The DELLA proteins function as negative regulators of the gibberellin (GA) signaling pathway. This article reports that RGL3 is a distinct DELLA protein that acts as an integrating factor that links GA and jasmonate signaling to enable adaptive regulation of plant resistance to pathogens.
Abstract
Gibberellins (GAs) are plant hormones involved in the regulation of plant growth in response to endogenous and environmental signals. GA promotes growth by stimulating the degradation of nuclear growth–repressing DELLA proteins. In Arabidopsis thaliana, DELLAs consist of a small family of five proteins that display distinct but also overlapping functions in repressing GA responses. This study reveals that DELLA RGA-LIKE3 (RGL3) protein is essential to fully enhance the jasmonate (JA)-mediated responses. We show that JA rapidly induces RGL3 expression in a CORONATINE INSENSITIVE1 (COI1)– and JASMONATE INSENSITIVE1 (JIN1/MYC2)–dependent manner. In addition, we demonstrate that MYC2 binds directly to RGL3 promoter. Furthermore, we show that RGL3 (like the other DELLAs) interacts with JA ZIM-domain (JAZ) proteins, key repressors of JA signaling. These findings suggest that JA/MYC2-dependent accumulation of RGL3 represses JAZ activity, which in turn enhances the expression of JA-responsive genes. Accordingly, we show that induction of primary JA-responsive genes is reduced in the rgl3-5 mutant and enhanced in transgenic lines overexpressing RGL3. Hence, RGL3 positively regulates JA-mediated resistance to the necrotroph Botrytis cinerea and susceptibility to the hemibiotroph Pseudomonas syringae. We propose that JA-mediated induction of RGL3 expression is of adaptive significance and might represent a recent functional diversification of the DELLAs.
INTRODUCTION
Bioactive gibberellins (GAs) are a class of tetracyclic diterpenoid plant hormones that modulate diverse developmental processes throughout the plant life cycle, such as seed germination, growth through cell elongation and division, and floral transition (Achard and Genschik, 2009). Typically, mutants deficient in GA synthesis or responses are dwarf or semidwarf in stature, whereas elevated GA concentrations or increased signaling result in taller plants. GA regulates growth by suppressing a group of nuclear growth–repressing DELLA proteins, a subset of the GRAS family of transcriptional regulators (Peng et al., 1997; Silverstone et al., 1998; Bolle, 2004; Cheng et al., 2004; Zentella et al., 2007; de Lucas et al., 2008; Feng et al., 2008; Cheminant et al., 2011). Binding of GA to its receptor GA INSENSITIVE DWARF1 (GID1) promotes the GID1–DELLA interaction, which in turn stimulates the interaction between DELLAs and the specific E3 ubiquitin ligase SCFSLY1/GID2 complex, leading to subsequent degradation of DELLAs by the 26S proteasome (Silverstone et al., 2001; McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004; Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006; Willige et al., 2007). The Arabidopsis thaliana genome encodes five DELLAs, GA INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3. Genetic analyses have shown that RGA and GAI are major repressors of vegetative growth (Dill and Sun, 2001; King et al., 2001). RGL2 mainly represses seed germination (Lee et al., 2002), whereas RGA, RGL1, and RGL2 are regulators of floral development (Cheng et al., 2004; Tyler et al., 2004). The biological function of the remaining Arabidopsis DELLA protein, RGL3, is less understood. RGL3 was recently proposed (with RGL2) to repress testa rupture during seed germination (Piskurewicz and Lopez-Molina, 2009). The functional diversification of DELLAs relies largely on their gene expression pattern (spatially and in term of regulatory responses) rather than on their molecular activity. Promoter swap experiments revealed that DELLAs could perform equivalent functions (Gallego-Bartolomé et al., 2010).
Recent advances revealed the role of DELLAs in many aspects of plant growth that are influenced by environmental cues, such as excess salt, light, or cold (Achard et al., 2006; Achard et al., 2008a; de Lucas et al., 2008; Feng et al., 2008; Cheminant et al., 2011). Moreover, DELLAs promote survival in an adverse environment (Achard et al., 2006; Achard et al., 2008a). Thus, it was proposed that DELLAs integrate environmental signals that enable plants to adapt their growth and development according to their surrounding environment. Furthermore, DELLAs are involved in response to biotic stress (Achard et al., 2008b; Navarro et al., 2008). DELLA activity promotes plant resistance to necrotrophs by potentiating jasmonate (JA) signaling and, reciprocally, increases plant susceptibility to virulent biotrophs by attenuating the salicylic acid (SA) pathway (Navarro et al., 2008). Thus, DELLAs modulate plant immune response by modulating the balance of JA/SA. Recently, the DELLA proteins have been demonstrated to interact with the JA ZIM-domain (JAZ) proteins, repressors of JA signaling (Hou et al., 2010; Yang et al., 2012). JAZ proteins bind and inhibit activity of a wide array of transcription factors, including the basic helix-loop-helix (bHLH) JASMONATE INSENSITIVE1 (JIN1/MYC2), MYC3, and MYC4 transcription factors, which regulate major aspects of JA-mediated gene expression (Chini et al., 2007; Thines et al., 2007; Gfeller et al., 2010; Fernández-Calvo et al., 2011). Besides the MYC proteins, JAZs interact with other members of the bHLHs, MYBs, and the ETHYLENE INSENSITIVE3 (EIN3)/EIN3-Like (EIL) family of transcription factors to repress a subset of JA-regulated responses, such as stamen and trichome development, anthocyanin synthesis, or defense against necrotrophic pathogens (Pauwels and Goossens, 2011; Qi et al., 2011; Song et al., 2011; Zhu et al., 2011). In the presence of jasmonyl-Ile (the active form of JA) bound to the E3 ubiquitin ligase SCFCOI1 complex (through the JA receptor/F-box protein COI1), JAZs interact with COI1, leading to their ubiquitylation and subsequent degradation by the 26S proteasome (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002; Yan et al., 2009). Upon degradation of JAZs, MYC2 and related transcription factors are released to promote JA-induced gene expression. The fact that DELLAs and MYC2 compete for binding to JAZ repressors provides a mechanism by which DELLAs could potentiate JA signaling. Consistent with this model, MYC2-dependent JA-responsive genes are upregulated in response to JA treatment in mutant backgrounds accumulating DELLAs (e.g., in the GA biosynthetic mutant ga1-3; Sun and Kamiya, 1994), whereas this upregulated expression is impaired under combined GA and JA treatment (Hou et al., 2010). However, although these observations suggest that DELLA activity modulates JA signaling via competitive binding to JAZs, the biological significance of this mechanism in a natural context remained unclear.
In this study, we present genetic and molecular data showing that the DELLA protein RGL3 is critical for the expression of JA-responsive genes in a biological context. In contrast with the other DELLA genes, we show that RGL3 is induced by JA in a MYC2-dependent fashion. Moreover, we show that RGL3 interacts with JAZs, thus suggesting that JA-induced RGL3 accumulation sequesters JAZ repressors, thereby releasing MYC2 transcriptional activity. Accordingly, overexpression of RGL3 activates MYC2-dependent JA-induced gene expression, whereas rgl3 mutation reduces it. In addition, phenotypic analyses show that the rgl3-5 mutant exhibits enhanced susceptibility to the necrotrophic fungus Botrytis cinerea and resistance to the hemibiotroph Pseudomonas syringae. Taken together, our findings show that RGL3 is a distinct negative regulator of GA responses that acts as an integrating factor that links GA and JA signaling to enable adaptive regulation of plant resistance to pathogens.
RESULTS
RGL3 and JA-Related Genes Are Coregulated and Induced by JA
To address the biological function of RGL3, we first surveyed its expression pattern using the ATTED-II Network Drawer, which predicts regulatory networks based on coexpressed genes determined from public microarray analyses (Obayashi et al., 2007). Bioinformatics study revealed that RGL3 is tightly coregulated with JA-related genes, including JAZs (JAZ2, JAZ3, JAZ6, JAZ9, and JAZ10), thus suggesting that JA regulates RGL3 expression (see Supplemental Figure 1 online). To confirm these data, we analyzed by quantitative RT-PCR (qRT-PCR) the induction kinetics of the five Arabidopsis DELLA genes (GAI, RGA, RGL1, RGL2, and RGL3) in 7-d-old seedlings treated with 50 μM of methyl jasmonate (MeJA). We found that JA transiently induced RGL3 (and to a lesser extent RGL1), with RGL3 reaching its maximum induction after 1 h of JA treatment (Figure 1A; see Supplemental Figure 2 online). DELLA proteins are subject to destabilization in the presence of GA (Achard and Genschik, 2009). Thus, we assessed whether JA-mediated transient induction of RGL3 was accompanied by an increase in RGL3 protein accumulation. To this end, we generated a translational fusion with the full-length RGL3 cDNA fused to the Green Fluorescent Protein (GFP) open reading frame downstream of the RGL3 promoter (pRGL3:RGL3-GFP). By protein gel blotting using an anti-GFP antibody, we found that JA rapidly enhanced the accumulation of RGL3-GFP protein in 7-d-old pRGL3:RGL3-GFP transgenic seedlings (Figure 1B). Furthermore, the level of RGL3-GFP accumulation was maintained over 24 h of JA treatment (Figure 1B). To visualize in planta the accumulation of RGL3 proteins, we next generated a line expressing a RGL3–β-glucuronidase (GUS) translational fusion driven by the RGL3 promoter (pRGL3:RGL3-GUS). In 3-week-old plants, trace levels of RGL3-GUS were visible only in the shoot apices (Figure 1C). In the presence of JA, RGL3-GUS expression was elevated throughout the plant, mainly in the petiole and the vasculature of the leaves. Thus, JA induces RGL3 expression rapidly and broadly within the plant.
Figure 1.
JA Enhances RGL3 Transcript and Protein Levels.
(A) Relative levels (mean ± sd) of GAI, RGA, RGL1, RGL2, and RGL3 gene transcripts (determined by qRT-PCR) in 7-d-old seedlings treated with 50 μM of MeJA for the time (15 min to 24 h) indicated. Similar results were obtained in three independent experiments.
(B) Immunodetection of RGL3-GFP in 7-d-old pRGL3:RGL3-GFP seedlings treated with 50 μM of MeJA for the time (15 min to 24 h) indicated (and mock treatment for controls). PSTAIRE serves as sample loading control. Similar results were obtained in three independent experiments.
(C) RGL3-GUS induction in 3-week-old pRGL3:RGL3-GUS plantlets after 2 h of JA-treatment (50 μM of MeJA) and control (mock treatment). Similar results were obtained in three independent experiments.
Next, to investigate whether JA modulated the GA-mediated degradation of RGL3 proteins, we examined the half-life of RGL3-GFP (in 7-d-old p35S:RGL3-GFP seedlings) in the presence of cycloheximide (CHX), a chemical blocking protein synthesis. As previously reported, the level of RGL3-GFP proteins was significantly reduced 2 h after CHX treatment, and the kinetic of RGL3-GFP degradation was accelerated in the presence of 50 μM of gibberellic acid (GA3) (see Supplemental Figure 3 online) (Feng et al., 2008). JA alone or combined with GA3 did slightly delay the kinetics of RGL3-GFP degradation in comparison with the controls, in the absence and presence of GA3, respectively (see Supplemental Figure 3 online) (Yang et al., 2012). However, we showed that JA did not statistically alter the content of the biologically active GA, GA4 (see Supplemental Figure 4 online). Thus, overall, these results indicate that JA rapidly enhances RGL3 protein accumulation essentially via induction of the gene transcripts.
RGL3 Is a Direct Target of MYC2
JA activates JA responses by stimulating the SCFCOI1-dependent degradation of the JAZ proteins, thereby releasing MYC2 to promote JA-induced gene expression (Chini et al., 2007; Thines et al., 2007). Here, we demonstrated that JA-induced RGL3 expression works via the COI1/MYC2-dependent signaling pathway (Figure 2). Loss-of-function coi1-1 confers stabilization of JAZs and impairs responsiveness to JA (Chini et al., 2007; Thines et al., 2007). We found that JA-mediated induction of RGL3 was abolished in the coi1-1 mutant, indicating that RGL3 is downstream of COI1 (Figure 2A). We next investigated the responsiveness of RGL3 to JA in the myc2/jin1-8 mutant. We found that RGL3 expression was partially insensitive to JA (Figure 2B). The Arabidopsis genome encodes two close MYC2 homologs, MYC3 and MYC4, which have an additive effect on the activation of the JA responses (Fernández-Calvo et al., 2011). As in the myc2 mutant, JA-mediated induction of RGL3 was significantly impaired in the myc3 mutant and to a lesser extent in the myc4 mutant. In addition, the responsiveness of RGL3 to JA was completely impaired in the triple myc2 myc3 myc4 mutant (Figure 2B). Altogether, these results indicate that MYC2/MYC3/MYC4 proteins act redundantly in the activation of RGL3 in response to JA.
Figure 2.
JA Induces the Expression of RGL3 in a COI1/MYC2-Dependent Manner.
(A) Time-course induction (mean ± sd) of RGL3 and MYC2 transcripts in 14-d-old wild-type (Col) and coi1-1 mutant seedlings treated with 50 μM of MeJA for the time indicated. Similar results were obtained in two independent experiments.Col, Col-0.
(B) Time-course induction (mean ± sd) of RGL3 transcripts in 7-d-old wild-type (Col), myc2/jin1-8, myc3, myc4, and triple myc2 myc3 myc4 mutant seedlings treated with 50 μM of MeJA for the time indicated. Similar results were obtained in two independent experiments.
MYC2/MYC3/MYC4 proteins are bHLH transcriptional factors that specifically bind to G-box (CACGTG) or G-box–like motifs (Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011). We found five G-box–related motifs (two CACATG and three CACGTT boxes) within the 1.5-kb promoter of RGL3 (Figure 3A). To investigate whether MYC2 binds directly to the RGL3 promoter, we performed chromatin immunoprecipitation (ChIP) with the jin1-8 pMYC2:MYC2-FLAG transgenic line described previously (Hou et al., 2010). After immunoprecipitation of protein–DNA complexes using an antibody against the FLAG epitope, enriched DNA sequences were amplified by quantitative PCR (qPCR) using primers that annealed near the G-box–like motifs present in the RGL3 promoter. A genomic fragment of ACTIN2 was also amplified as a negative control, and the promoter of JAZ1 was amplified as a positive control (Hou et al., 2010). We performed ChIP assays on plants that had been treated for 4 h with 50 μM of MeJA to induce MYC2-FLAG expression (and on controls without JA treatment). As shown in Figure 3A, we found an enrichment of the F3 region of the RGL3 promoter, indicating that RGL3 is a direct target of MYC2. To confirm the specificity of the FLAG antibody, we also performed ChIP assays on nontransgenic jin1-8 mutant, and we did not find an enrichment of the same promoter region by qPCR (see Supplemental Figure 5 online).
Figure 3.
MYC2 Regulates RGL3 Expression through a Direct Association with Its Promoter.
(A) MYC2 interacts with the RGL3 promoter according to ChIP analysis. (Top) Schematic representation of the RGL3 promoter, including potential MYC2 binding sites (white and black triangles) and DNA fragments used for ChIP (F1, F2, and F3) and for EMSA. (Bottom) Fold enrichment of indicated RGL3 promoter fragments and of JAZ1. Chromatin of transgenic plants expressing MYC2-FLAG (jin1-8 pMYC2:MYC2-FLAG) treated with 50 μM of MeJA for 4 h and untreated controls (Mock) were subjected to ChIP with anti-FLAG antibodies followed by qPCR. Data represent means ± sd of triplicate determinations. The experiment was repeated twice with several technical replicates.
(B) EMSA showing that MBP-MYC2 fusion protein binds to the DNA probe of RGL3 in vitro. 32P-radiolabeled probes were incubated with MBP-MYC2 or MBP purified proteins, and the free and bound probes were separated in an acrylamide gel. As indicated, unlabeled probes were used as competitors. Similar results were obtained in two independent experiments. Mut, mutated unlabeled probe in which the CACATG motif was deleted.
To validate further the interaction of MYC2 with the RGL3 promoter, we next conducted a DNA electrophoretic mobility shift assay (EMSA) with a recombinant maltose binding protein (MBP)-MYC2 protein produced in Escherichia coli and affinity-purified. We found that MBP-MYC2 fusion proteins (but not MBP alone) were able to bind a DNA probe containing the CACATG G-box–like motif located near the F3 region of the RGL3 promoter (Figure 3B). Furthermore, unlabeled DNA probes competed for binding in a dose-dependent manner, but the DNA probes containing the mutant form of the CACATG motif did not (Figure 3B). Together, these results reveal that MYC2 regulates RGL3 expression through a direct association with its promoter.
RGL3 Interacts with JAZ1 and JAZ8 and Modulates MYC2-Dependent Gene Expression
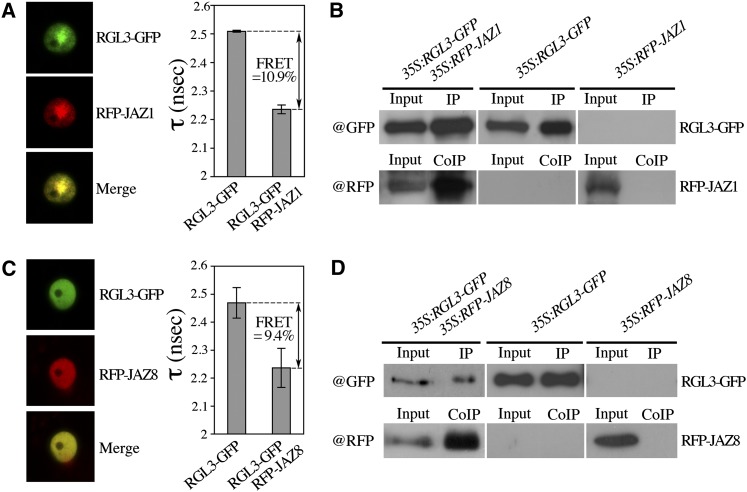
Recent studies reported the interaction of the DELLA proteins with JAZ proteins, providing a mechanism by which DELLAs contribute to JA signaling (Hou et al., 2010; Yang et al., 2012). By fluorescence lifetime imaging microscopy (FLIM) analysis of nuclei in transiently transformed Nicotiana benthamiana cells, we showed the direct interaction of RGL3 with two phylogenetically distant JAZ isoforms, JAZ1 and JAZ8 (a JAZ resistant to JA-mediated degradation; Shyu et al., 2012) with a fluorescence resonance energy transfer of 10.9 and 9.4%, respectively (Figures 4A and 4C). We also showed the direct interaction of RGL3 with JAZ1 and JAZ8 by yeast two-hybrid assays (see Supplemental Figure 6 online). This latest result contrasts with those from Yang et al. (2012), who did not observe an interaction between RGL3 and JAZ8 in yeast. Moreover, coimmunoprecipitation studies further confirmed an in vivo interaction of RGL3 with JAZ1 and with JAZ8 (Figures 4B and 4D). Taken together, our results demonstrate that RGL3 interacts in planta with various JAZ proteins and therefore suggest that JA-mediated induction of RGL3 transcripts is critical to enhance the JA responses.
Figure 4.
RGL3 Interacts with JAZ1 and JAZ8.
(A) and (C) Fluorescence lifetime analyses (in nanoseconds [nsec]) of RGL3-GFP alone or together with RFP-JAZ1 or RFP-JAZ8, and mean (±se) fluorescence resonance energy transfer (FRET) value (%) in N. benthamiana agroinfiltrated leaves.
(B) and (D) Coimmunoprecipitation studies. Total protein extract from N. benthamiana agroinfiltrated leaves with p35S:RGL3-GFP and p35S:RFP-JAZ1 or p35S:RFP-JAZ8 were immunoprecipitated with anti-GFP antibody. The coimmunoprecipitated proteins were detected by anti-RFP antibody. The experiment was repeated twice with several technical replicates. CoIP, coimmunoprecipitated proteins; IP, immunoprecipitated proteins.
To investigate further the biological function of RGL3, we identified a new rgl3 mutant allele (SALK_082546), hereafter called rgl3-5 (see Supplemental Figure 7 online). We speculated that lack of RGL3 activity would reduce or delay the induction of MYC2-dependent gene expression. To test this hypothesis, we compared the kinetics of induction of typical MYC2-regulated genes (including two MYC2 direct targets, LOX2 and TAT1; Hou et al., 2010) in 3-week-old wild-type and rgl3-5 plants treated with JA. We found that the rgl3-5 mutant exhibited reduced induction levels of VSP2, TAT1, and LOX2 transcripts in comparison with the wild type (Figure 5A). Thus, RGL3 is essential to fully enhance JA-induced MYC2-dependent gene expression. Consistent with this result, overexpression of RGL3 (in p35S:RGL3-GFP transgenic line) constitutively enhanced the expression of VSP2, TAT1, and LOX2 in comparison with the wild type (Figure 5B). Thus, JA enhances MYC2 transcriptional activity by a dual mechanism. First, JA stimulates the SCFCOI1-dependent degradation of the JAZs, thus releasing MYC2 transcriptional activity. Second, JA (in a MYC2-dependent fashion) enhances the accumulation of RGL3, which in turn sequesters JAZs into inactive complexes unable to restrain MYC2 transcriptional activity (Hou et al., 2010). This second regulatory mechanism is likely to play a prominent role in modulating the activity of the JAZs that are relatively resistant to JA-mediated degradation, such as JAZ8 (Shyu et al., 2012).
Figure 5.
RGL3 Enhances JA-Responsive Gene Expression.
(A) Time-course induction (mean ± sd) of VSP2, TAT1, and LOX2 transcripts in 3-week-old wild-type (Col) and rgl3-5 mutant plants treated with 50 μM of MeJA for the time indicated. Similar results were obtained in three of four biological replicates.Col, Col-0.
(B) Relative transcript levels of RGL3, VSP2, TAT1, and LOX2 in 6-week-old wild-type (Ler) and a line overexpressing RGL3 (p35S:RGL3-GFP). Data (mean ± sd) are represented as fold change in gene expression (RGL3ox/Ler). Similar results were obtained in two independent experiments.
RGL3 Modulates JA-Dependent Plant Defense Responses
The JA signaling pathway performs a critical role in plant defense. JA is the major signaling molecule implicated in plant resistance to necrotrophic pathogens (Lorenzo and Solano, 2005). For example, mutants impaired in JA production exhibit enhanced susceptibility to the necrotrophic fungus B. cinerea (Stintzi et al., 2001; Kunkel and Brooks, 2002). To address the biological role of RGL3 in JA-mediated plant defense responses, we first examined the expression pattern of RGL3 in rosette leaves of 6-week-old plants inoculated with B. cinerea spores over a time-course experiment. We found that RGL3 was broadly induced and accumulated throughout the leaves 24 to 48 h after inoculation (Figures 6A and 6B). Next, we assessed whether RGL3 modulates JA-dependent plant defense responses. To this end, we inoculated leaves of 6-week-old wild-type and rgl3-5 mutant plants with B. cinerea spores and examined disease symptoms 2 d after inoculation. rgl3-5 mutant plants were found to be more susceptible to B. cinerea, because they presented larger lesions compared with wild-type infected plants (Figure 6C). Consistent with this, the increased severity of the disease symptoms in rgl3-5 mutant plants correlated with increased pathogen content (Figure 6D). Finally, we monitored the kinetics of JA-responsive gene induction (including the pathogen-responsive PLANT DEFENSIN1.2 [PDF1.2] and ETHYLENE RESPONSE FACTOR1 [ERF1] genes; Zhu et al., 2011), in wild-type and rgl3-5 mutant plants inoculated with B. cinerea. All the JA-responsive genes tested displayed decreased induction rates in rgl3-5 infected plants compared with wild-type infected plants (Figure 6E). Thus, RGL3 enhances JA-dependent plant defense responses to necrotrophic pathogens.
Figure 6.
RGL3 Enhances JA-Dependent Plant Defense Responses to the Necrotroph B. cinerea.
(A) to (E) Six-week-old rosette leaves were inoculated with B. cinerea spores at a concentration of 2 × 105 spores/mL.
(A) Time-course induction (mean ± sd) of RGL3 and MYC2 transcripts in wild-type inoculated leaves. Similar results were obtained in three independent experiments.
(B) RGL3-GUS induction in pRGL3:RGL3-GUS infected leaves at 0, 1, 2, and 3 d after inoculation.
(C) Graphical representation of disease symptoms (diameter of lesions) at 2 d after inoculation (mean ± se). At least 25 leaves per genotype were inoculated in each experiment. Experiments were repeated at least five times with similar results.
(D) Quantification of in planta growth of B. cinerea in wild-type and rgl3-5 infected leaves at 0 to 3 d after inoculation. qPCR was used to analyze the relative genomic DNA level of B. cinerea CUTINASE A gene compared with Arabidopsis ACTIN2. Data represent means ± sd of triplicate determinations. Similar results were obtained in three independent experiments.
(E) Time-course induction (mean ± sd) of TAT1, LOX2, ERF1, and PDF1.2 transcripts in wild-type and rgl3-5 inoculated leaves. Similar results were obtained in three of four biological replicates.
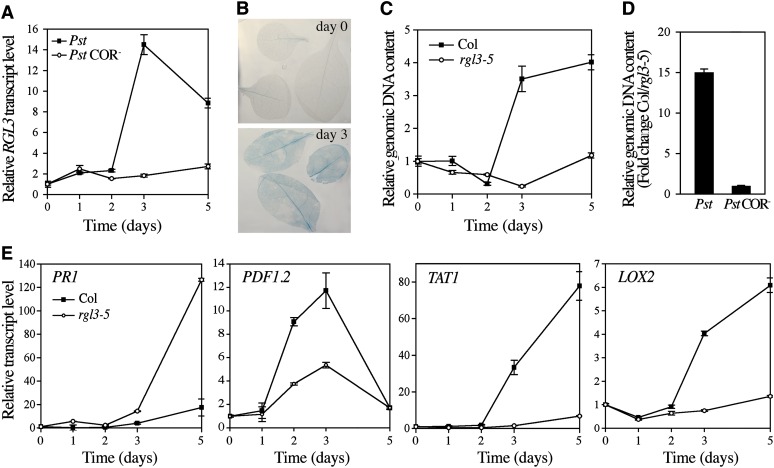
We also analyzed the biological function of RGL3 in response to infection by the hemibiotrophic pathogen P. syringae pv tomato DC3000 (Pst). Contrary to necrotrophic pathogens, which activate the JA pathway and trigger a strong defense response, Pst produces a JA-mimicking phytotoxin, coronatine (COR), which can induce a set of JA-responsive genes to promote susceptibility (Laurie-Berry et al., 2006). As expected, we found that Pst induced RGL3 expression in leaves 3 d after inoculation (Figures 7A and 7B). Conversely, RGL3 was not induced by a P. syringae strain defective in COR synthesis (Pst COR−), thus indicating that P. syringae uses the COR-dependent pathway to modulate RGL3 expression (Figure 7A). We then analyzed whether the loss-of-function rgl3 mutation altered resistance to P. syringae. Infected rgl3-5 plants displayed an ∼15-fold reduction in bacterial growth at 3 d after inoculation compared with wild-type infected plants (Figures 7C and 7D). Consistent with this, no significant difference in bacterial growth was observed between rgl3-5 and wild-type infected plants with the COR-deficient Pst COR− strain (Figure 7D).
Figure 7.
RGL3 Enhances Plant Susceptibility to the Hemibiotrophic Pathogen P. syringae.
(A) to (E) Six-week-old rosette leaves were inoculated by spray with Pst and Pst COR− (as indicated) at a bacterial concentration of 2 × 108 colony-forming units/mL.
(A) Time-course induction (mean ± sd) of RGL3 transcripts in wild-type inoculated leaves. Similar results were obtained in three independent experiments.
(B) RGL3-GUS induction in pRGL3:RGL3-GUS leaves inoculated with Pst at 0 and 3 d after inoculation.
(C) Quantification of in planta growth of Pst in wild-type and rgl3-5 infected leaves at 0 to 5 d after inoculation. qPCR was used to analyze the relative genomic DNA level of P. syringae oprf gene compared with Arabidopsis ACTIN2. Data represent mean ± sd of triplicate determinations. Similar results were obtained in three independent experiments.
(D) Relative growth of Pst and Pst COR− assessed at 3 d after inoculation. Data (mean ± sd; calculated as described in [C]) are represented as fold change of genomic DNA content (Col/rgl3-5). Similar results were obtained in three independent experiments.Col, Col-0.
(E) Time-course induction (mean ± sd) of PR1, PDF1.2, TAT1, and LOX2 transcripts in wild-type and rgl3-5 leaves inoculated with Pst. Similar results were obtained in three of four independent experiments.
SA is the main signaling molecule involved in plant resistance to P. syringae (Kunkel and Brooks, 2002). Because the SA pathway is antagonistic to the JA pathway (Robert-Seilaniantz et al., 2011), we tested whether the increased resistance of the rgl3-5 mutant to Pst correlated with a hyperinduction of SA-dependent defense genes and a suppression of JA-dependent gene expression, respectively. We found that SA-dependent PATHOGENESIS-RELATED1 (PR1) transcripts displayed stronger induction in infected leaves of rgl3-5 versus the wild type, indicating that RGL3 represses the SA pathway (Figure 7E). Conversely, the induction of JA-dependent gene expression, including PDF1.2, TAT1, and LOX2, was severely compromised in rgl3-5 infected leaves in comparison with wild-type infected leaves (Figure 7E). Altogether, these results indicate that RGL3 modulates plant defense to pathogens via its positive regulation of JA signaling.
DISCUSSION
DELLA proteins mediate GA signaling in a wide range of plant species. Studies of loss-of-function alleles of GAI, RGA, RGL1, and RGL2 in Arabidopsis have shown that GAI and RGA function redundantly as negative regulators of GA responses affecting stem elongation (Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001), and RGL2 functions as a main repressor of seed germination (Lee et al., 2002), whereas RGA, RGL1, and RGL2 together are important for modulating GA-regulated floral development (Cheng et al., 2004; Tyler et al., 2004). The RGL3 amino acid sequence is related to GAI/RGA/RGL1/RGL2 throughout its entire length (Lee et al., 2002), suggesting that RGL3 also regulates GA responses. Accordingly, a recent study revealed that GA stimulates the degradation of TAP-tagged RGL3 protein and that MG132 (a 26S proteasome inhibitor) prevents GA-mediated degradation of RGL3 (Feng et al., 2008). Moreover, RGL3 (like the other DELLAs) interacts with SLY1, the F-box component of the SCFSLY1 complex that specifically ubiquitinates DELLAs (Tyler et al., 2004). However, because RGL3 is expressed at low levels, it was suggested that RGL3 plays only a minor role in the control of GA-regulated responses (Tyler et al., 2004; Piskurewicz and Lopez-Molina, 2009). Therefore, no clear function has yet been attributed to RGL3.
To study the function of RGL3, we first surveyed its expression pattern in microarray data sets from various growth and stress conditions. We found that RGL3 was coregulated with several JAZ members, suggesting that JA regulates RGL3 expression. Consistent with this observation, our expression studies have further confirmed that JA induced the expression of RGL3 via the COI1/MYC2-dependent signaling pathway (Figures 1 and 2). Collectively, these results indicate that RGL3 acts downstream of MYC2/MYC3/MYC4 in the JA signaling pathway. Interestingly, a previous work provided genetic evidence showing that GA acts through JA to promote stamen filament growth in Arabidopsis (Cheng et al., 2009). Thus, the hierarchical relationship between GA and JA signaling pathways might be different depending on the tissue or the developmental stage.
Our findings raised the possibility that MYC2 directly regulates RGL3 expression, like other primary JA-responsive genes, such as LOX2, TAT1, JAZ1, or JAZ3 (Chung et al., 2008; Hou et al., 2010). Indeed, ChIP experiments indicated that MYC2 directly associates with a G-box–related motif CACATG present in the promoter of RGL3 (Figure 3). It is noteworthy that the CACATG motif is not the preferred MYC2 core binding site (which is CACGTG), but several lines of evidence indicate that MYC2 can bind efficiently to this G-box–related hexamer and regulate the expression of the corresponding genes (Dombrecht et al., 2007; Chen et al., 2011).
JA treatment also weakly induced RGL1 expression (Figure 1). Sequence analysis of the RGL1 promoter revealed one G-box–related motif (CACGTT) located 1447 bp upstream of the translational start codon. However, we did not detect an interaction of MYC2 with this promoter region by ChIP assays (data not shown), suggesting that the induction of RGL1 is indirect.
Previous reports showed that the DELLA proteins interact with the repressors of MYC2 activity, JAZs (Hou et al., 2010; Yang et al., 2012). We also showed that RGL3 interacts in vivo with JAZ1 and JAZ8, a JAZ resistant to JA-mediated degradation (Figure 4). Thus, according to our current knowledge about the JA signaling pathway, JA-mediated accumulation of RGL3 protein should enhance the expression of JA-responsive genes via competitive binding to the JAZs. Indeed, we showed that the MYC2 direct targets are reduced in rgl3-5 mutants and enhanced in a transgenic line overexpressing RGL3 (Figure 5). Therefore, we propose a model in which RGL3 enhances JA action (Figure 8). In the absence of JA, JAZs repress MYC2 activity and thus the expression of MYC2-regulated genes. In the presence of JA, the JAZs containing the conserved LPIAR degron motif (such as JAZ1; Shyu et al., 2012) are ubiquitylated and destabilized by the proteasome, and MYC2 is released from the negative effect of this subgroup of JAZ proteins. In turn, JAZs are positively regulated by MYC2 by a feedback mechanism conferring a transitory character to JA. However, MYC2 also positively regulates RGL3 accumulation, which leads to the release of MYC2 by competitive binding to JAZs. Given the ability of RGL3 to interact with JAZ8, the induction of RGL3 by JA provides an efficient mechanism by which JA may repress the activity of the JAZs that are resistant to the action of JA (Figure 8). Taken together, our results indicate that RGL3 plays a critical role in modulating JA action by enhancing the responsiveness of the plant to JA. RGL3 is destabilized in the presence of GA (see Supplemental Figure 3 online); therefore, the importance of RGL3 in this response is inversely proportional to the amount of bioactive GAs accumulated in the plant. This is consistent with other findings reporting that pretreatment with exogenous GA compromised JA signaling responses (Navarro et al., 2008; Hou et al., 2010).
Figure 8.
Model of RGL3-Mediated Regulation of JAZ1 and JAZ8 by JA/MYC2.
In the absence of JA, the JAZs sequester the transcription factor MYC2, and thereby repress the expression of early JA-responsive genes. Binding of JA to the JA-receptor/F-box COI1 promotes the polyubiquitylation and subsequent destruction of degradable JAZ1 (but not of nondegradable JAZ8), thus releasing the JAZ1-mediated inhibition of MYC2 target genes. Moreover, JAZs are regulated by a feedback mechanism creating a transitional character of the JA signaling. JA-induced MYC2 expression also leads to RGL3 accumulation, which competes with MYC2 binding to JAZs, leading to the release of MYC2, which in turn enhances the expression of JA-responsive genes. In the presence of GA, RGL3 is immediately degraded by the proteasome, allowing the accumulation of JAZs free to interact with MYC2.
[See online article for color version of this figure.]
To investigate further the role of RGL3 in modulating JA action in a natural context, we analyzed JA-regulated defense responses in wild-type and rgl3 loss-of-function mutant plants. Plant defense is mediated by complex signaling networks in which the plant hormones JA and SA play key roles (Robert-Seilaniantz et al., 2011). In general, JA-mediated signaling pathways elevate necrotroph resistance, whereas the SA pathway is associated with enhanced resistance to biotrophs. Moreover, these two pathways are mostly antagonistic. Thus, elevated necrotroph resistance is often correlated with increased biotroph susceptibility and vice versa. As an example of JA-dependent resistance, we analyzed the infection by the necrotrophic fungus B. cinerea, and as an example of JA-mediated susceptibility, we analyzed the infection by the hemibiotroph Pst. Consistent with our model, rgl3-5 mutation enhanced susceptibility to B. cinerea and resistance to P. syringae (Figures 6 and 7). Collectively, these results indicate that RGL3 contributes substantially to JA-regulated defense responses. Interestingly, we have already shown that RGL3 is induced by various environmental stresses, such as cold (Achard et al. 2008a). Therefore, it is likely that the contribution of RGL3 to plant fitness increases when the environmental conditions are less favorable for plant growth.
Previous studies have indicated the existence of two branches in the JA signaling pathway that are antagonistically regulated by MYC2 and ERF1 (Lorenzo et al., 2004). Whereas MYC2 is responsible for the activation of wound/insect-responsive genes, such as LOX2 and TAT1, and the repression of pathogen-responsive genes, such as PDF1.2, ERF1 exerts antagonistic effects. Interestingly, ERF1, PDF1.2, TAT1, and LOX2 transcripts all displayed decreased induction rates in rgl3-5 infected plants compared with wild-type infected plants (Figures 6 and 7). The JAZ proteins interact with various transcription factors, including EIN3, a positive regulator of ERF1 and PDF1.2 (Zhu et al., 2011); therefore, RGL3 may enhance the activity of EIN3 (or other JAZ-interacting factors) via competitive binding to JAZs. Further analyses will be required to assess these possibilities.
Gene duplications are considered to be the major source for variation and the acquisition of novel functions (Ohno, 1970). Unlike in monocots, DELLA genes are often duplicated in dicots. A recent report proposed that functional diversification of Arabidopsis DELLAs relied on changes in their expression patterns (Gallego-Bartolomé et al., 2010). Thus, JA-mediated induction of RGL3 expression could represent a novel function with adaptive significance. Bioinformatics studies performed on publicly available microarray resources (www.bar.utoronto.ca) indicate that some Arabidopsis accessions, including ecotypes Canary Islands (Can-0) and San Feliu (Sf-2), express RGL3 at higher levels in comparison with ecotype Columbia (Col-0) seedlings, suggesting that these accessions are hyperresponsive to JA. Consistent with this, we found that JA induces RGL3 expression to a substantially higher level in Can-0 and Sf-2 than in Col-0 and that the basal level of primary JA-responsive gene transcripts is also higher and is induced to higher levels by JA in Sf-2 than in Col-0 (see Supplemental Figure 8 online). Interestingly, the Sf-2 accession was recently reported to be naturally more resistant to various pathogens, including the necrotrophic fungus Plectosphaerella cucumerina (Ahmad et al., 2011). Results from our study raise the possibility that DELLA gene duplication events (that probably occurred after the divergence between dicots and monocots) increase the plasticity and the adaptation of plants to their surrounding growth conditions.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 is the genetic background for all mutant lines. rgl3-5 (SALK_082546) and jin1-8 (SALK_061267) mutant lines were obtained from the Nottingham Arabidopsis Stock Centre resource center. The mutants coi1-1, myc3, myc4, myc2 myc3 myc4, and the transgenic line jin1-8 pMYC2:MYC2-FLAG have been previously described (Xie et al., 1998; Hou et al., 2010; Fernández-Calvo et al., 2011). Arabidopsis Sf-2 and Can-0 accessions were obtained from the Nottingham Arabidopsis Stock Centre resource center. Plants were grown on soil (12-h light/12-h dark; 22°C) or on plates containing 1× Murashige and Skoog medium (Duchefa Biochemical), 0.5% Suc, and 1% agar under a 16-h photoperiod at 22°C. For hormone treatments, 7-d-old seedlings grown in Murashige and Skoog liquid medium or 3-week-old plantlets grown on soil were treated with 50 μM of MeJA (Duchefa Biochemical) or 50 μM of GA3 (Sigma-Aldrich) for the time indicated.
Plasmid Construction and Plant Transformation
pRGL3:RGL3 fragment was PCR amplified from ecotype Landsberg erecta (Ler) genomic DNA with appropriate primers encompassing 1.5-kb promoter region of RGL3. Subsequently, the fragment was inserted into pDONR207 (Invitrogen) by Gateway cloning methods and then recombined with the plant binary vectors pMDC204 (Curtis and Grossniklaus, 2003) and pBGWFS7 (Karimi et al., 2002) to generate the pRGL3:RGL3-GUS and pRGL3:RGL3-GFP constructs, respectively. RGL3 coding sequence was PCR amplified from Col-0 genomic DNA, subsequently inserted into pDONR207 and recombined with pB7FWG2 (Karimi et al., 2002) to generate p35S:RGL3-GFP. The N-terminal part of the DELLA proteins is subject to autoactivation in yeast two-hybrid assays (de Lucas et al., 2008); therefore, only the C-terminal domain of RGL3 (amino acids 140 to 523) was cloned and recombined with pGBT9 (Clontech) to generate BD-RGL3. The JAZ1 and JAZ8 cDNAs were amplified by RT-PCR, subsequently inserted into pDONR207, and recombined with pB7WGR2 (Karimi et al., 2002) to generate p35S:red fluorescent protein (RFP)-JAZ1 and p35S:RFP-JAZ8 or with pGADT7 (Clontech) to generate AD-JAZ1 and AD-JAZ8. Finally, MYC2 coding sequence was amplified from Col-0 genomic DNA, inserted into pDONR207, and recombined with the Escherichia coli expression vector pHMGWA (Busso et al., 2005) to express His-tagged MBP-MYC2 protein. Primers used for the cloning are listed in Supplemental Data Set 1 online. The plant binary vectors were introduced into Agrobacterium tumefaciens GV3101 strain by electroporation, and Arabidopsis Ler plants were transformed by floral dip.
Gene Expression Analyses
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen) following the user manual. First, 2 μg of total RNA were treated with two units of DNase I (Promega) and then were reverse transcribed in a total volume of 40 μL with 2 μM of oligo(dT)20, 0.5 mM of deoxynucleotide triphosphate, 5 mM of DTT, and 200 units of Superscript III reverse transcriptase (Invitrogen). qRT-PCR was performed using gene-specific primers in a total volume of 6 μL of SYBR Green Master mix (Roche) on a Lightcycler LC480 apparatus (Roche) according to manufacturer’s instructions. The GAPDH and TIP41-like genes were used as internal reference genes. The relative expression level of each gene was calculated using Lightcycler 480 software, release 1.5.0 SP3, and averaged over three replicates. qRT-PCR analyses were performed on at least two biological repeats. qRT-PCR primers used are listed in Supplemental Data Set 1 online.
Immunoblot Analyses and Protein Half-Life Determination
pRGL3:RGL3-GFP seedlings that were 7 d old and inoculated with 50 μM of MeJA for the time indicated were ground in 2× SDS-PAGE buffer and then boiled for 5 min. After centrifugation, the protein extracts were fractionated on a 10% SDS-PAGE gel and blotted onto membrane. Immunoblots were performed using a 10,000-fold dilution of anti-GFP-horseradish peroxidase (HRP) antibodies (MACS Miltenyi Biotec). Signals were detected by film (within linear range of detection) using the Luminata Forte Western HRP Substrate (Millipore). The blot was subsequently stripped with 0.2 N of Gly, pH 2.5, and reprobed with anti-cdc2 (PSTAIRE) antibody (Santa Cruz Biotechnology) for loading control. The immunoblot assay was repeated three times.
For the determination of protein half-life, 7-d-old p35S:RGL3-GFP were incubated with 100 μM of CHX (Fluka Analytical) to inhibit protein synthesis. A total of 50 μM of GA3 and/or 50 μM of MeJA were added with CHX when indicated. After incubation for 0 to 180 min, seedlings were frozen in liquid nitrogen, and total proteins were extracted as described above. Immunoblotting was performed with anti-GFP-HRP in the same fashion as described above.
GUS Analyses
Histochemical detection of GUS activity was performed on 3-week-old (MeJA induction assay) or 6-week-old (pathogen assays) pRGL3:RGL3-GUS plants using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) as a substrate. Plants were placed in 80% acetone on ice for 10 min, then infiltrated in X-Gluc buffer solution (500 μg/mL of X-Gluc; 50 mM of sodium phosphate, pH 7; 10 mM of EDTA; 0.01% Triton X-100) for 10 min and incubated at 37°C overnight.
ChIP Assays
ChIP assays were performed on 7-d-old jin1-8 pMYC2:MYC2-FLAG seedlings as previously described (Berr et al., 2010). Briefly, chromatin was immunoprecipitated with anti-FLAG antibodies (Sigma-Aldrich) together with protein A magnetic beads (Millipore). ChIP experiments using mouse IgG were performed as negative controls. The resulting ChIP DNA was subjected to qPCR analysis. Enrichment of promoter regions was averaged over three replicates and normalized using ACTIN2. Enrichment of a JAZ1 promoter fragment was used as positive control. ChIP-qPCR analyses were performed on two independent biological repeats with several technical repeats. qPCR primers used are listed in Supplemental Data Set 1 online.
EMSA Assays
The His-tagged MBP-MYC2 recombinant protein was expressed in the Rosetta 2 (DE3) pLysS (Novagen) E. coli strain by induction with 0.25 mM of isopropyl-β-d-thiogalactopyramoside for 4 h at 20°C. The recombinant MBP-MYC2 protein was purified binding onto a Ni-NTA column (His60 Ni Superflow; Clontech) and eluted with 500 mM of imidazole. Elution buffer was replaced by EMSA buffer (15 mM of HEPES-KOH, pH 7.5; 40 mM of KCl; 0.1 mM of dithiothreitol; 10% glycerol) by filtration through a Sephadex-G25 HiTrap column (GE Healthcare Life Sciences). Oligonucleotide probe containing the G-box–like motif was end-filled labeled with the Klenow enzyme (Fermentas) in the presence of 32P-dCTP. The sequence of the oligonucleotides is indicated in Supplemental Data Set 1 online. The EMSA reaction was performed with 1 ng of 32P-labeled probe, 2 μg of poly(dI-dC), and 100 ng of MBP-MYC2 fusion proteins (or MBP as control) and was incubated at room temperature for 20 min. The binding reactions were analyzed by electrophoresis on 6% native acrylamide gels in 0.5× TBE buffer. After drying, the gels were subjected to autoradiography and were exposed at −80°C overnight.
Yeast Two-Hybrid Assays
Interaction assays in yeast were performed following the Clontech small-scale LiAc yeast transformation procedure. Yeast strain AH109 was cotransformed with BD-RGL3 (amino acids 140 to 523) and AD-JAZ1 or AD-JAZ8 or empty vector, and interaction tests were surveyed on selective media lacking Leu, Trp, Ade, and His (Clontech).
Coimmunoprecipitation Assays
Coimmunoprecipitation assays of RGL3 and JAZ1 or JAZ8 proteins were performed on Nicotiana benthamiana agroinfiltrated leaves with p35S:RGL3-GFP and p35S:RFP-JAZ1 or p35S:RFP-JAZ8 constructs. Total proteins were extracted in Lysis buffer containing 50 mM of Tris-HCl, pH 8, 150 mM of NaCl, 1% Triton X-100, supplemented with EDTA-free protease inhibitors (Roche), and then incubated for 2 h at 4°C with 50 μL of anti-GFP antibody conjugated to micro beads (μMACS GFP-tagged beads; Miltenyi Biotec). μMACS GFP-tagged beads were retained and washed onto a magnetic column system (M columns; Miltenyi Biotec) to recover the immunoprotein complexes according to the manufacturer’s instructions. RGL3 and JAZ1 or JAZ8 proteins bound to the beads were resolved by SDS-PAGE and detected by anti-GFP-HRP (Miltenyi Biotec) or anti-RFP antibody (Clontech) followed by a goat anti-rabbit-HRP antibody (Invitrogen).
Observation of GFP/RFP Fluorescence and FLIM Analyses
Confocal microscopy images were obtained with a Zeiss LSM510 inverted confocal laser microscope with 40× objectives. The excitation wavelength for GFP and RFP detection was 488 and 561 nm, respectively. Fluorescence was determined on N. benthamiana agroinfiltrated leaves with p35S:RGL3-GFP and p35S:RFP-JAZ1 or p35S:RFP-JAZ8 constructs.
FLIM was performed using a Nikon TE2000 microscope connected to a LiFA FLIM system. Fluorescence lifetime was measured using the LiFLIM software version 1.2.8. on N. benthamiana agroinfiltrated leaves expressing RGL3-GFP (control) and coexpressing RGL3-GFP and RFP-JAZ1 or RFP-JAZ8 proteins. At least 60 nuclei per condition were analyzed.
Pathogen Inoculation and Analyses
Pathogen inoculation assays were performed on 6-week-old plants (12-h light/12-h dark). Botrytis cinerea (5 × 105 spores/mL in one-half potato dextrose broth [Duchefa Biochemical]) inoculation was performed by placing 5-μL droplets onto fully expanded leaves. Disease symptoms were scored 2 d after inoculation. RNA and DNA analyses were conducted on 8-mm leaf disks centered on the infection site of at least 30 leaves per genotype. For in planta quantification of pathogen growth, DNA content of B. cinerea was compared with plant DNA by comparing the abundance of the fungal CUTINASE gene to the Arabidopsis-specific ACTIN2 DNA by qPCR analysis as described in Berr et al. (2010). The bacterial strains used in this study were Pseudomonas syringae pv tomato DC3000 (Pst) and Pst COR−. Bacterial inoculations were performed by spraying at a concentration of 2 × 108 colony-forming units/mL. RNA and DNA analyses were conducted on whole mature leaves from six different plants. Quantification of bacterial growth was determined by relative quantification of bacteria and plant DNA by comparing the abundance of P. syringae oprf gene with plant ACTIN2 by means of qPCR analysis as in Brouwer et al. (2003). Primer sequences are detailed in Supplemental Data Set 1 online. Work with pathogens was repeated at least five times, and similar results were obtained.
GA4 Quantification
GA4 were extracted from 3-week-old plants by adding 2 mL of ice-cold acetone to 600 mg of ground fresh whole plant material. The mixture was agitated at 4°C for 20 min at 1300 rpm and then centrifuged at 4°C for 20 min at 16,000g. The supernatant was separated and completely evaporated under argon on ice. After resuspension with 130 μL of methanol, GA4 contents were measured by ultra performance liquid chromatography coupled to tandem mass spectrometry as described in Supplemental Methods 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g01570 (RGA), At1g14920 (GAI), At1g66350 (RGL1), At3g03450 (RGL2), At5g17490 (RGL3), At1g32640 (MYC2/JIN1), At5g46740 (MYC3), At4g17880 (MYC4), At2g39940 (COI1), At1g19180 (JAZ1), At1g30135 (JAZ8), At5g24770 (VSP2), At4g23600 (TAT1), At3g45140 (LOX2), AT3G23240 (ERF1), AT5G44420 (PDF1.2), and At2g14610 (PR1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Coexpression Analysis of RGL3.
Supplemental Figure 2. JA Induces RGL3 Expression.
Supplemental Figure 3. Effect of GA and JA on RGL3-GFP Protein Abundance.
Supplemental Figure 4. Effect of JA on GA4 Levels.
Supplemental Figure 5. Negative Control of ChIP Experiments.
Supplemental Figure 6. RGL3 Interacts with JAZ1 and JAZ8 in Yeast.
Supplemental Figure 7. Characterization of rgl3-5 Mutant.
Supplemental Figure 8. Natural Variation of RGL3 Expression.
Supplemental Methods 1. GA4 Extraction and Quantification.
Supplemental Data Set 1. DNA Primer Sequences Used in the Study.
Supplementary Material
Acknowledgments
We thank Roberto Solano for the myc3, myc4, and myc2 myc3 myc4 mutants, John G. Turner for coi1-1, Hao Yu for the jin1-8 pMYC2:MYC2-FLAG line, Thierry Heitz for B. cinerea spores and Pst bacterial strain, Lionel Navarro for Pst COR− strain, and Alexandre Berr for technical help with ChIP and pathogen growth analyses. This article was supported by Bayer.
AUTHOR CONTRIBUTIONS
M.W., R.B., P.G., and P.A. designed the research. M.W., J.-M.D., S.C., T.R., N.B., D.H., R.B., P.G., and P.A. performed different aspects of the research and contributed to the analysis of the data. M.W., N.B., D.H., R.B., P.G., and P.A. wrote the article.
Glossary
- GA
gibberellin
- JA
jasmonate
- SA
salicylic acid
- bHLH
basic helix-loop-helix
- MeJA
methyl jasmonate
- GA3
gibberellic acid
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- CHX
cycloheximide
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- FLIM
fluorescence lifetime imaging microscopy
- Can-0
ecotype Canary Islands
- Sf-2
ecotype San Feliu
- Col-0
ecotype Columbia
- Ler
Landsberg erecta
- HRP
horseradish peroxidase
- qRT-PCR
quantitative RT-PCR
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-d-glucuronide
- RFP
red fluorescent protein
- Pst
Pseudomonas syringae pv tomato DC3000
- COR
coronatine
- qPCR
quantitative PCR
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008a). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P. (2008b). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Van Hulten M., Martin J., Pieterse C.M., Van Wees S.C., Ton J. (2011). Genetic dissection of basal defence responsiveness in accessions of Arabidopsis thaliana. Plant Cell Environ. 34: 1191–1206 [DOI] [PubMed] [Google Scholar]
- Berr A., McCallum E.J., Alioua A., Heintz D., Heitz T., Shen W.H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Brouwer M., Lievens B., Van Hemelrijck W., Van den Ackerveken G., Cammue B.P., Thomma B.P. (2003). Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiol. Lett. 228: 241–248 [DOI] [PubMed] [Google Scholar]
- Busso D., Delagoutte-Busso B., Moras D. (2005). Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343: 313–321 [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D.E., Cao D., Luo D., Harberd N.P., Peng J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Gfeller A., Liechti R., Farmer E.E. (2010). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3: cm4. [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- King K.E., Moritz T., Harberd N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B.N., Brooks D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Laurie-Berry N., Joardar V., Street I.H., Kunkel B.N. (2006). The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Pol. Plant Microbe Interact. 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Solano R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8: 532–540 [DOI] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. (New York: Springer)
- Pauwels L., Goossens A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Lopez-Molina L. (2009). The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal. Behav. 4: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Weber H., Reymond P., Browse J., Farmer E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Kamiya Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.