from the cdka:1 mutant, it has been shown that initiation of autonomous endosperm by CC nuclear divisions was correlated with the failure of karyogamy of the CC and sperm nucleus (Aw et al., 2010), although in that study the migration of the sperm cell nucleus after entry into the CC appeared to be unaffected in the mutants.
Molecular Identification of the Genes for Double Fertilization within the glc Deletion
To understand the molecular basis for the failure of CC fertilization in glc mutants, we embarked on the molecular isolation of the gene(s) responsible. A segment of 215 kb was reported to be deleted from chromosome #1 in the glc mutant (Ngo et al., 2007; Figure 2A). To examine the possibility that the glc phenotype is caused by loss of function of a single gene, we obtained and screened knockout lines in 25 out of the ∼55 annotated genes residing in the deletion. However, we did not observe the glc phenotype in any of these gene knockouts (list of all knockout lines screened here is found in Supplemental Table 2 online).
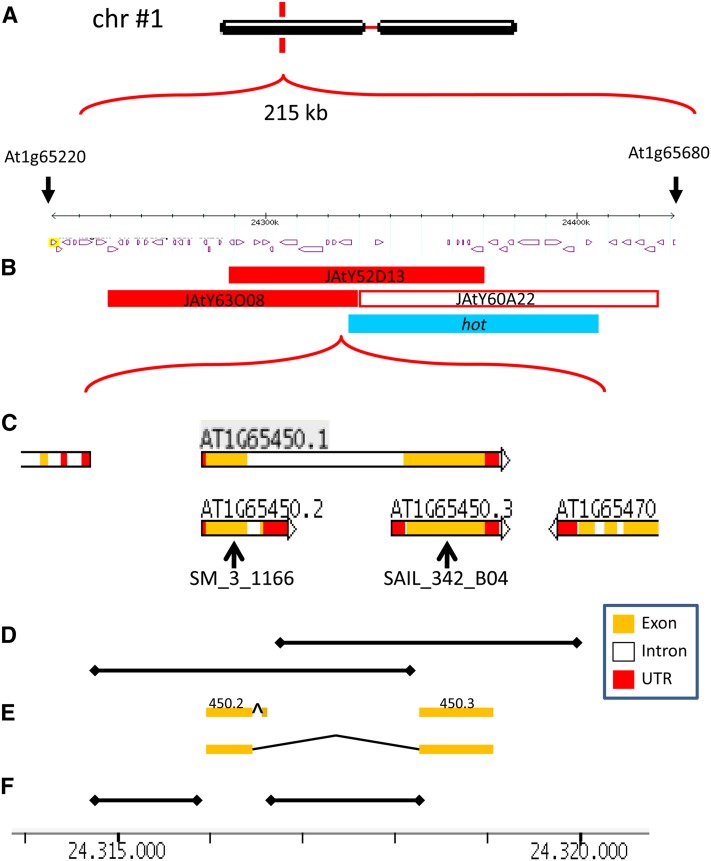
Figure 2.
Fine Mapping of the glc Deletion Region.
(A) The 215-kb deleted region on glc chromosome 1 (Ngo et al., 2007).
(B) Exact position of the JAtY BACs used in this study and of the reported hot deletion mutant (Kaya et al., 2000).
(C) Close-up of the right border of BAC JAtY 63O08 and gene model of At1g65450, adapted from The Arabidopsis Information Resource. Black arrows indicate localization of T-DNA in mutant lines.
(D) Genomic DNA regions cloned in this study indicated by horizontal black lines. UTR, untranslated region.
(E) cDNAs cloned in this study are indicated by orange bars.
(F) Intergenic regions (putative promoters) cloned in the study are indicated by horizontal black lines. Bar at the base of the figure is in kilobases and shared by (C) to (F).
We then attempted to narrow down the region responsible for the glc phenotype, using complementation by BAC clones that overlie the deletion interval. Mutant heterozygotes were transformed with the following BAC clones (JAtY, John Innes Center) that together covered the deleted region: JAtY63O08 (At1g65290 to At1g65470), JAtY52D13 (At1g65380 to At1g65540), and JAtY60A22 (At1g65480 to At1g65660) (Figure 2B). Seeds were collected from successful glc BAC transformed plants and were plated on plates with kanamycin (kan) to determine the kan resistance/kan sensitivity (kanr/kans) segregation ratio to assay whether the BACs could rescue the glc defect.
The kanr/kans segregation ratio in glc heterozygotes was 0.58:1, in a range typical of gametophytic mutations (Moore et al., 1997; Pagnussat et al., 2005). This ratio increased to 2.35:1 and 2.41:1 in glc T2 plants transformed with the BACs JatY63O08 and JAtY52D13, respectively, in a statistically significant fashion (Table 1). The kanr/kans segregation ratio remained unchanged (0.56:1) when glc plants were transformed with BAC JAtY60A22 (Table 1). The failure of this BAC to complement glc is consistent with the absence of the glc phenotype in the reported deletion mutant hot, whose entire deletion overlaps ∼85% of JAtY60A22 (Kaya et al., 2000; blue bar in Figure 2B). Since the BACs JatY63O08 and JAtY52D13 are overlapping (Figure 2B, closed red bars), we concluded that one or more genes residing in this 35-kb region of overlap must play a role in the glc phenotype.
Table 1. Complementation of glc kanr/kans Distorted Ratio by JatY BACs.
| Genotype | kanr | kans | Ratio kanr/kans | P Value (Fisher’s Exact Test) |
|---|---|---|---|---|
| glc; JatY52D13 | 304 (70. 7%) | 126 (29.3%) | 2.41:1 | 5.11e−26 |
| glc; JatY63O08 | 214 (70.2%) | 91 (29.8%) | 2.35:1 | 7.89e−21 |
| glc; JatY60A22 | 161 (36.1%) | 285 (63.9%) | 0.56:1 | 0.84 (N.S.) |
| glc (self) | 195 (36.7%) | 335 (63.3%) | 0.58:1 | N.A. |
glc plants were transformed with JatY binary vectors containing genomic DNA segments from the glc deleted region. The segregation of the linked nptll gene conferring kanr was scored in T2 seedlings grown on kan-MS medium. N.A., not applicable; N.S., not significant.
At1g65450.2 and At1g65450.3 Complement the glc Mutant
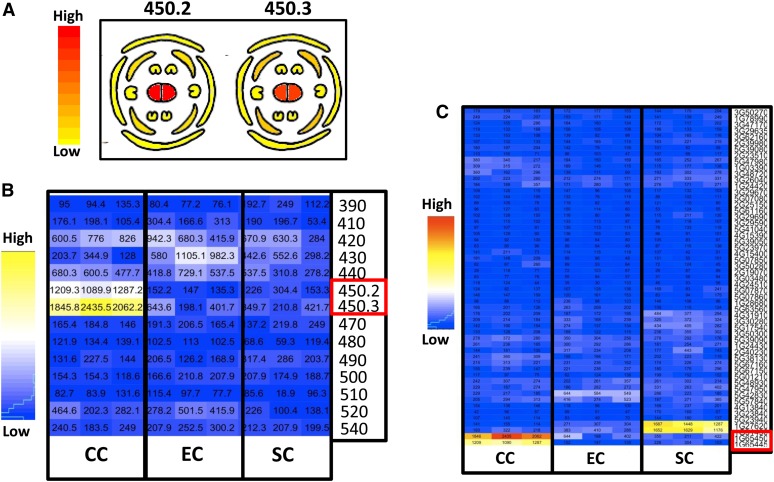
Expression data in female reproductive tissues of the 10 genes residing in the overlapping region of JAtY63O08 and JAtY52D13 were derived from the Bio Array Resource (BAR) microarray database and showed that two small neighboring unknown transferases, At1g65450.2 and At1g65450.3 (formerly annotated as At1g65445 and At1g65450, respectively), were the only ones to be highly and specifically expressed in pistils of mature nonpollinated flower (Figure 3A; data adapted from BAR; http://bar.utoronto.ca). More importantly, the recently published Arabidopsis female gametophyte transcriptome (Wuest et al., 2010) enabled us to extract specific expression profiles of these genes in the different embryo sac cell types: EC, CC, and SC. It was found that most of the genes residing in the overlapping region (and in the entire deleted region; data presented only for genes in JAtY63008) had a low expression in all three cell types, except for At1g65450.2 and At1g65450.3, which were highly expressed in the CC (Figure 3B). Furthermore, expression of these two genes could not be found in the recently published seed development transcriptome (SeedGenes database, http://seedgenenetwork.net; Le et al., 2010) either in the early endosperm (preglobular stage) or other seed developmental stages, thereby restricting the role of these genes to earlier stages. Based on this information, these two genes were considered good candidates for being affected in the glc mutant.
Figure 3.
Tissue-Specific Expression Profiles of At1g65450 and Other Arabidopsis BAHD Members.
(A) Gene expression in mature nonfertilized flower. Cross-section cartoons are presented, obtained from the BAR microarray database.
(B) Gene expression in the different embryo sac cell types of mature unfertilized ovules, extracted from the Arabidopsis female gametophyte transcriptome (Wuest et al., 2010). Shown are three replicates per cell type (CC, EC, and SC).
(C) Gene expression of 54 Arabidopsis BAHD members in the different embryo sac cell types of mature unfertilized ovules using data extracted from Wuest et al. (2010). Shown are three replicates per cell type (CC, EC, and SC) derived from the Arabidopsis female gametophyte transcriptome, as in (B). Red boxes in (B) and (C) indicate 450.2 and 450.3.
Both genes were cloned into the binary vector pCAMBIA1300 (see Figure 2C for gene models and Figure 2D for the cloned genomic regions) and transformed into glc heterozyotes. Seeds were collected from transformed glc/GLC plants and were plated on plates with kan to assess their kanr/kans transmission ratio. Both genes successfully complemented the kanr/kans segregation ratio distortion typical of glc/GLC plants (Table 2) and rescued glc/GLC semisterility in mature siliques (Figure 4, Table 3). Therefore, we concluded that both genes can complement the fertility function that is disrupted in glc mutants.
Table 2. Complementation of glc kanr/kans Distorted Ratio by Genomic DNA of 450.2 and 450.3.
| Genotype | kanr | kans | Ratio kanr/kans | P Value (Fisher’s Exact Test) |
|---|---|---|---|---|
| glc; 450.2 | 263 (72.5%) | 98 (27.5%) | 2.68:1 | 1.47e−25 |
| glc; 450.3 | 218 (68.8%) | 99 (31.2%) | 2.20:1 | 4.76e−19 |
| glc (self) | 152 (35.8%) | 273 (74.2%) | 0.56:1 | N.A. |
glc plants were transformed with pCAMBIA1300 binary vector cloned with genomic DNA of 450.2 or 450.3. The segregation of the linked nptll gene conferring kanr was scored in T2 seedlings grown on kan-MS medium. N.A., not applicable.
Figure 4.
Rescue of the glc Semisterility Phenotype by Genomic DNA of At1g65450.2 (450.2) and At1g65450.3 (450.3).
Opened siliques of wild-type (wt; Ler), glc, and glc plants transformed with genomic DNA of 450.2 and 450.3. White arrows indicate aborted ovules (more aborted ovules are present underneath the seeds). At least 10 siliques per genotype were used for the measurements presented in Table 3.
Table 3. Complementation of glc Semisterility Phenotype by Genomic DNA of 450.2 and 450.3.
| Genotype | Wild-Type–Looking Seeds | Aborted Ovules | Total | Seed Set (%) | P Value (Fisher’s Exact Test) |
|---|---|---|---|---|---|
| glc; 450.2 (HYGR+) | 374 | 131 | 505 | 74.1 | 1.26e−16 |
| glc; 450.3 (HYGR+) | 395 | 149 | 544 | 72.6 | 2.49e−15 |
| glc (self) | 288 | 293 | 580 | 49.7 | N.A. |
| glc;450.2 (HYGR-) | 233 | 284 | 517 | 45.1 | 0.13 (N.S.) |
| glc;450.3 (HYGR-) | 290 | 279 | 569 | 51 | 0.68 (N.S.) |
glc plants were transformed with pCAMBIA1300 binary vector cloned with genomic DNA of 450.2 or 450.3. Seed set was scored in at least 10 siliques of T2 plants, which were genotyped for the presence/absence of the HYG resistance gene, which is linked to the genomic cloned insert. N.A., not applicable; N.S., not significant.
It is important to note that the large-scale screen of T-DNA knockout lines mentioned above included knockouts of At1g65450.2 and At1g65450.3, corresponding to T-DNA insertion lines SM_3_1166 and SAIL_342_B04, respectively. In both cases, the insertions are in the middle of coding exons (Figure 2C); however, both lines exhibited a normal seed set and individuals homozygous for the insertion could be isolated, suggesting that 450.2 and 450.3 are genetically redundant.
Furthermore, no homozygous glc plants could be isolated among the rescued glc transformants, implying that another gene or genes within the deletion are required to restore viability in glc homozygotes. For example, loss-of-function mutants of the gene AGL23, which lies within the glc deletion interval, is known to result in seedling lethality when homozygous (Colombo et al., 2008). Another possible reason for not obtaining glc/glc homozygotes might arise from the 135-kb segment duplicated elsewhere in the genome (Ngo et al., 2007), potentially affecting the expression of other unknown genes.
The At1g65450 Locus Contains a Unique Dual Transcript Organization with an Alternately Spliced Transcript Encoding a Member of the BAHD Superfamily
As mentioned above, At1g65450.2 and At1g65450.3 were previously annotated as two separate genes that encode two small transferases of unknown function. However, we noticed that BLAST searches of each one aligns with the N-terminal and C-terminal regions, respectively, of most known acyl-transferase proteins of the BAHD superfamily (see amino acid sequence in Figure 5). These enzymes use acyl-CoA and are considered to be involved in secondary metabolism, which has been demonstrated for some of the BAHD members (D’Auria 2006). In Arabidopsis, at least 61 BAHD members were identified (Yu et al., 2009). At the amino acid level, the BAHDs are highly divergent, showing 10 to 30% similarity, yet they share several highly conserved regions, specifically the HXXXD motif that is located near the enzyme’s catalytic center and the C-terminal DFGWG motif. Manipulation of these motifs has been shown to result in a significant reduction in enzymatic activity (D’Auria, 2006).
Figure 5.
Amino Acid Sequences of At1g65450 cDNAs.
The amino acid sequences of the three At1g65450 cDNAs (Ler). At1g65450.1 product (50.5 kD) corresponds to the combined cDNA, while the products of At1g65450.2 (18.7 kD) and At1g65450.3 (32.3 kD), which are derived from the nonoverlapping 5′ and 3′ mature transcripts, are shown aligned. Highlighted in yellow are regions of similarity between the amino acid sequences of At1g65450.2 and At1g65450.3 (asterisk = identity, period = hydrophobicity or size similarity, and colon = both hydrophobicity and size similarity). The conserved BAHD motifs (bold blue text) are found only in At1g65450.1 and At1g65450.3 products. There are residues (bold red text) that are unique to At1g65450.1 and At1g65450.3. The extra amino acids in the At1g65450.3 product contain a putative canonical NLS sequence (underlined) that is absent from the other products due to alternative splicing. The extra residues in the At1g65450.1 product are a short stretch of amino acids linking the front and back ends of the combined product.
Indeed, the latest version of the Arabidopsis genome, TAIR10, has reannotated these as a single gene, At1g65450, containing two small exons separated by one long intron (see gene model in Figure 2C). Nevertheless, the known full-length cDNAs for this gene consist mostly of its shorter variants with the expected untranslated regions for two separate transcripts rather than truncated ESTs of the full gene (see Supplemental Figure 4 online). This indicates that the truncated variants are transcribed and may actually be more abundant than the combined sequence, therefore supporting the former annotation. Currently, a combination of both annotations is presented by TAIR10 (see Figure 2C for gene models that were taken from TAIR10 website).
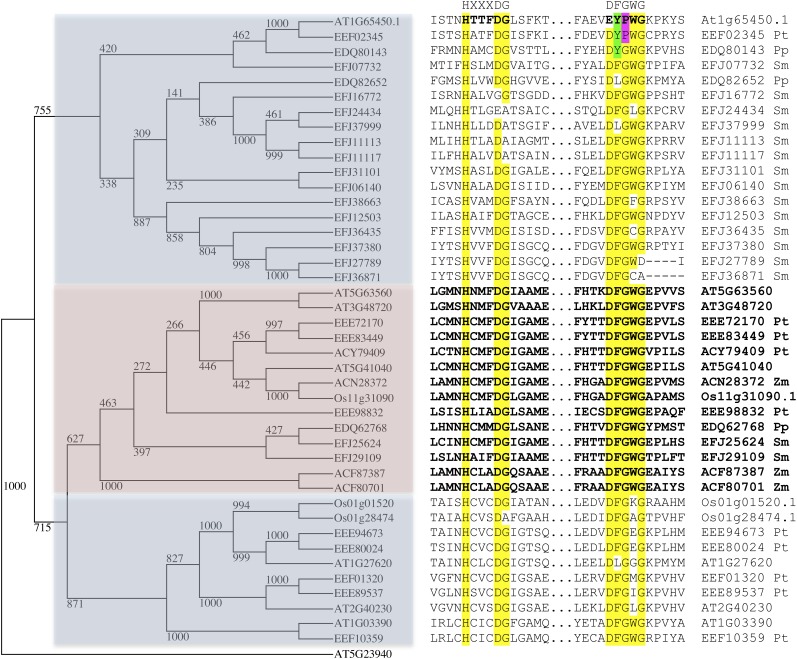
Interestingly, embryo sac–specific gene expression profiles of 54 Arabidopsis BAHD members in general showed very low expression, except for At1g65450.2 (450.2) and At1g65450.3 (450.3), which were the only ones highly expressed in the CC (Figure 3C). These data are consistent with a specific role for these two genes in the CC of the mature unfertilized ovule. In line with the former annotation, 450.2 and 450.3 were recently regarded as partial polypeptides that are lacking the two essential HXXXD and DFGWG motifs associated with functional BAHD genes and were therefore excluded from a large-scale phylogenetic analysis of BAHDs performed in Arabidopsis and poplar (Populus spp; Yu et al., 2009). Nevertheless, we could successfully identify both motifs in the middle and the C-terminal end of At1g65450.1 (450.1) and in the N-terminal and C-terminal ends of 450.3 (Figures 5 and 6). It is important to note that we could detect several BAHD members from a variety of plants in which the motif diverged from the canonical conserved DFGWG motif. In the case of At1g65450, it is represented by EYPWG. A very similar change, DYPWG, occurred in the poplar BAHD EEF02345 protein, which groups into the same clade as At1g65450 (Figure 6).
Figure 6.
Alignment of At1g65450.1 Amino Acid Sequence to Members of Clade I of the BAHD Transferase Family.
Two subsections of an amino acid sequence alignment (ClustalW), containing the HXXXDG and DFGWG motifs, is shown (right panel) with At1g65450.1 (top) and other members of clade I of the BAHD transferase family (Yu et al., 2009). The sequences used were for the plants Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), Populus trichocarpa (Pt), Zea mays (Zm), Oryza sativa (Os), and Arabidopsis (At), and only those that were complete. A rooted cladogram (left panel), with At5g23940 as the outgroup, consists of members that represent a subtree from the BAHD transferase family and was made using a t-coffee alignment of complete protein sequences (available as Supplemental Data Set 1 online). It shows three clades correlated with variation in both conserved motifs. The divergence of the DFGWG motif, which has occurred for At1g65450 and a related poplar sequence, have residue differences for positions 2 and 3 highlighted (green and purple).
cDNAs of All three Isoforms Can Rescue the glc Defect When Expressed in the CC
To determine if the combined sequence is capable of rescuing glc and to rule out the possibility that the overlapping intergenic region (or intron) shared by the genomic DNA pieces of 450.2 and 450.3 is responsible for the rescue of glc, RNA was isolated from wild-type pistils and cDNAs were synthesized. All three isoforms discussed above were identified and successfully cloned, thus confirming the dual nature of At1g65450 expression. The complicated transcriptional structure of this locus made it difficult to pinpoint the native promoters. Since the embryo sac expression profiles of this gene were high in the CC (Figure 3B), we chose to use the reported DD65 CC-specific promoter (Steffen et al., 2007) for expression of the cloned cDNAs in the CC. The DD65 promoter was cloned and fused to a GUS reporter and indeed showed high and specific GUS activity in the CC (see Supplemental Figure 5 online). The DD65 promoter was then swapped in place of the original 35S promoter of the pB7W2 binary vector, into which the above cDNAs had been cloned. Seeds were collected from successfully transformed glc plants and plated onto plates containing kan to assess the kanr/kans segregation ratio. All three isoforms successfully rescued the distorted kanr/kans segregations ratio typical of glc/GLC plants (Table 4), indicating that both the combined sequence and its two truncated variants can functionally complement glc.
Table 4. Complementation of glc kanr/kans Distorted Ratio by cDNAs of 450.1, 450.2, and 450.3 Operating under the DD65 CC-Specific Promoter.
| Genotype | kanr | kans | Ratio kanr/kans | P Value (Fisher’s Exact Test) |
|---|---|---|---|---|
| glc; 450.1 | 159 (73.9%) | 56 (26.1%) | 2.8:1 | 4.88e−15 |
| glc; 450.2 | 140 (72.9%) | 52 (27.1%) | 2.7:1 | 1.4oe−13 |
| glc; 450.3 | 141 (73.8%) | 50 (26.2%) | 2.8:1 | 4.61e−14 |
| glc (self) | 75 (36.2%) | 132 (63.8%) | 0.57:1 | N.A. |
The pB7WG2 binary vector was modified and its original 35S promoter was swapped with the DD65 CC-specific promoter, then cloned with cDNAs of 450.1, 450.2, and 450.3 and transformed to glc plants. The segregation of the linked nptll gene conferring kanr was scored in T2 seedlings grown on kan-MS medium. N.A., not applicable.
To find out if the glc effect on pollen tube guidance was rescued as well, emasculated flowers form glc/GLC plants transformed with 450.3 were pollinated with pollen of expressing the LAT52:GUS pollen-specific marker. At 8 HAP, 86.3% of the ovules were reached by pollen tubes that burst at the micropylar end, which is not significantly different from wild-type ovules at the same time (see Supplemental Figure 1A online). Interestingly, even though 450.2 and 450.3 are not identical, the amino acid alignment of both shows regions of similarity (Figure 5). Again, no homozygous glc plants could be isolated among the rescued glc transformants, suggesting that another gene(s) within the deletion might be required for viability when homozygous.
Intracellular Localization and Promoter Analysis
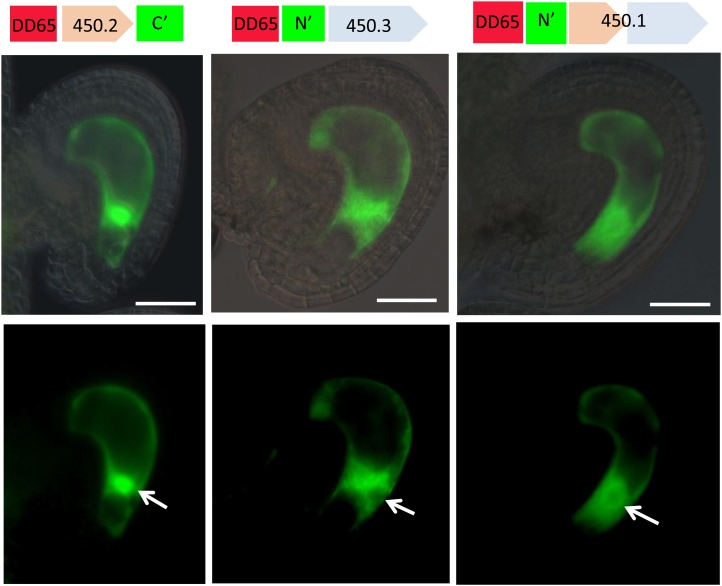
Most BAHDs are known to be localized to the cytoplasm (D’Auria, 2006), but functions of this family have not been characterized in detail. Since the phenotype we describe suggests a communication problem between the CC and the externally located sperm cell, we explored the possibility of plasma membrane localization. Alternately, a clear nuclear localization using the 450.1 and 450.3 constructs would have suggested a nuclear role, such as involvement in gene expression through acetylation. GFP translational fusions of the complete protein and its subisoforms were constructed; the different cDNAs were cloned into N- and C-terminal GFP translational fusion constructs driven by the DD65 CC-specific promoter. Since the constructs used were driven by the DD65 promoter, we were expecting to observe GFP in the CC.
This was indeed the case for 450.2 C-terminal, 450.3 N-terminal, and 450.1 N-terminal constructs. In these cases, the GFP was located in the cytoplasm and was highly concentrated in the nucleus only in the 450.2 construct (Figure 7). The nuclear GFP signal in the 450.3 and 450.1 constructs was considerably reduced compared with the cytoplasm, suggesting that the fusions might be excluded from the nucleus. However, due to the low signal that is detectable with 450.1 and 450.3, it is possible that the fusion proteins are still able to enter the nucleus.
Figure 7.
GFP Translational Fusions of the Different Gene Products from At1g65450 in Wild-Type Unfertilized Ovules.
Provided are fluorescence micrographs (bottom panel) that were merged with their bright-field images (top panel). The construct used is given on top of each pair of vertical images. White arrows indicate position of the nucleus. Bar = 50 μm.
To examine whether the GFP translational fusions are functional, they were introduced into the glc mutant background. Since direct transformation of the constructs into glc plants was not possible (due to having the same selective marker, i.e., kanr), glc/GLC served as a pollen donor and was crossed to pistils of wild-type transformants expressing the 450.2 C-terminal and the 450.3 N-terminal constructs mentioned above. F1 heterozygous plants that were genotyped as glc/GLC and expressed GFP in their ovules were isolated and their seed set was assessed. Partial but significant rescue of the glc/GLC semisterility phenotype was observed for both the 450.2 C-terminal and the 450.3 N-terminal constructs, thus indicating that both GFP fusions are biologically functional (Table 5).
Table 5. Complementation of glc Semisterility Phenotype by GFP Translational Fusions of 450.1, 450.2, and 450.3 cDNAs.
The unusual gene organization resulting in the 450.2 and 450.3 transcripts raised the question of where the promoters might reside for initiation of these transcripts. To characterize their putative promoters, genomic DNA from two intergenic regions was cloned. The first isolated fragment, from here on called the 450.2 promoter, resides between At1g65440 and the first exon of At1g65450. The second isolated fragment, that we call the 450.3 promoter, is the long intron of 450.1 that corresponds to the intergenic region between the 450.2 and 450.3 loci (see regions in Figure 2F). Both fragments were successfully cloned into the binary vector pKGWFS7, and their GUS expression was determined in mature nonfertilized ovules of wild-type transformants. In both cases, GUS activity could be detected in the embryo sac. With the 450.2 promoter, strong GUS activity was observed throughout the embryo sac, whereas the GUS activity driven by the 450.3 promoter was weaker and mainly restricted to the SC (see Supplemental Figures 6A to 6C online). In both cases, strong GUS activity was also observed in sporophytic tissues, such as the funiculus and the placenta for the 450.2 promoter and the micropylar end of integuments and placenta for the 450.3 promoter (see Supplemental Figures 6A to 6C online). These observations differ from the specific CC expression of the At1g65450 isoforms deduced from the microarray profiles (Figure 3B). However, it is very likely that in addition to the 5′ end promoter sequences used, the differential gene expression found in the embryo sac cell types requires enhancer activity of elements located up/downstream of the gene, particularly in the case of the 450.3 gene. Since the isolated putative promoters were likely incomplete and were investigated out of their native chromosomal context, we were not expecting to observe an exact match between the GUS activity and the actual expression pattern of these genes. Still, both DNA fragments were found to have promoter activity in the embryo sac. Importantly, the GUS activity resulting from the 450.3 promoter suggests that the intron between the two exons of 450.1 can act as a promoter for the 450.3 gene and therefore can explain the presence of the 450.3 transcripts in wild-type plants.
DISCUSSION
Fertilization of the CC Requires Specific Factors That Are Not Essential for the EC
Double fertilization is initiated with the discharge of the two sperm cells carried by the pollen tube into one of the two SCs. For many years, the exact fate of the sperm cells in the SC between discharge and cell fusion was obscure. A recent live-cell imaging study revealed details of the dynamics of the two sperm cells in the Arabidopsis SC prior to fertilization (Hamamura et al., 2011). This study also reported that the two sperm cells share an equal capacity to fuse with either female gamete, EC, or CC, a conclusion consistent with previous studies using genetic mutants (Pagnussat et al., 2005; Ingouff et al., 2007). Yet, the factors required for fusion of these two functionally identical sperm cells with the female gametes are not well understood. In Arabidopsis, the absence of the sperm-specific factor GCS1/HAP2 resulted in a failure of the sperm cells to fuse with either of the female gametes (Mori et al., 2006, von-Besser et al., 2006). The sperm cells also failed to fuse with either of the female gametes in the absence of ANK6, which is normally expressed in both the female gametophyte and the sperm cell (Yu et al., 2010). However, to our knowledge, no female gamete-specific factor (e.g., unique to either the EC or the CC) has been identified so far.
Here, we report that in glc ovules, uncoupling of the two gamete fusion events has occurred, with one sperm cell remaining excluded from the CC (Figure 1), while a sexual embryo develops from the fusion product of the EC with the other sperm cell. Moreover, we also report that transcripts of At1g65450.1 and its two other isoforms, which were able to rescue glc, are highly expressed in the CC of the unfertilized ovule. As rescue of the glc mutant could be performed using just the CC-specific promoter DD65, we conclude that expression of these genes within the CC is sufficient for fertilization. Differences between EC and CC fertilization have been previously described using the tetraspore mutant that results in additional sperm cells within a pollen tube (Scott et al., 2008). It was found that whereas there is a block against polyspermy for EC fertilization, a significant fraction of the CC had undergone polyspermy (i.e., multiple fertilizations by different sperm cells). From that study, it was not clear whether the CC was passively receptive to any sperm cells in the vicinity, and therefore permitted additional sperm cell fusion, or whether the CC was actively expressing functions required for fertilization that promoted sperm cell fusion. Here, we show that the mere presence of a released sperm cell in the degenerating synergid is not sufficient for fusion with the CC. Rather, the CC is an active player in the process, expressing CC-specific factors that promote fusion with the sperm cell. These factors must include the products of at least one of the isoforms of the At1g65450 gene. The available expression databases show that At1g65450.1 transcripts are not detectable in the endosperm at later stages, consistent with a fertilization-specific role for the gene product(s).
Prior to fertilization, the pollen tube is attracted and guided to the micropylar pole of the embryo sac. The direct role that the SCs play in this process has been demonstrated in Torenia fornieri (Okuda et al., 2009), maize (Zea mays; Márton et al., 2005), and Arabidopsis (Kasahara et al., 2005). Chen et al. (2007) reported that an Arabidopsis knockout of a CC-expressed, putative transcription factor called CCG resulted in defective micropylar pollen tube guidance, which led to sterility of the ovules harboring ccg mutant embryo sac. The effect of glc on pollen tube attraction and the fact that the late pollen tube arrival in glc was rescued by the 450.3 isoform driven by a CC-specific promoter is consistent with a proposed role for the CC in pollen tube attraction, at least in Arabidopsis.
Possible Functions of At1g65450 and Its Isoforms in the CC
In this study, we found that the product of At1g65450.1 is a member of the BAHD superfamily. Previous studies regarded the N-terminal (At1g65450.2) and C-terminal (At1g65450.3) products as partial polypeptides lacking the two essential HXXXD and DFGWG BAHD motifs (Yu et al., 2009). Yet, these two BAHD motifs were identified in the complete At1g65450.1 protein and the product of the C-terminal isoform At1g65450.3 (Figures 5 and 6). The BAHD acyl-transferases form a large family of acetyl-CoA using plant proteins that are considered to play biosynthetic roles using a wide range of substrates in secondary metabolism. Still, only few examples of fully biochemically characterized BAHDs exist (D’Auria, 2006). Interestingly, among these studies are reports of the involvement of BAHDs in the production of floral scent compounds in petals of several species, such as Clarkia breweri, rose (Rosa hybrida), and petunia (Petunia hybrida), during flower development (Dudareva et al., 1998; Shalit et al., 2003; Boatright et al., 2004). Therefore, it is possible that At1g65450 might be involved in the production of a secondary metabolite that plays a role in communication of the sperm with the CC.
The GFP translational fusions of At1g65450.1 and its two other isoforms were subcellularly localized to the cytoplasm, consistent with the general prediction for cytoplasmic localization of the BAHDs (D’Auria, 2006). Since the nuclear presence of both 450.1 and 450.3 GFP fusions was significantly reduced compared with the cytoplasm (Figure 7), it seems that the native isoforms do not have nuclear functions. However, the 450.2 isoform appeared to be concentrated in the nucleus. In general, nuclear pore complexes allow passive diffusion of proteins smaller than 40 kD (Lange et al., 2007). This might be the explanation for the 450.2 nuclear localization since its molecular size together with the GFP is about ∼46 kD, which might still be at the upper limit of molecules allowed passive entry to the nucleus. BAHD proteins are considered to be lacking any transit/signal peptide or nuclear localization signals (NLSs) (D’Auria, 2006). However, we located a putative classic SV40-like NLS sequence (RKKKSKV; Lange et al., 2007) at the C-terminal end of the 450.2 isoform, which might also explain its nuclear localization. The putative NLS is located in the small exon at the 3′ end of 450.2; this sequence is alternatively spliced out of the complete 450.1 product, which was not observed to be concentrated in the nucleus.
The N-terminal 450.2 isoform is lacking the two BAHD-like motifs, whereas the C-terminal 450.3 isoform does contain the two motifs but is nevertheless missing ∼40% of the complete protein. It is therefore questionable whether they can exhibit the characteristic BAHD activity. Even the full-length product At1g65450.1 exhibits differences from the canonical DFGWG motif of the BAHD superfamily. One explanation for these atypical BAHD-like isoforms is that the different variants of At1g65450 may still retain properties of the BAHD family, such as interaction with other cellular components to carry out their functions. These might be positive functions, in which they promote synthesis of a signaling molecule. Alternatively, the isoforms might be required to act as inhibitors of other proteins that negatively regulate CC fusion with the sperm cell (e.g., by blocking CC–sperm cell communication). In this model, the truncated isoforms may retain sufficient recognition/binding activity to perform the inhibitory function and thus permit fertilization. The alternate modes of action in which At1g65450 isoforms might operate in CC–sperm cell interaction are summarized in Supplemental Figure 7 online.
The unique dual nature of At1g65450 expression and the presence of transcripts and full-length cDNAs for all three isoforms raise interesting questions about the evolution of this complex locus. Alternative splicing can explain the presence of the 450.1 and 450.2 products but not the 450.3 isoform. We presented evidence that the sequence immediately upstream of the transcript for the 450.3 isoform can act as a promoter, which might be responsive to distal enhancers for 450.1 (see Supplemental Figures 6B and 6C online). The redundancy of the 450.2 and 450.3 isoforms, that both can rescue glc, combined with the similarities observed in their aligned amino acid sequences raises the possibility that the full-length BAHD protein 450.1 might actually be composed of two similar non-BAHD transferases. For example, a tandem duplication could give rise to two small neighboring independent transferases, followed by sequence divergence and subsequent incorporation into a single transcript through splicing, while retaining both promoters.
Implications for the Evolution of Double Fertilization
The fertilization process in glc mutant embryo sacs has features reminiscent of fertilization in gymnosperms. Gymnosperm pollen also contains two sperm cells, but only one participates in fertilization (Fernando et al., 2010). For example, in the conifers, the pollen tube enters the archegonium of the female gametophyte, where the sperm cells are released. However, only a single sperm will fertilize the EC; the second sperm cell can persist for some time within the female gametophyte but will eventually disintegrate, similar to the fate of the second sperm cell in the glc mutant. From an evolutionary perspective, the CC is a recent cell type found only in the angiosperm female gametophyte, and the nutritive function of the endosperm replaces that of the bulk of the cells of the female gametophyte in gymnosperms. Two hypotheses have been proposed for the origin of the CC and the endosperm. The first is that it arose from a gymnosperm female gametophyte cell that acquired gametic attributes (i.e., the capability to participate in fertilization). The second is that the endosperm arose by duplication of EC fertilization, as a sacrificial, altruistic embryo that is now exclusively nourishing its twin (reviewed in Baroux et al., 2002). The findings from this study have implications for both hypotheses. According to the first hypothesis, the interaction of the second sperm occurred with a cell whose identity was distinct from that of the EC; therefore, it acquired a novel function that permitted fertilization through a process that might be similar or different from that of the EC. This novel function would be provided in part through expression of the genes identified in this study. According to the second hypothesis, a duplication of the EC-sperm cell fertilization occurred, suggesting that the ancestral CC began with the same capacity for fertilization as the EC. In this case, we would need to postulate that as the ancestral CC acquired a new identity and functions, it also developed pathways for fertilization that differentiate it from the EC, including a requirement for the products of specific genes such as At1g65450 to permit fusion with the sperm cell.
METHODS
Plant Materials and Growth Conditions
The LAT52:GUS pollen-specific marker was a gift from Sheila McCormick (University of California, Berkeley; Twell et al., 1990). The embryo sac cell-specific GFP markers lines, DD31 (SC), DD45 (EC), and DD65 (CC), were a gift from Gary Drews (University of Utah; Steffen et al., 2007). The sperm-specific RFP marker HTR10 was a gift from Fred Berger (Temasek Lifesciences Laboratories, Singapore; Ingouff et al., 2007). The T-DNA knockout lines SM_3_1166 and SAIL_342_B04 were obtained from the ABRC and Nottingham Arabidopsis Stock Centre, respectively. The rest of the T-DNA lines screened in this study can be found in Supplemental Table 2 online. These mutants were genotyped according to the instructions found at http://signal.salk.edu. To genotype glc, primers that bridge the junction between the Ds element and the genomic flanking region (Ngo et al., 2007) were used (5′ to 3′): TAAGGAAGCTCAAGGGGAATC and CGTTCCGTTTTCGTTTTTTACC. Seed sterilization and growth conditions of the plants were performed as described (Pagnussat et al., 2005).
Genomic DNA and RNA Extractions and RT-PCR
Genomic DNA was extracted from rosette leaves (accession Landsberg erecta [Ler]) as described (Capron et al., 2008). RNA extraction (from nonpollinated Ler pistils) and RT-PCR were done as described (Ron et al., 2010).
Plasmids
BACs JAtY52D13, JAtY60A22, and JAtY63O08 were obtained from the JAtY Clone Library at the John Innes Centre (http://jicgenomelab.co.uk/libraries.html). To amplify the genomic DNAs of At1g65450.2 and At1g65450.3 (see cloned regions in Figure 2C), the following primers (5′ to 3′) were used: GGTTTGCGACGAAGCGATTAAGGT and GTCCATCGAACGTGGTATGGTTTG for the former and AGGTCATTAGTCGAGGACCATG and CATCCATCCCTCAACTTGGCTA for the latter. These genes were then cloned into the binary vector pCAMBIA1300. To amplify the DD65 promoter we used (5′ to 3′) CGAGCTCTTAGTCAGCAAAATCAAAATT and CGACTAGTCAAAACACAAACACTTCTTAT. The amplified product was then cloned into the binary vector pCAMBIA1300. The different cDNAs of At1g65450 (see regions in Figure 2D) were cloned into pB7W2 (Gateway; Invitrogen). For At1g65450.1, the following primers were used (5′ to 3′): CACCGGTTCTTCGTACCAAGAATCTCCA and TCAGCACAGAGTGTCTTCAAACCA. For At1g65450.2, we used (5′ to 3′) CACCGGTTCTTCGTACCAAGAATCTCCA and TTATACACTCAAAACTTTTGATTT. For At1g65450.3, we used (5′ to 3′) CACCATGGGAATCTCAACAAACCATACC and TCAGCACAGAGTGTCTTCAAACCA. For N-terminal GFP translational fusions, the same cDNAs were cloned into pK7FWG2 (Gateway; Invitrogen). For C-terminal GFP translational fusions, the same primers were used, but excluded the first three nucleotides (stop codon) from the 5′ of the reverse primer. These cDNAs were cloned into pK7WFG2 (Gateway; Invitrogen). For putative promoter analysis, the following primers were used to amplify intergenic regions (Figure 2E): CACCCAGTGTGAATTTTTCATA and CATTTTGCAACCCTTCTCTCTC for the 450.2 promoter and CACCACAAAGAAGCTTCCTTA and CATTGCAAATCCACCGCACTT for the 450.3 promoter. The amplified DNAs were then cloned into pKGWFS7 (Gateway; Invitrogen).
In all the Gateway destination vectors used here, the original 35S promoter was removed by SacI-SpeI double digestion and replaced by DD65 promoter, which has gone through the same double digest. The destinations backbone and the DD65 promoter bands were cut from the gel, and the DNAs were extracted by NucleoSpin Extract II kit (Macherey-Nagel) and ligated overnight using T4 ligase (New England Biolabs).
Transformation of Agrobacterium tumefaciens and Arabidopsis thaliana
The vectors were electroporated into Agrobacterium strains AGL1 or GV3101, which were later grown with the appropriate antibiotics. Transformation into Arabidopsis plants was performed by the floral dipping method (Zhang et al., 2006).
Microscopy
Fluorescent and GUS signals were observed under a Zeiss Axioplan imaging 2 microscope equipped with epifluorescence illumination. Filter sets for GFP (green), RFP (Texas Red), and differential interference contrast were used. In some cases, a MRC-1024 confocal microscope was used (Bio-Rad). Images were captured with a CCD Zeiss camera using the AxioVision 4.8 software and were later assembled with the Adobe Photoshop software package. GUS staining and ovule Hoyer’s clearing was performed as published (Stangleland and Salehian, 2002); in the case of LAT52:GUS, we used the protocol published by Johnson et al. (2004).
Bioinformatics and Statistics
Amino acid sequences of related proteins used here were obtained from GenBank (National Center for Biotechnology Information) using homology with the transferase gene family. For phylogenetic analysis, these sequences were aligned using the t-coffee alignment software, version 9.02 (Notredame et al., 2000), and the resulting alignment was not edited. A rooted cladogram was produced using the PHYLIP package. Specifically, the protpars algorithm was used with 1000 alignments (made using seqboot) with the sequence order jumbled one time for each. The demonstration of conservation of the motifs HXXXDG and DFGWG, for which the emphasis is on function (rather than sequence evolution), was more clearly displayed (Figure 6, right panel) using a ClustalW alignment. Fisher’s exact tests were performed using the fisher.test of the core statistics package (stats) in the R statistical environment.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: At1g65450.1, 2206339; At1g65450.2, 6530296489; and At1g65450.3, 6530296490.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pollen Tube Guidance and Burst at the Embryo Sac Micropylar End of Wild-Type, glc/GLC, and glc/GLC;450.3 Ovules.
Supplemental Figure 2. Sperm Localization in glc Ovules 24 h after Pollination.
Supplemental Figure 3. Complete Z-Stack Cross-Section Confocal Images of HTR10:RFP in glc Ovule Expressing DD65 Central Cell Marker.
Supplemental Figure 4. The Known Population of cDNAs of At1g65450 and Its Variants.
Supplemental Figure 5. Central Cell–Specific GUS Activity in Wild-Type Ovule Driven by DD65 Promoter Fusion.
Supplemental Figure 6. Putative Promoter Analysis.
Supplemental Figure 7. Alternative Modes of Action of At1g65450 Isoforms in Central Cell–Sperm Cell Interaction.
Supplemental Table 1. Presence of Sperm Nuclei in the Embryo Sac 24 h after Pollination.
Supplemental Table 2. List of T-DNA Knockout Lines Screened in This Study.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Tree Shown in Figure 6.
Supplementary Material
Acknowledgments
We thank Gary Drews for the DD embryo sac marker lines, Frédéric Berger for the sperm-specific HTR10:RFP marker line, and Sheila McCormick for the LAT52:GUS pollen-specific marker. We thank Charles Gasser and Marissa Simon for predicting the dual gene annotation ahead of the TAIR10 release. We also thank Mily Ron, Aneesh Panoli, Liza Conrad, Ryan Kirkbride, Yael Golani, Dudy Bar-Zvi, and Naomi Melamed-Book for helpful discussions and the two anonymous reviewers for their critical reading. Y.L. was partially supported by Vaadia-BARD Postdoctoral Fellowship Award FI-392-07 from BARD, the U.S.–Israel Binational Agricultural Research and Development Fund. This work was also supported by a National Science Foundation grant IOS-1051951 to V.S. Work in U.G.’s laboratory was supported by the University of Zurich and the Swiss National Science Foundation.
AUTHOR CONTRIBUTIONS
V.S. and Y.L. designed the research. Y.L., C.J., and X.S. performed research. S.E.W., Q.A.N., and U.G. performed microarray data analysis. Y.L. and C.J. analyzed data. Y.L. wrote the article.
Glossary
- EC
egg cell
- CC
central cell
- SC
synergid cells
- GUS
β-glucuronidase
- HAP
h after pollination
- RFP
red fluorescent protein
- Kanr/Kans
kanamycin resistance/kanamycin sensitivity
- BAR
Bio Array Resource
- BAHD
to be defined
- GFP
green fluorescent protein
- NLS
nuclear localization signal
- Ler
Landsberg erecta
References
- Aw S.J., Hamamura Y., Chen Z., Schnittger A., Berger F. (2010). Sperm entry is sufficient to trigger division of the central cell but the paternal genome is required for endosperm development in Arabidopsis. Development 137: 2683–2690 [DOI] [PubMed] [Google Scholar]
- Baroux C., Spillane C., Grossniklaus U. (2002). Evolutionary origins of the endosperm in flowering plants. Genome Biol. 3: 1026.1–1026.5
- Berger F., Hamamura Y., Ingouff M., Higashiyama T. (2008). Double fertilization—Caught in the act. Trends Plant Sci. 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Boatright J., Negre F., Chen X., Kish C.M., Wood B., Peel G., Orlova I., Gang D., Rhodes D., Dudareva N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A., Gourgues M., Neiva L.S., Faure J.E., Berger F., Pagnussat G., Krishnan A., Alvarez-Mejia C., Vielle-Calzada J.P., Lee Y.R., Liu B., Sundaresan V. (2008). Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Li H.J., Shi D.Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Masiero S., Vanzulli S., Lardelli P., Kater M.M., Colombo L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dudareva N., D’Auria J.C., Nam K.H., Raguso R.A., Pichersky E. (1998). Acetyl-CoA:benzylalcohol acetyltransferase—An enzyme involved in floral scent production in Clarkia breweri. Plant J. 14: 297–304 [DOI] [PubMed] [Google Scholar]
- Fernando D.D., Quinn C.R., Brenner E.D., Owens J.N. (2010). Male gametophyte development and evolution in extant gymnosperms. Intl. J. Plant Dev. Biol. 4: 47–63 [Google Scholar]
- Gross-Hardt R., Kägi C., Baumann N., Moore J.M., Baskar R., Gagliano W.B., Jürgens G., Grossniklaus U. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamura Y., Saito C., Awai C., Kurihara D., Miyawaki A., Nakagawa T., Kanaoka M.M., Sasaki N., Nakano A., Berger F., Higashiyama T. (2011). Live-cell imaging reveals the dynamics of two sperm cells during double fertilization in Arabidopsis thaliana. Curr. Biol. 21: 497–502 [DOI] [PubMed] [Google Scholar]
- Ingouff M., Hamamura Y., Gourgues M., Higashiyama T., Berger F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., Preuss D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Sato S., Tabata S., Kobayashi Y., Iwabuchi M., Araki T. (2000). hosoba toge toge, a syndrome caused by a large chromosomal deletion associated with a T-DNA insertion in Arabidopsis. Plant Cell Physiol. 41: 1055–1066 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. (2007). Classical nuclear localization signals: Definition, function, and interaction with importin α. J. Biol. Chem. 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márton M.L., Cordts S., Broadhvest J., Dresselhaus T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- Moore J.M., Calzada J.P., Gagliano W., Grossniklaus U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62: 35–47 [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71 [DOI] [PubMed] [Google Scholar]
- Ngo Q.A., Moore J.M., Baskar R., Grossniklaus U., Sundaresan V. (2007). Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134: 4107–4117 [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.-J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.-F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Ron M., Saez M.A., Williams L.E., Fletcher J.C., McCormick S. (2010). Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.D. (1993). The egg cell: Development and role in fertilization and early embryogenesis. Plant Cell 5: 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandaklie-Nikolova L., Palanivelu R., King E.J., Copenhaver G.P., Drews G.N. (2007). Synergid cell death in Arabidopsis is triggered following direct interaction with the pollen tube. Plant Physiol. 144: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Armstrong S.J., Doughty J., Spielman M. (2008). Double fertilization in Arabidopsis thaliana involves a polyspermy block on the egg but not the central cell. Mol. Plant 1: 611–619 [DOI] [PubMed] [Google Scholar]
- Shalit M., Guterman I., Volpin H., Bar E., Tamari T., Menda N., Adam Z., Zamir D., Vainstein A., Weiss D., Pichersky E., Lewinsohn E. (2003). Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/Citronellol acetyltransferase in developing rose petals. Plant Physiol. 131: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangleland B., Salehian Z. (2002). An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol. Biol. Rep. 20: 107–114 [Google Scholar]
- Steffen J.G., Kang I.H., Macfarlane J., Drews G.N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713 [DOI] [PubMed] [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., Preuss D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yu F., et al. (2010). ANK6, a mitochondrial ankyrin repeat protein, is required for male-female gamete recognition in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 22332–22337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.H., Gou J.Y., Liu C.J. (2009). BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: Bioinformatics and gene expression. Plant Mol. Biol. 70: 421–442 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.