Heat shock protein101 (HSP101) is essential for rescue of proteins damaged by heat stress. However, reduced function of a protein involved in mitochondrial gene expression was found, unlike many mitochondrial mutations, to reduce oxidative stress and allow survival of heat stress in the absence of HSP101. Understanding control of oxidative stress by mitochondria is important to engineering plants with stress tolerance.
Abstract
The molecular chaperone heat shock protein101 (HSP101) is required for acquired thermotolerance in plants and other organisms. To identify factors that interact with HSP101 or that are involved in thermotolerance, we screened for extragenic suppressors of a dominant-negative allele of Arabidopsis thaliana HSP101, hot1-4. One suppressor, shot1 (for suppressor of hot1-4 1), encodes a mitochondrial transcription termination factor (mTERF)–related protein, one of 35 Arabidopsis mTERFs about which there is limited functional data. Missense (shot1-1) and T-DNA insertion (shot1-2) mutants suppress the hot1-4 heat-hypersensitive phenotype. Furthermore, shot1-2 suppresses other heat-sensitive mutants, and shot1-2 alone is more heat tolerant than the wild type. SHOT1 resides in mitochondria, indicating it functions independently of cytosolic/nuclear HSP101. Microarray analysis suggests altered mitochondrial function and/or retrograde signaling in shot1-2 increases transcripts of other HSPs and alters expression of redox-related genes. Reduced oxidative damage is the likely cause of shot1 thermotolerance, indicating HSP101 repairs protein oxidative damage and/or reduced oxidative damage allows recovery in the absence of HSP101. Changes in organelle-encoded transcripts in shot1 demonstrate that SHOT1 is involved in organelle gene regulation. The heat tolerance of shot1 emphasizes the importance of mitochondria in stress tolerance, and defining its function may provide insights into control of oxidative damage for engineering stress-resistant plants.
INTRODUCTION
Plants have evolved many different strategies to cope with heat stress, including long-term adaptations in life cycle or morphology, short-term heat avoidance strategies (e.g., leaf orientation and transpirational cooling), and rapid cellular acclimation mechanisms. It has long been known that plants can survive extreme heat stress if first acclimated by either exposure to sublethal temperatures or by a gradual increase to normally lethal temperatures (Vierling, 1991). This acquired thermotolerance is dependent on the induction of heat shock proteins (HSPs) during the acclimation treatment. Many HSPs protect plants from heat stress by either preventing irreversible protein denaturation (e.g., small HSPs) or rescuing heat-denatured proteins (e.g., Hsp70 and Hsp100/Casein lytic protease type B [ClpB]). In addition to causing protein denaturation, heat stress can also disrupt membrane integrity and homeostasis of metabolic processes and lead to oxidative stress (Vierling, 1991; Dat et al., 1998; Alfonso et al., 2001; Larkindale and Knight, 2002; Sangwan et al., 2002). Thus, other mechanisms besides enhanced protein quality control by HSPs must contribute to thermotolerance. Larkindale et al. (2005) showed that, indeed, there are other genes involved in thermotolerance by testing heat sensitivity of various mutants with defects in aspects of cellular function, including hormone signaling, reactive oxygen species (ROS) metabolism and signaling, and fatty acid metabolism.
One of the key HSPs essential for acquired thermotolerance in Arabidopsis thaliana and other plants is HSP101, which is a member of the Hsp100/ClpB chaperones in the AAA+ (for ATPases associated with various cellular activities) family of proteins. Using energy from ATP, Hsp100/ClpB chaperones play an important role in protecting organisms from severe heat stress by resolubilizing protein aggregates and aiding the refolding of denatured proteins (Parsell et al., 1994; Glover and Lindquist, 1998). The critical role of this protein in the acquisition of thermotolerance in Arabidopsis was revealed in a screen for heat-sensitive mutants, in which the first mutant isolated (hot1) was found to have a mutation in the gene encoding HSP101 (Hong and Vierling, 2000). The HSP101 protein contains two AAA modules, each composed of a nucleotide binding domain, sensor motifs, and a smaller C-terminal domain. The two AAA modules are separated by a coiled-coil domain, a key structural feature of the Hsp100/ClpB protein disaggregases (Doyle and Wickner, 2009). While there have been many biochemical studies on mechanistic aspects of the Hsp100/ClpB proteins, it remains largely unknown whether this family of proteins removes all aggregated proteins or rescues certain critical proteins in vivo.
We have taken a genetic approach to find HSP101-interacting proteins or other factors that are involved in thermotolerance. We screened for suppressor mutants in an ethyl methanesulfonate (EMS)–mutagenized M2 population of a semidominant mutant allele of HSP101 (hot1-4). The hot1-4 allele carries a mutation (A499T) in the unique Hsp100/ClpB coiled-coil domain, and hot1-4 is considered dominant negative because it is more heat sensitive than a T-DNA protein null allele of HSP101 (hot1-3) (Lee et al., 2005). In this report, an extragenic suppressor (suppressor of hot1-4 1 [shot1]) is shown to have a mutation in a gene encoding a mitochondrial transcription termination factor (mTERF)–related protein. Genetic analyses and localization studies revealed that SHOT1 works independently of HSP101, and the loss-of-function mutants of SHOT1 are more heat tolerant than the wild type. Reduced oxidative damage correlated with lowered ROS accumulation in shot1 mutants indicates that protection from oxidative damage associated with heat stress is a critical determinant of thermotolerance. Furthermore, the shot1 mutations provide direct genetic evidence that the chaperone function of HSP101 is not sufficient to counteract all oxidative damage. Finally, changes in the levels of mitochondrial transcripts suggest that the mTERF encoded by SHOT1 is involved in regulating expression of mitochondrial-encoded genes.
RESULTS
The SHOT1 Gene Encodes an mTERF-Related Protein
The sensitivity of Arabidopsis plants to heat stress can be quantitatively measured in dark-grown seedlings by the amount of hypocotyl elongation after heat stress. This hypocotyl elongation assay was used to screen for suppressors of the heat-sensitive, semidominant HSP101 mutant allele, hot1-4 (Lee et al., 2005). Dark-grown, 2.5-d-old hot1-4 seedlings are blocked in hypocotyl elongation after 2 h of 38°C heat treatment, while the wild type continues to grow. EMS-mutagenized hot1-4 M2 seeds were screened for shot mutants under this condition. Intragenic suppressors were analyzed and published previously (Lee et al., 2005). Four extragenic suppressors were also identified; here, we report detailed analysis of the first of these suppressors, shot1.
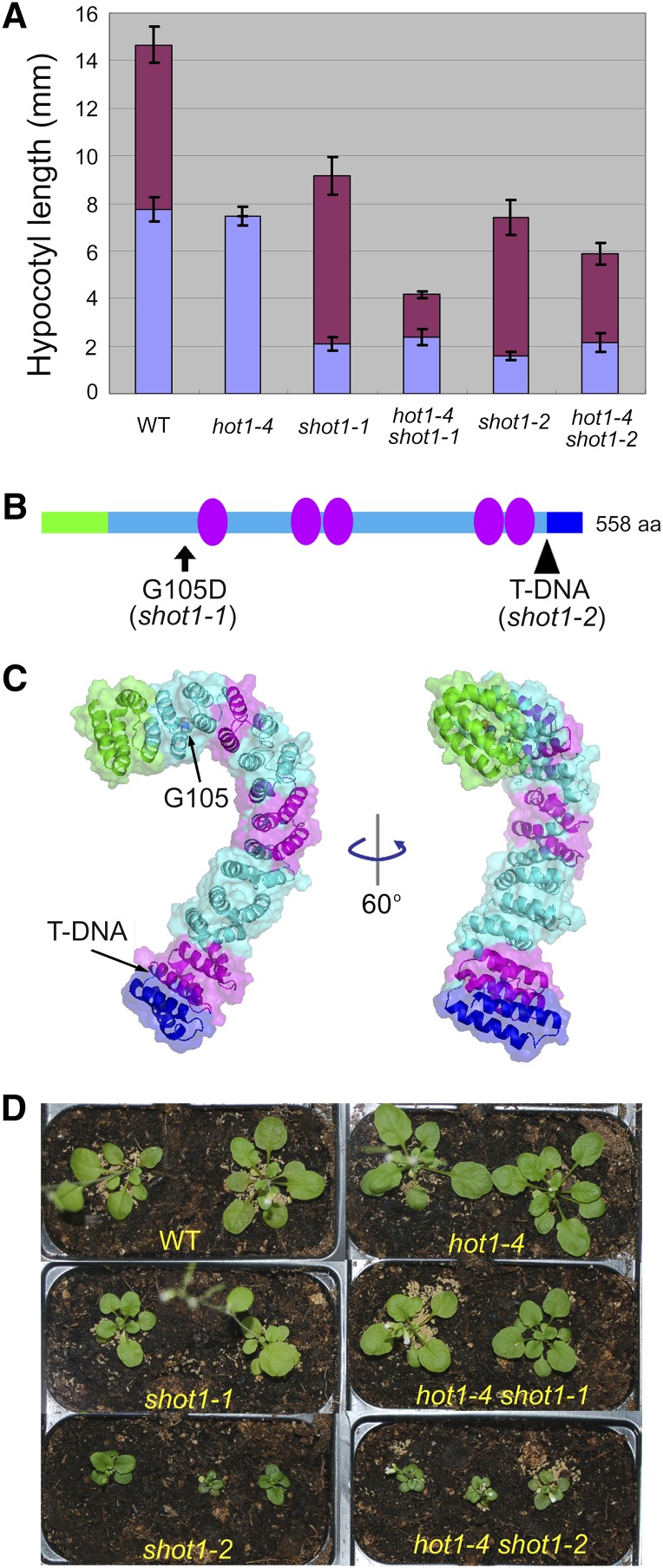
In addition to suppressing the defect in hypocotyl elongation after heat stress, the hot1-4 shot1-1 mutant has a short hypocotyl phenotype under optimal growth conditions (Figure 1A). We determined that the short hypocotyl phenotype cosegregates with the suppressing phenotype as a single recessive trait (see Supplemental Figure 1A online) and therefore used the short hypocotyl phenotype for map-based cloning of the mutation. After localization to a segment of chromosome 3 (see Supplemental Figure 1B online), sequencing of genes in the mapped region revealed shot1-1 to be a guanine-to-adenine transition converting a Gly to Asp at residue 105 in an mTERF-related protein (At3g60400) (Figure 1B; see Supplemental Figures 1B and 1C online).
Figure 1.
Mutations in an mTERF-Related Gene Suppress the Heat-Sensitive Phenotype of a hot1-4 Mutant.
(A) Hypocotyl growth before (purple bar) and after (maroon bar) heat treatment. Seedlings were grown on plates in the dark for 2.5 d and heat-acclimated at 38°C for 90 min followed by 2 h at 22°C (abbreviated AC), then heat-shocked at 45°C for 1 h (AC > 45°C/1 h). Growth after heat stress was measured 2.5 d later. Error bars indicate sd; n = 12. WT, the wild type.
(B) Location of the mutations on the SHOT1 protein structure. A T-DNA allele, shot1-2, truncates the C terminus, removing the region shown in blue. Green indicates the putative targeting peptide. Magenta ovals represent an mterf motif. aa, amino acids.
(C) A SHOT1 model was generated using the I-TASSER server. Arrows point to the locations of the shot1-1 and shot1-2 mutations. The same color scheme is used as in (B).
(D) Plants were grown on soil for 6 weeks under long-day conditions (16 h day/8 h night).
The SHOT1 protein is predicted to contain five mterf motifs and an N-terminal transit peptide of ∼60 amino acids (Figures 1B and 1C). When the SHOT1 protein sequence was used to query the SUBA database (http://suba.plantenergy.uwa.edu.au/), which contains prediction programs for subcellular localization, seven out of eight programs predicted mitochondrial localization (Heazlewood et al., 2007). Babiychuk et al. (2011) experimentally determined the localization of 28 of 35 Arabidopsis mTERF proteins using green fluorescent protein (GFP) fusions driven by the 35S promoter in both transient assays and transgenic plants. Their study concluded that SHOT1 is a mitochondrial protein. We confirmed this result by making a translational fusion of SHOT1 to the N terminus of GFP driven by the 35S promoter. Stable transgenic plants in the hot1-4 shot1-1 mutant background were generated. T3 homozygous plants carrying the mTERF-GFP fusion complemented the heat-tolerant phenotype of hot1-4 shot1-1, indicating that the transgene is functional and that the correct gene was identified by map-based cloning (see Supplemental Figure 2A online). Protoplasts were isolated from the complemented transgenic plants to examine subcellular localization (see Supplemental Figure 2B online). To discriminate GFP fluorescence from chlorophyll autofluorescence and MitoTracker fluorescence, microscopy was performed with spectral imaging followed by linear unmixing, revealing GFP fluorescence restricted to mitochondria. These results, along with data from Babiychuk et al. (2011), support the conclusion that SHOT1 is a mitochondrion-localized mTERF.
The founding member of the mTERF family is human MTERF1, which was originally identified as a factor responsible for transcription termination by binding to a specific site downstream of the MT-RNR2 gene encoding the mitochondrial 16S rRNA, thereby regulating the ratio of mitochondrial rRNA to mRNA (Kruse et al., 1989). This family of proteins shares an ∼30–amino acid mterf motif, which is composed of two α helices followed by a 310 helix (Yakubovskaya et al., 2010). The shot1-1 mutation is located upstream of the first mterf motif. A structural model of the SHOT1 protein was created using I-TASSER (Figure 1C) (Roy et al., 2010). Although SHOT1 does not share significant amino acid sequence similarity with MTERF1, I-TASSER identified MTERF1 as the closest structural homolog, and the SHOT1 model shows an all-helical repeat protein comparable to MTERF1. This type of helical repeat arrangement is typical of RNA and DNA binding proteins containing repeats of armadillo, HEAT, and PUM/PUF motifs (Groves and Barford, 1999; Lu et al., 2009), suggesting that SHOT1 is also likely to have nucleic acid binding properties.
A Second shot1 Allele Is a Stronger Suppressor of the HSP101 Mutant
We next identified a second allele of shot1, designated shot1-2, in the Columbia-0 (Col-0) background (GABI-Kat line 040C09) carrying a T-DNA insertion after the final mterf motif, which would result in elimination of the last 44 amino acids of the predicted protein (Figures 1B and 1C). We found by RT-PCR that the shot1-2 mutant produced a truncated transcript (see Supplemental Figure 1C online) and noticed that the T-DNA starts with an in-frame stop codon. Therefore, the shot1-2 mutant could produce a partially functional protein.
We then created the double mutant hot1-4 shot1-2 to determine if shot1-2 would also suppress the hot1-4 thermotolerance defect. shot1-2 would not be expected to suppress hot1-4 if suppression by the missense shot1-1 mutation was allele specific, due to direct physical interaction between HSP101 and SHOT1. However, after heat stress (AC > 45°C/1 h: acclimation at 38°C for 1.5 h and 22°C for 2 h, followed by 45°C for 1 h), shot1-2 suppressed the heat-sensitive phenotype of hot1-4 even more strongly than the shot1-1 mutation (maroon bars in Figure 1A). Thus, it is unlikely that there is direct interaction between the SHOT1 and hot1-4 proteins, consistent with their different compartmentalization within the cell, and the extent of suppression suggests that shot1-2 is a stronger allele than shot1-1. Like shot1-1, shot1-2 seedlings also have a short hypocotyl phenotype in the presence or absence of the hot1-4 mutation (purple bars in Figure 1A).
Both mutations in the SHOT1 gene make plants grow slowly, leading to small stature (Figure 1D), with shot1-2 showing a more severe growth reduction, consistent with the conclusion that this is a stronger allele. We could complement the growth phenotype (see Supplemental Figure 3A online) and the heat stress phenotype (see Supplemental Figure 3B online) caused by the shot1-2 mutation by transformation with wild-type SHOT1 genomic DNA, providing independent confirmation that both phenotypes are caused by the same gene. For further analyses, we used mainly the stronger shot1-2 mutant allele.
The hot1-4 allele has a dominant-negative phenotype, which means that the hot1-4 mutant protein is toxic and makes plants more sensitive to heat stress than an HSP101 protein null allele (i.e., hot1-3) (Lee et al., 2005). It is possible that the suppressing phenotype could result from reducing the dominant-negative effect of the hot1-4 mutant protein by downregulating the level of the toxic hot1-4 protein. Protein blot analysis of HSP101 protein levels, either after the heat treatment (AC > 45°C/1 h) or after recovery at 22°C, did not show a significant difference between plants with and without the shot1 mutations (see Supplemental Figures 3C and 3D online). The results indicate that the shot1 mutations provide thermotolerance by a mechanism that is independent from the mechanism involving the chaperone function of HSP101.
The shot1-2 Mutation Is a General Suppressor of Heat Sensitivity
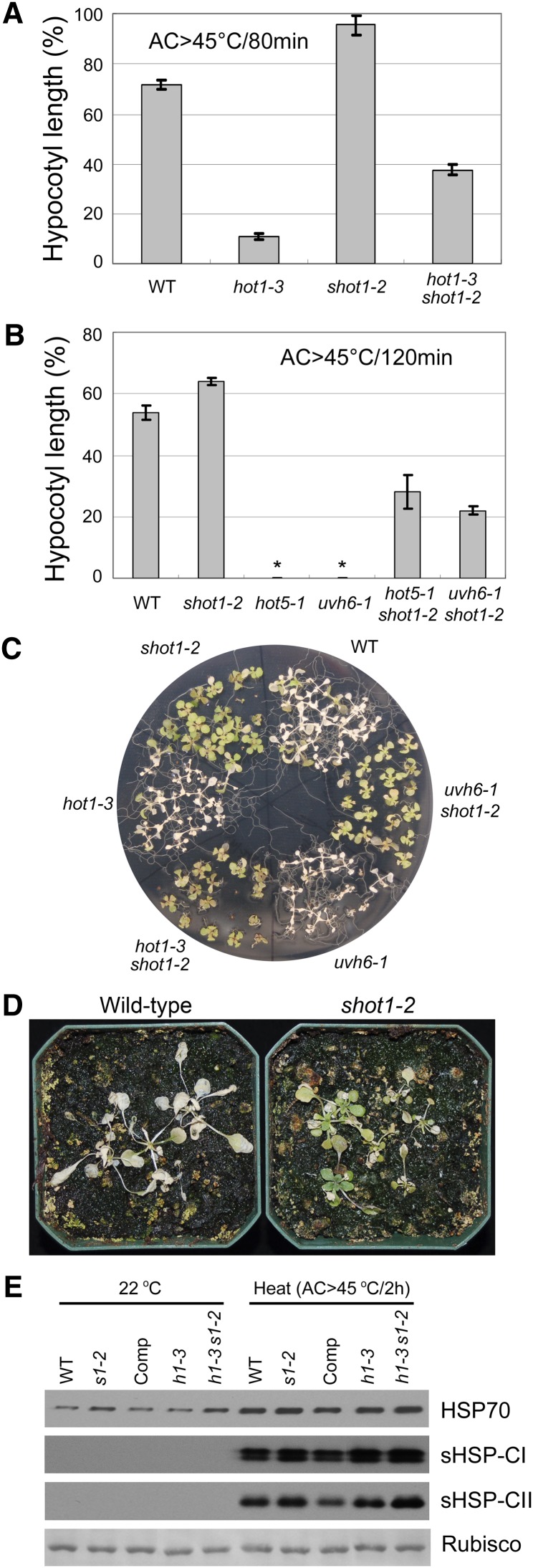
To test whether the suppressing phenotype is dependent in any way on HSP101, a double mutant with the HSP101 protein null allele hot1-3 was made. The double mutant hot1-3 shot1-2 also was more thermotolerant than the hot1-3 single mutant, when assayed both for hypocotyl elongation (Figure 2A) and survival of heat-treated light-grown seedlings (Figure 2C). Thus, the shot1-2 mutation can suppress heat sensitivity in the absence of functional HSP101 protein.
Figure 2.
The shot1-2 Mutant Can Suppress hot1-3 and Other Heat-Sensitive Mutants.
(A) The hypocotyl elongation assay shows partial suppression of heat sensitivity of hot1-3 by shot1-2. Hypocotyl elongation 2.5 d after heat treatment (AC > 45°C/80min) was measured and expressed as a percentage of growth during the same time period for seedlings that had not been heat treated. Error bars indicate se; n = 12. WT, the wild type.
(B) The hypocotyl elongation assay of seedlings heat-stressed at 45°C for 120 min after acclimation (AC > 45°C/120min) shows partial suppression of the heat sensitivity of hot5-1 and uvh6-1 by shot1-2. Asterisk indicates no growth after heat treatment. Error bars indicate se; n = 12.
(C) Ten-day-old seedlings were heat-treated (AC > 45°C/3.5 h). Photograph was taken after 7 d.
(D) Sixteen-day-old wild-type and 28-d-old shot1-2 plants were heat-treated at 37°C for 17 d and returned to 22°C for 8 d before photographing.
(E) The shot1-2 mutation causes a minor increase in cytosolic HSP70s and small HSPs. Total protein was extracted from 10-d-old seedlings with heat treatment (AC > 45°C/2 h) or without heat treatment (22°C control). Coomassie blue–stained ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit bands are shown as a loading control. Comp, complemented line; h1-3, hot1-3; h1-4, hot1-4; s1-1, shot1-1; s1-2, shot1-2.
It is clear that other pathways besides those involving HSPs contribute to thermotolerance (Larkindale et al., 2005). Therefore, we sought to determine if the shot1-2 mutation could suppress two other heat-sensitive mutants, UV hypersensitive6-1 (uvh6-1) and sensitive to hot temperatures5-1 (hot5-1), which impact very different aspects of cellular function. The uvh6-1 mutant was identified as a UV-sensitive mutant (Liu et al., 2003) but was later found to be defective in thermotolerance (Larkindale et al., 2005). The UVH6 gene encodes a homolog of human Xeroderma Pigmentosum D (XPD), which is an integral part of the basal transcription factor BTF2/TFIIH complex and belongs to the RAD3/XPD subfamily of helicases (Winkler et al., 2000). The HOT5 gene encodes S-nitrosoglutathione reductase, which metabolizes the nitric oxide adduct S-nitrosoglutathione (Lee et al., 2008). hot5 mutants have increased nitric oxide species that are associated with heat sensitivity. The hypocotyl elongation assay shows that the heat-sensitive phenotype of both uvh6-1 and hot5-1 is suppressed by the shot1-2 mutation (Figure 2B). Suppression is also observed with light-grown seedlings in the case of uvh6-1 (Figure 2C). The hot5-1 mutant was not tested in this assay because this allele is not sensitive to heat when grown in the light (Lee et al., 2008). Remarkably, the shot1-2 single mutant was more tolerant to heat stress than the wild type (Figures 2A to 2C), indicating that the shot1-2 mutation is a general suppressor of heat sensitivity. Enhanced thermotolerance of the shot1-2 mutant was also observed when soil-grown plants were subjected to prolonged, chronic heat stress at 37°C (Figure 2D).
Many studies have reported that increases in HSP expression could enhance heat tolerance of diverse organisms, including plants (Solomon et al., 1991; Wissing and Jäättelä, 1996; Queitsch et al., 2000). Therefore, we checked the protein level of some major HSPs, other than HSP101, known to be important for thermotolerance. The shot1-2 single and hot1-3 shot1-2 double mutants, when compared with wild-type and hot1-3 plants, respectively, show a minor increase in cytosolic HSP70s and small HSPs under the heat stress conditions used to assay heat sensitivity (Figure 2E). Cytosolic HSP70s show a minor increase in shot1-2 plants even without heat stress, implying that shot1-2 mutant plants may exhibit a stress response even without any heat treatment.
The Transcriptome of the shot1-2 Mutant Shows Enrichment of Stress-Responsive Genes
It is well established that enhancement of the protein quality control network through increased HSP expression is a major component of the thermotolerance mechanism. However, even though the shot1 mutants show a small increase in some HSPs (Figure 2E), this subtle difference appears unlikely to account for shot1-2 thermotolerance. Therefore, to identify other mechanisms that could enhance shot1 thermotolerance, we compared the transcriptional profile of the shot1-2 single mutant with that of wild-type plants both with and without heat treatment.
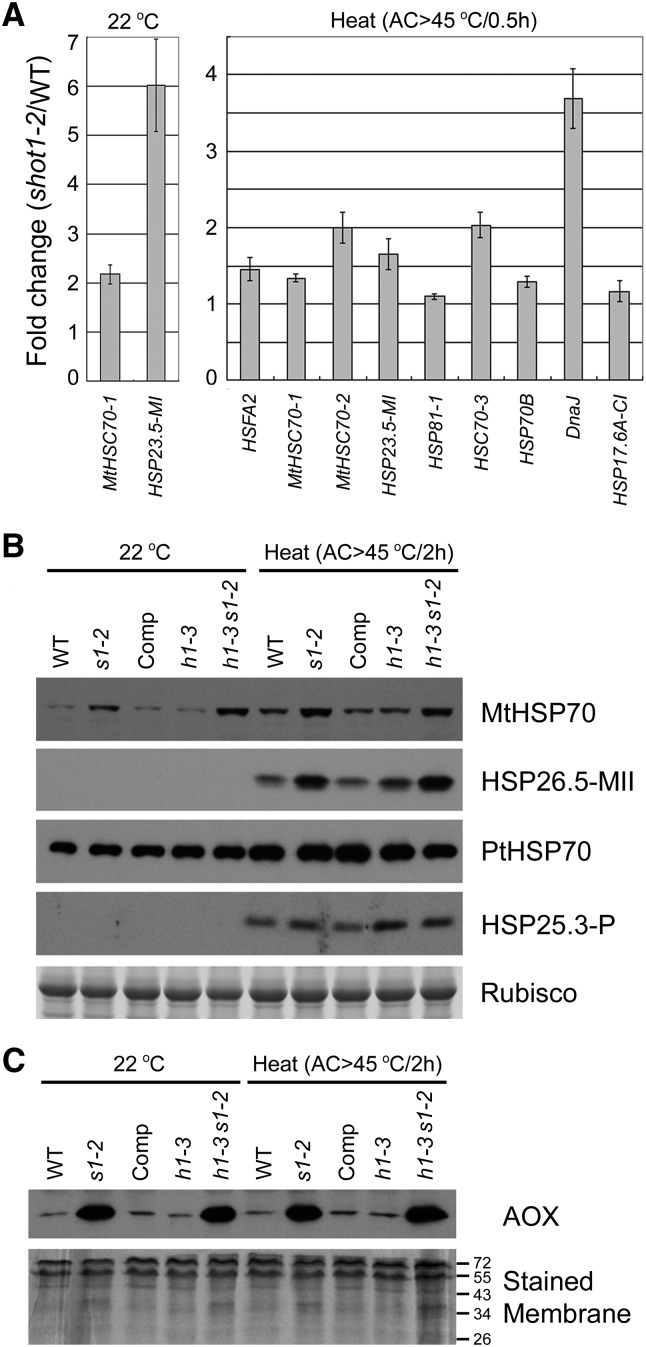
Microarray analysis was performed using Arabidopsis oligonucleotide arrays with the two-color dye method to compare the wild type to the mutant (Zanetti et al., 2005; Zhang et al., 2008). RNA samples were prepared from 10-d-old, plate-grown seedlings sampled with or without heat treatment (AC > 45°C/30 min). The microarray data can be found in Supplemental Data Sets 1 and 2 online. For a general overview, genes that were both differentially regulated more than 0.5-fold log2 ratio and that had a false discovery rate–adjusted P value of <0.05 were selected for analysis. This cutoff value resulted in 700 and 484 differentially regulated genes for the 22°C control and heat treatment, respectively. Gene Ontology (GO) enrichment analyses performed with these gene subsets are shown for control samples in Supplemental Table 1 online and heat stress samples in Supplemental Table 2 online. The shot1-2 mutant showed significant differences in transcripts of genes responsive to stress, more specifically oxidative stress, in both conditions. A similar observation was made relative to genes with redox-related GO terms in the GO category of molecular function. Almost twice as many genes related to antioxidant activity were downregulated in shot1-2 under control conditions compared with the wild type. Genes with glutathione S-transferase activity showed the opposite pattern, with nine out of 10 upregulated in shot1-2. The fact that the expression pattern of many stress-responsive genes is altered may indicate that the mutant plants experience stress even under normal growth conditions, as is suggested by the increase in cytosolic HSP70 proteins without heat treatment (Figure 2E). Another noticeable GO term was, not surprisingly, differences in genes responsive to heat. Supplemental Table 3 online lists the 19 heat stress–responsive genes that changed in both control and heat stress conditions. Specific HSP transcripts were increased in the shot1-2 mutant although the difference in expression level compared with the wild type was for the most part small, with a maximum of ∼2.7-fold.
From the microarray data, it appeared that two possible mechanisms could contribute to higher thermotolerance in the shot1-2 mutant. The first mechanism is enhancement of protein quality control through increased expression of HSPs other than HSP101. The second mechanism is reduced oxidative stress.
Mitochondrial HSPs and Alternative Oxidases Are Increased in the shot1-2 Mutant
We observed a small fold increase in transcripts of cytosolic HSP70s in the microarray experiment, which correlates well with the immunoblot analysis in Figure 2E. In addition, the transcripts of mitochondrial HSP70s and mitochondrial small HSPs were shown to be higher in shot1-2 than in the wild-type, with or without heat stress. To confirm results of the microarray experiments, quantitative RT-PCR (qRT-PCR) was performed on these and other HSP genes (Figure 3A). The microarray data were readily confirmed, with the exception of transcript levels for HSP81-1 and HSP17.6A-CI, which were not significantly different between wild-type and mutant plants (P > 0.05). Related to the potential role of SHOT1 in mitochondria, transcripts of HSPs targeted to mitochondria were increased, but not transcripts of HSPs targeted to plastids. This differential mRNA expression was accompanied by related increases in protein levels, as shown in Figure 3B; mitochondrial HSPs, but not plastidic HSPs, increased in shot1-2 mutants. These results indicate that the mutation causes an indirect effect on the level of mitochondrial HSPs. The increased mitochondrial HSPs may enhance thermotolerance by maintaining protein quality in mitochondria.
Figure 3.
Mitochondrial HSPs and AOXs Are Upregulated in the shot1-2 Mutant.
(A) Increased expression of HSPs determined in microarray experiments was confirmed by qRT-PCR. Values represent the mean and se of at least two biological replicates with three technical replicates. WT, the wild type.
(B) Protein blot analysis shows that mitochondrial HSP70s and a mitochondrial small HSP are increased in shot1-2 mutants. Comp, complemented shot1-2 line; h1-3, hot1-3; s1-2, shot1-2. Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit bands are shown as a loading control.
(C) AOX is increased in shot1-2 mutants. Stained membrane is shown as a loading control.
In the microarray experiment, we also found that the Alternative Oxidase 1a (AOX1a) transcript was increased 1.6-fold in shot1-2. AOX genes have been described as markers for many different stresses (Vanlerberghe and McLntosh, 1996; Clifton et al., 2005; Zarkovic et al., 2005). In the mitochondrial electron transport chain, AOX proteins play a key role in preventing ROS production by accepting electrons from the reduced quinone pool. To determine if this small change in AOX mRNA level was reflected at the protein level, protein blot analysis was performed. The AOX protein level was highly increased in plants with the shot1-2 mutation (Figure 3C), with the increase in protein being much more dramatic than that of the transcript. The increase in AOX protein even without the heat treatment indicates that the shot1-2 mutation triggers stress signaling, as is also indicated by the increased HSP levels. AOX proteins show no significant further increase with heat treatment.
To determine the potential contribution of AOX to the increased thermotolerance of shot1, we tested transgenic plants that either under- or overexpressed AOX1a protein (see Supplemental Figure 4 online). The overexpression line had even more AOX protein than the shot1-2 mutant, but it was as heat sensitive as the wild type. In addition, over- or underexpression of AOX in the shot1-2 background did not affect the shot1-2 heat stress phenotype, suggesting that the changes in AOX1a protein level in the shot1-2 mutant alone cannot confer thermotolerance.
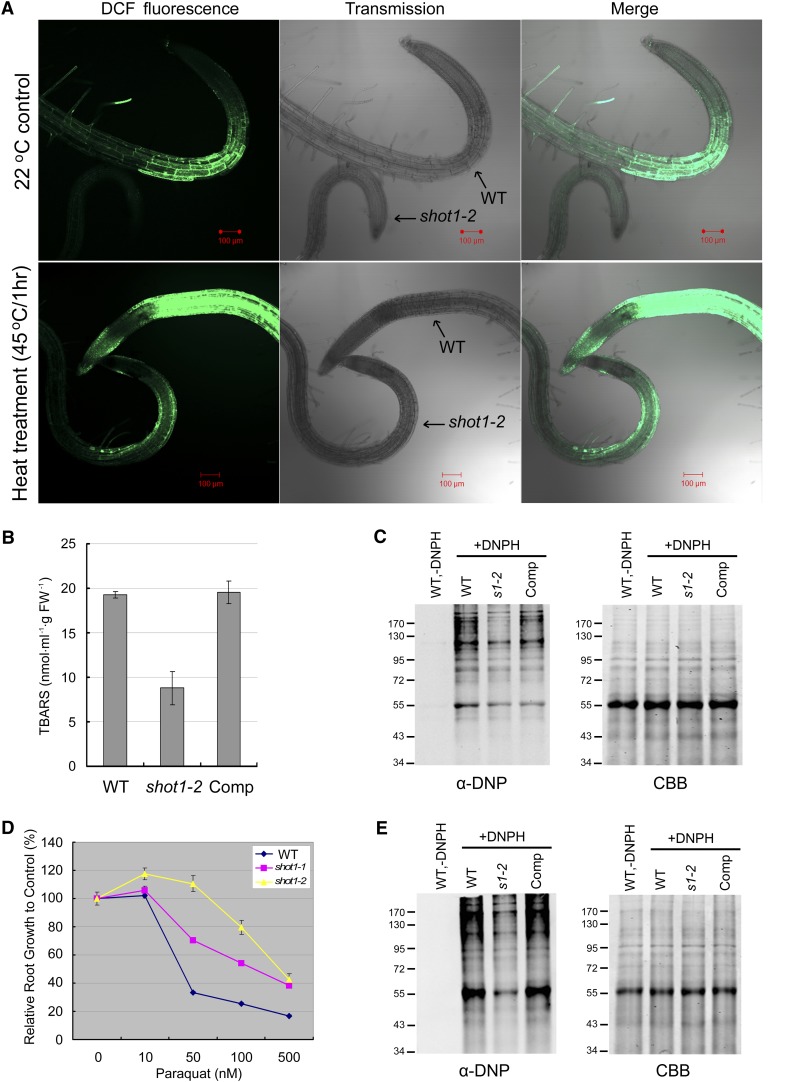
The shot1-2 Mutant Accumulates Less ROS, Has Less Oxidative Damage, and Is More Tolerant to Paraquat
The induction of AOX protein and GO analysis of microarray data led us to look at the oxidative stress aspect of heat stress. We hypothesized that shot1-2 is more tolerant to heat stress because it can better cope with the oxidative stress that occurs with heat treatment. We first examined ROS levels using the fluorescent dye 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA). Plate-grown seedlings were treated at 22 or 45°C for 1 h and then incubated with H2DCFDA before being observed by confocal microscopy (Figure 4A). While the ROS signal is clearly increased by the heat treatment, there was consistently significantly less ROS in the shot1-2 mutant than in the wild type. This result predicts there should be also less oxidative damage in the shot1-2 mutant. To test this prediction, we measured both lipid and protein oxidation after heat treatment. In accordance with our expectation, shot1-2 mutant plants showed less lipid and protein oxidation than the wild type (Figures 4B and 4C). These data further suggest that the shot1-2 mutant would be more tolerant to oxidative stress. Tolerance to oxidative stress was measured by root growth after transferring seedlings to paraquat-containing medium. Root growth of the shot1 mutants was indeed more tolerant to paraquat (Figure 4D), and the shot1-2 mutant was more tolerant than shot1-1, again indicating that shot1-2 is a stronger allele. Consistent with the sensitivity to paraquat, shot1-2 had less protein oxidation than the wild type when treated with paraquat as shown in Figure 4E.
Figure 4.
The shot1-2 Mutant Has Less ROS and Oxidative Damage and Is More Resistant to the Oxidative Agent Paraquat.
(A) Roots of 5-d-old seedlings were stained with the ROS marker H2DCFDA (DCF). WT, the wild type.
(B) Twelve-day-old plants were heat-treated (AC > 45°C/3 h) and harvested 2 d later for the thiobarbituric acid-reactive-substances assay. Values represent averages and se of three biological replicates. FW, fresh weight.
(C) The level of carbonylated proteins is higher in the wild type than shot1 after heat stress. Proteins were extracted from plants after heat treatment (AC > 45°C/2 h). Four micrograms of total proteins was derivatized with (+DNPH) or without DNPH (−DNPH, negative control) and separated on 10% SDS-PAGE gel for either immunoblot analysis with anti-DNP antibody (left panel) or Coomassie blue staining (right panel; CBB). The wild-type sample without derivatization (−DNPH) is shown as a negative control. Comp, complemented shot1-2 line; DNPH, 2,4-dinitrophenylhydrazine.
(D) Relative root growth as a percentage of growth without paraquat is shown as a measure of paraquat sensitivity. Error bars indicate se of 12 seedlings.
(E) The level of carbonylated proteins is higher in the wild type than shot1 after paraquat treatment. Proteins were extracted from plants after 4 h of treatment with 100 µM paraquat. Four micrograms of total proteins was derivatized with (+DNPH) or without DNPH (−DNPH, negative control) and separated on 10% SDS-PAGE gel for either immunoblot analysis with anti-DNP antibody (left panel) or Coomassie blue staining (right panel). The wild-type sample without derivatization (−DNPH) is shown as a negative control.
To test the possibility that reduced ROS in shot1-2 correlated with increased ability to eliminate ROS, we checked the activities of the antioxidant enzymes, superoxide dismutase, and catalase. While shot1-2 consistently showed a minor increase in mitochondria-localized manganese superoxide dismutase, a minor decrease in catalase activity was also detected in shot1-2 (see Supplemental Figure 5 online). These minor changes in the antioxidant enzyme activities would not appear to explain the significant alteration in ROS level. Thus, shot1-2 may have reduced ROS due to decreased production of ROS rather than increased removal of ROS by these enzymes.
In summary, shot1 mutants show reduced accumulation of ROS and have better tolerance to oxidative stress, which likely contributes to their heat stress tolerance.
Other Mutants Compromised in Mitochondrial Function Are More Sensitive to Heat Stress
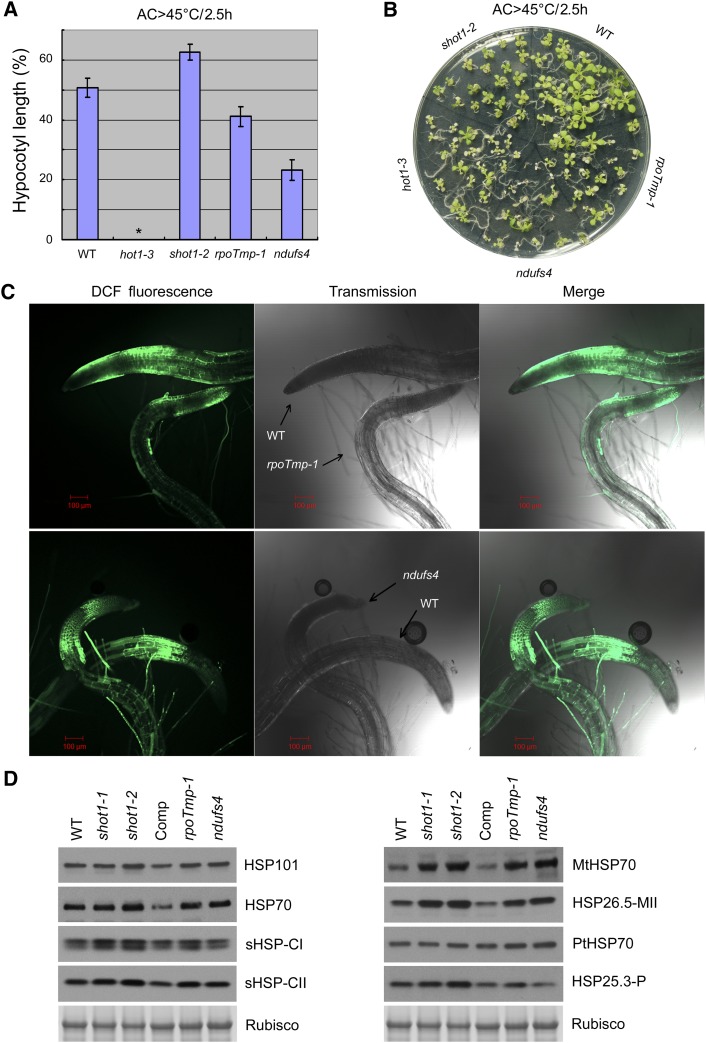
Mutants impaired in mitochondrial function tend to have slow growth rate and wrinkly rosette leaves (de Longevialle et al., 2007; Kühn et al., 2009). The shot1-2 mutant shares these phenotypes, which along with the mitochondrial localization of the protein and altered AOX levels, indicate that shot1 mutants may also be impaired in mitochondrial function. This led us to ask if the response of shot1 mutants to heat stress is common to other mutants with mitochondrial dysfunction and if these mutants also have enhanced tolerance to heat stress. We selected two mutants, rpoTmp-1 (for RNA polymerase T-phage type in mitochondria and chloroplasts) and ndufs4 (for NADH dehydrogenase [ubiquinone] iron-sulfur protein 4) to address these questions. The RPOTmp gene encodes a phage-type RNA polymerase that performs gene-specific transcription in mitochondria and is targeted to both mitochondria and plastids. The loss-of-function mutant rpoTmp-1 is known to have a reduced abundance of respiratory chain complexes I and IV (Kühn et al., 2009). The NDUFS4 gene encodes NADH dehydrogenase (ubiquinone) iron-sulfur protein subunit 4, which is a component of respiratory chain complex I (Meyer et al., 2009). The ndufs4 null mutation prevents assembly of complex I without affecting other components of respiratory complexes. frostbite1, another loss-of-function allele of the same gene, was shown to be sensitive to freezing and to have increased ROS level (Lee et al., 2002).
In thermotolerance assays with both hypocotyl elongation and light-grown seedlings, the rpoTmp-1 and ndufs4 mutants showed more sensitivity to heat stress, in contrast with the shot1-2 mutant (Figures 5A and 5B). These results demonstrate that mitochondrial dysfunction does not necessarily result in the same heat-tolerant phenotype. In addition, the rpoTmp-1 and ndufs4 mutants had more ROS production under heat treatment than the shot1-2 mutant (Figure 5C). However, the levels of HSPs were quite similar to the shot1 mutants with heat treatment (Figure 5D). In particular, the mitochondrial HSPs were notably increased in the rpoTmp-1 and ndufs4 mutants, similar to the shot1 mutants. These data indicate that low ROS levels in the shot1-2 mutant may be more critical for providing thermotolerance than the increased level of mitochondrial HSPs.
Figure 5.
Mitochondrial Dysfunction Does Not Lead to Heat Tolerance.
(A) Hypocotyl elongation after heat stress (AC > 45°C/2.5 h) relative to no heat treatment was measured to determine heat sensitivity of shot1 and two other mitochondrial mutants. Error bars indicate se of 12 seedlings. Asterisk indicates no growth after heat treatment. WT, the wild type.
(B) Heat tolerance test on light-grown seedlings of the same genotypes tested in (A).
(C) ROS accumulation in roots of 5-d-old seedlings after heat treatment at 45°C for 1 h detected with H2DCFDA (DCF).
(D) Immunoblot analysis of HSPs from shot1 and other mitochondrial mutants. Protein was extracted from light-grown seedlings after heat treatment (AC > 45°C/2 h). Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit bands are shown as a loading control. Comp, complemented shot1-2 line.
Mutation of a Chloroplast mTERF Does Not Confer Heat Tolerance
We noted that soldat10 is a mutation in an Arabidopsis plastid-localized mTERF protein and that it shows parallels to shot1 in that it was isolated as a suppressor of the flu mutant, which is highly sensitive to singlet oxygen–induced cell death. Furthermore, like shot1, soldat10 was subsequently found to be more stress tolerant even in the wild-type background (Meskauskiene et al., 2009). We therefore tested soldat10 mutant seedlings for heat tolerance. However, unlike shot1, the soldat10 mutant was not more heat tolerant than the wild type (see Supplemental Figure 6 online). Comparison of the 257 genes upregulated in shot1 compared with the 201 genes reported to be upregulated in soldat10 also showed only eight common upregulated genes (see Supplemental Table 4 online). Thus, the mechanisms leading to the different stress protection by these two mTERF mutants are distinct and may partly reflect differences in responses to alterations in chloroplasts compared with mitochondria.
Mitochondrial Genes Are Upregulated and Chloroplast Genes Are Downregulated in the shot1-2 Mutant
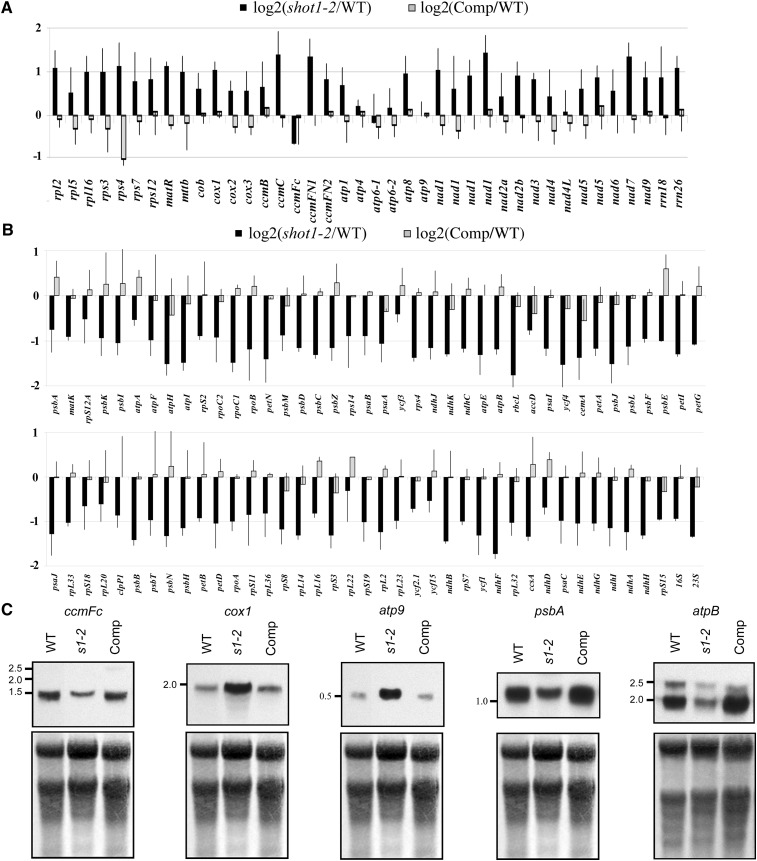
In mitochondria, mTERF family proteins are involved in transcription termination as well as mitochondrial DNA replication, transcription initiation, and translational control (Kruse et al., 1989; Martin et al., 2005; Roberti et al., 2006; Hyvärinen et al., 2007; Wenz et al., 2009; Cámara et al., 2011). Since the SHOT1 protein contains no defined motifs other than the mterf motif, and it is readily modeled based on MTERF1 with features consistent with nucleic acid binding, we considered that SHOT1 may have a biochemical function similar to homologs in other organisms. To gain insight into potential roles of the SHOT1 protein in nucleic acid interactions in mitochondria, we assayed the level of transcripts using qRT-PCR. The shot1-2 mutant and the line complemented with wild-type SHOT1 genomic DNA were compared with the wild type. As shown in Figure 6A, the shot1-2 mutation caused an overall twofold increase in levels of mitochondrial transcripts when normalized against a number of housekeeping genes. There were almost no changes in the transcripts of atp4, atp6-1, atp6-2, atp9, and nad4L. The only downregulated transcript in the shot1-2 mutant was the ccmFC transcript, which encodes a subunit of a protein complex reported to be involved in cytochrome c maturation (Rayapuram et al., 2008).
Figure 6.
Most Mitochondrion-Encoded Genes Are Upregulated and Plastid-Encoded Genes Are Downregulated in the shot1-2 Mutant.
(A) Transcript abundance of all protein-encoding and rRNA genes of the mitochondrial genome from shot1-2 (black bars) and shot1-2 complemented with SHOT1 (Comp; gray bars) was compared with the wild type (WT) by qRT-PCR and normalized against the geometric mean of five cytosolic housekeeping transcripts (YSL8, RPL5B, UBC, ACTIN2/ACTIN8, and TUB6). Error bars are sd based on three biological replicates.
(B) Transcript abundance of all protein-encoding and rRNA genes of the chloroplast genome from shot1-2 (black bars) and shot1-2 complemented with SHOT1 (Comp; gray bars) was compared with the wild type by qRT-PCR and normalized against the geometric mean of six cytosolic housekeeping transcripts (YSL8, RPL5B, UBC, ACTIN2/ACTIN8, POR, and RRN18S). Genes are sorted according to their physical location on the chloroplast chromosome. Error bars are sd based on two biological replicates.
(C) RNA gel blot analysis of organelle-encoded genes. Methylene blue–stained membrane is shown as a loading control. Comp, complemented shot1-2 line; s1-2, shot1-2.
In contrast with mitochondrial transcripts, the shot1-2 mutation had the opposite effect on plastid transcripts, showing an overall twofold decrease with no significant variation between different genes (Figure 6B). To confirm the qRT-PCR results, specific genes were chosen for RNA gel blot analysis (Figure 6C). The transcript of the mitochondrial gene cox1 was increased, but ccmFC was decreased in the shot1-2 mutant as expected based on the qRT-PCR data. The ccmFC transcript, which contains an intron, was correctly spliced in the mutant, despite the reduced transcript levels. Inconsistent with the qRT-PCR data, the atp9 transcript was increased in the shot1-2 mutant, which may reflect the choice of primers for the qRT-PCR measurement. Changes in transcripts of the plastid genes psbA and atpB confirmed the qRT-PCR data, decreasing in the shot1-2 mutant. Thus, the shot1-2 mutation influences transcript levels of the two organelles in opposite ways and may have a specific role in determining the levels of mitochondrial ccmFC RNA.
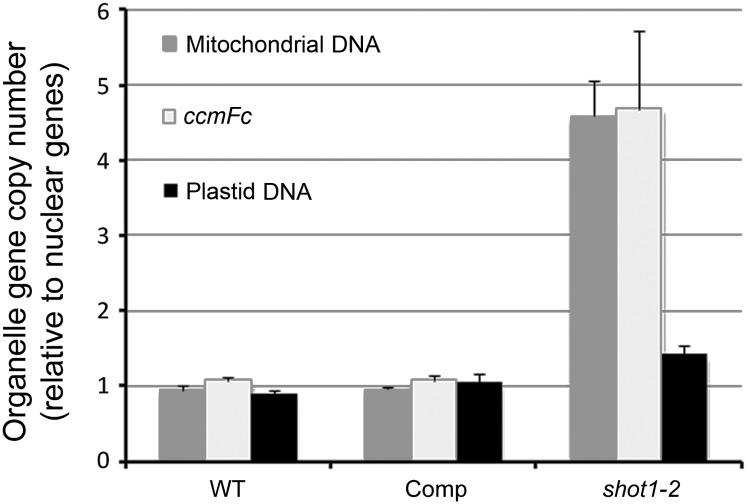
To check if the shot1-2 mutation also causes alterations in organelle genome copy numbers, qPCR was performed to compare copy numbers of organelle genes in the shot1-2 mutant and the wild type. Five representative genes of the organellar and nuclear genome were selected for this experiment (see Supplemental Table 5 online). Values for the plastid and mitochondrial gene amplifications were normalized to the nuclear gene amplifications. Mitochondrial DNA levels in shot1-2 were ∼4.5-fold higher than in the wild type and complemented line (Figure 7). The observed general increase in mitochondrial gene transcripts may reflect this increased gene copy number. However, the copy number of ccmFC gene was also increased, in contrast with the observed transcript decrease. Plastidic DNA levels in shot1-2 were only slightly higher than in the wild type and the complemented line. In summary, SHOT1 function appears to be required for coordination of nuclear and mitochondrial gene copy number.
Figure 7.
Organelle Gene Copy Numbers in shot1-2.
Five single-copy genes from each genome (nuclear, plastid, and mitochondrial) were amplified by quantitative PCR. Each reaction was performed with two biological replicates and three technical replicates. The concentrations obtained for all five amplification products belonging to the same compartment were averaged (geometric mean), and the value of the plastid and mitochondrial amplifications was normalized to the value of the nuclear gene amplifications. The bars in the graph represent the mean and ±sd from two biological samples. Comp, complemented shot1-2 line; WT, the wild type.
DISCUSSION
The Thermotolerance Pathway Involving the mTERF Protein, SHOT1, Acts Parallel to the Pathway of Protein Quality Control by HSP101
We set out to find interacting partners of HSP101 or other factors/pathways involved in heat tolerance mechanisms by genetic suppressor screening. Surprisingly, the first extragenic suppressor isolated from this screen, shot1, bypassed the requirement for HSP101 in thermotolerance, providing direct genetic evidence for the importance of other mechanisms in heat tolerance. Partial suppression of heat sensitivity by shot1 mutations in the hot1 mutant background indicates that cells can recover in the absence of the protein quality control function of HSP101, provided that other damage is limited or cells are otherwise primed for protection from stress. We conclude that this is primarily the result of reduced ROS accumulation in shot1 but that stress tolerance is further enhanced by other factors, including increased expression of specific HSPs, particularly those localized to mitochondria.
It is of significant interest to have found a mechanism that bypasses the activity of HSP101 in thermotolerance. This suggests that one or more critical substrates of HSP101 may not be damaged in the absence of heat-induced oxidative stress, implying HSP101 is involved in repair of oxidatively damaged proteins. In Saccharomyces cerevisiae, Hsp104 is found associated with protein aggregates formed by oxidative stress and is implicated in preventing segregation of oxidatively damaged proteins into daughter cells on division (Erjavec et al., 2007). However, there are no direct data demonstrating that Hsp100/ClpB proteins repair oxidative damage. It is also possible, and not mutually exclusive, that the increased levels of other chaperones in shot1 can prevent or repair some damage to proteins that are normally HSP101 substrates. However, the fact that the shot1 mutation does not exhibit wild-type thermotolerance indicates that much of the heat-induced damage remains unrepaired in the absence of HSP101 function. We suggest that cells can survive failure to rescue heat-denatured proteins as long as they are not also overloaded with oxidatively damaged cellular components.
According to Genevestigator expression data (Zimmermann et al., 2004), the level of SHOT1 mRNA is very low (close to the detection limit) and is not very responsive to different environmental conditions. We also saw no significant change in expression of the gene with heat stress treatment as determined by qRT-PCR. Thus, we conclude that SHOT1 is not likely to be a regular factor involved in heat stress signaling or protection in wild-type plants. Altogether, our data support the hypothesis that mutations in SHOT1 indirectly increase thermotolerance due to adaptation of plants to defective mitochondrial function. The critical role of mitochondrial metabolism in tolerance to a variety of stresses is being increasingly recognized (Dutilleul et al., 2003; Atkin and Macherel, 2009; Jacoby et al., 2011; Millar et al., 2011) and is further supported by our data. Studies of the molecular function of shot1 and the resulting impact on mitochondrial metabolism will be important to defining the mechanism of stress tolerance.
Reduced Oxidative Damage Is Likely the Major Contributor to the Enhanced Thermotolerance of shot1 Mutants
The overall growth phenotype of shot1 mutants is similar to mutants with mitochondrial dysfunction, such as rpoTmp (Kühn et al., 2009), ndufs4 (Meyer et al., 2009), and otp43 mutants (de Longevialle et al., 2007), suggesting that shot1 mutants have problems in the key mitochondrial function of ATP production by oxidative phosphorylation. One common molecular response of the rpoTmp, ndufs4, and shot1 mutants is an increased level of specific HSPs, especially mitochondrial HSP70s and small HSPs, both with or without heat treatment (Figure 5D). Maize (Zea mays) respiratory-deficient NCS (nonchromosomal stripe) mutants with permanent mitochondrial dysfunction also exhibit constitutive expression of both cytosolic and mitochondrial HSPs (Kuzmin et al., 2004). Transcripts of two closely related mitochondrial small HSPs, HSP23.5-MI and HSP23.6-MI, are induced by many stresses, including heat and hydrogen peroxide, and are reported to localize both to mitochondria and chloroplasts (Van Aken et al., 2009b). In the same study, MtHSC70-2 was found to be induced most by heat and moderately by UV and osmotic stress. Because they are heat induced, mitochondrial HSPs are proposed to have a heat-protective function. However, there is a lack of genetic and functional evidence demonstrating their role in thermotolerance. The fact that rpoTmp-1 and ndufs4 mutants show similar levels of HSP accumulation as the shot1 mutants, yet are sensitive to heat stress, indicates that these HSPs alone cannot account for the heat tolerance of shot1.
Another common molecular response between mutants that are affected in mitochondrial electron transport (ETC) and shot1 mutants is the accumulation of AOX proteins, which are components of alternative ETC. AOX is a well-documented marker for stress responses and has a role in suppressing accumulation of ROS (Van Aken et al., 2009a). Other components of alternative ETC include nicotinamide adenine dinucleotide dehydrogenases, which can accept electrons from NAD(P)H without generating a proton gradient (Rasmusson et al., 2004). One of the key nicotinamide adenine dinucleotide dehydrogenases that is coexpressed with AOX1a is NDB2 (Clifton et al., 2005). In our microarray data, the NDB2 transcript is also increased 1.57-fold in shot1-2 (see Supplemental Table 6 online). Uncoupling proteins (UCPs) are another component of alternative ETC that can dissipate the proton gradient without producing ATP. UCP1 has been shown to be involved in reducing oxidative stress damage (Brandalise et al., 2003; Sweetlove et al., 2006). Transcripts of both UCP1 and UCP2 are increased 1.43- and 1.53-fold in shot1-2, respectively (see Supplemental Table 6 online). Therefore, the whole alternative ETC circuit may be induced in shot1-2.
The heat sensitivity of rpoTmp-1, ndufs4, and AOX1a transgenic plants indicates that heat sensitivity is not simply inversely correlated with the level of AOX proteins; increased expression of AOX in ndufs4 and the AOX1a overexpression line does not lead to increased heat tolerance. AOX is known to ameliorate accumulation of ROS and oxidative damage caused by KCN, a potent inhibitor of cytochrome c oxidase (Umbach et al., 2005). Therefore, we could expect that increased expression of AOX would lead to better heat stress tolerance, but this is not the case. These confounding results from plants with increased AOX protein levels may indicate that there are other levels of regulation in the activity of alternative ETC. The activity of AOX is regulated by many factors, such as redox state of the Cys residues in AOX and the availability of pyruvate (Day and Wiskich, 1995; Umbach et al., 2006). Furthermore, unlike shot1-2, AOX1a transgenic plants did not show significant changes in other components of alternative ETC (Umbach et al., 2005). Further study of the redox state and metabolic environment is needed to understand how increased AOX may contribute to the thermotolerance of shot1 mutants.
There is one clear difference between shot1 mutants and the dysfunctional mitochondrial ETC mutants that are heat sensitive. The rpoTmp-1 and ndufs4 mutants had more ROS than shot1 mutants (Figures 4A and 5C). Many reports imply a connection between oxidative stress and heat stress. ROS increases upon heat shock (Dat et al., 1998; Volkov et al., 2006; Kristiansen et al., 2009), heat stress causes oxidative damage (Larkindale et al., 2005), and specific HSPs are induced by oxidative stress (Banzet et al., 1998; Desikan et al., 2001; Volkov et al., 2006; Swindell et al., 2007). Even though the mechanism of reduced ROS in shot1 mutants remains unknown, it is associated with reduced oxidative damage and enhanced heat tolerance. The slow growth rate of rpoTmp-1, ndufs4, and shot1 mutants is likely due to altered metabolism resulting from defective oxidative phosphorylation, but this phenotype does not necessarily lead to reduced ROS and heat tolerance; in this regard, shot1 appears unique. The specific metabolic alteration in shot1 mitochondria remains to be determined, but at the morphological level, we found no evidence of major structural differences between wild-type and shot1-2 mitochondria (see Supplemental Figure 7 online).
SHOT1 Is Required for Normal Mitochondrial Gene Regulation and Alters Nuclear Gene Expression
Concerning its biochemical function, SHOT1 can be modeled to have structural features like MTERF1 and other classes of nucleic acid binding proteins. We experimentally showed that it impacts both transcript levels and copy number of mitochondrial genes (Figures 6A and 7). Although we also saw an effect of shot1 on levels of chloroplast transcripts, this is likely an indirect effect explained by the extensive connection of these two organelles through metabolic pathways and retrograde signaling to the nucleus (Schönfeld et al., 2004; Umbach et al., 2005; Noctor et al., 2007). Transcripts of mitochondrial genes were increased approximately twofold, with the exception of the cytochrome c maturation factor ccmFC. We checked the accumulation of cytochrome c by immunoblot analysis and, contrary to our expectation, shot1-2 showed an increased level of protein (see Supplemental Figure 8 online). It remains to be resolved whether cytochrome c in shot1-2 is functional because CcmFC is proposed to be involved in the assembly of heme with apocytochromes, not in protein accumulation. Regulation of gene expression in organelles by mTERF proteins has been suggested by recent analyses of plastid-targeted mTERFs, SINGLET OXYGEN DEATH ACTIVATOR10, and BELAYA SMERT/RUGOSA2. Loss-of-function mutants of these proteins showed reduction of 16S and 23S rRNAs, which ultimately resulted in reduced plastid translation activity (Meskauskiene et al., 2009; Babiychuk et al., 2011; Quesada et al., 2011). Relative to the shot1-2 mutation, which leads to increased mitochondrial gene transcripts and decreased plastidic gene transcripts, the rug2-1 mutant showed the opposite phenotype (Quesada et al., 2011). In addition, although soldat10 is more resistant to light stress, it is not more resistant to heat stress, and exhibits very different changes in nuclear gene expression than shot1. Determining which of the transcript effects are direct or indirect, whether they result from transcriptional or posttranscriptional mechanisms, and their link to each mTERF phenotype will require further biochemical studies. However, the fact that mTERF proteins from other organisms bind nucleic acids to regulate steps of mitochondrial gene expression supports the hypothesis that plant mTERFs also mediate steps in both plastid and mitochondrial gene expression.
The origin of the shot1 phenotype clearly resides in mitochondria but no doubt requires the observed alterations in nuclear gene expression. Therefore, the metabolic status of shot1 mitochondria has to be communicated to the nucleus, either by a specific retrograde signal or by more general changes in the balance of cellular metabolites. Metabolic alterations in shot1-2 are evident as it has increased transcript levels for genes involved in glycolysis and mitochondrial ETC but decreased transcript levels for chloroplast ETC genes (see Supplemental Figure 9 online). In addition, two-thirds of differentially regulated genes with antioxidant activity were downregulated in the shot1-2 mutant. It is possible that the shot1-2 mutant does not require increased antioxidant activity because it is already producing less ROS. In contrast with antioxidant activity, genes with glutathione S-transferase activity are mostly upregulated in shot1-2. Overexpression of tobacco (Nicotiana tabacum) glutathione S-transferase has been shown to reduce oxidative damage leading to enhanced growth of transgenic tobacco seedlings during stress (Roxas et al., 1997, 2000). AOX has been suggested as the primary target of the retrograde signal from mitochondrial stress (Zarkovic et al., 2005; Rhoads and Subbaiah, 2007; Van Aken et al., 2009a, 2009b), and AOX1a is also found to be regulated by ABI4, which is involved in retrograde signaling from chloroplasts (Koussevitzky et al., 2007; Giraud et al., 2009). Recent articles comparing the transcriptomic responses of mitochondrial mutants or plants treated with ETC inhibitors (Schwarzländer et al., 2012), as well as comparisons of phenotypes of different respiratory complex I mutants (Juszczuk et al., 2012), also highlight the complexity of responses to different mitochondrial dysfunctions. Defining the cause of phenotypic differences between diverse mitochondrial mutants should provide insight into the proposed, but currently unknown, retrograde signaling mechanisms from mitochondria to the nucleus. Furthermore, understanding how the absence of shot1 function can reduce ROS and limit oxidative damage may make it possible to separate the slow growth phenotype from the enhanced thermotolerance with the goal of increasing plant stress tolerance.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana mutants were in the Col-0 background, as described previously, unless otherwise indicated: hot1-3 (Hong and Vierling, 2000), hot1-4 (Lee et al., 2005), hot5-1 (Lee et al., 2008), uvh6-1 (Liu et al., 2003), soldat10 (Landsberg erecta background; Meskauskiene et al., 2009), rpoTmp-1 (Kühn et al., 2009), and ndufs4 (Meyer et al., 2009). Transgenic plants with altered expression of AOX1a protein were provided by James Siedow (Duke University) (Umbach et al., 2005).
Seeds were surface sterilized and plated on nutrient medium (Hong and Vierling, 2000) containing 0.5% (w/v) Suc or planted directly on soil. Seeds were stratified at 4°C for 2 to 3 d. Plants were grown in controlled growth chambers (∼100 µmol m–2 s–1) on a 22°C/18°C, 16-h-day/8-h-night cycle.
For thermotolerance assays, 2.5-d-old dark-grown and 10- to 12-d-old light-grown seedlings were treated according to Hong and Vierling (2000). Soil-grown plants were also tested for chronic heat stress at 37°C for 17 d and returned to 22°C. Sixteen-day-old wild-type and 28-d-old shot1-2 plants were compared to reflect the developmental delay in shot1-2.
SDS-PAGE and Immunoblot Analysis
Total protein extracts were prepared by grinding plant tissues in a 1.5-mL microtube with SDS sample buffer (60 mM Tris-HCl, pH 8.0, 60 mM DTT, 2% SDS, 15% Suc, 5 mM ε-amino-N-caproic acid, and 1 mM benzamidine). Protein concentration was measured with a Coomassie Brilliant Blue binding assay (Hong and Vierling, 2000). For AOX and cytochrome c, mitochondria were enriched by differential centrifugation according to Kühn et al. (2009). Protein samples were separated by SDS-PAGE and blotted onto nitrocellulose membrane for immunoblot analysis. Protein blots were stained with Memcode reversible protein stain (Thermo Scientific) to check transfer efficiency. After the stain was removed, blots were probed with antibodies against HSP101, HSP70s (cytosolic HSP70s), MtHSP70s (mitochondrial HSP70s), PtHSP70s (plastidic HSP70s), small HSPs (HSP17.6C-I and II, HSP26.5-MII, and HSP25.3-P), AOX, or cytochrome c. Anti-HSP101 antibody was raised against a recombinant protein of the N-terminal 145 amino acids of HSP101 protein in rabbits. Anticytosolic, mitochondrial, and plastidic HSP70 antibodies were purchased from Agrisera. Antibodies against small HSPs were raised against a full-length recombinant protein in rabbits. Anti-AOX antibody was kindly provided by James Whelan (University of Western Australia). Anti-pigeon cytochrome c antibody was purchased from Millipore. Blots were incubated with secondary antibodies conjugated with horseradish peroxidase and visualized by enhanced chemiluminescence (Amersham International).
Identification of shot1 Mutant Alleles
The hot1-4 shot1-1 mutant was isolated from a pool of EMS-mutagenized hot1-4 seeds in a screen for suppressors of hot1-4 (Lee et al., 2005). When the original mutant was backcrossed to hot1-4, it was found that the short hypocotyl phenotype cosegregated with the hot1-4 suppressing phenotype. The mutant was also crossed with Col-0 to separate the shot1-1 mutation from hot1-4. Homozygosity of the single shot1-1 mutant was confirmed after crossing back to hot1-4. The single shot1-1 mutant in the Col-0 background was then crossed with Landsberg erecta for mapping. The short hypocotyl phenotype was tracked for genetic mapping. A total of 1053 F2 plants that showed the short hypocotyl phenotype were selected and scored for molecular markers. When the region was narrowed, the open reading frames of all the candidate genes were sequenced.
The shot1-2 mutant was obtained by screening plants from GABI-Kat line 040C09 using PCR primers: P1, 5′-ATTGAAGAATCTGCCTTATGTGCT-3′; MK1-2, 5′-CCTCTCACTTCATAATCTGTGAAG-3′; GABI-T, 5′-CCCATTTGGACGTGAATGTAGACAC-3′.
The genomic SHOT1 DNA construct for the complementation of the shot1 mutations was generated by amplifying the appropriate DNA fragment with the primers MK5-1, 5′-caccTCAGAATCAATGTGTGGCTG-3′ and MK5-2, 5′-ATCTTACCCTTCCTATATCGTCGG-3′, and cloning the fragment into the entry vector pENTR/D-TOPO (Invitrogen). The fragment was then inserted into the pMDC123 binary vector carrying Basta resistance (Curtis and Grossniklaus, 2003) by Gateway LR cloning (Invitrogen). Transformation of Arabidopsis was done by floral dipping (Clough and Bent, 1998).
Microarray Array Analysis
Five samples were prepared from 10-d-old seedlings grown on agar plates with or without heat treatment (AC > 45°C/30 min) using an RNeasy plant mini kit (Qiagen) according to the manufacturer’s protocol. RNA quality and quantity were checked with an Agilent 2100 bioanalyzer. Four biological replicates of the wild-type and shot1-2 samples from each treatment (control and heat stress; 16 RNA samples total) were used in a two-color, dye swap hybridization experiment. Hybridization on a long-oligonucleotide array chip was performed by the Galbraith lab (University of Arizona) according to the Arabidopsis microarray methods (http://ag.arizona.edu/microarray/methods.html) (Zanetti et al., 2005; Zhang et al., 2008). The wild type and mutant, either control or heat stress samples, were directly compared on each of four slides (eight slides total), swapping the dye colors to control for dye effects. Normalization of expression data, analysis using a linear model, and Benjamini and Hochberg false discovery rate correction for multiple comparisons were performed using the LIMMA package (Smyth and Speed, 2003). The data were then further sorted and filtered using Microsoft Excel. The microarray data have been deposited at ArrayExpress (E-MEXP-3322). For GO enrichment analysis, genes with the false discovery rate–adjusted P < 0.05 and log2 fold ratio >0.5 were selected and analyzed using AgriGO, which is publicly accessible at http://bioinfo.cau.edu.cn/agriGO/ (Du et al., 2010). Transcript levels of metabolic pathways were analyzed using MAPMAN (http://mapman.gabipd.org/web/guest/mapman) (Thimm et al., 2004).
Validation of Microarray Data by qRT-PCR
Total RNA was extracted as described in the microarray analysis with additional RNase-free DNase (DNA-free; Ambion) treatment steps according to the manufacturer’s protocol. One microgram of total RNA was reverse-transcribed using oligo(dT) primers with Superscript III reverse transcriptase (Invitrogen). qRT-PCR analysis was performed using iTaq SYBR green supermix (Bio-Rad) and the Applied Biosystems 7300 Real-Time PCR System according to the manufacturer’s instructions. The primer sequences are provided in the Supplemental Table 7 online. Two biological replicates with three technical replicates were run for HSFA2, MtHSC70-1, and HSP23.5-MI. Three biological replicates with three technical replicates were run for all other genes.
Analysis of Organelle Transcripts by qRT-PCR and RNA Gel Blotting
The qRT-PCR analysis was conducted on three independent biological replicates of RNA from the shot1-2 mutant, shot1-2 complemented with genomic DNA, and the wild type. Total RNA was extracted as described in validation of microarray data. The absence of DNA contamination was confirmed by PCR. Reverse transcription was performed with random hexanucleotide primers and Superscript III reverse transcriptase (Invitrogen) as described previously (Chateigner-Boutin et al., 2008). qRT-PCR analysis was conducted in 384-well plates with a Light-Cycler 480 (Roche) using the LightCycler 480 SYBR I Master mix (Roche). Primer pairs for the mitochondrial transcriptome and the chloroplast transcriptome are described in Supplemental Tables 8 and 9 online, respectively, as are the primer pairs used to measure the accumulation of cytosolic transcripts (housekeeping genes).
For RNA gel blot analysis, total RNA extracted from 12-d-old seedlings was separated on 1.2% agarose-formaldehyde gels and transferred onto Hybond N+ nylon membrane (GE Healthcare). Template DNAs for generation of biotinylated complementary RNA (cRNA) probes were amplified from wild-type genomic DNA using Ex-Taq polymerase (Takara) with the primer pairs in Supplemental Table 7 online. The cRNA probes were made using the Ambion MAXIscript T7 kit (Applied Biosystems) and biotin-14-CTP (Invitrogen) according to the manufacturer’s instructions. For the detection of cRNA probes, the chemiluminescent nucleic acid detection module kit (Thermo Scientific) was used.
Estimation of Mitochondrial and Chloroplast Genome Copy Numbers
Total DNA was extracted from 10-d-old plate-grown seedlings using the Qiagen DNeasy plant kit according to the manufacturer’s instructions. Five single-copy genes from each genome (nuclear, plastid, and mitochondrial) were amplified by quantitative PCR (see Supplemental Table 5 online). The same primer sets were used as in the qRT-PCR experiment for the organelle genes. The total DNA concentration in the standard curve ranged between 2 and 500 pg per 5 μL reaction, and the concentration of the samples was ∼50 pg per 5-μL reaction. Each reaction was performed in triplicate (technical repeats) on two biological samples each for the wild type, shot1-2 complemented with SHOT1, and the shot1-2 mutant. The qPCR was performed as described for the organelle transcript analysis. The concentrations obtained for all five amplification products belonging to the same compartment were averaged (geometric mean), and the value of the chloroplast and mitochondrial amplifications was normalized to the value for the nuclear genome.
Microscopy Detection of ROS
Five-day-old plate-grown seedlings were either kept at 22°C or treated at 45°C for 1 h and incubated with 5 µM H2DCFDA (Invitrogen) in 10 mM MES/KOH buffer, pH 5.7, for 30 min. The roots of seedlings were visualized under a confocal microscope (LSM model 510 META microscope; Zeiss). H2DCFDA signals were excited at 488 nm and visualized using band-pass filters at 505 to 570 nm for H2DCFDA.
Thiobarbituric Acid-Reactive-Substances Assay and Detection of Oxidized Proteins
Twelve-day-old plants grown on plates were treated at 45°C for 3 h after heat acclimation (38°C for 1.5 h, followed by 22°C for 2 h). Plant tissue samples were harvested 2 d later. Estimation of lipid oxidation was performed according to Hodges et al. (1999).
Ten- to 12-d-old plate-grown plants were treated with heat stress or with 100 µM paraquat solution for 4 h at 22°C in light. Plants were ground in liquid nitrogen and then extraction buffer (100 mM Tris-HCl, pH 7.5, 0.3 M Suc, 1 mM EDTA, 1 mM PMSF, and 2% 2-mercaptoethanol) was added at a ratio of 100 mg tissue to 300 μL buffer. Protein extracts were centrifuged at 1000g for 10 min to remove cell debris and quantified with a Coomassie Brilliant Blue dye binding assay (Hong and Vierling, 2000). Oxidized proteins were detected using an OxyBlot protein oxidation detection kit (Chemicon International) according to the manufacturer’s instructions.
Measurement of Paraquat Sensitivity
Plants were grown vertically for 4 d on nutrient medium containing 0.5% (w/v) Suc and then transferred to plates containing different concentrations of paraquat. Root growth was measured after 3 d and compared with growth on plates without paraquat.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or the GenBank/EMBL data libraries under accession numbers provided in Supplemental Tables 3 to 9 online. The microarray data can be found at ArrayExpress under accession number E-MEXP-3322.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Segregation of the shot1 Mutation, Map-Based Cloning, and RT-PCR in shot1 Mutants.
Supplemental Figure 2. SHOT1-GFP Localizes to Mitochondria.
Supplemental Figure 3. Complementation Analysis and HSP101 Protein Level in shot1 Mutants.
Supplemental Figure 4. Heat Stress Phenotype of Transgenic Plants with Altered AOX1a Protein Level.
Supplemental Figure 5. Plants with the shot1-2 Mutation Have Slightly Higher Manganese Superoxide Dismutase Activity but Lower Catalase Activity.
Supplemental Figure 6. The Chloroplast mTERF Mutant soldat10 Does Not Show Increased Heat Tolerance.
Supplemental Figure 7. Mitochondria Appear Normal in shot1-2.
Supplemental Figure 8. Immunoblot Analysis of Cytochrome c Level in the shot1 Mutants.
Supplemental Figure 9. Changes in Transcript Levels of Genes Involved in Glycolysis, Mitochondrial ETC, and Chloroplastic ETC of 22°C-Grown Plants.
Supplemental Table 1. GO Analysis of Differentially Regulated Transcripts in 22°C-Grown Wild-Type and shot1-2 Plants.
Supplemental Table 2. GO Analysis of Differentially Regulated Transcripts in Heat-Stressed Wild-Type and shot1-2 Plants.
Supplemental Table 3. Heat-Responsive Genes That Are Differentially Regulated in shot1-2.
Supplemental Table 4. Genes Upregulated in Both shot1-2 and soldat10.
Supplemental Table 5. List of Genes and Primer Sequences Used in Estimation of Mitochondrial and Plastid Genome Copy Numbers.
Supplemental Table 6. Changes in Transcript Levels of Mitochondrial ETC Components.
Supplemental Table 7. List of Primer Sequences Used in the qRT-PCR for the Validation of Microarray Experiments and Generation of Biotinylated Complementary RNA Probes.
Supplemental Table 8. List of Primer Sequences Used in the qRT-PCR of Mitochondrial Transcripts.
Supplemental Table 9. List of Primer Sequences Used in the qRT-PCR of Plastid Transcripts.
Supplemental Data Set 1. Microarray Data Comparing the Transcriptome of shot1-2 with That of the Wild Type under Normal Growth Conditions.
Supplemental Data Set 2. Microarray Data Comparing the Transcriptome of shot1-2 with That of the Wild Type under Heat Stress Conditions.
Supplementary Material
Acknowledgments
We thank Patricia Jansma for assistance with confocal microscopy and William Day for imaging mitochondria with transmission electron microscopy. We thank Jeremy D. Edwards for advice and programming for statistical analysis of the microarray data. We also thank Carol Dieckmann, Frans Tax, and Eman Basha for critically reading the article. This work was supported by the Korea Science and Engineering Foundation Grant C00120 to M.K., the Australian Research Council Grant CE0561495 to I.S., and the U.S. Department of Energy Grant DE-FG02-99ER20338 to E.V.
AUTHOR CONTRIBUTIONS
M.K. designed and performed research, analyzed data, and wrote the article. U.L. designed and performed research. I.S. analyzed data. C.C.d.F.-S. performed research and analyzed data. E.V. designed research, analyzed data, and wrote the article.
Glossary
- HSP
heat shock protein
- ROS
reactive oxygen species
- EMS
ethyl methanesulfonate
- mTERF
mitochondrial transcription termination factor
- GFP
green fluorescent protein
- Col-0
Columbia-0
- GO
Gene Ontology
- qRT-PCR
quantitative RT-PCR
- H2DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- ETC
electron transport
- cRNA
complementary RNA
References
- Alfonso M., Yruela I., Almárcegui S., Torrado E., Pérez M.A., Picorel R. (2001). Unusual tolerance to high temperatures in a new herbicide-resistant D1 mutant from Glycine max (L.) Merr. cell cultures deficient in fatty acid desaturation. Planta 212: 573–582 [DOI] [PubMed] [Google Scholar]
- Atkin O.K., Macherel D. (2009). The crucial role of plant mitochondria in orchestrating drought tolerance. Ann. Bot. (Lond.) 103: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E., Vandepoele K., Wissing J., Garcia-Diaz M., De Rycke R., Akbari H., Joubès J., Beeckman T., Jänsch L., Frentzen M., Van Montagu M.C.E., Kushnir S. (2011). Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet N., Richaud C., Deveaux Y., Kazmaier M., Gagnon J., Triantaphylidès C. (1998). Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J. 13: 519–527 [DOI] [PubMed] [Google Scholar]
- Brandalise M., Maia I.G., Borecký J., Vercesi A.E., Arruda P. (2003). Overexpression of plant uncoupling mitochondrial protein in transgenic tobacco increases tolerance to oxidative stress. J. Bioenerg. Biomembr. 35: 203–209 [DOI] [PubMed] [Google Scholar]
- Cámara Y., et al. (2011). MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A.-L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L.F., Lurin C., Small I., León P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K.L., Sappl P.G., Elhafez D., Millar A.H., Day D.A., Whelan J. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J.F., Lopez-Delgado H., Foyer C.H., Scott I.M. (1998). Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 116: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day D.A., Wiskich J.T. (1995). Regulation of alternative oxidase activity in higher plants. J. Bioenerg. Biomembr. 27: 379–385 [DOI] [PubMed] [Google Scholar]
- de Longevialle A.F., Meyer E.H., Andrés C., Taylor N.L., Lurin C., Millar A.H., Small I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., A-H-Mackerness S., Hancock J.T., Neill S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.M., Wickner S. (2009). Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem. Sci. 34: 40–48 [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38 (Web Server issue): W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C., Garmier M., Noctor G., Mathieu C., Chétrit P., Foyer C.H., de Paepe R. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., Nyström T. (2007). Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21: 2410–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. (1998). Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Groves M.R., Barford D. (1999). Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9: 383–389 [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis subcellular database. Nucleic Acids Res. 35 (Database issue): D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges D.M., DeLong J.M., Forney C.F., Prange R.K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Hong S.-W., Vierling E. (2000). Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen A.K., Pohjoismäki J.L.O., Reyes A., Wanrooij S., Yasukawa T., Karhunen P.J., Spelbrink J.N., Holt I.J., Jacobs H.T. (2007). The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 35: 6458–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby R.P., Taylor N.L., Millar A.H. (2011). The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci. 16: 614–623 [DOI] [PubMed] [Google Scholar]
- Juszczuk I.M., Szal B., Rychter A.M. (2012). Oxidation-reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction. Plant Cell Environ. 35: 296–307 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kristiansen K.A., Jensen P.E., Møller I.M., Schulz A. (2009). Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H2DCFDA and confocal laser microscopy. Physiol. Plant. 136: 369–383 [DOI] [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., Attardi G. (1989). Termination of transcription in human mitochondria: Identification and purification of a DNA binding protein factor that promotes termination. Cell 58: 391–397 [DOI] [PubMed] [Google Scholar]
- Kühn K., Richter U., Meyer E.H., Delannoy E., de Longevialle A.F., O’Toole N., Börner T., Millar A.H., Small I.D., Whelan J. (2009). Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin E.V., Karpova O.V., Elthon T.E., Newton K.J. (2004). Mitochondrial respiratory deficiencies signal up-regulation of genes for heat shock proteins. J. Biol. Chem. 279: 20672–20677 [DOI] [PubMed] [Google Scholar]
- Larkindale J., Hall J.D., Knight M.R., Vierling E. (2005). Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Knight M.R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Lee H., Xiong L., Zhu J.K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U., Wie C., Escobar M., Williams B., Hong S.-W., Vierling E. (2005). Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell 17: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U., Wie C., Fernandez B.O., Feelisch M., Vierling E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Hong S.-W., Escobar M., Vierling E., Mitchell D.L., Mount D.W., Hall J.D. (2003). Arabidopsis UVH6, a homolog of human XPD and yeast RAD3 DNA repair genes, functions in DNA repair and is essential for plant growth. Plant Physiol. 132: 1405–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Dolgner S.J., Hall T.M.T. (2009). Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 19: 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Cho J., Cesare A.J., Griffith J.D., Attardi G. (2005). Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 123: 1227–1240 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Würsch M., Laloi C., Vidi P.-A., Coll N.S., Kessler F., Baruah A., Kim C., Apel K. (2009). A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J. 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Meyer E.H., Tomaz T., Carroll A.J., Estavillo G., Delannoy E., Tanz S.K., Small I.D., Pogson B.J., Millar A.H. (2009). Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Whelan J., Soole K.L., Day D.A. (2011). Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Noctor G., De Paepe R., Foyer C.H. (2007). Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 12: 125–134 [DOI] [PubMed] [Google Scholar]
- Parsell D.A., Kowal A.S., Singer M.A., Lindquist S. (1994). Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478 [DOI] [PubMed] [Google Scholar]
- Queitsch C., Hong S.-W., Vierling E., Lindquist S. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Sarmiento-Mañús R., González-Bayón R., Hricová A., Pérez-Marcos R., Graciá-Martínez E., Medina-Ruiz L., Leyva-Díaz E., Ponce M.R., Micol J.L. (2011). Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 68: 738–753 [DOI] [PubMed] [Google Scholar]
- Rasmusson A.G., Soole K.L., Elthon T.E. (2004). Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu. Rev. Plant Biol. 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Rayapuram N., Hagenmuller J., Grienenberger J.M., Bonnard G., Giegé P. (2008). The three mitochondrial encoded CcmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J. Biol. Chem. 283: 25200–25208 [DOI] [PubMed] [Google Scholar]
- Rhoads D.M., Subbaiah C.C. (2007). Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Roberti M., Bruni F., Loguercio Polosa P., Manzari C., Gadaleta M.N., Cantatore P. (2006). MTERF3, the most conserved member of the mTERF-family, is a modular factor involved in mitochondrial protein synthesis. Biochim. Biophys. Acta 1757: 1199–1206 [DOI] [PubMed] [Google Scholar]
- Roxas V.P., Lodhi S.A., Garrett D.K., Mahan J.R., Allen R.D. (2000). Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 41: 1229–1234 [DOI] [PubMed] [Google Scholar]
- Roxas V.P., Smith R.K., Jr, Allen E.R., Allen R.D. (1997). Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15: 988–991 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan V., Örvar B.L., Beyerly J., Hirt H., Dhindsa R.S. (2002). Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31: 629–638 [DOI] [PubMed] [Google Scholar]
- Schönfeld C., Wobbe L., Borgstädt R., Kienast A., Nixon P.J., Kruse O. (2004). The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 279: 50366–50374 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., König A.C., Sweetlove L.J., Finkemeier I. (2012). The impact of impaired mitochondrial function on retrograde signalling: A meta-analysis of transcriptomic responses. J. Exp. Bot. 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Solomon J.M., Rossi J.M., Golic K., McGarry T., Lindquist S. (1991). Changes in hsp70 alter thermotolerance and heat-shock regulation in Drosophila. New Biol. 3: 1106–1120 [PubMed] [Google Scholar]
- Sweetlove L.J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N.L., Baxter C.J., Eickmeier I., Fernie A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103: 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell W.R., Huebner M., Weber A.P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Umbach A.L., Fiorani F., Siedow J.N. (2005). Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach A.L., Ng V.S., Siedow J.N. (2006). Regulation of plant alternative oxidase activity: A tale of two cysteines. Biochim. Biophys. Acta 1757: 135–142 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Giraud E., Clifton R., Whelan J. (2009a). Alternative oxidase: A target and regulator of stress responses. Physiol. Plant. 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Zhang B., Carrie C., Uggalla V., Paynter E., Giraud E., Whelan J. (2009b). Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G.C., McLntosh L. (1996). Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol. 111: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. (1991). The roles of heat-shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 579–620 [Google Scholar]
- Volkov R.A., Panchuk I.I., Mullineaux P.M., Schöffl F. (2006). Heat stress-induced H(2)O (2) is required for effective expression of heat shock genes in Arabidopsis. Plant Mol. Biol. 61: 733–746 [DOI] [PubMed] [Google Scholar]
- Wenz T., Luca C., Torraco A., Moraes C.T. (2009). mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 9: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Winkler G.S., Araújo S.J., Fiedler U., Vermeulen W., Coin F., Egly J.-M., Hoeijmakers J.H.J., Wood R.D., Timmers H.T.M., Weeda G. (2000). TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J. Biol. Chem. 275: 4258–4266 [DOI] [PubMed] [Google Scholar]
- Wissing D., Jäättelä M. (1996). HSP27 and HSP70 increase the survival of WEHI-S cells exposed to hyperthermia. Int. J. Hyperthermia 12: 125–138 [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E., Mejia E., Byrnes J., Hambardjieva E., Garcia-Diaz M. (2010). Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141: 982–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.E., Chang I.F., Gong F., Galbraith D.W., Bailey-Serres J. (2005). Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic J., Anderson S.L., Rhoads D.M. (2005). A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol. Biol. 57: 871–888 [DOI] [PubMed] [Google Scholar]
- Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. (2008). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 247: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.