This article examines the crosstalk between gibberellin responses, which result in degradation of DELLAs, and the microRNA-regulated SQUAMOSA PROMOTER BINDING LIKE (SPL) transcription factors, which activate miR172 and MADS box transcription factors. The authors find that DELLA binds to SPLs and interfere with SPL transcriptional activation of miR172 and MADS box genes, thereby delaying flowering.
Abstract
Gibberellin (GA), a diterpene hormone, plays diverse roles in plant growth and development, including seed germination, stem elongation, and flowering time. Although it is known that GA accelerates flowering through degradation of transcription repressors, DELLAs, the underlying mechanism is poorly understood. We show here that DELLA directly binds to microRNA156 (miR156)-targeted SQUAMOSA PROMOTER BINDING–LIKE (SPL) transcription factors, which promote flowering by activating miR172 and MADS box genes. The interaction between DELLA and SPL interferes with SPL transcriptional activity and consequently delays floral transition through inactivating miR172 in leaves and MADS box genes at shoot apex under long-day conditions or through repressing MADS box genes at the shoot apex under short-day conditions. Our results elucidate the molecular mechanism by which GA controls flowering and provide the missing link between DELLA and MADS box genes.
INTRODUCTION
The shoot apical meristem (SAM) of plants continuously produces lateral organs. Based on the identity and morphological traits of the lateral organs, the life cycle of a plant can be divided into two major phases: vegetative and reproductive. The SAM produces leaves during the vegetative phase, whereas it gives rise to flowers in the reproductive phase (Poethig, 2003). The switch from vegetative to reproductive growth, also known as the floral transition, is controlled by both endogenous and exogenous cues, such as age, temperature, photoperiod, and hormones. Molecular and genetic analyses have revealed that the multiple floral inductive cues are integrated via a set of floral-promoting MADS box genes, including APETALA1 (AP1), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), FRUITFULL (FUL), and plant-specific transcription factor LEAFY (LFY) (Amasino, 2010; Lee and Lee, 2010; Srikanth and Schmid, 2011).
In Arabidopsis thaliana, the onset of flowering is accelerated by long-day conditions and delayed by short-day conditions. Seasonal changes in daylength are perceived in leaves and transduced to CONSTANS (CO), which activates the expression of FLOWERING LOCUS T (FT) in the vascular tissues of the leaves (Samach et al., 2000; An et al., 2004; Kobayashi and Weigel, 2007). The FT protein, as the output of the photoperiodic cue, moves from the leaves to the shoot apex, where it binds to the 14-3-3 protein and the transcription factor FD to activate the expression of MADS box genes (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Taoka et al., 2011). In addition to being activated by CO, the expression of FT is negatively regulated by other transcriptional regulators, such as FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), and TEMPRANILLO (TEM) (Searle et al., 2006; Castillejo and Pelaz, 2008; Li et al., 2008).
Under noninductive short-day conditions, two pathways play critical roles in flowering: one is dependent on the biosynthesis of the plant hormone gibberellin (GA) (Mutasa-Göttgens and Hedden, 2009); another is mediated by microRNA156 (miR156), which targets a group of transcription factors called SQUAMOSA PROMOTER BINDING–LIKEs (SPLs) (Cardon et al., 1999; Rhoades et al., 2002).
The miR156–SPL interaction constitutes an evolutionarily conserved, endogenous cue for both vegetative phase transition and flowering (Huijser and Schmid, 2011). The age-dependent decrease in miR156 results in an increase in SPLs that promote juvenile to adult phase transition and flowering through activation of miR172, MADS box genes, and LFY (Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009). Interestingly, SPLs not only act as the upstream activators of the floral-promoting MADS box genes but also serve as their downstream targets. The expression of three miR156-targeted SPLs, namely SPL3, SPL4, and SPL5, is highly induced by photoperiod (Schmid et al., 2003). More recently, SPL3 has been shown to be directly regulated by SOC1 (Jung et al., 2012), and the transcript level of SPL4 is reduced in the SAM of the soc1 ful double mutant (Torti et al., 2012). This interlocking feed-forward loop might contribute to a rapid and irreversible transition from vegetative to reproductive development.
GA is essential for floral induction in short-day conditions, because the plants that harbor the mutation in a GA biosynthetic gene, such as GA1, fail to flower (Wilson et al., 1992). In long-day conditions, the effect of GA on flowering is less pronounced. However, the analyses of the GA receptor mutants indicate that GA also plays an important role in flowering in long-day conditions (Griffiths et al., 2006). Recent studies have demonstrated that the GA response is mediated by the ubiquitin-proteasome pathway (Harberd, 2003; Schwechheimer and Willige, 2009). By binding to a nuclear receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1), GAs regulate gene expression by promoting the degradation of the transcriptional regulator DELLA proteins, including REPRESSOR OF GA1-3 1 (RGA), GA INSENSITIVE (GAI), RGA-LIKE1 (RGL1), and RGL2 (Murase et al., 2008). The degradation of DELLA proteins is mediated by 17 amino acids, called the DELLA motif (Dill et al., 2001). The Arabidopsis gai-1 mutant, which carries a deletion of the DELLA motif, is insensitive to GA-induced proteolysis and delays flowering (Peng et al., 1997; Dill et al., 2001). Although it is known that GA promotes flowering through activating MADS box genes and LFY (Blazquez et al., 1998; Moon et al., 2003; Eriksson et al., 2006; Achard et al., 2007), the underlying mechanism is largely elusive. Interestingly, several studies have revealed that DELLA exerts its biological functions through interacting with other transcription factors. For example, DELLA regulates hypocotyl elongation by interacting with PHYTOCHOME INTERACTING FACTORs (PIFs) (de Lucas et al., 2008), contributes to plant defense by interacting with JASMONATE ZIM-DOMAIN (JAZ) (Hou et al., 2010; Yang et al., 2012), and participates in secondary metabolism by interacting with MYC2 (Hong et al., 2012).
Here, we demonstrate the existence of crosstalk between GA and miR156 age pathways. DELLA regulates flowering partially through a direct interaction with miR156-targeted SPL transcription factors. The DELLA–SPL interaction inhibits SPL transcriptional activation of MADS box genes and miR172.
RESULTS
RGA Represses Flowering both in Leaves and at Shoot Apices
Under normal conditions, DELLAs are subjected to GA-induced proteolysis. To reveal the role of GA in flowering, we expressed RGAd17, the GA-insensitive form of RGA (Dill et al., 2001), from its own upstream regulatory sequence (ProRGA:RGAd17). ProRGA:RGAd17 phenocopied the GA-deficient mutant, developed small dark green leaves, and delayed flowering (see Supplemental Figure 1 online). Both phloem and SAM have been shown to play critical roles in the floral induction. To understand where RGA regulates flowering, we generated transgenic plants in which RGAd17 was expressed from either a phloem-specific promoter, SUC2 (Truernit and Sauer, 1995), or a meristem-specific promoter, FD (Abe et al., 2005).
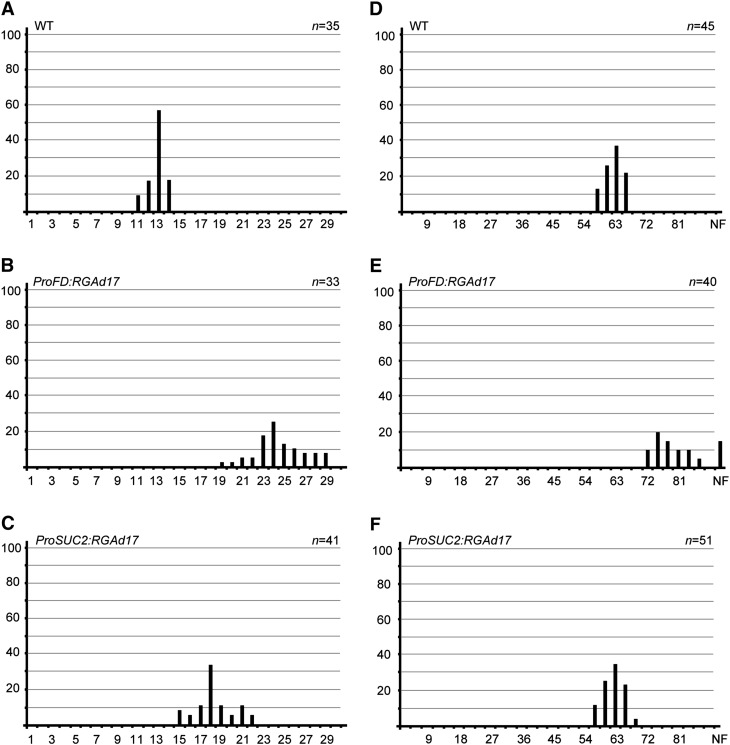
In long-day conditions, the wild-type plants began to flower with ∼12 leaves. Both ProSUC2:RGAd17 and ProFD:RGAd17 exhibited a late flowering phenotype, producing more than 20 leaves at the time of bolting (Figures 1A to 1C). In short-day conditions, the flowering of the wild type was greatly delayed, because of the absence of photoperiodic input (Figure 1D). ProSUC2:RGAd17 plants flowered nearly at the same time as the wild type, whereas ProFD:RGAd17 severely blocked the floral transition (Figures 1E and 1F). Six out of 40 T1 ProFD:RGAd17 plants failed to flower. Taken together, these results indicate that RGA regulates flowering via two distinct mechanisms: it suppresses flowering both in the leaves and at the shoot apices in long-day conditions and delays flowering at the shoot apices in short-day conditions.
Figure 1.
RGA Represses Flowering Both in Leaves and at Shoot Apices.
Flowering time of wild-type (WT), ProSUC2:RGAd17, and ProFD:RGAd17 plants under long-day ([A] to [C]) or short-day conditions ([D] to [F]). Flowering frequency of T1 transgenic lines is shown as a histogram, with the y axis indicating percentage of plants that flower with a given number of leaves. The x axis indicates the number of leaves. NF, never flowering.
RGA Represses Flowering through FT and miR172 in Leaves under Long-Day Conditions
To assess whether the late flowering phenotype of ProSUC2:RGAd17 in long-day conditions was caused by a low amount of FT, we performed quantitative real-time PCR. To facilitate the expression analyses, we chose one representative T3 line of ProFD:RGAd17 and ProSUC2:RGAd17. Both of these flowered late under long-day or short-day conditions (see Supplemental Figure 2 online).
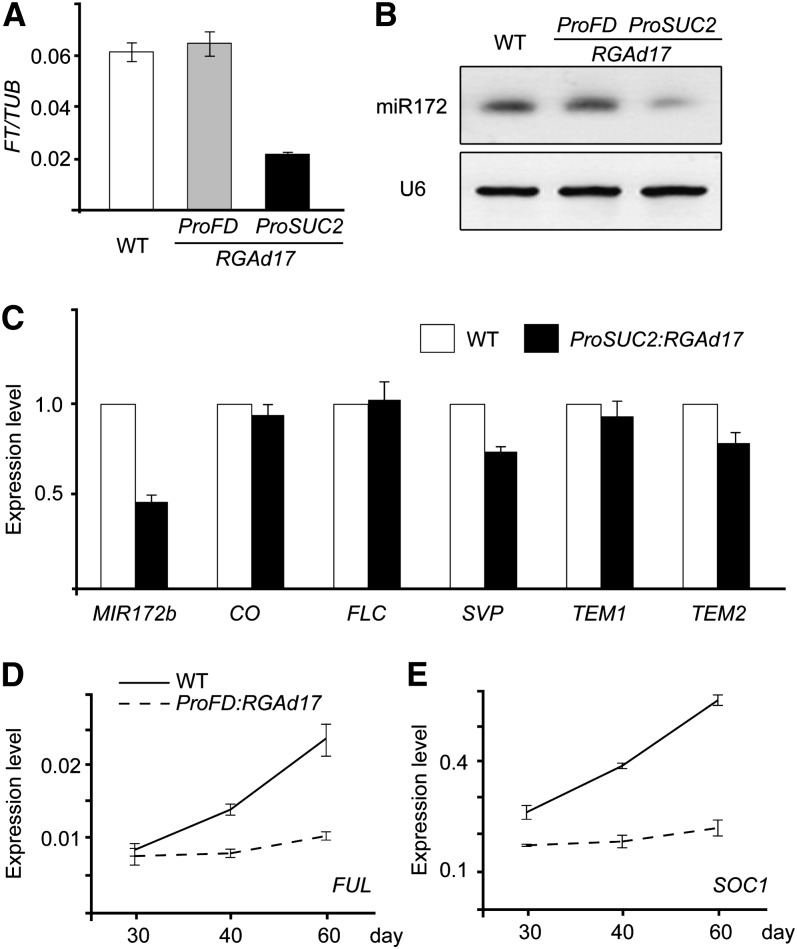
The leaves of wild-type, ProFD:RGAd17, and ProSUC2:RGAd17 plants were collected at zeitgeber time 16, when FT shows the highest expression level (Kobayashi et al., 1999). Compared with wild-type and ProFD:RGAd17 plants, the transcript level of FT was markedly less in ProSUC2:RGAd17 (Figure 2A), indicating that RGA is able to repress FT in the vascular tissue of the leaves. In agreement with this finding, it has been shown that GA was able to induce FT expression in long-day conditions (Hisamatsu and King, 2008; Porri et al., 2012).
Figure 2.
RGA Represses Flowering through FT and MADS Box Genes.
(A) Expression of FT normalized to β-TUBULIN-2 (TUB) in the wild type (WT) and plants expressing RGAd17 under the FD or SUC2 promoters.
(B) Expression of miR172 by small RNA gel blot. The amount of U6 was monitored as loading control.
(C) Expression of MIR172b and FT regulators.
The leaves of wild-type and ProSUC2:RGAd17 plants were used for expression analyses by quantitative real-time–PCR. Plants were grown in long-day conditions for 14 d, and the leaves were harvested at zeitgeber time 16. Expression was normalized to that of β-TUBULIN-2. Expression in the wild type was set as 1 for each gene. Two biological replicates were performed with similar results. Error bars represent ±se (n = 3).
(D) and (E) Expression of FUL (D) and SOC1 (E). The shoot apices of short-day–grown wild-type and ProFD:RGAd17 plants were harvested at different time points and subjected to quantitative real-time PCR analyses. Error bars represent ±se (n = 3).
Recent studies have demonstrated that miR172, which is activated by miR156-targeted SPLs, targets AP2-like transcription factors that negatively control FT expression in leaves (Mathieu et al., 2009; Yant et al., 2010). Overexpression of SCHLAFMUTZE (SMZ) or SCHNARCHZAPFEN (SNZ), two miR172-targed AP2-like genes, results in a decrease in FT expression and a late flowering phenotype in long-day conditions (Mathieu et al., 2009). To test whether DELLA represses FT through the SPL-miR172-AP2 module, we analyzed the level of miR172. Compared with wild-type and ProFD:RGAd17 plants, the level of mature miR172 was much lower in the leaves of ProSUC2:RGAd17 plants (Figure 2B). Consistent with this, the accumulation of the primary transcript of MIR172b, one of the five coding genes of miR172, was accordingly less (Figure 2C).
We then examined the expression of miR172-targeted AP2-like genes, including AP2, SMZ, SNZ, TARGET OF EAT1 (TOE1), TOE2, and TOE3 (Rhoades et al., 2002). The transcript levels of all these genes except those of SMZ were not greatly changed (see Supplemental Figure 3 online), which is probably because miR172 controls its targeted genes mainly through the translational inhibition (Aukerman and Sakai, 2003; Chen, 2004; Schwab et al., 2005).
In addition to the miR172-AP2 module, the expression of FT is regulated by other transcriptional regulators, such as CO, FLC, SVP, and TEM (Searle et al., 2006; Castillejo and Pelaz, 2008; Li et al., 2008). The expression of these genes was not significantly altered in ProSUC2:RGAd17 in comparison with wild-type and ProFD:RGAd17 plants (Figure 2C).
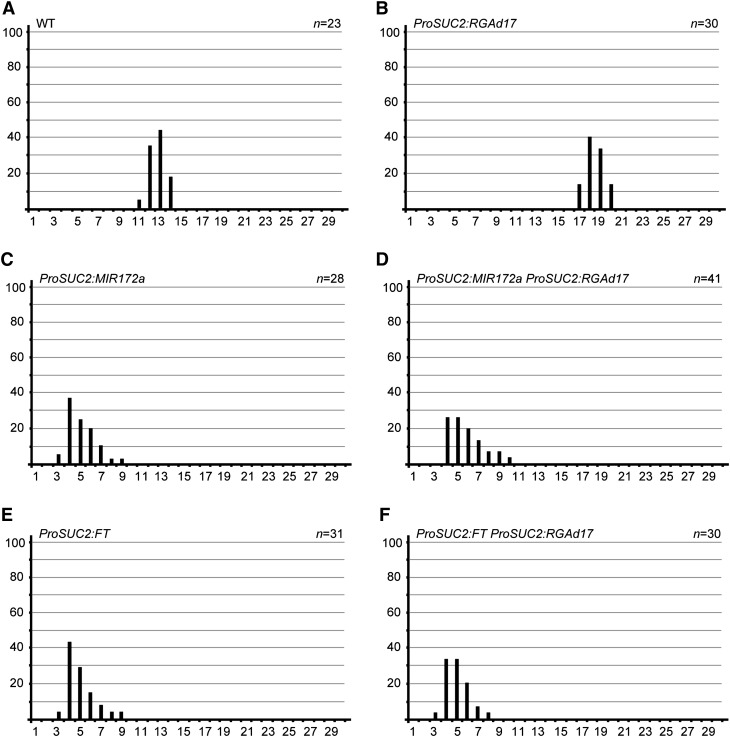
To confirm that RGA suppresses flowering through the miR172-AP2-FT module in leaves, we expressed MIR172a or FT from the SUC2 promoter in wild-type and ProSUC2:RGAd17 plants. Both ProSUC2:FT and ProSUC2:MIR172a flowered earlier than the wild type and were sufficient to suppress the late flowering phenotype of ProSUC2:RGAd17 under long-day conditions (Figure 3; see Supplemental Figure 4 online).
Figure 3.
RGA Represses Flowering through miR172-AP2-FT in Leaves.
Flowering time of wild-type (WT), ProSUC2:RGAd17, ProSUC2:MIR172a, ProSUC2:FT, ProSUC2:RGAd17 ProSUC2:MIR172a, and ProSUC2:RGAd17 ProSUC2:FT plants under long-day conditions. Flowering frequency of T1 transgenic lines is shown as a histogram, with the y axis indicating percentage of plants that flower with a given number of leaves. The x axis indicates the number of leaves.
RGA Represses Flowering through MADS Box Genes at Shoot Apex
We have shown that ProFD:RGAd17 delays flowering at the shoot apex under both long-day and short-day conditions (Figure 1). At the shoot apex, the transition from the vegetative to the reproductive phase is executed by the MADS box genes, such as FUL and SOC1. We extracted the RNAs from the shoot apices of wild-type and ProFD:RGAd17 plants of different ages in short-day conditions. In 20-d-old seedlings, FUL and SOC1 transcripts accumulated to a similar level in ProFD:RGAd17 plants as in the wild type (Figures 2D and 2E). As the plants grew, the expression of FUL and SOC1 was gradually increased in the wild type. However, we did not observe the same increase in both genes in ProFD:RGAd17 plants (Figures 2D and 2E). Under long-day conditions, the expression of FUL and SOC1 was also decreased in ProFD:RGAd17 in comparison with the wild type (see Supplemental Figure 5 online). These results indicate that RGA blocks the activation of MADS box genes at the shoot apices. Consistent with this, it has been shown that the activation of SOC1 is attenuated in the ga1-3 mutant and that overexpression of SOC1 rescues the flowering phenotype of the ga1-3 plants in short-day conditions (Moon et al., 2003).
Overexpression of miR156 Reduces the GA Response in Flowering
To understand the genetic interaction between GA and miR156, we studied the GA response of the wild type, the miR156 overexpression line (Pro35S:MIR156), in which miR156 was expressed from the 35S promoter (Schwab et al., 2005), and the miR156 target mimicry line (Pro35S:MIM156), which reduces miR156 activity (Franco-Zorrilla et al., 2007; Wang et al., 2008; Todesco et al., 2010). miR156 has been shown to affect leaf initiation rate (Wang et al., 2008); therefore, we measured the flowering time by counting both the number of leaves and the number of days until the plants started to flower.
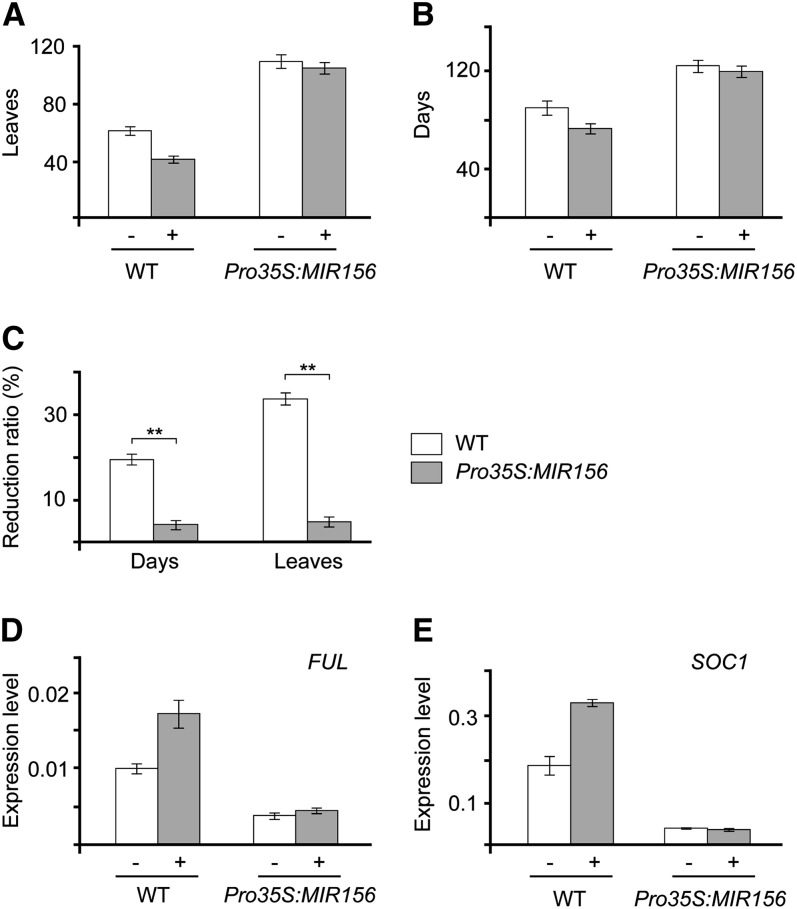
Under long day conditions, 5-d-old seedlings were sprayed once with 50 μM of gibberellic acid (GA3). The photoperiod pathway plays a predominant role in long-day conditions; therefore, the flowering response to GA was largely masked (see Supplemental Figure 6 online). We performed the same GA treatment assay under short-day conditions. As shown in Figure 4, application of GA3 was sufficient to accelerate flowering in the wild-type plants. The number of leaves or days was accordingly decreased by 33 or 19%, respectively (Figures 4A to 4C). By contrast, Pro35S:MIR156 significantly reduced GA sensitivity. The GA3-treated Pro35S:MIR156 plants flowered almost as late as the mock-treated plants (Figures 4A and 4B; see Supplemental Figure 7 online). We only observed 4.4% reduction in the number of days and 3.7% reduction in the number of leaves (Figure 4C).
Figure 4.
Pro35S:MIR156 Reduces the GA Response.
(A) and (B) GA response of wild-type (WT) and Pro35S:MIR156 plants under short-day conditions. We sprayed 7-d-old seedlings with 50 μM of GA3 (+) or ethanol (mock, −). The number of leaves (A) and the days to flowering (B) were counted.
(C) The reduction ratio in response to GA. The reduction ratio was calculated as (number of leaves/days [mock] − number of leaves/days [GA3] / number of leaves/days [mock]). **, Student’s t test, P < 0.01.
(D) and (E) Expression of SOC1 and FUL in 50-d-old GA3-treated wild-type and Pro35S:MIR156 plants in short-day conditions. The shoot apices were collected 6 h after treatment. Error bars represent ±se (n = 3).
To understand whether the change in GA response of Pro35S:MIR156 plants under short-day conditions is caused by a reduction in MADS box genes, we analyzed the expression of SOC1 and FUL. We sprayed 50-d-old short-day–grown plants with 50 μM of GA3, and their shoot apices were collected after 6 h. The expression of SOC1 and FUL was elevated in the GA3-treated wild-type plants but not in Pro35S:MIR156 plants (Figures 4D and 4E).
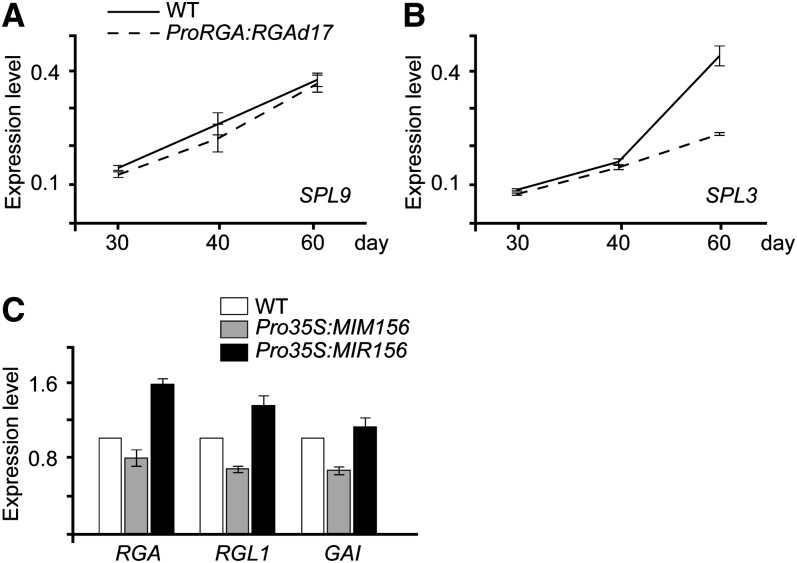
Expression of SPL and DELLA
In the Arabidopsis genome, miR156-targeted SPLs can be divided into two groups, represented by SPL3 and SPL9 (Guo et al., 2008; Xing et al., 2010). To determine whether RGA regulates the transcription of SPLs, we analyzed the mRNAs of SPL3 and SPL9 in wild-type and Pro35S:RGAd17 plants at different time points in short-day conditions. There was no significant change in SPL9 transcript levels between wild-type and Pro35S:RGAd17 plants (Figure 5A). SPL3 exhibited a distinct expression pattern: its mRNAs gradually increased in wild-type plants but increased less in Pro35S:RGAd17 plants (Figure 5B). In agreement with this finding, a recent report has shown that SPL3 level was repressed by paclobutrazol (PAC), a GA biosynthesis inhibitor (Jung et al., 2012). Compared with the wild type, the expression of DELLAs, including RGA, RGL1, and GAI, was not greatly altered in either Pro35S:MIR156 or Pro35S:MIM156 plants (Figure 5C).
Figure 5.
Expression of SPLs and DELLAs.
(A) and (B) Expression of SPL3 (A) and SPL9 (B) in wild-type (WT) and ProRGA:RGAd17 plants. Error bars represent ±se (n = 3). The shoot apices of short-day–grown plants were harvested at different time points.
(C) Expression of DELLAs in wild-type, Pro35S:MIM156, and Pro35S:MIR156 plants. We used 20-d-old plants grown in short-day conditions for expression analyses. Error bars represent ±se (n = 3).
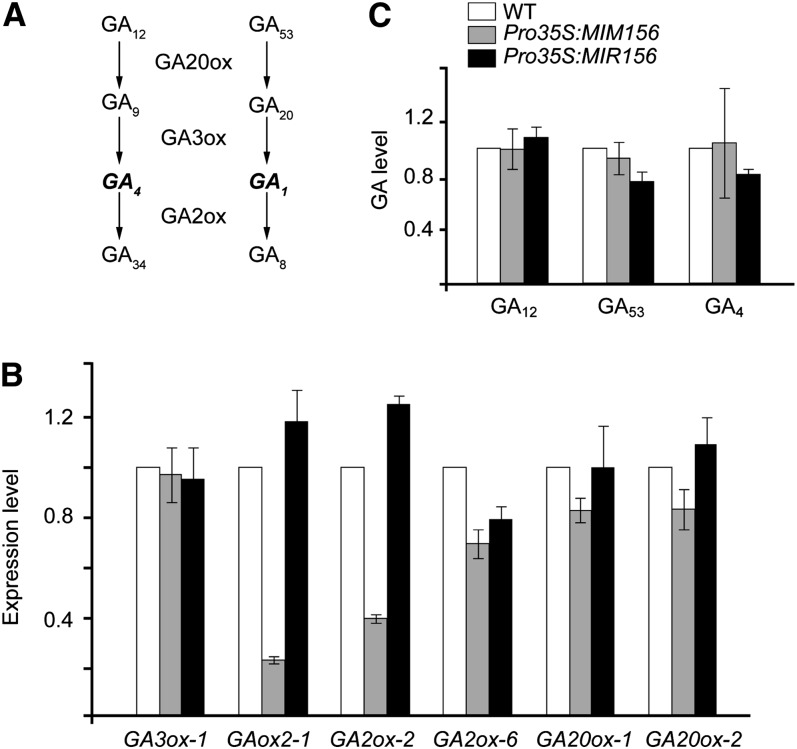
Expression of GA Biosynthetic and Catabolic Genes
To understand whether SPL affects GA biosynthesis, we monitored the expression of several GA biosynthetic genes that are highly expressed in leaves, including GA2-oxidase-1 (GA2ox-1), GA2ox-2, GA2ox-6, GA3-oxidase-1 (GA3ox-1), GA20-oxidase-1 (GA20ox-1), and GA20ox-2. GA3ox and GA20ox are responsible for the biosynthesis of bioactive GA4, whereas GA2ox catalyzes the deactivation of GA4 by oxidation (Figure 6A) (Eriksson et al., 2006; Yamaguchi, 2008). Compared with the wild type, the expression of the genes encoding GA2ox-1, GA3ox-1, GA20ox-1, and GA20ox-2 was not changed in either Pro35S:MIR156 or Pro35S:MIM156 plants. The transcript levels of GA2ox-2 and GA2ox-6 were moderately decreased in Pro35S:MIM156 and increased in Pro35S:MIR156 plants (Figure 6B). To test whether the change in GA2ox expression results in an increase in bioactive GA, we measured the content of GAs, including GA4, GA53, and GA12. As shown in Figure 6C, Pro35S:MIM156 and Pro35S:MIR156 plants accumulated the same amount of GA4, one of the bioactive forms of GA, as wild-type plants (Figure 6C).
Figure 6.
Expression of GA Biosynthetic and Catabolic Genes.
(A) GA biosynthetic and catabolic pathway. The bioactive forms of GA are labeled in bold italic.
(B) Expression of GA3ox, GA20ox, and GA2ox in 15-d-old plants grown in short-day conditions. Error bars represent ±se (n = 3).
(C) GA measurement. The level in the wild type (WT) was set to 1. Error bars represent ±se (n = 3).
Genetic Interaction between Age and GA Pathway
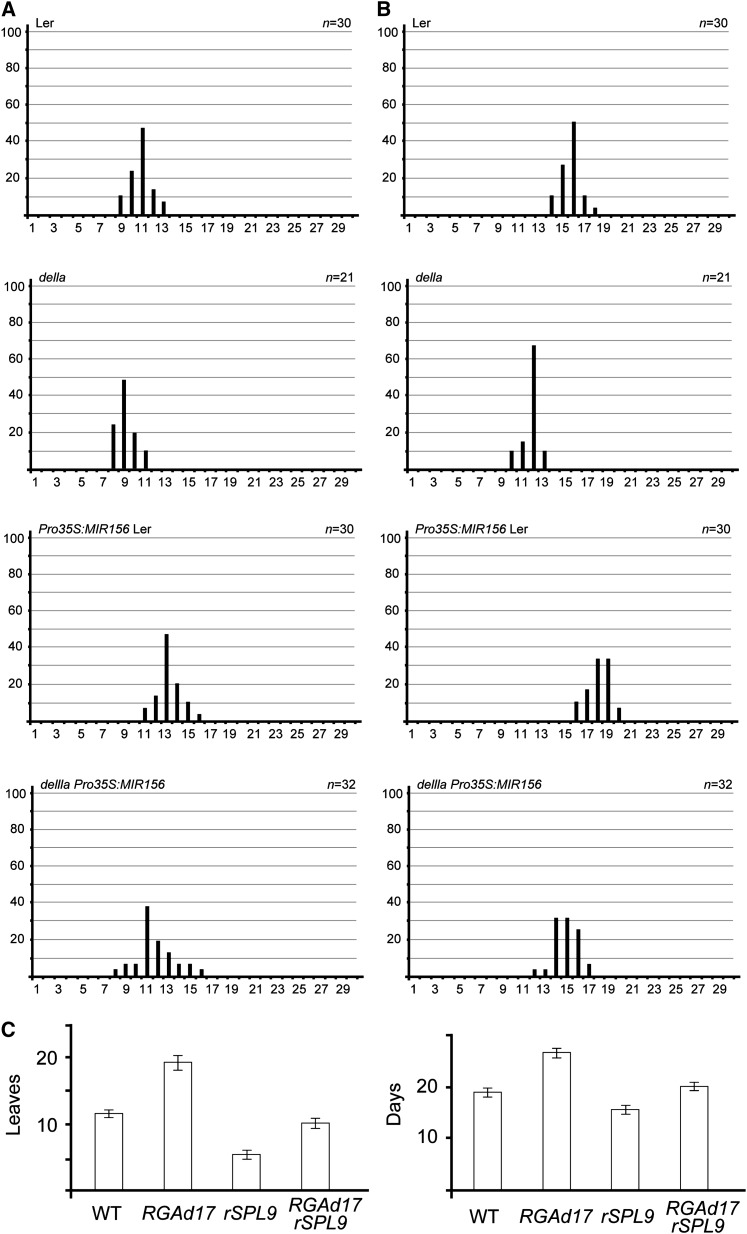
To further elucidate the genetic interaction between DELLA and SPL, we overexpressed miR156 in the della pentuple mutant (Landsberg erecta [Ler] background) (Feng et al., 2008). The della mutant flowered earlier than the wild type (Ler) under long-day conditions. Overexpression of MIR156 resulted in a delay of flowering in both the wild type (Ler) and della mutant background (Figures 7A and 7B; see Supplemental Figure 8 online).
Figure 7.
Genetic Interaction between GA and miR156.
(A) and (B) Flowering time of della and della Pro35S:MIR156 under long-day conditions. The x axis indicates the number of leaves (A) or the number of days (B). Flowering frequency of della homozygous plants and T1 della Pro35S:MIR156 transgenic lines are shown as a histogram, with the y axis indicating percentage of plants that flower with a given number of leaves.
(C) Flowering time of wild-type (WT), ProRGA::RGAd17, ProSPL9:rSPL9, and ProRGA:RGAd17 ProSPL9:rSPL9 plants under long-day conditions. Homozygous ProRGA:RGAd17, ProSPL9:rSPL9, and ProRGA:RGAd17 ProSPL9:rSPL9 plants were used for flowering time measurements.
SPL9 and SPL15 play a dominant role within miR156-targeted SPLs. The spl9 spl15 double mutant shows a similar but weak phenotype as the miR156 overexpression line (Schwarz et al., 2008; Wang et al., 2008). Under long-day conditions, ProSPL9:rSPL9 plants, where the miR156-resistant form of SPL9 (rSPL9) was expressed under its own regulatory sequence (Wang et al., 2008), promoted flowering in long-day conditions (Figure 7C). We crossed ProSPL9:rSPL9 to ProRGA:RGAd17. ProSPL9:rSPL9 ProRGA:RGAd17 plants developed the same small dark green leaves as Pro35S:RGAd17 and flowered earlier than ProRGA:RGAd17 (Figure 7C; see Supplemental Figure 9 online). Taken together, our genetic and expression analyses indicate that miR156-targeted SPLs are essential for the floral induction by GAs and that DELLA represses flowering partially through miR156-targeted SPLs.
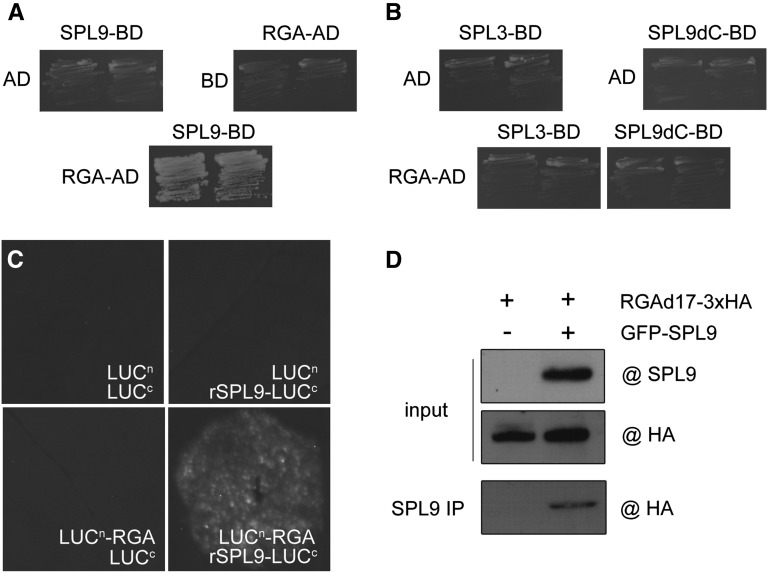
RGA Binds Directly to SPLs
Because of lack of a canonical DNA binding domain, DELLA regulates plant development and physiology by interacting with other transcription factors, such as PIFs, SCL3, MYC2, and JAZ (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Zhang et al., 2011; Hong et al., 2012; Yang et al., 2012). Given the fact that both SPLs and RGA regulate the floral transition through the same downstream targets, such as miR172, FUL, and SOC1 (Wang et al., 2009; Wu et al., 2009), we suspected that RGA might directly bind to SPL. To test this hypothesis, we performed yeast two-hybrid assays. A strong interaction was observed when RGA was fused to GAL4 activation domain and SPL9 was fused to GAL4 DNA binding domain (BD) (Figure 8A). This interaction was compromised when the C-terminal domain of SPL9 was deleted (SPL9dC) (Figure 8B). Consistent with this finding, an SPL3 mutant that only harbors the SBP DNA binding domain also failed to interact with RGA (Gandikota et al., 2007) (Figure 8B), suggesting that the C-terminal domain of SPL9 is responsible for its interaction with RGA. Yeast two-hybrid assays further demonstrated the widespread interactions between DELLAs and miR156-targeted SPLs (see Supplemental Figure 10 online).
Figure 8.
RGA Directly Interacts with SPL9.
(A) Yeast two-hybrid assay. SPL9 was fused to GAL4 BD and RGA to GAL4 activation domain. Interactions were examined on SD/-Leu/-Trp/-His plates supplemented with 15 mM of 3-amino-1,2,4-triazole. Two independent clones are shown.
(B) SPL9 binds to RGA through its C-terminal. SPL3 and SPL9dC were fused to BD.
(C) BiFC analyses. N. benthamiana leaves were infiltrated with agrobacteria. The combinations of LUCc-rSPL9 with LUCn and LUCc with RGA-LUCn were used as negative controls.
(D) CoIP analyses. Soluble protein extract was immunoprecipitated with anti-SPL9 antibody. RGAd17-3xHA proteins were detected by immunoblot with anti-HA antibody.
To examine the interaction between SPL9 and RGA in vivo, we used a bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana (Chen et al., 2008). rSPL9 was in-frame fused to the N-terminal half of firefly luciferase (LUC) (rSPL9-LUCn), and RGA was fused to the C-terminal half of LUC (LUCc-RGA). Luminescence was detected when the leaves were infiltrated with LUCc-rSPL9/RGA-LUCn, but not in those infiltrated with LUCc-rSPL9/LUCn or LUCc/RGA-LUCn (Figure 8C).
To further confirm the direct interaction between SPL9 and RGA, we performed a coimmunoprecipitation (CoIP) experiment using an N. benthamiana transient expression assay. Hemagglutinin (HA)-tagged RGAd17 and green fluorescent protein (GFP)-tagged rSPL9 were transiently expressed in the leaves of N. benthamiana. Protein extract of RGAd17-3xHA or RGAd17-3xHA GFP-rSPL9 was immunoprecipitated by the antibody against SPL9. In the immunoprecipitation fraction, RGAd17-3xHA was readily detected in the sample of RGAd17-3xHA GFP-rSPL9 but not in that of RGAd17-3xHA (Figure 8D).
Interaction between DELLA and SPL Interferes with SPL Transcriptional Activity
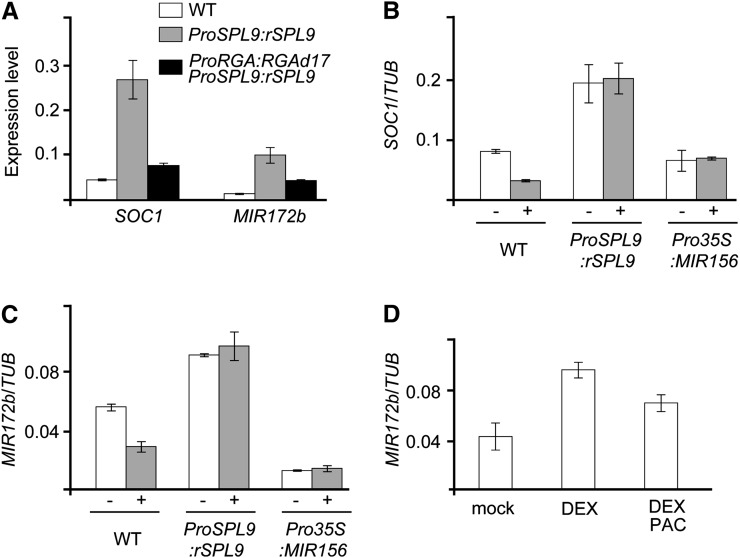
To understand whether DELLA interferes with SPL transcriptional activity, we examined the expression of SOC1 and MIR172b in ProSPL9:rSPL9 ProRGA:RGAd17 plants. Consistent with previous reports (Wang et al., 2009; Wu et al., 2009), the expression of SOC1 and MIR172b was upregulated in ProSPL9:rSPL9 in comparison with the wild type (Figure 9A). If DELLA interferes with the transcriptional activity of SPL, one would expect that the activation of SOC1 and MIR172b by SPL would be compromised by the increased level of RGA. Indeed, the transcripts of SOC1 and MIR172b accumulated to the same level in ProSPL9:rSPL9 ProRGA:RGAd17 as in the wild type (Figure 9A).
Figure 9.
RGA Impairs the Activation of miR172 and SOC1 through SPL9.
(A) RGA interferes with the activation of SOC1 and MIR172b. We analyzed 30-d-old short-day–grown wild-type (WT), ProSPL9:rSPL9, and ProRGA:RGAd17 ProSPL9:rSPL9 plants. Error bars represent ±se (n = 3).
(B) and (C) Expression of SOC1 (B) and MIR172b (C) in the PAC-treated wild-type, ProSPL9:rSPL9, and Pro35S:MIR156 plants. We collected 20-d-old short-day–grown seedlings 2 d after treatment. Error bars represent ±se (n = 3).
(D) Inducible expression of MIR172b. We sprayed 10-d-old long-day–grown ProSPL9:rSPL9-GR seedlings with DEX, DEX + PAC, or ethanol (mock) for 12 h. Error bars represent ±se (n = 3).
Next, we studied the sensitivity of wild-type, ProSPL9:rSPL9, Pro35S:MIR156 plants to PAC. To verify the treatment efficiency, we examined the expression of NOD26-LIKE INTRINSIC PROTEIN (NOD26, At4g19030) and LIPID TRANSFER PROTEIN3 (LTP3, At5g59320), both of which are the direct targets of the DELLA-PIF3 module (de Lucas et al., 2008; Feng et al., 2008). After 2 d of the treatment with PAC, the expression of NOD26 and LTP3 was greatly decreased (see Supplemental Figure 11 online). We observed a similar reduction in SOC1 and MIR172b transcripts in the PAC-treated wild-type plants, whereas the expression of both genes was insensitive to PAC in ProSPL9:rSPL9 and Pro35S:MIR156 plants (Figures 9B and 9C).
To further confirm these results, we used an inducible system in which rSPL9 fused to the rat glucocorticoid receptor (GR) was expressed under its regulatory sequence (ProSPL9:rSPL9-GR) (Wang et al., 2009). Treatment with the GR ligand dexamethasone (DEX) resulted in a threefold increase in MIR172b transcripts after 12 h (Figure 9D). By contrast, we only observed a 1.5-fold induction in MIR172b when 50 μM of PAC was coapplied. Taken together, these observations indicate that a high level of RGA impairs the activation of MADS box genes and miR172 through SPL.
DISCUSSION
Integration of Flowering Time Pathways
Forward and reverse genetics have identified five flowering pathways in Arabidopsis, including photoperiod, vernalization, GA, autonomous, and age pathways (Amasino, 2010; Srikanth and Schmid, 2011). Elucidation of how these pathways are integrated is of great importance in understanding how plants flower in response to diverse developmental and environmental signals. Previous results have shown that vernalization and autonomous pathways converge at FLC, which encodes a MADS box–type flowering repressor (Simpson, 2004). Interestingly, FLC could also inactivate FT in leaves, providing a molecular link between vernalization and photoperiod pathways (Searle et al., 2006).
The age pathway is governed by miR156, the level of which gradually decreases as age increases (Wu and Poethig, 2006; Wang et al., 2009). The integration between photoperiod and age pathway has been extensively studied. In leaves, miR156-SPL acts in parallel with CO, both of which are positive regulators of FT. SPL promotes flowering through the miR172-AP2-FT signaling cascade (Mathieu et al., 2009; Wang et al., 2009; Yant et al., 2010). At the shoot apex, the photoperiod pathway acts upstream of SPL. The FT–14-3-3–FD complex activates the expression of SPL3 and SPL4 (Jung et al., 2012).
Our results reveal that age and GA pathways are integrated through a direct physical interaction between SPL and DELLA. The binding of DELLA to SPLs attenuates SPL transcriptional activities toward FT and MADS box genes, subsequently blocking the floral transition. It will be interesting to see how the age pathway is integrated into vernalization and autonomous pathways. Indeed, a recent study in Arabis alpina, a perennial herb, has revealed that the vernalization response of this perennial plant is age-dependent (Wang et al., 2011). Given the fact that the transcript level of FLC is not altered in either Pro35S:MIM156 or Pro35S:MIR156 Arabidopsis plants (Wang et al., 2009), it is unlikely that the age pathway regulates vernalization through modulating FLC expression.
DELLA Represses Flowering via Distinct Mechanisms
Our results demonstrate that DELLA regulates flowering via two distinct mechanisms (see Supplemental Figure 12 online). Under short-day conditions, miR156-targeted SPLs play a major role in flowering by activating MADS box genes at the shoot apex. DELLAs delay the floral transition through interfering with the transcriptional activities of SPL. Under long-day conditions, in addition to a similar role at the shoot apex, the interaction between DELLA and SPL leads to a decrease of miR172. As a result, the increased level of AP2-like transcription factors represses flowering through inactivating FT.
Pro35S:MIR156 still responds to GA; therefore, additional flowering targets of DELLA must exist. Indeed, a recent report has shown that simultaneous inactivation of two GA-responsive GATA transcription factors, GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE/CYTOKININ-RESPONSIVE GATA FACTOR1 (GNL), partially rescues the flowering defect of ga1-3 plants in short-day conditions (Richter et al., 2010). Another potential flowering target of DELLA is SVP, which encodes a MADS box–type floral repressor. The expression of SVP is reduced in the GA-treated plants and increased in a GA-deficient mutant (Li et al., 2008). Moreover, because the level of SPL3 was decreased in the shoot apices of ProFD:RGAd17 plants, we could not exclude the possibility that DELLA controls flowering through modulating the expression of SPLs. Indeed, a recent study has suggested that GA could promote flowering through SPL3 (Porri et al., 2012).
DELLA and SPL in Vegetative Phase Transition
In addition to a role in flowering, GA is essential for the expression of adult phase traits (Chien and Sussex, 1996; Poethig, 2003). In Arabidopsis, juvenile leaves only develop trichomes on their adaxial (upper) sides, whereas adult leaves produce trichomes on their adaxial and abaxial (lower) sides. The GA-deficient mutant delays the appearance of abaxial trichomes, whereas exogenous application of GA accelerates the formation of abaxial trichomes (Telfer et al., 1997).
miR156-targeted SPLs exert a similar role as that of GA in vegetative phase transition. The SPL level is correlated with abaxial trichome production (Wu and Poethig, 2006; Wu et al., 2009). DELLA directly binds to SPL; therefore, it is plausible that GA could promote abaxial trichome formation through releasing the inhibition of DELLA on SPL. However, a recent study has shown that application of GA3 induces abaxial trichome formation in the miR156 overexpression line, suggesting that GA is able to promote abaxial trichome formation independent of SPL (Schwarz et al., 2008). Whether GA regulates the display of adult vegetative phase traits through SPL awaits further investigation.
METHODS
Plant Materials
Arabidopsis thaliana plants, ecotypes Columbia and Ler, and Nicotiana benthamiana were grown at 21°C in long days (16-h light/8-h dark) or short days (8-h light/16-h dark). Pro35S:MIR156, Pro35S:MIM156, ProSPL9:rSPL9, and ProSPL9:rSPL9-GR have been described elsewhere (Wang et al., 2008; Wang et al., 2009). The della mutant (N16298) was ordered from the Nottingham Arabidopsis Stock Centre (www.Arabidopsis.info). For GA and PAC treatment, 50 μM of GA3 (Sigma-Aldrich) and 50 μM of PAC were used. For DEX induction experiment, 10 μM of DEX or 10 μM of DEX plus 50 μM of PAC was used.
Constructs and Plant Transformation
For RGAd17 constructs, RGAd17 was cloned into the binary constructs behind the RGA, SUC2, or FD promoter (Wang et al., 2008). For FT and MIR172a constructs, the coding region of FT and a 410-bp fragment harboring the stem loop of MIR172a was PCR amplified and cloned into the binary construct behind the pSUC2 promoter. For yeast two-hybrid constructs, the cDNAs of SPL2, SPL3, SPL9, SPL9dC, SPL10, and SPL11 were PCR amplified and cloned into pGBKT7 or pGADT7 (Clontech). RGA was cloned into pGADT7. The pGBKT7 series of GAI, RGA, RGL1, RGL2, and RGL3 constructs was generated as described in de Lucas et al. (2008). BiFC constructs were generated as described elsewhere (Chen et al., 2008; Gou et al., 2011). For CoIP constructs, RGAd17 and rSPL9 was PCR amplified and cloned into the binary constructs with 3xHA or GFP tag. The oligonucleotide primers for these constructs are given in Supplemental Table 1 online. The constructs were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90) by the freeze-thaw method. Transgenic plants were generated by the floral dipping method (Clough and Bent, 1998) and were screened with 0.05% glufosinate (Basta) on soil or 50 μg/mL of hygromycin on one-half–strength Murashige and Skoog plate.
Flowering Time Measurement and Expression Analyses
Flowering time was measured by counting the total number of leaves (rosette and cauline leaves) and the number of days to flower (when the floral buds are visible).
Total RNA was extracted with TRIzol reagent (Invitrogen). A total of 1 μg of total RNA was DNase I–treated and used for cDNA synthesis with oligo (dT) primer and RevertAid reverse transcriptase (Fermentas). Quantitative real-time–PCR was performed with SYBR-Green PCR Mastermix (TaKaRa), and amplification was real-time monitored on Mastercycler Realplex2 (Eppendorf). Quantitative real-time PCR primers for FT, MIR172b, SOC1, FUL, SPL3, SPL9, and TUB have been described elsewhere (Wang et al., 2009; Wu et al., 2009). Other oligonucleotide primers are given in Supplemental Table 1 online.
Yeast Two-Hybrid Assay
Plasmids were transformed into yeast strain AH109 (Clontech) by the lithium chloride–polyethyleneglycol method according to the manufacturer’s manual (Clontech). The transformants were selected on SD/-Leu/-Trp plates. The interactions were tested on SD/-Leu/-Trp/-His plate with 3-amino-1,2,4,-triazole (Figures 8A and 8B) or SD/-Ade/-Leu/-Trp/-His plate (see Supplemental Figure 10B online). At least 10 individual clones were analyzed.
BiFC Analysis
BiFC assays were performed as described in Gou et al. (2011). A. tumefaciens was resuspended in infiltration buffer at OD600 = 0.8. Pro35S:P19-HA (Papp et al., 2003) was coinfiltrated to inhibit gene silencing. A total of 1 mM of luciferin was infiltrated before LUC activity was monitored after 3 d.
CoIP Analysis
SPL9 antibody was raised against the peptide QHQYLNPPWVFKDNC, corresponding to the residues 299 to 312 of SPL9 (Willget Biotech). Agrobacteria-infiltrated N. benthamiana leaves were used for CoIP analyses. The soluble proteins were extracted in the extraction buffer (100 mM of Tris-HCl, 5 mM of EDTA, 100 mM of NaCl, 0.2% Nonidet P-40, 1.0% Triton X-100, pH 7.5). Immunoprecipitation was performed with anti-SPL9 antibody. RGAd17-3xHA fusion proteins were detected by immunoblot with anti-HA-horseradish peroxidase antibody (Roche).
GA Measurement
We harvested 2.0-g 40-d-old short-day–grown plants. Extraction and measurement of GA was performed according to a published protocol (Qi et al., 2011).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AP2 (At4g36920); CO (At5g15840); FLC (At5g10140); FUL (At5g60910); FT (At1g65480); GAI (At1g14920); GA20ox-1 (At4g25420); GA20ox-2 (At5g51810); GA2ox-1 (At1g78440); GA2ox-2 (At1g30040); GA2ox-6 (At1g02400); GA3ox-1 (At1g15550); MIR172a (At2g28056); MIR172b (At5g04275); RGA (At2g01570); RGL1 (At1g66350); RGL2 (At3g03450); RGL3 (At5g17490); SOC1 (At2g45660); SPL2 (At5g43270); SPL3 (At2g33810); SPL9 (At2g42200); SPL10 (At1g27370); SPL11 (At1g27360); TEM1 (At1g25560); TEM2 (At1g68840); TOE1 (At2g28550); TOE2 (At5g60120); TOE3 (At5g67180); TUB (At5g62690); SMZ (At2g39250); SNZ (At3g54990); SVP (At2g22540).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotype of ProRGA:RGAd17 Plants under Long-Day Conditions.
Supplemental Figure 2. Flowering Time of the Wild Type, ProSUC2:RGAd17, and ProFD:RGAd17.
Supplemental Figure 3. Expression of miR172 Targets.
Supplemental Figure 4. Flowering Time of ProSUC2:MIR172a ProSUC2:RGAd17 and ProSUC2:FT ProSUC2:RGAd17.
Supplemental Figure 5. Expression of SOC1 and FUL at the Shoot Apex in Long-Day Conditions.
Supplemental Figure 6. GA Response in Long-Day Conditions.
Supplemental Figure 7. GA Response of the Wild Type and Pro35S:MIR156 in Short-Day Conditions.
Supplemental Figure 8. Phenotype of della Pro35S:MIR156 Double Mutant.
Supplemental Figure 9. Phenotype of ProRGA::RGAd17 ProSPL9::rSPL9 Double Mutant.
Supplemental Figure 10. Interaction between DELLAs and SPLs in Yeast.
Supplemental Figure 11. Expression of NOD26 and LTP3 in Response to PAC.
Supplemental Figure 12. A Model for Integration of GA and miR156 Pathway.
Supplemental Table 1. Oligonucleotide Primer Sequences.
Supplementary Material
Acknowledgments
We thank Xiao-Ya Chen and Detlef Weigel for their generous scientific support. This article was supported by the National Natural Science Foundation of China (31222029), Recruitment Program of Global Expects (China), and initiation grants from the National Key Laboratory of Plant Genetics, Shanghai Institute of Plant Physiology, and Shanghai Institutes for Biological Sciences to J.-W.W. and by grants from National Nature Science Foundation (91017013 and 31070327) to Q.-Y.F. Work in the Schmid lab on the regulation on flowering time is supported by the Max Planck Institute for Developmental Biology and by grants from the Deutsche Forschungsgemeinschaft.
AUTHOR CONTRIBUTIONS
J.-W.W. and M.S. designed the research. S.Y., V.C.G., Y.-C.Z., D.H., T.-Q.Z., Y.-H.H., and J.-W.W. performed the experiments. Y.-Q.F., M.S., and J.-W.W. analyzed the data. S.W. and J.-W.W. wrote the article.
Glossary
- GA
gibberellin
- SAM
shoot apical meristem
- GA3
gibberellic acid
- PAC
paclobutrazol
- Ler
Landsberg erecta
- BD
binding domain
- BiFC
bimolecular fluorescence complementation
- CoIP
coimmunoprecipitation
- HA
hemagglutinin
- GFP
green fluorescent protein
- GR
glucocorticoid receptor
- DEX
dexamethasone
- LUC
luciferase
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Achard P., Baghour M., Chapple A., Hedden P., Van Der Straeten D., Genschik P., Moritz T., Harberd N.P. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010). Seasonal and developmental timing of flowering. Plant J. 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez M.A., Green R., Nilsson O., Sussman M.R., Weigel D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104 [DOI] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J.C., Sussex I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A., Jung H.S., Sun T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.Y., Zhu Q.H., Gu X., Ge S., Yang J., Luo J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- Harberd N.P. (2003). Botany. Relieving DELLA restraint. Science 299: 1853–1854 [DOI] [PubMed] [Google Scholar]
- Hisamatsu T., King R.W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59: 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jung J.H., Ju Y., Seo P.J., Lee J.H., Park C.M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D. (2007). Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Lee J., Lee I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Lin M.K., Belanger H., Lee Y.J., Varkonyi-Gasic E., Taoka K., Miura E., Xoconostle-Cázares B., Gendler K., Jorgensen R.A., Phinney B., Lough T.J., Lucas W.J. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E., Hedden P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Ray A., van der Winden J., Matzke M., Matzke A.J. (2003). Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Qi W., Sun F., Wang Q., Chen M., Huang Y., Feng Y.Q., Luo X., Yang J. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157: 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Richter R., Behringer C., Müller I.K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Willige B.C. (2009). Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. (2004). The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman K.M., Poethig R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Albani M.C., Vincent C., Bergonzi S., Luan M., Bai Y., Kiefer C., Castillo R., Coupland G. (2011). Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wilson R.N., Heckman J.W., Somerville C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Salinas M., Höhmann S., Berndtgen R., Huijser P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.