This work shows that SKIP, a spliceosomal component, is critical for clock function in Arabidopsis thaliana. Loss of SKIP function impairs splicing on a genomic scale, and skip shows multiple defects in circadian clock function, including altered period, temperature compensation, and light sensitivity.
Abstract
Circadian clocks generate endogenous rhythms in most organisms from cyanobacteria to humans and facilitate entrainment to environmental diurnal cycles, thus conferring a fitness advantage. Both transcriptional and posttranslational mechanisms are prominent in the basic network architecture of circadian systems. Posttranscriptional regulation, including mRNA processing, is emerging as a critical step for clock function. However, little is known about the molecular mechanisms linking RNA metabolism to the circadian clock network. Here, we report that a conserved SNW/Ski-interacting protein (SKIP) domain protein, SKIP, a splicing factor and component of the spliceosome, is involved in posttranscriptional regulation of circadian clock genes in Arabidopsis thaliana. Mutation in SKIP lengthens the circadian period in a temperature-sensitive manner and affects light input and the sensitivity of the clock to light resetting. SKIP physically interacts with the spliceosomal splicing factor Ser/Arg-rich protein45 and associates with the pre-mRNA of clock genes, such as PSEUDORESPONSE REGULATOR7 (PRR7) and PRR9, and is necessary for the regulation of their alternative splicing and mRNA maturation. Genome-wide investigations reveal that SKIP functions in regulating alternative splicing of many genes, presumably through modulating recognition or cleavage of 5′ and 3′ splice donor and acceptor sites. Our study addresses a fundamental question on how the mRNA splicing machinery contributes to circadian clock function at a posttranscriptional level.
INTRODUCTION
The circadian clock plays critical role in diverse aspects of plant growth and development and in coordination of the biological activities with daily environmental cycles (McClung, 2006; Harmer, 2009). Circadian clocks are composed of negative feedback loops, although the specific core clock components recruited to form circadian oscillators vary among taxa (Bell-Pedersen et al., 2005; Wijnen and Young, 2006; McClung and Gutiérrez, 2010; Zhang and Kay, 2010).
In Arabidopsis thaliana, as in most other eukaryotes studied to date, a circadian clock includes multiple interlocked feedback loops. The initially described central loop is based on reciprocal regulation between CIRCADIAN CLOCK ASSOCIATED1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY) and TIMING OF CAB EXPRESSION1 (TOC1) (Harmer, 2009). In this loop, CCA1 and LHY repress TOC1 expression through direct promoter binding. Similarly, TOC1 binds directly to specific regions of the CCA1 and LHY promoters as a transcriptional repressor (Harmer, 2009; Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012). This central loop integrates with two other morning and evening loops to establish the basic architecture of the plant circadian clock. The expression of PSEUDORESPONSE REGULATOR7 (PRR7) and PRR9, two homologs of TOC1, is directly activated by CCA1 and LHY, and in turn PRR7 and PRR9 suppress the expression of CCA1 and LHY in the morning loop. Mutations in PRR7 and PRR9 lead to long circadian period, whereas mutations in CCA1, LHY, and TOC1 shorten circadian period (Harmer, 2009).
TOC1, PRR5, GIGANTEA (GI), LUX ARRHYTHMO (LUX; also called PHYTOCLOCK1), and possibly other circadian components play essential roles in the night-specific loop (Hazen et al., 2005; Onai and Ishiura, 2005; Más and Yanovsky, 2009; Harmer, 2010; McClung and Gutiérrez, 2010; Nakamichi et al., 2010; Pruneda-Paz and Kay, 2010; Wang et al., 2010). LUX, in an evening complex with EARLY FLOWERING3 (ELF3) and ELF4, has been recently shown to link the evening and morning loops by negatively regulating PRR9 expression via direct promoter binding (Helfer et al., 2011; Nusinow et al., 2011; Chow et al., 2012). Limiting light input, either by reducing fluence rate and abundance of photoreceptors, or blocking photoreceptor cascades slows the pace of the oscillator (Millar et al., 1995; Somers et al., 1998). Many other clock components, including REVEILLE8 (Farinas and Mas, 2011; Rawat et al., 2011), TIME FOR COFFEE (Hall et al., 2003; Ding et al., 2007), GI (Park et al., 1999; Tseng et al., 2004; Mizoguchi et al., 2005), and XAP5 CIRCADIAN TIMEKEEPER (Martin-Tryon and Harmer, 2008) have been identified, although how these contribute to clock function remains incompletely understood.
In addition, posttranslational regulatory mechanisms, including regulated protein modification by phosphorylation and proteasomal degradation, are also a theme seen in clocks of most taxa (Harms et al., 2004; Guo et al., 2009; Mehra et al., 2009). Proteasome-mediated degradation of TOC1 and its homolog PRR5 are triggered through interaction with the cytosolic F-box protein ZEITLUPE (ZTL) (Más et al., 2003; Kiba et al., 2007; Kim et al., 2007). Phosphorylation also regulates activity and abundance of clock components. Phosphorylation of CCA1, mediated at least in part by Casein Kinase2, is required for its normal function (Sugano et al., 1998; Daniel et al., 2004). TOC1 and each of the PRRs exhibit cycling levels of phosphorylation, although the responsible kinases are unknown (Fujiwara et al., 2008). Phosphorylation of PRR3 promotes its interaction with TOC1, which stabilizes TOC1 by sequestering it from ZTL (Para et al., 2007; Fujiwara et al., 2008). Interaction of PRR5 with TOC1 promotes the nuclear localization of TOC1, which is necessary for its function (Wang et al., 2010).
Pre-mRNA processing (5′ capping, splicing, and 3′ polyadenylation) is an essential step in eukaryotic gene expression that not only affects the mature mRNA level but is intimately interconnected with both transcription itself and downstream mRNA metabolic events including mRNA export and turnover (Moore and Proudfoot, 2009). Pre-mRNA splicing takes place within the spliceosome. Components of the splicing complex include several small nuclear ribonucleoproteins (snRNPs), numerous Ser/Arg-rich (SR) proteins, and other non-snRNP proteins. The SR proteins act as splicing factors in either constitutive or alternative splicing (Deckert et al., 2006; Behzadnia et al., 2007; Bessonov et al., 2008). The spliceosome is highly dynamic during splicing progression, guided by consensus sequences in the pre-mRNA to form sequential complexes (Wahl et al., 2009).
Splicing has been employed by plants to modulate gene expression and development (Lorković et al., 2000; Reddy, 2007). Alternative processing of the tobacco (Nicotiana tabacum) N gene and of the Arabidopsis Flowering Time Control Locus A pre-mRNA is an important regulatory stage in the control of disease resistance and of the floral transition (Dinesh-Kumar and Baker, 2000; Jordan et al., 2002; Macknight et al., 2002; Quesada et al., 2003). Dysfunction of the plant-specific SR45 protein leads to splicing deficiency and late flowering, as well as to abnormal leaf morphological defects (Ali et al., 2007; Tanabe et al., 2009; Zhang and Mount, 2009). Alternative splicing is also emerging as an important mechanism to regulate clock gene expression, including the period gene in Drosophila melanogaster (Cheng et al., 1998), the frequency gene in Neurospora crassa (Colot et al., 2005), and the CCA1 gene in Arabidopsis (Filichkin et al., 2010). Recently, temperature-dependent alternative splicing has been shown to contribute to temperature compensation of the plant circadian clock (James et al., 2012). PROTEIN ARGININE METHYL TRANSFERASE5 (PRMT5) has been shown to contribute to circadian period determination in Arabidopsis, at least in part through the regulation of alternative splicing of PRR9 (Hong et al., 2010; Sanchez et al., 2010). PRMT5, a type II protein Arg methyltransferase, methylates various nonhistone substrates, such as heterogeneous nuclear ribonucleoproteins, snRNP, SmD1, D3, and LSm4, and reduced methylation of SmD1 and LSm4 is thought to elongate the circadian period of prmt5 mutants (Deng et al., 2010; Sanchez et al., 2010).
In this work, we characterize a novel mutant with a defective (long) circadian period and show that the mutation disrupts the function of a gene encoding the Arabidopsis homolog of mammalian Ski-interacting protein (SKIP). Our studies reveal that in Arabidopsis SKIP is a component of a splicing complex and plays a pivotal role in pre-mRNA splicing of several circadian oscillator genes through direct pre-mRNA binding. Mutations of SKIP compromise splice site choice and, hence, alter genome-wide splicing patterns. Splicing defects in clock genes, including PRR7 and PRR9, contribute to a lengthened period of the circadian clock. Our studies reveal a critical role for the SKIP splicing factor in the regulation of splicing and in the determination of circadian period.
RESULTS
Mutation of EIP1 Confers Circadian Defects in Arabidopsis
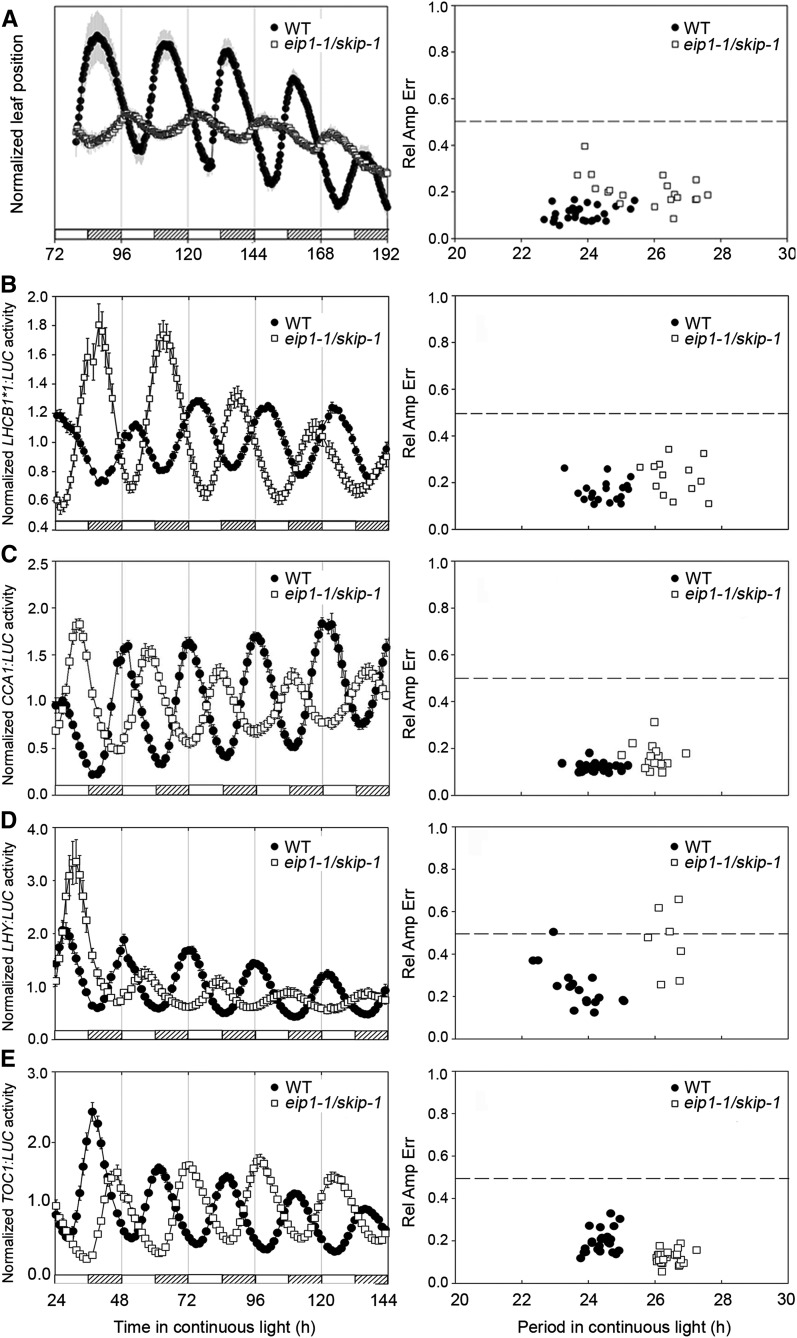
To find new components in the floral transition pathway, we performed a genetic screen in a T-DNA–mutagenized background for early flowering mutants. From this, we isolated a recessive mutant that exhibits an early flowering phenotype under both long-day (LD) and short-day photoperiod conditions, which we have designated as early flowering and insensitive to photoperiod1-1 (eip1-1) (see Supplemental Table 1 online). Because some mutants affecting circadian clock function exhibit a photoperiod-insensitive phenotype in terms of flowering, we tested eip1-1 for altered rhythmicity and observed a lengthened (by ∼2.4 h) period of circadian leaf movement in continuous white light (LL) (Figure 1A; see Supplemental Table 2 online). We then tested the expression of the circadian clock-controlled LHCB1*1 (CAB2) gene, encoding a light-harvesting chlorophyll a/b binding protein, and observed a similar long period in eip1-1 under LL (Figure 1B; see Supplemental Table 2 online). Thus, EIP1 is necessary for circadian period determination of multiple circadian clock-controlled output rhythms.
Figure 1.
Effects of SKIP (EIP1) Mutation on Circadian Clock Outputs.
Mean traces (±se) of cotyledon movement (A) or of luciferase activity from seedlings transformed with LHCB1*1:LUC (B), CCA1:LUC (C), LHY:LUC (D), and TOC1:LUC (E) and scatterplots of RAE versus period length in eip1-1/skip-1 (n = 20) and the wild type (WT; n = 30). Seedlings were entrained at 22°C in 12-h-light/12-h-dark photocycles for 7 d before release to LL (150 µmol m−2 s−1) at ZT = 0. RAE is a measure of the strength of the oscillation, with RAE = 0 corresponding to a perfect sine wave and RAE = 1 defining the lower limit of statistically significant rhythmicity. One-way analysis of variance was used for statistical analysis of period length, and in each case the period of eip1-1/skip-1 was longer than that of the wild type (P < 0.001). Hatched bars represent subjective night, and blank bars represent subjective day.
Next, we monitored the effect of EIP1 mutation on the expression pattern of three core oscillator genes, CCA1, LHY, and TOC1. In both LL and continuous dark (DD) free-running conditions, eip1-1 lengthens the period in transcription of the three oscillator genes, as measured with Promoter:LUCIFERASE (LUC) assays (Figures 1C to 1E; see Supplemental Figure 1 and Supplemental Table 2 online). The elongated period of CCA1, LHY, and TOC1 expression was further confirmed at the level of steady state mRNA abundance by RNA gel blot analysis and quantitative RT-PCR (qRT-PCR) (see Supplemental Figure 2 online). Thus, these results indicate that EIP1 is critical for the function of the Arabidopsis circadian clock.
EIP1 Encodes a Conserved SNW Domain–Containing Protein, SKIP, and Its Expression Is Global and Constitutive
EIP1 (At1g77180) was isolated through a map-based cloning approach (the T-DNA was not linked to the mutation) and a 22-bp deletion was detected in eip1-1 (see Supplemental Figure 3A online). At1g77180 previously has been identified as SKIP on the basis of altered abiotic stress responses (Lim et al., 2010); therefore, we rename EIP1 as SKIP and eip1-1 as skip-1. The skip-1 mutation did not abolish the expression of SKIP (see Supplemental Figure 4 online). The SKIP transcript in skip-1 has a 22-nucleotide deletion that results in a frame shift and is predicted to result in a truncated protein that retains the whole SNW and nuclear localization sequence domains and may retain partial function (see Supplemental Figure 3D online). A T-DNA insertion allele, skip-2, in which the T-DNA inserted into the nuclear localization sequence domain and therefore is predicted to result in a null phenotype was viable as a heterozygote but was severely dwarfed and infertile when homozygous (see Supplemental Figure 3D online). These results suggest that SKIP is an essential gene and that skip-1 is a weak allele, but skip-2 is a strong and presumably null allele. Because skip-1 is inherited recessively, we do not favor the alternate hypothesis that skip-1 is a neomorphic allele. SKIP encodes a 613–amino acid SNW/SKIP domain protein that is phylogenetically conserved from fungi to humans (see Supplemental Figures 3B and 3C online) and is an ortholog of SKIP in humans, Prp45 in yeast, and Bx42 in Drosophila (Saumweber et al., 1990; Albers et al., 2003; Zhang et al., 2003; Brès et al., 2005; Scott and Plon, 2005; Gahura et al., 2009).
The circadian clock defects in skip-1 were completely complemented by transforming SKIP genomic DNA into skip-1 plants (see Supplemental Figure 5A online), which confirms that SKIP is responsible for the circadian clock defects in skip-1. We performed green fluorescent protein (GFP)-tagged SKIP reporter, SKIP promoter-driven β-glucuronidase (GUS), and qRT-PCR assays to explore the subcellular localization and the expression pattern of SKIP. The results show that SKIP is a nuclear-localized protein with a broad expression pattern (Figures 2A to 2C).
Figure 2.
Expression, Subcellular Localization, and Circadian Regulation of SKIP.
(A) Expression of SKIP as determined by confocal microscopy of SKIP:GFP-SKIP/skip-1 transgenic lines. Samples were collected at ZT6 (6 h after dawn). Bars = 40 µm.
(B) SKIP expression in plants expressing SKIP:GUS. CL, cauline leaf; RL, rosette leaf. Bars = 1 mm.
(C) Expression of SKIP in seedlings and different organs by qRT-PCR assay. Samples were collected at ZT6. The values are the mean ± sd from three biological independent experiments.
(D) Expression of SKIP under free-running conditions detected by RNA gel blots. Seedlings were entrained at 22°C in 12-h-light/12-h-dark photocycles for 7 d before release to LL (150 µmol m−2 s−1) at ZT = 0. Samples were harvested after 24 h in LL and every 3 h thereafter for the next 24 h. Total RNA was assayed by RNA gel blot hybridization with SKIP-specific probes. Two biological replicates were performed with similar results, and the result from one of the experiments is shown.
(E) Expression of SKIP under free-running conditions measured by qRT-PCR. Abundance of SKIP total mRNA was measured from seedlings entrained at 22°C in 12-h-light/12-h-dark photocycles for 7 d before release to LL (150 µmol m−2 s−1) and harvested at ZT = 24 through qRT-PCR. The values are the mean and sd from three biological replicates. There was no significant difference over time among the expression level of SKIP as determined by Tukey's multiple comparison test (P < 0.05).
Not all genes associated with the circadian clock exhibit circadian control of expression. We did not observe cycling abundance of SKIP steady state mRNA by RNA gel blot and qRT-PCR analysis (Figures 2D and 2E).
SKIP Promotes Light Input and Affects the Sensitivity of the Clock to Light Resetting
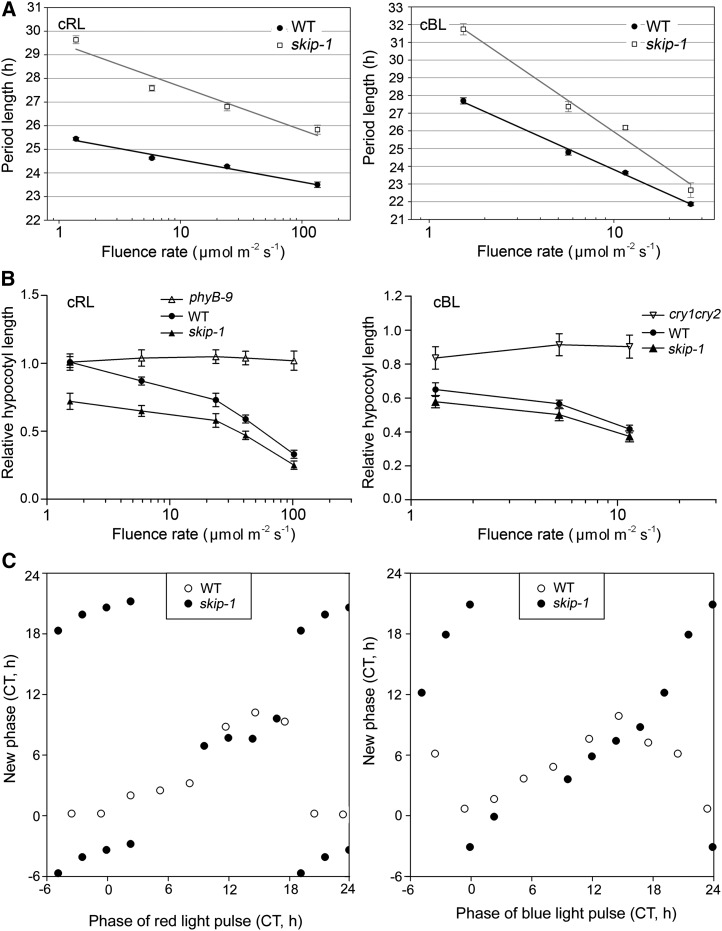
Light and temperature are the most extensively studied input signals of the clock system. Light/dark cycles mediate circadian entrainment through associated photoreceptors, which regulate the abundance and/or activity of clock components. Therefore, we examined the function of SKIP in light input to the clock. The period of circadian rhythms in plants and day-active animals is inversely proportional to the environmental light intensity, known as Aschoff’s rule (Aschoff, 1979). We observed that skip-1 is hypersensitive to both red and blue light compared with the wild type because the mutant exhibited a steeper slope in a plot of period versus fluence rate (Figure 3A; see Supplemental Table 3 online); analysis of covariance showed slopes for skip-1 to be significantly steeper than those of the wild type for both red (P < 0.0015) and blue (P < 0.0001) light. Like many clock mutants, skip-1 also showed defects in photomorphogenesis, with a shorter hypocotyl in constant red (cRL) or constant blue (cBL) light conditions (Figure 3B). These phenotypes were completely rescued in skip-1 complementation lines (see Supplemental Figure 5B online). Therefore, SKIP negatively regulates the inhibition of hypocotyl elongation in response to both red and blue light. These data reveal that SKIP plays a role in red and blue light input to the clock and in photomorphogenesis.
Figure 3.
SKIP Mediates Light Input to and Resetting the Circadian Clock.
(A) Fluence response curves of CCA1:LUC period in skip-1 and wild-type (WT) plants under cRL or cBL conditions.
(B) Hypocotyl elongation is hypersensitive to light inhibition in cRL or cBL light in skip-1 relative to the wild type. The values are mean ± se from three independent experiments.
(C) PTCs. Data from Supplemental Figure 6 and Supplemental Table 4 online are replotted as PTCs in which the new acrophase (peak phase) of CCA1:LUC expression is plotted against the time of pulses of red or blue light. The slope of the curve is closer to 1 than to 0; hence, this represents type 1 (weak) resetting.
The circadian clock is able to modulate its own sensitivity to environmental stimuli, such as dawn or dusk and temperature changes. Thus, the ability of a given stimulus (e.g., light pulse) to shift the phase of the clock varies according to the time of day (Johnson, 1999). To confirm the function of SKIP in light signal transduction to the clock, we developed phase transition curves (PTCs) and phase response curves (PRCs) to pulses of red (30 min at 100 µmol m−2 s−1) or blue (30 min at 25 µmol m−2 s−1) light given to dark-adapted skip-1 and wild-type plants by monitoring the expression of CCA1. We represented the PRC data as PTCs, in which the new phase, converted to circadian time (CT; which represents phase normalized to an arbitrary 24-h day), was plotted against the CT of the light pulse (Nagoshi et al., 2004; Locke et al., 2005). The PTCs showed that skip-1 was more sensitive to light pulses in the late night/early morning than the wild type (Figure 3C; see Supplemental Table 4 online), indicating that skip-1 is hypersensitive to both red and blue light and SKIP negatively regulates clock sensitivity to light resetting. As can be seen in the PRCs, skip-1 exhibited stronger (by ∼3 h) phase advances to pulses given in the subjective day and stronger (by ∼3.5 h) phase delays to red light pulses given in the subjective night (see Supplemental Figure 6 and Supplemental Table 4 online). For blue light pulses, skip-1 exhibited stronger (by ∼2.4 h) phase advances to pulses given in the subjective day and dramatically greater phase delays to pulses given in the subjective night (see Supplemental Figure 6 and Supplemental Table 4 online).
SKIP Is Required for Maintaining Temperature Compensation
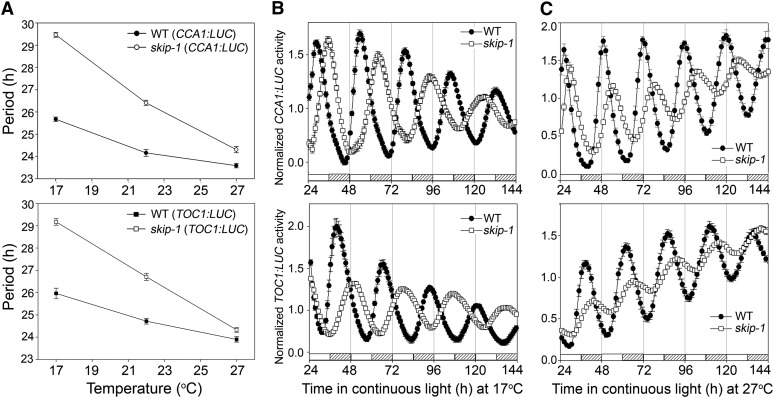
Organisms are able to maintain a circadian period close to 24 h across a broad range of physiological temperatures, which is known as temperature compensation and is one of hallmarks of circadian clocks (Zimmerman et al., 1968). The lengthened period phenotype of skip-1 was temperature sensitive and gradually disappeared with increasing temperature from 17 to 27°C (Figure 4). The period length in skip-1 was ∼3.5 h longer than that of the wild type at 17°C but very close to the wild type at 27°C (Figure 4). Thus, skip-1 has partially lost temperature compensation, indicating that SKIP plays a critical role in maintaining temperature compensation of the circadian clock.
Figure 4.
SKIP Is Involved in Maintaining Temperature Compensation of the Clock.
(A) Period length of CCA1:LUC (top panel) and TOC1:LUC (bottom panel) at 17, 22, and 27°C for the wild type (WT; closed symbols) and skip-1 (open symbols). The values are the mean ± sd from three biological independent experiments.
(B) Luciferase activity (mean ± se) of two independent transgenic lines for the wild type (closed symbols) and skip-1 (open symbols) carrying CCA1:LUC (top panel) or TOC1:LUC (bottom panel) at 17°C in continuous light. White and gray bars at bottom indicated subjective day and night, respectively.
(C) Luciferase activity (mean ± se) of two independent transgenic lines for the wild type (closed symbols) and skip-1 (open symbols) carrying CCA1:LUC (top panel) or TOC1:LUC (bottom panel) at 27°C in continuous light. White and gray bars at bottom indicated subjective day and night, respectively.
SKIP Is a Splicing Factor and One of the Components of the Spliceosome in Planta
It is well documented that SKIP acts as a splicing factor in both mammals and yeast (Albers et al., 2003; Bessonov et al., 2008; Chen et al., 2011). To investigate further the molecular mechanism by which SKIP affects circadian clock function in Arabidopsis, we asked whether SKIP functions as a splicing factor.
We first tested the ability of SKIP to complement the yeast prp45(1-169) mutant, which is defective in cell division and exhibits temperature sensitivity of growth due to a defect in pre-mRNA splicing (Gahura et al., 2009). ADH1:SKIP was able to rescue the cell division and temperature-dependent growth arrest defects in prp45(1-169) (Figures 5A and 5B).
Figure 5.
At-SKIP Is Able to Complement Morphologic, Temperature-Sensitive, and Splicing Efficiency Defects of prp45(1-169) in Yeast.
(A) Cell morphologies of PRP45, prp45(1-169), and prp45(1-169)+SKIP.
(B) The growth of cells from PRP45, prp45(1-169), and prp45(1-169)+SKIP at 30 and 37°C.
(C) Schematic of ACT1-CUP1 pre-mRNA and its mutation sites as used in (D). Mutations of the ACT1-CUP1 pre-mRNA in the 5′SS, BPS, or the 3′SS region of the ACT1 intron are shown. gAG is the mutated 3′SS of UAG. A3C represents the 5′SS GUA- to-GUC mutation. BS-C and BS-G is the 259-bp mutation of branch site from A to C and G, respectively.
(D) SKIP rescues the prp45(1-169) splicing deficiency phenotype as observed through primer extension analysis. The abundances of pre-mRNA (P; top), mRNA (M; middle), and lariat intermediate (LI; bottom) were estimated with Quantity One. The first-step efficiency (gray bars) and the second step efficiency (black bars) were calculated with (M+LI)/(P+M+LI) and M/(M+LI), respectively. Three biologically independent experiments were conducted, similar results were obtained, and the results from one replicate are shown.
To examine further whether SKIP acts as a splicing factor like Prp45 in yeast, we introduced the plasmids expressing an unmutated ACT1-CUP1 fusion construct or an ACT1-CUP1 fusion construct containing mutations in the 5′ splice site (5′SS), branch point site (BPS), or the 3′ splice site (3′SS) region of the ACT1 intron as indicated (Figure 5C) into prp45(1-169). Levels of pre-mRNA, lariat-exon 2 intermediate, and mature mRNA are analyzed by primer extension. Consistent with a previous report (Gahura et al., 2009), the splicing efficiency of unmutated ACT1-CUP1 fusion template in the prp45(1-169) appears the same as in the wild type (PRP45) (Figure 5D, lanes 1 and 2), and the splicing efficiency is not altered by transforming prp45 (1-169) with SKIP (Figure 5D, lanes 1 to 3). However, the splicing efficiency for the mutated templates, including 3′SS UAG to gAG, 5′SS GUA to GUc, and BPS (UACUAAC to UACUAcC or UACUAgC), was compromised in prp45(1-169) but was partially rescued by introducing the SKIP gene into prp45(1-169) (Figure 5D, lanes 4 to 15). That SKIP is able to rescue the cell division, temperature-sensitive, and splicing efficiency defects of prp45(1-169) in yeast indicates that SKIP is functionally conserved and possesses splicing activity in yeast.
Pre-mRNA is spliced to generate mature mRNA within a ribonucleoprotein complex termed the spliceosome. To address whether SKIP is a component of the spliceosome in planta, we employed fluorescence resonance energy transfer (FRET) to test for colocalization and physical interaction of SKIP with the spliceosome components U1-70K (Golovkin and Reddy, 1996) and SR45 (Ali et al., 2007) in Nicotiana benthamiana. SKIP colocalized in nuclear speckles with the spliceosome components U1-70K and SR45 (Figures 6A and 6B). No FRET signal was detected in cells cotransformed with GFP-SKIP and U1-70K-mCherry (Figure 6A), indicating that these proteins do not interact closely. However, a strong FRET emission was observed in nuclear speckles of cells coexpressing GFP-SKIP and SR45-mCherry proteins (Figure 6B). The results were further confirmed by photobleaching. In agreement with the above results, FRET between GFP-SKIP and SR45-mCherry was detectable before bleaching the SR45-mCherry energy acceptor (Figure 6C, the prebleach panel), but enhanced fluorescent intensity of GFP-SKIP was observed after photodestruction of SR45-mCherry (Figure 6C, the postbleach panel), indicating energy transfer between GFP-SKIP and SR45-mCherry (Figure 6C). Increased GFP:SKIP fluorescence intensity after photobleaching of the mCherry acceptor is quantified in Figure 6D. These results indicate that SKIP is a component of the spliceosome in which it is closely associated with SR45.
Figure 6.
SKIP Is a Component of the Spliceosome and Interacts with SR45.
(A) Colocalization of SKIP with U1-70K in vivo by sensitized emission assay. Bar = 5 μm.
(B) Colocalization and interaction of SKIP with SR45 in vivo by sensitized emission assay. Bar = 5 μm.
(C) Colocalization and interaction of SKIP with SR45 in vivo by photobleaching assay. Images from GFP, mCherry, and FRET channels before and after SR45-mCherry photobleaching using illumination at 543 nm. Bar = 5 μm.
(D) Emission of donor and acceptor images with acceptor photobleaching FRET. The fluorescent intensity of pre- and postbleach images from donor and acceptor was calculated with Image J, and it was normalized to the nonbleached region. Five independent photobleaching results are shown.
Mutation of SKIP Causes Genome-Wide Splicing Defects in Arabidopsis
To investigate further the function of SKIP in mRNA metabolism, we performed ultra-high-throughput RNA sequencing (RNA-seq) analysis using a mutant carrying the strong skip-2 loss-of-function allele. About 14.47 and 15.76 million reads with an average length of 42 bp from the wild type and skip-2, respectively, were aligned to the Arabidopsis reference genome (TAIR 9) (see Supplemental Table 5 online). Among them, ∼11.95 and 14.76 million reads for the wild type and skip-2, respectively, were mapped to unique loci in the genome (see Supplemental Table 5 online). An additional 0.34 and 0.48 million reads for the wild type and skip-2, respectively, were aligned to annotated exon-intron junctions (see Supplemental Table 5 online). Compared with the wild type, in skip-2, we identified ∼2979 novel splicing events, which could be sorted into four categories: intron retention (IR), alternative 5′ or 3′ splice site (A5′SS or A3′SS), and exon skipping (ES) (Figure 7A; see Supplemental Data Set 1 online). Although examples of both IR and ES were detected, the majority of the altered splicing events involved the use of alternative donor and acceptor splice sites, indicating that SKIP is probably involved in splice site recognition.
Figure 7.
Genome-Wide Effects of SKIP Mutation on Alternative Splicing.
(A) Frequency of new splicing events detected by RNA-seq in skip-2.
(B) Pre-mRNAs with IR splicing defects in skip-2 detected by RNA-seq. One gene from each chromosome was randomly picked as representative. Arrows indicate retained intron in skip-2. WT, the wild type.
(C) to (F) Validation of the selected genes with IR (C), A5′SS (D), A3′SS (E), or ES (F) splicing defects by RT-PCR in the wild type, skip-1 and skip-2.
(G) The frequency distribution of nucleotides at consensus 5′ and 3′SSs. Sequence logos illustrate consensus sequences for splice sites in the wild type (top logos), skip-2 (middle logos), and for novel splice sites detected in skip-2 (bottom logos).
We validated these observations by detecting representative IR, A5′SS, A3′SS, and ES novel splicing events in skip-2 through RT-PCR using primers flanking them (see Supplemental Table 6 online). The IR, A5′SS, A3′SS, and ES events sampled from each chromosome could be detected in skip-1 and skip-2, but not in the wild type (Figures 7B to 7F; see Supplemental Figure 7 online).
Consensus sequences at the 5′SS and 3′SS are important for accurate splicing of pre-mRNAs in plants (Reddy, 2007). Although the majority of splice sites used in skip-2 conformed to consensus, the frequency distribution of the nucleotides at 5′SS and 3′SS from the novel splicing events detected in skip-2 differed from consensus, with obvious decreases in the frequency of both the dominant G and T at +1 and +2 positions of 5′ alternative donor sites and of both the A and G at −2 and −1 positions of 3′ alternative acceptor sites, consistent with a role of SKIP protein in 5′SS and 3′SS recognition or cleavage (Figure 7G; see Supplemental Data Set 2 online).
To identify biological pathways in which SKIP is involved, we conducted Gene Ontology analysis of genes undergoing abnormal splicing in skip-2 mutants through the agriGO Web server (Du et al., 2010). The results show that SKIP may be involved in multiple biological processes, including many aspects of metabolism, development, and reproduction (see Supplemental Figure 8A online). All the categories observed were overrepresented relative to the percentage in the genome (see Supplemental Figure 8B online).
SKIP Is Involved in the Pre-mRNA Splicing of Oscillator Genes in Arabidopsis
To determine whether SKIP directly regulates the expression of clock genes through splicing, we measured alternative splicing of PRR7 and PRR9 by an electrophoretic approach. In skip-1, aberrantly spliced variants of PRR7 and PRR9 were increased compared with the wild type (see Supplemental Figures 9A and 9B online).
To determine whether patterns of alternative splicing of PRR7 and PRR9 mRNAs are regulated by the circadian clock, we further performed qRT-PCR to detect the expression of the multiple PRR7 and PRR9 splicing isoforms. The skip-1 mutation decreased the accumulation of the fully spliced PRR7_a isoform and increased the accumulation of PRR7_b and PRR7_c aberrantly spliced variants (Figures 8A to 8C). Similarly, the accumulation of the fully spliced PRR9_a isoform was decreased, whereas the accumulation of PRR9_b, PRR9_c, PRR9_d, and PRR9_e incompletely spliced isoforms was elevated in skip-1 compared with the wild type (Figures 8D to 8H). These splicing defects in skip-1 resulted in an ∼50% reduction in the peak level of PRR7_a and a slight reduction in the peak level of PRR9_a, the only isoforms capable of being translated to give functional PRR7 and PRR9 proteins (Figures 8A and 8D). The splicing defects also reduced the proportion of PRR7_a and PRR9_a relative to total PRR7 and PRR9 transcripts in skip-1 (Figures 8A to 8H; see Supplemental Figures 9C and 9D online). These results suggest that the decreased peak levels of PRR7_a and, to a lesser extent, PRR9_a and possibly the enhanced accumulation of the aberrantly spliced PRR7 and PRR9 isoforms in skip-1 contribute to the lengthened period phenotype, which is consistent with the lengthened period seen in prr7 and prr9 loss-of-function mutants (Michael et al., 2003; Farré et al., 2005; Salomé and McClung, 2005).
Figure 8.
SKIP Is Required for Regulating the Alternative Splicing and Function of PRR7 and PRR9.
Splicing variants of PRR7 ([A] to [C]) and PRR9 ([D] to [H]) as detected by qRT-PCR. Seedlings in continuous light were entrained in 12-h-light/12-h-dark photocycles for 7 d and transferred to LL prior to harvest at the indicated times. The values are the mean and sd from three biological independent experiments. WT, the wild type.
(I) RNA-IP analysis reveals that SKIP binds to the pre-mRNAs of PRR7 and PRR9 but not to the pre-mRNA of APX3. Detection of SKIP-associated pre-mRNA by RT-PCR. For Input, NA (no antibody), GFP-IP RT (+), and GFP-IP RT (−), the left lane is the wild type and the right lane is a complemented SKIP:GFP-SKIP/skip-1 T3 transgenic line. GFP-IP RT (+) or (−) is the PCR performed with the templates directly eluted from RNA-IP against GFP with reverse transcription [RT(+)] or without reverse transcription [RT(−)].
(J) Genetic analysis of circadian period length in the wild type and in single, double, and triple mutants of prrr7-3, prr9-1, and skip-1 through detecting the LUC activity of CCA1:LUC (*Tukey's multiple comparison test, P < 0.05).
Splicing of pre-mRNA requires recruitment to the spliceosome. Therefore, we asked if pre-mRNAs associate with SKIP in planta using the circadian oscillator genes PRR7 and PRR9 as examples. We took advantage of the SKIP:GFP-SKIP transgenic line in the skip-1 background and applied antibody against GFP to immunoprecipitate GFP-SKIP. The immunoprecipitates of GFP-SKIP were then reverse transcribed into cDNA and further probed by RT-PCR with primers specific to PRR7 and PRR9, using no immunoprecipitation (input) and immunoprecipitation without anti-GFP antibody (NA) samples as controls. Pre-mRNA for each of these clock genes was detected in the input and the immunoprecipitation sample from the SKIP:GFP-SKIP transgenic line and confirmed by sequencing (Figure 8I). The results support a role for the SKIP-containing spliceosome in splicing of the PRR7 and PRR9 pre-mRNAs.
To determine whether SKIP modulates circadian period solely through alternative splicing of PRR7 and PRR9, we generated double or triple mutants of prr7-3, prr9-1, and skip-1 and assessed circadian rhythmicity with CCA1:LUC. The periods of prr7-3 skip-1 and prr9-1 skip-1 double mutants were significantly longer than those of prr7-3, prr9-1, and skip-1 single mutants but were significantly shorter than that of the prr7-3 prr9-1 double mutant, which suggests that the skip-1 mutation partially compromises the expression of both PRR7 and PRR9, but the defect is not as severe as a complete loss of PRR7 and PRR9 function. This is consistent with the reduced but still detectable levels of PRR7_a and PRR9_a mRNAs (Figure 8J). Moreover, there must be circadian clock targets in addition to PRR7 and PRR9 because the period of the prr7-3 prr9-1 skip-1 triple mutant was significantly longer than that of the prr7-3 prr9-1 double mutant (Figure 8J). Consistent with this prediction, we observed skip-1 splicing defects for several other clock genes, including ARR4, CCA1, LHY, and TOC1, although not obvious for FIO1, TEJ, and ARR3 under the same condition (see Supplemental Figures 9E and 9F online). We conclude that the splicing defects in other clock genes besides PRR7 and PRR9 may also contribute to the elongated period or other circadian defects of skip-1. It was observed that the splicing patterns for LHY, PRR9, TEJ, and FIO1 in the wild type are quite dissimilar from those in skip-2 determined with RNA-seq (see Supplemental Figure 9E online). We attribute this to low expression in the wild type; the period difference between the wild type and skip-2 resulted in sampling at the peak of expression in skip-2 but at the trough of expression in the wild type.
We initially identified skip-1 on the basis of a flowering time defect, so we also assessed splicing of the transcripts of flowering time genes, including SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), CONSTANS, FLOWERING LOCUS T (FT), ELF3, ELF4, and GI. Among these genes, we observed obvious splicing defects (IR) for ELF3 and GI in skip-1 compared with the wild type (see Supplemental Figures 9E and 9F online), suggesting that SKIP may affect both circadian clock and flowering time through alternative splicing. Loss of ELF3 function through IR would be consistent with the early flowering phenotype (Zagotta et al., 1996) characteristic of skip mutants, although loss of GI function would be expected to confer late flowering (Rédei, 1962; Koornneef et al., 1991). However, the abundance of the mature fully spliced transcript was not obviously reduced in skip-1 for either ELF3 or GI, suggesting that these transcripts are unlikely to be the primary SKIP targets of relevance for the early flowering phenotype seen in skip-1 mutants.
DISCUSSION
To date, most studies of the circadian clock network in plants have emphasized transcriptional and posttranslational regulatory mechanisms. Recent genome-wide gene expression analyses have suggested that mRNA splicing is under the control of the circadian clock (Hazen et al., 2009), and it has been demonstrated that pre-mRNAs of several clock genes, including CCA1, LHY, PRR7, and PRR9, undergo alternative splicing (Filichkin et al., 2010; Sanchez et al., 2010; James et al., 2012). PRMT5 has been shown to be important for correct alternative splicing of PRR9 (Hong et al., 2010; Sanchez et al., 2010), but splicing components necessary for correct alternative splicing of these and other clock genes remain only incompletely known. Our study identifies SKIP as a splicing factor important for full splicing of PRR7 and PRR9 and other clock gene pre-mRNAs.
SKIP, a Splicing Factor, Is Required for Normal Function of the Clock
We identified SKIP, a single-copy gene in Arabidopsis, through a forward genetic analysis on the basis of a partial loss-of-function early-flowering and photoperiod-insensitive phenotype. Because other mutations with similar phenotypes often affect circadian clock function, we assessed circadian rhythms in wild-type and skip-1 plants and found that defects in SKIP lengthen circadian period of both clock-controlled output rhythms as well as of transcription of the core oscillator genes CCA1, LHY, and TOC1 (Figure 1). Thus, SKIP is critical for circadian period determination in Arabidopsis. Furthermore, the skip-1 long period phenotype exhibits temperature sensitivity, indicating that SKIP is required for temperature compensation of circadian period length (Figure 4). In addition, SKIP also modulates clock entrainment to light input and negatively regulates the inhibition of hypocotyl elongation in response to red and blue light (Figure 3). Taken together, our results suggest that SKIP is important for clock function in the Arabidopsis.
SKIP Acts as a Spliceosomal Component Important for Alternative Splicing
SKIP is evolutionarily conserved, with close homologs in yeast and mammals. SKIP plays important roles as a transcriptional coactivator/corepressor in mammals (Scott and Plon, 2005; Brès et al., 2009). However, a recent proteomic analysis of the spliceosome identified SKIP as part of the Prp19-related complex or Nineteen Complex in yeast (Bessonov et al., 2008). Consistent with this observation, human SKIP acts as a splicing factor to specifically regulate alternative splicing and expression of p21 (Chen et al., 2011). SKIP interacts with the 3′SS recognition factor U2AF65 and recruits it to the p21 mRNA in vivo, and downregulation of SKIP induces a rapid repression of p21 expression via reduced splicing of the first or second p21 introns, resulting in subsequent p53-mediated apoptosis (Chen et al., 2011).
The yeast SKIP homolog, Prp45, is a component of the spliceosomal Nineteen Complex and functions as a splicing factor (Gahura et al., 2009). Weak prp45 alleles cause defects in the splicing of actin and other genes, resulting in temperature-sensitive growth, while strong prp45 alleles are lethal (Figueroa and Hayman, 2004; Gahura et al., 2009).
In plants, the molecular mechanisms by which SKIP functions are incompletely established. We have shown that SKIP rescues both alternative splicing and temperature-sensitive growth defects in prp45-mutated yeast (Figure 5), indicating that SKIP possesses similar properties to Prp45 as a splicing factor. Consistent with this, in planta SKIP colocalizes with the spliceosomal protein U1-70K and interacts physically with SR45 (Figure 6), a second component of the plant spliceosome (Ali et al., 2007; Tanabe et al., 2009). Therefore, we conclude that SKIP is a component of the Arabidopsis spliceosome.
Consistent with this role, we observe genome-wide defects in pre-mRNA splicing in skip mutants (Figure 7). Our analysis implicates SKIP in multiple biological processes, including postembryonic development, protein metabolism, response to stress, reproductive development, and response to abiotic stimuli, which is consistent with an important role for SKIP in pre-mRNA splicing and for roles for alternative splicing in multiple biological processes (see Supplemental Figure 8 online). Others have established that SKIP is involved in abscisic acid signaling, as well as in salt and osmotic tolerance in Arabidopsis and rice (Oryza sativa), although these studies attributed the effects of SKIP misexpression primarily to roles in transcriptional regulation (Hou et al., 2009; Lim et al., 2010). Thus, we cannot rule out the possibility that SKIP regulates circadian clock function at the transcriptional level as well.
Consistent with its role in pre-mRNA splicing, aberrant splicing events in skip-2 include examples of IR and ES, and, most commonly, alternative 5′ donor and 3′ acceptor splice site selection (Figure 7; see Supplemental Figures 7 and 9 online). The nucleotide sequences of these novel 5′SS and 3′SSs diverge considerably from consensus, indicating that SKIP plays an important role in the utilization of splice donor and acceptor sites that diverge from consensus (Figure 7G), although this role could be associated with splice site recognition, cleavage, or both. Consistent with a role in cleavage, SKIP is a component of splicing complexes B and C, which catalyze both 5′SS and 3′SS cleavage (Bessonov et al., 2008), and SKIP interacts with the 3′SS recognition factor U2AF65 to regulate splicing and expression of p21 gene in mammals (Chen et al., 2011).
SKIP Is Essential for Normal Function of the Circadian Clock through Modulating the Alternative Splicing of Clock Genes
The roles for splicing factors in circadian clock control are only incompletely elucidated. SKIP associates with the pre-mRNAs of PRR7 and PRR9 and is necessary for their clock-regulated splicing (Figure 8; see Supplemental Figure 9 online). Failure to fully splice PRR7 and PRR9 mRNAs may contribute to the long period seen in the skip-1, although there must be additional clock-relevant defects in the skip-1 because a triple prr7 prr9 skip-1 exhibits a longer period than does the prr7 prr9 double mutant (Figure 8J; see Supplemental Figures 9E and 9F online). PRR7, PRR9, LHY, and CCA1 all are important for proper temperature compensation of the Arabidopsis circadian clock (Salomé and McClung, 2005; Gould et al., 2006; Salomé et al., 2010), and alternative splicing of LHY and PRR7 has been established as critical for proper temperature compensation (James et al., 2012). The temperature-sensitive period lengthening of skip-1 may result from a temperature-sensitive splicing defect of one or more of these targets and suggests SKIP as a component of the temperature-sensitive alternative splicing mechanism.
In conclusion, our findings uncover a general role of a splicing factor, SKIP, for proper function of the circadian clock and provide a molecular link between the circadian clock and alternative splicing.
METHODS
Plant Materials and Growth Conditions
All plant materials used in this study were in the Columbia-0 ecotype (Col-0) background of Arabidopsis thaliana. Seeds were sterilized and placed on Murashige and Skoog medium with 0.3% agar and 1% Suc. After stratification in the dark at 4°C for 2 d, plates were transferred to white light (70 µmol m−2 s−1) in a Percival CU36L5 growth chamber (Percival Scientific). Plants for flowering time determination were grown under various light–dark photocycles (LD) with cool white fluorescent light (100 µmol m−2 s−1) at 22°C for the day and 18°C for the night.
For hypocotyl length measurement, sterilized and stratified seeds were planted on Murashige and Skoog medium with 0.3% agar and 1% Suc plates under white light for 12 h before transfer into cRL (670 nm) or cBL (470 nm) at 22°C under various fluence rates, provided with a Percival E30LED chamber. Hypocotyl length was measured after 6 d in cRL and cBL.
Leaf Movement and Bioluminescence Measurement
Leaf movement was measured as described previously (Kim et al., 2010). For LUC activity assays, 7- to 10-d-old seedlings entrained to LD (12 h light/12 h dark) were released into LL or DD conditions for LUC measurement (Plautz et al., 1997). Circadian rhythms were assayed with BRASS 2.1.4 (Southern and Millar, 2005), which employs fast Fourier transform nonlinear least squares (Plautz et al., 1997). The strength of a circadian rhythm is expressed as relative amplitude error (RAE). An ideal cosine wave is defined as RAE = 0 and RAE = 1 defines the statistically detectable limit of rhythmicity.
For temperature compensation assays, seedlings were entrained in 12-h-light/12-h-dark (LD) cycles at 22°C for 7 to 10 d before transferring to LL at 12, 22, and 27°C for LUC activity measurement.
For fluence response curves, CCA1:LUC transgenic seedlings were entrained to LD cycles at 22°C for 7 d before transfer to cRL or cBL at indicated fluence rates. On the first day in LL, seedlings were transferred to 96-well microplates (Perkin-Elmer) for LUC activity measurement; microplates were transferred manually to the Packard TopCount at 3-h intervals. The response of period to fluence rate of red and blue light was analyzed by linear regression followed by analysis of covariance using GraphPad Prism software.
PRCs were generated with CCA1:LUC seedlings entrained to 12-h-light/12-h-dark cycles at 22°C for 7 d before transferring to DD. After one whole day in DD, pulses (30 min) of red (100 µmol m−2 s−1) or blue (25 µmol m−2 s−1) light were given to individual plates at 3-h intervals scanning one complete circadian cycle (24 h). Free-running rhythms of CCA1:LUC were monitored in DD. Acrophase (peak phase) was determined by Fourier transform nonlinear least squares. Changes in acrophase relative to seedlings not exposed to light pulse are plotted against the CT of the light pulses. For CT, phase is normalized for period length to an arbitrary 24-h day, with CT0 defined as dawn (lights on) of the entraining cycle.
Constructs of firefly LUC reporter gene driven by the CCA1, LHY, and TOC1 promoters (Salomé and McClung, 2005) and LHCB1*1 (CAB2):LUC in pZP222 vector (−199/+1) (Anderson et al., 1994) were introduced into skip-1 through transformation.
Positional Cloning of the EIP1 Gene
The eip1-1 mutation was isolated by map-based positional cloning (Lukowitz et al., 2000). To generate a mapping population, eip1-1 was crossed to Landsberg erecta (Ler), and 467 eip1-1 plants with early flowering phenotype in the F2 population were scored by PCR for molecular markers cosegregating with the eip1-1 mutation. The eip1-1 mutation was initially anchored to the lower arm of chromosome I between AthATPASE and K14-1 simple sequence length polymorphism (see Supplemental Figure 3A online). A high-resolution map was generated using simple sequence length polymorphism or cleaved amplified polymorphic sequences markers by taking advantage of the polymorphism list between Col-0 and Ler (TAIR; http://www.Arabidopsis.org/). Two recombinants, L12B-57 and L12B-108, flanking a 25-kb region in which there are 13 open reading frames, were obtained between N5-7 and N5-4 cleaved amplified polymorphic sequences markers. The open reading frames in the region were sequenced and a 22-bp deletion was detected in the SKIP (At1g77180) locus, which results in a frame shift to truncate the SKIP protein (see Supplemental Figure 3B online).
Alignment and Molecular Phylogenetic Analysis
Amino acid sequences of At-SKIP and its homologs from other organisms were aligned using Jalview software through ClustalW (Thompson et al., 1994). GenBank accessions of SKIP homologs are the following: Homo sapiens, NP_036377, SKIP (Dahl et al., 1998); Mus musculus, NP_079783 (Barry et al., 2003); Drosophila melanogaster, NP_511093, Bx42 (Saumweber et al., 1990); Caenorhabditis elegans, NP_505950 (direct submission); Oryza sativa japonica, NP_001048184, Os-SKIP (Tanaka et al., 2008); Schizosaccharomyces pombe, NP_588213 (Wood et al., 2002); Saccharomyces cerevisiae, NP_00937, Prp45 (Albers et al., 2003); and Neurospora crassa OR74A, XP_961188 (Galagan et al., 2003). The corresponding molecular phylogenetic tree was built by maximum likelihood using MEGA 4 (Tamura et al., 2007). The alignment and other parameters used to generate the tree are available as Supplemental Data Set 3 online.
Complementation Test in Arabidopsis
Both genomic and cDNA constructs of SKIP were tested for the ability to complement the skip-1 phenotype. A 5770-bp genomic fragment spanning the SKIP locus (2704 bp upstream of the ATG, 1842 bp coding region, and 1201 bp downstream of the TAA) and a SKIP cDNA fused to GFP were cloned into the pCambia1300 binary vector. The resulting SKIP:SKIP and SKIP:GFP-SKIP plasmids were introduced into skip-1 by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Flowering time, leaf movement, and hypocotyl length were determined in the T2 or T3 generations of transgenic lines (SKIP:SKIP/skip-1). Complemented T3 lines (SKIP:GFP-SKIP/skip-1) were used for RNA immunoprecipitation (RNA-IP).
Gene Expression Pattern Assays
SKIP:GUS transgenic (T3) lines were used to determine the expression pattern of SKIP via histochemical GUS reporter activity (Jefferson et al., 1987). Transgenic (T3) lines carrying an N-terminal fusion of GFP to SKIP, driven by the SKIP promoter (SKIP:GFP-SKIP), were generated to determine the tissue, organ, and temporal expression patterns and subcellular localization of SKIP.
For RT-PCR and qRT-PCR, total RNA was extracted from seedlings using TaKaRa RNAiso Plus. After RNase-free DNase I (RQ1 RNase-Free DNase; Promega) treatment, 3 µg of RNA was used for the first-strand cDNA synthesis (RevertAid first-strand cDNA synthesis kit; Fermentas). TaKaRa SYBR Premix Ex Taq and a 7500 Fast Real-Time PCR instrument (Applied Biosystems) were used for qRT-PCR.
For the electropherogram analysis, the forward primers of PRR7 and PRR9 were labeled with carboxyfluorescein. After RT-PCR, the PCR products are applied to ABI3730 sequencer for electrophoresis. Collected data were analyzed with GeneMapper ID v3.2 software.
For RNA gel blot hybridization, 10 µg of total RNA was separated on 1.2% agarose denaturing formaldehyde gels and transferred to nylon membranes. Hybridization probes were radiolabeled and hybridization was done in Church’s buffer (Church and Gilbert, 1984) at 65°C overnight with gentle rotation. The blot was washed sequentially with 2× SSC, 1× SSC, and 0.5× SSC, twice each for 10 min at 65°C with gentle rotation. Images were visualized using a phosphor imager (Amersham Biosciences; Storage Phosphor Screen), and the intensities were quantified using ImageQuant software.
Complementation Analysis in Yeast
The BamHI-SalI fragment of SKIP from Arabidopsis was cloned under the yeast ADH1 promoter in pRS416 as ADH1:SKIP. Rescue of the morphological defects in prp45(1-169) was scored in EGY48 (MATα ura3-1 leu2-3, 112 trp1-1 his3-1, 115 ade2-1 can1-100), KAY02 [prp45(1-169)-HA:kanMX6] (Siatecka et al., 1999; Gahura et al., 2009), or KAY02 transformed with ADH1:SKIP plasmid strains and grown at 30 or 37°C on the appropriate selective media for 3 d.
Primer extension was conducted as described previously with slight modification (Siatecka et al., 1999; Gahura et al., 2009). Extension products were separated in an 8% polyacrylamide/8 M urea gel, and the results were visualized by autoradiography.
To determine the band intensity in Figure 5D, we analyzed the band with Quantity One software. The Volume Analysis Report was selected to analyze the band intensity. The mean value in the chosen sizeable box represents the mean intensity of the specific band.
RNA-IP
The RNA-IP assay was conducted as described previously (Wierzbicki et al., 2008; Zheng et al., 2009) with slight modification. Ten-day-old whole seedlings of transgenic complementation lines harboring SKIP:GFP-SKIP in the skip-1 background grown under 12-h-light/12-h-dark conditions were harvested at Zeitgeber time 0 (ZT0) and ZT8 (dawn and 8 h after dawn) to detect the association of SKIP with the pre-mRNA of PRR7 and PRR9. After cross-linking in 1% formaldehyde, a series preimmunoprecipitation treatment and immunoprecipitation with GFP antibody, the immunoprecipitation products were eluted with immunoprecipitation elution buffer. The associated RNAs were quantified by RT-PCR with primer pairs crossing the intron-exon junctions of PRR7 and PRR9 pre-mRNAs after reversing cross-link.
Confocal Microscopy and FRET Analysis
FRET was conducted as described previously (Más et al., 2000). 35S:GFP-SKIP in pCambia1300 binary vector and 35S:SR45-mCherry or 35S:U1-70K-mCherry in pCambia2300 binary vector were transiently expressed in Nicotiana benthamiana leaves infiltrated with Agrobacterium strains carrying the appropriate binary plasmids. pCambia1300-P19 plasmid was coinfiltrated to suppress RNA interference. After 3 d of growth in a greenhouse at 26°C under 16 h light/8 h dark, infiltrated leaves were examined by confocal microscopy.
To test for interaction of SKIP and SR45 or U1-70K, 2- to 3-week-old N. benthamiana leaves were transformed with GFP-SKIP, SR45-mCherry, or U1-70K-mCherry alone or with GFP-SKIP cotransformed with either SR45-mCherry or U1-70K-mCherry. Both imaging sensitized emission and acceptor photobleaching approaches for FRET measurement were applied. Three days after transformation, sensitized emission FRET was performed with an Olympus FluoView FV1000 laser scanning confocal microscope. Images in donor (excitation 488 nm; emission 500 to 530 nm), acceptor (excitation 543 nm; emission 560 to 630 nm), and FRET (excitation 488 nm; emission 560 to 630 nm) channels were captured. The energy transfer efficiency and distance between the two interacted proteins were calculated as follows:
where PFRET is precision FRET (correction done FRET image); DSBT is donor spectral bleed-through; ASBT is acceptor spectral bleed-through; a is donor channel image of donor excited, with donor only dyed; b is acceptor channel image of donor excited, with donor only dyed; c is acceptor channel image of donor excited, with acceptor only dyed; d is acceptor channel image of acceptor excited, with acceptor only dyed; e is donor channel image of donor excited, with donor and acceptor dyed; f is acceptor channel image of donor excited, with donor and acceptor dyed; g is acceptor channel image of acceptor excited, with donor and acceptor dyed.
For acceptor photobleaching FRET, the fluorescence of the GFP and RFP channels were scanned as for sensitized emission FRET before and after photobleaching. Bleaching of the acceptor fluorescence signal was performed using a 543-nm beam at maximum intensity for 4 s.
mRNA Sequencing and Bioinformatics Analysis of RNA-Seq Data
For mRNA sequencing, total RNA was isolated using TRI reagent (Ambion). mRNA was extracted from total RNA using Dynabeads mRNA DIRECT kit (Invitrogen). First- and second-strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen) and random hexamers primers. DNA Sample Prep Master Mix Set 1 (New England Biolabs) was used to add adapters to the ends of the cDNA. Then, the products of the ligation reaction were purified on a gel to select a size range of templates for downstream enrichment by PCR amplification. After enrichment, the library was sequenced thoroughly.
An overview of the RNA-seq analysis pipeline for reads mapping, expression quantification, detection of differential expression, data visualization, and downstream bioinformatics analysis is summarized in Supplemental Figure 10 online. In brief, the Arabidopsis genome build and annotated gene set were downloaded from The Arabidopsis Information Resource (ftp://ftp.Arabidopsis.org/home/tair/Sequences/). After removing reads containing sequencing adapters, we mapped reads to the Arabidopsis TAIR9 genome build with BOWTIE (Langmead et al., 2009), allowing up to two mismatches. Reads that failed to be mapped to the genome build were aligned by de novo mapping using BLAT (Kent, 2002) against the same genome build to find splice junctions.
The gene locus expression levels were measured in the reads per kilobase of exon per million mapped reads unit and were calculated based on mapping outputs using ERANGE (Mortazavi et al., 2008). The cutoff value of determining significant expression or translation was 1.0 RPKM based on spike-in controls and reproducibility (Jiao and Meyerowitz, 2010). Differential expression analysis was performed using edgeR using raw mapping counts from BOWTIE and BLAT mapping (Robinson et al., 2010). Data were modeled as negative binomial distributed because our data set contains biological replicates instead of technique replicates. The multiple testing errors were addressed using the false discovery rate. Differential expression cutoff was set as above twofold changes in expression and false discovery rate–adjusted P < 0.001. To alleviate the bias in the detection of differential expression influenced by the transcript length (Young et al., 2010), we added the expression ratio cutoff (twofold).
The raw data and processed data of high-throughput mRNA sequencing were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32216).
Novel Splicing Event Detection and Analysis
Splicing events identified by de novo mapping using BLAT were compared with the gene annotation in TAIR9. IR is considered as the complete retention of an intron in a transcript, which has >1.0 RPKM, the same detection criteria for exons and transcripts (Jiao and Meyerowitz, 2010). Alternative 5′ or 3′SS, ES, or unclassified splicing (with novel 5′ and 3′SSs) is defined by the existence of a de novo mapping identified read, which represents a splicing event not annotated in TAIR9. Alternative splicing events classification is according to Matlin et al. (2005).
Nucleotide frequencies around novel and known splicing sites were counted and visually displayed using sequence logos (Schneider and Stephens, 1990).
Gene Ontology Analysis
GO analysis was conducted through the website GO Analysis Toolkit and Database for Agricultural Community (Du et al., 2010).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SKIP (At1g77180), PRR7 (At5g02810), PRR9 (At2g46790), SR45 (At1g16610), U1-70K (At3g50670), TEJ (At2g31870), ARR4 (At1g10470), CCA1 (At2g46830), LHY (At1g01060), TOC1 (At5g61380), FIO1 (At2g21070), ARR3 (At1g59940), SOC1 (At2g45660), FT (At1g65480), ELF3 (At2g25930), ELF4 (At2g40080), and GI (At1g22770). SKIP homologs are as follows: H. sapiens, NP_036377, M. musculus, NP_079783, Drosophila, NP_511093, C. elegans, NP_505950, O. sativa japonica, NP_001048184, S. pombe, NP_588213, S. cerevisiae, NP_00937, and N. crassa OR74A, XP_961188.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effects of eip1-1 on Clock Gene Expression in Constant Dark.
Supplemental Figure 2. Circadian Expression of CCA1, LHY, and TOC1 in the Wild Type and skip-1.
Supplemental Figure 3. EIP1 Encodes a Conserved SNW Domain–Containing Protein, SKIP.
Supplemental Figure 4. skip-1 Mutation Does Not Abolish the Expression of SKIP Partial Transcript.
Supplemental Figure 5. skip-1 Phenotypes Are Rescued by SKIP Genomic DNA.
Supplemental Figure 6. Phase Response Curve of CCA1:LUC Expression to Pulses of Red or Blue Light in the Wild Type and skip-1.
Supplemental Figure 7. Validation of Intron Retention Splicing Defects in skip-2 Detected by RNA-Seq through RT-PCR.
Supplemental Figure 8. Enriched Gene Ontology Terms (False Detection Rate <0.01) in Genes with Splicing Defects in skip-2.
Supplemental Figure 9. Expression of PRR7 and PRR9 Splicing Isoforms and Splicing Alteration of Circadian Associated and Flowering Time Control Genes in skip Detected through RNA Sequencing and RT-PCR.
Supplemental Figure 10. The Bioinformatic Pipeline for RNA Sequencing.
Supplemental Table 1. Early Flowering and Photoperiod-Insensitive Phenotypes in eip1-1.
Supplemental Table 2. Circadian Period Lengthening in eip1-1/skip-1 under LL Condition.
Supplemental Table 3. CCA1:LUC Rhythm under Differential Intensity and Spectrum of Light Conditions.
Supplemental Table 4. Phase Responses to Red Light and Blue Light Pulses.
Supplemental Table 5. Results of Ultra-High-Throughput RNA Sequencing Analysis.
Supplemental Table 6. Primers Used in the Studies.
Supplemental Data Set 1. Number of New Splicing Events Belonging to Four Alternative Splicing Types in skip-2.
Supplemental Data Set 2. The Frequency Distribution of Nucleotides at 5 and 3 Prime Splicing Site.
Supplemental Data Set 3. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 3C.
Supplementary Material
Acknowledgments
We thank Petr Folk for critical reading of the article, Chen Zhan and Ping Wu for their technical assistance with FRET and GFP fluorescence, Yuelin Zhang for his suggestion on positional cloning, Lilin Du for the pRS416 plasmid, and Zhiyu Peng for his help on RNA-seq analysis. We acknowledge the Arabidopsis Genome Initiative and the ABRC for providing us with the polymorphism between Arabidopsis Col-0 and Ler ecotypes and the T-DNA insertion line, respectively. This work was supported by grants from Ministry of Science and Technology of China 973 projects (2012CB910900 and 2012CB114200; L.M.), by the Hebei Province key laboratory program (L.M.), by the National Science Foundation of China (X.X. and Y.C.), and by the National Science Foundation (IOS-0950703 and IOS1025965; C.R.M.).
AUTHOR CONTRIBUTIONS
L.M., X.X, X.W., Q.X., F.W., Y.C., and C.R.M. designed the study. X.W. and H.W. identified the mutant, cloned the gene, provided the SKIP-related constructs, and generated all the transgenic lines. X.X., Q.X., and X.W. performed circadian clock analysis. F.W. and L.L. performed FRET. F.W. performed RIP experiments. F.W., O.G., and F.P. performed yeast complementation and primer extension assay. X.W., Y.Y., and F.W. performed oscillator gene pre-mRNA splicing assay. Y.W., Y.J., X.W., and S.M. performed the RNA-seq analysis and validation. L.M., X.X., C.R.M., X.W., Q.X., and F.W. wrote the article. All authors discussed the results and commented on the article.
Glossary
- snRNP
to be defined
- LD
long-day
- LL
continuous white light
- DD
continuous dark
- qRT-PCR
quantitative RT-PCR
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- cRL
constant red light
- cBL
constant blue light
- PTC
phase transition curve
- PRC
phase response curve
- CT
circadian time
- 5′SS
5′ splice site
- BPS
branch point site
- 3′SS
3′ splice site
- FRET
fluorescence resonance energy transfer
- RNA-seq
RNA sequencing
- IR
intron retention
- A5′SS
alternative 5′ splice site
- A3′SS
alternative 3′ splice site
- ES
exon skipping
- Col-0
Columbia-0
- RAE
relative amplitude error
- Ler
Landsberg erecta
- RNA-IP
RNA immunoprecipitation
- ZT
Zeitgeber time
References
- Albers M., Diment A., Muraru M., Russell C.S., Beggs J.D. (2003). Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9: 138–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali G.S., Palusa S.G., Golovkin M., Prasad J., Manley J.L., Reddy A.S. (2007). Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.L., Teakle G.R., Martino-Catt S.J., Kay S.A. (1994). Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 6: 457–470 [DOI] [PubMed] [Google Scholar]
- Aschoff J. (1979). Circadian rhythms: Influences of internal and external factors on the period measured in constant conditions. Z. Tierpsychol. 49: 225–249 [DOI] [PubMed] [Google Scholar]
- Barry J.B., Leong G.M., Church W.B., Issa L.L., Eisman J.A., Gardiner E.M. (2003). Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues. J. Biol. Chem. 278: 8224–8228 [DOI] [PubMed] [Google Scholar]
- Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Lührmann R. (2007). Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C.L., Urlaub H., Lührmann R. (2008). Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- Brès V., Gomes N., Pickle L., Jones K.A. (2005). A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 19: 1211–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V., Yoshida T., Pickle L., Jones K.A. (2009). SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell 36: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang L., Jones K.A. (2011). SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 25: 701–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Gvakharia B., Hardin P.E. (1998). Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol. Cell. Biol. 18: 6505–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B.Y., Helfer A., Nusinow D.A., Kay S.A. (2012). ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colot H.V., Loros J.J., Dunlap J.C. (2005). Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol. Biol. Cell 16: 5563–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R., Wani B., Hayman M.J. (1998). The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 16: 1579–1586 [DOI] [PubMed] [Google Scholar]
- Daniel X., Sugano S., Tobin E.M. (2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C.L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26: 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Baker B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97: 1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Millar A.J., Davis A.M., Davis S.J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38(Web Server issue): W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B., Mas P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Figueroa J.D., Hayman M.J. (2004). The human Ski-interacting protein functionally substitutes for the yeast PRP45 gene. Biochem. Biophys. Res. Commun. 319: 1105–1109 [DOI] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S.S., Salomé P.A., McClung C.R., Somers D.E. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gahura O., Abrhámová K., Skruzný M., Valentová A., Munzarová V., Folk P., Půta F. (2009). Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J. Cell. Biochem. 106: 139–151 [DOI] [PubMed] [Google Scholar]
- Galagan J.E., et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868 [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (1996). Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8: 1421–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.D., Locke J.C., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., Hall A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Cheng P., Yuan H., Liu Y. (2009). The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138: 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Bastow R.M., Davis S.J., Hanano S., McWatters H.G., Hibberd V., Doyle M.R., Sung S., Halliday K.J., Amasino R.M., Millar A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S. (2010). Plant biology in the fourth dimension. Plant Physiol. 154: 467–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harms E., Kivimäe S., Young M.W., Saez L. (2004). Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19: 361–373 [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Naef F., Quisel T., Gendron J.M., Chen H., Ecker J.R., Borevitz J.O., Kay S.A. (2009). Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Xie K., Yao J., Qi Z., Xiong L. (2009). A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. USA 106: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- James A.B., Syed N.H., Bordage S., Marshall J., Nimmo G.A., Jenkins G.I., Herzyk P., Brown J.W., Nimmo H.G. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.H. (1999). Forty years of PRCs—What have we learned? Chronobiol. Int. 16: 711–743 [DOI] [PubMed] [Google Scholar]
- Jordan T., Schornack S., Lahaye T. (2002). Alternative splicing of transcripts encoding Toll-like plant resistance proteins - What’s the functional relevance to innate immunity? Trends Plant Sci. 7: 392–398 [DOI] [PubMed] [Google Scholar]
- Kent W.J. (2002). BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Salomé P.A., Fujiwara S., Somers D.E., McClung C.R. (2010). Characterization of pseudo-response regulators in plants. Methods Enzymol. 471: 357–378 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G.H., Zhang X., Chung M.S., Lee D.J., Woo Y.M., Cheong H.S., Kim C.S. (2010). A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 185: 103–113 [DOI] [PubMed] [Google Scholar]
- Locke J.C., Southern M.M., Kozma-Bognar L., Hibberd V., Brown P.E., Turner M.S., Millar A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1: 2005.0013. [DOI] [PMC free article] [PubMed]
- Lorković Z.J., Wieczorek Kirk D.A., Lambermon M.H., Filipowicz W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5: 160–167 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon E.L., Harmer S.L. (2008). XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell 20: 1244–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Devlin P.F., Panda S., Kay S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Más P., Kim W.Y., Somers D.E., Kay S.A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Más P., Yanovsky M.J. (2009). Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 12: 574–579 [DOI] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W. (2005). Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 6: 386–398 [DOI] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R., Gutiérrez R.A. (2010). Network news: Prime time for systems biology of the plant circadian clock. Curr. Opin. Genet. Dev. 20: 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A., Baker C.L., Loros J.J., Dunlap J.C. (2009). Post-translational modifications in circadian rhythms. Trends Biochem. Sci. 34: 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar A.J., Straume M., Chory J., Chua N.H., Kay S.A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wright L., Fujiwara S., Cremer F., Lee K., Onouchi H., Mouradov A., Fowler S., Kamada H., Putterill J., Coupland G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Proudfoot N.J. (2009). Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004). Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K., Ishiura M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Kay S.A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Macknight R., Dean C., Simpson G.G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22: 3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Rédei G.P. (1962). Supervital mutants of Arabidopsis. Genetics 47: 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., Weigel D., McClung C.R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Saumweber H., Frasch M., Korge G. (1990). Two puff-specific proteins bind within the 2.5 kb upstream region of the Drosophila melanogaster Sgs-4 gene. Chromosoma 99: 52–60 [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.L., Plon S.E. (2005). CHES1/FOXN3 interacts with Ski-interacting protein and acts as a transcriptional repressor. Gene 359: 119–126 [DOI] [PubMed] [Google Scholar]
- Siatecka M., Reyes J.L., Konarska M.M. (1999). Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 13: 1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Southern M.M., Millar A.J. (2005). Circadian genetics in the model higher plant, Arabidopsis thaliana. Methods Enzymol. 393: 23–35 [DOI] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Green R.M., Wang Z.Y., Tobin E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 95: 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanabe N., Kimura A., Yoshimura K., Shigeoka S. (2009). Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus. Plant Mol. Biol. 70: 241–252 [DOI] [PubMed] [Google Scholar]
- Tanaka T., et al. Rice Annotation Project (2008). The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 36(Database issue): D1028–D1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.S., Salomé P.A., McClung C.R., Olszewski N.E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., Lührmann R. (2009). The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang L., Fujiwara S., Somers D.E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H., Young M.W. (2006). Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Wood V., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 Erratum. Nature 421: 94. [DOI] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zhang C., Dowd D.R., Staal A., Gu C., Lian J.B., van Wijnen A.J., Stein G.S., MacDonald P.N. (2003). Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278: 35325–35336 [DOI] [PubMed] [Google Scholar]
- Zhang E.E., Kay S.A. (2010). Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 11: 764–776 [DOI] [PubMed] [Google Scholar]
- Zhang X.N., Mount S.M. (2009). Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 150: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W.F., Pittendrigh C.S., Pavlidis T. (1968). Temperature compensation of the circadian oscillation in Drosophila pseudoobscura and its entrainment by temperature cycles. J. Insect Physiol. 14: 669–684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.