Arabidopsis thaliana GROWTH-REGULATING FACTOR7 (GRF7) was identified as a transcriptional repressor of DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2A and other osmotic stress–responsive genes under normal growth conditions. This work proposes a mechanism in which GRF7 minimizes adverse effects on plant growth by repressing the expression of stress-responsive genes under favorable conditions.
Abstract
Arabidopsis thaliana DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) functions as a transcriptional activator that increases tolerance to osmotic and heat stresses; however, its expression also leads to growth retardation and reduced reproduction. To avoid these adverse effects, the expression of DREB2A is predicted to be tightly regulated. We identified a short promoter region of DREB2A that represses its expression under nonstress conditions. Yeast one-hybrid screening for interacting factors identified GROWTH-REGULATING FACTOR7 (GRF7). GRF7 bound to the DREB2A promoter and repressed its expression. In both artificial miRNA-silenced lines and a T-DNA insertion line of GRF7, DREB2A transcription was increased compared with the wild type under nonstress conditions. A previously undiscovered cis-element, GRF7-targeting cis-element (TGTCAGG), was identified as a target sequence of GRF7 in the short promoter region of DREB2A via electrophoretic mobility shift assays. Microarray analysis of GRF7 knockout plants showed that a large number of the upregulated genes in the mutant plants were also responsive to osmotic stress and/or abscisic acid. These results suggest that GRF7 functions as a repressor of a broad range of osmotic stress–responsive genes to prevent growth inhibition under normal conditions.
INTRODUCTION
Unlike animals, plants cannot avoid environmental stresses by moving to a different location. Instead, they have developed complicated mechanisms for sensing and responding to environmental stresses, such as drought, high salinity, and extreme temperatures. A number of genes have been reported that allow plants to tolerate and overcome these unfavorable circumstances (Thomashow, 1999; Zhu, 2002; Bartels and Sunkar, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006; Mittler and Blumwald, 2010).
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) is one such gene in Arabidopsis thaliana. Harboring an ethylene-responsive element binding factor/APETALA2-type DNA binding domain, DREB2A functions as a transcriptional activator by interacting with a cis-element, dehydration responsive element/C-repeat (A/GCCGAC) in the promoters of downstream genes (Liu et al., 1998; Sakuma et al., 2002). DREB2A is induced by osmotic and high-temperature stresses; however, overexpression of full-length DREB2A does not result in the activation of downstream genes (Liu et al., 1998; Sakuma et al., 2006b). By contrast, overexpression of a constitutively active form of DREB2A (DREB2A CA) that lacks a negative regulatory domain confers enhanced tolerance to dehydration and high temperature in plants (Sakuma et al., 2006a, 2006b). Furthermore, T-DNA insertion mutant plants of DREB2A were shown to be less tolerant of high temperature (Sakuma et al., 2006b). Genome-wide transcriptomic analyses revealed that DREB2A regulates a number of genes involved in osmotic and/or heat stress tolerance, with many of those genes harboring more than one dehydration responsive element sequence in their promoter region (Sakuma et al., 2006a, 2006b).
The overexpression of DREB2A CA also adversely affects plant growth. Transgenic plants overexpressing DREB2A CA showed growth retardation, dwarfism, and diminished reproduction (Sakuma et al., 2006a). Therefore, multiple regulatory steps that prevent DREB2A activity under nonstress conditions are expected to exist, and several reports have begun to elucidate these steps. Posttranslational regulation of DREB2A involving its degradation under nonstress conditions was reported; overproduced DREB2A was not stable under nonstress conditions but was stabilized under heat stress conditions (Sakuma et al., 2006a, 2006b). By contrast, DREB2A CA stably accumulated in the nucleus even under nonstress conditions (Sakuma et al., 2006a). Subsequent research on DREB2A-INTERACTING PROTEIN1 and DRIP2 revealed their roles in the rapid degradation of DREB2A through 26S proteasome-dependent proteolysis under nonstress conditions (Qin et al., 2008). Recently, reports on Arabidopsis MEDIATOR25, a newly described DREB2A-interacting cofactor protein, suggested another method of regulation of DREB2A activation (Elfving et al., 2011; Blumberg et al., 2012).
DREB2A expression is also regulated at the transcriptional level. When Arabidopsis plants are exposed to abiotic stress conditions, such as heat or high osmolarity, the transcript level of DREB2A increases rapidly and reaches up to 250-fold of the basal transcript level (Liu et al., 1998; Sakuma et al., 2006b). The expression patterns of DREB2A in response to heat and osmotic stresses are different from each other, which suggests the presence of two separate transcriptional regulation mechanisms in Arabidopsis, one heat stress specific and the other osmotic stress specific (Sakuma et al., 2006b). Recently, three heat shock transcription factors, HsfA1a, HsafA1b, and HsfA1d, were demonstrated to play critical roles in the heat stress–induced expression of DREB2A via a heat shock element in the promoter region (Liu et al., 2011; Yoshida et al., 2011). In the osmotic stress response, an abscisic acid–responsive element (ABRE) and a coupling element 3-like sequence in the promoter region were identified as essential cis-elements (Kim et al., 2011). The ABRE binding proteins/factors (AREBs/ABFs) were found to be major transcription factors that activate the transcription of DREB2A via the ABRE sequence in response to osmotic stress; however, the transcription factors that recognize the coupling element 3-like sequence have not been identified (Kim et al., 2011).
Despite remarkable advances in recent years, we still have little information on how plants regulate the expression of DREB2A to bring about the precise and robust responses to the rapidly changing environment. In this article, we report a region of the DREB2A promoter that represses the activity of the promoter. We isolated GROWTH-REGULATING FACTOR7 (GRF7) via yeast one-hybrid screening as a protein that binds to this region of the DREB2A promoter and represses DREB2A promoter activity under nonstress conditions. Microarray analysis showed that GRF7 negatively regulates the expression of a number of osmotic stress–responsive genes under nonstress conditions, which might sustain the growth of Arabidopsis plants under normal growth conditions by minimizing the adverse effects of the stress-related genes, including DREB2A.
RESULTS
Region S Represses DREB2A Promoter Activity
In a previous report, we performed a truncation analysis of the DREB2A promoter under dehydration stress conditions (Kim et al., 2011). Arabidopsis plants containing various promoter-GUS (for β-glucuronidase) constructs were exposed to controlled dehydration conditions, and the transcript levels of endogenous DREB2A and GUS were measured by quantitative RT-PCR (qRT-PCR). The absence of the region between −314 and −271 bp from the transcription initiation site significantly enhanced promoter activity before the stress treatment (Figure 1; Kim et al., 2011). We hypothesized that this region has an important role in the negative regulation of DREB2A expression under nonstress conditions and termed it Region S (Suppression) (Figure 1A). To validate the role of Region S, a construct containing 1.8 kb of the DREB2A promoter without Region S (N0ΔS) was prepared, and its promoter activity was evaluated and compared with that of other intact or truncated promoter constructs (Figure 1B). The transcript levels of GUS were significantly higher in plants harboring the constructs without Region S (N3 and N0ΔS) than in those with constructs containing Region S (N2 and N0) under nonstress conditions (Figure 1B). The absence of Region S had only a moderate impact on the promoter activity after 5 h of dehydration (Figure 1B).
Figure 1.
Activity of Region S in the DREB2A Promoter.
(A) Schematic diagram of the DREB2A promoter-GUS constructs used in this study and the sequence information for Region S. Numbers indicate locations relative to the transcription start site.
(B) qRT-PCR analysis of various promoter-GUS constructs. The left panel presents the schematic structure of the promoter in each construct. The right panel shows the relative GUS transcript levels before (closed bar) and after (open bar) dehydration treatment. Data represent the means of triplicate biological repeats, and the error bars indicate sd. Asterisks denote significant differences (multiple t test with Bonferroni correction; *P < 0.05).
GRF7 Directly Interacts with Region S and Represses DREB2A Promoter Activity
To identify transcription factors that bind to and regulate Region S, we searched for known cis-elements in Region S using internet-based cis-element databases. However, no cis-element was reliably predicted. Therefore, yeast one-hybrid (Y1H) screening was performed using a cDNA library composed of quantitatively equalized Arabidopsis transcription factors (Mitsuda et al., 2010). From the screening of more than 4 × 104 colonies, we obtained 18 positive colonies, eight of which contained the coding region of GRF7 (see Supplemental Table 1 online). Other candidates included single members of the MYB and IAA transcription factor families, but we failed to validate their binding affinities with Region S in further trials (see Supplemental Table 1 online). Then we examined the interaction of all nine Arabidopsis GRF family members (GRF1 to GRF9; Kim et al., 2003; see Supplemental Figure 1 and Supplemental Data Set 1 online) with Region S in yeast cells, but only GRF7 displayed a positive signal (Figure 2A). To verify the capability of GRF7 to interact with Region S in planta, we performed chromatin immunoprecipitation (ChIP) assays using transgenic Arabidopsis plants expressing GFP-fused GRF7 under the cauliflower mosaic virus (CaMV) 35S promoter (35Spro:GFP-GRF7) (see Supplemental Figure 2 online). Significant enrichment of the DREB2A promoter fragments was observed in the 35Spro:GFP-GRF7 plants compared with the negative control, supporting the idea that GRF7 could localize to the DREB2A promoter in planta (see Supplemental Figure 2 online).
Figure 2.
GRF7 as a Candidate Interactor with Region S.
(A) Y1H analysis using Region S as bait and GRF family members as prey. Representative growth status of yeast cells is shown on SD/-UHL agar media with or without 3-AT from triplicate independent trials. Numbers on the top side of each photograph indicate relative densities of the cells. DPI, days postinoculation.
(B) Transient expression assay using Arabidopsis mesophyll protoplasts. The top panel shows the schematic structures for the effector and reporters. The bottom panel shows the mean activity ratios of GUS and luciferase (LUC) from quadruplicate biological repeats. Values represent GUS/LUC ratios relative to mean values obtained using the empty effector plasmid (VEC) for each reporter construct. The error bars indicate sd. Asterisks denote significant difference compared with the VEC sample (multiple t test with Bonferroni correction; *P < 0.05).
To establish a role for GRF7 in the regulation of DREB2A expression through Region S, its activity was evaluated by transient expression in Arabidopsis mesophyll protoplasts (Figure 2B). Full-length GRF7 driven by the CaMV 35S promoter was used as an effector, and two DREB2A promoter-GUS constructs were used as reporters (Figure 2B). The promoter activity of the N0 construct was decreased by 39.5% when it was cotransfected with GRF7, whereas no significant difference was observed for the N0ΔS construct (Figure 2B).
Expression Profile of GRF7
The spatial expression pattern of GRF7 was analyzed (Figure 3). Histochemical GUS staining showed that a 3-kb region of the GRF7 promoter was not active in dry seeds (Figure 3B). In 1- and 2-week-old seedlings, the staining was mainly detected in developing tissues and at veins in leaf blades and petioles of true leaves, though rarely at cotyledons (Figures 3C to 3E). In reproductive tissues of 5-week-old plants, GUS staining was mainly observed around the inflorescence meristem, the pistil of buds, and the replum of siliques and only rarely in other floral organs or maturing siliques (Figures 3F to 3H). We examined the subcellular localization pattern of GRF7 using the 35Spro:GFP-GRF7 transgenic Arabidopsis. Fluorescence signals were detected in cellular nuclei, suggesting that GRF7 is a nuclear-localized protein (Figure 3J).
Figure 3.
Spatial Expression Pattern of GRF7.
(A) to (H) Histochemical GUS assay using 3 kb of the GRF7 promoter region.
(A) Schematic view of the GRF7pro:GUS construct.
(B) to (H) GUS assays in dry seeds (B), a 1-week-old seedling (C), developing leaves (D), a mature leaf of 2-week-old seedlings (E), inflorescence apex (F), a developing silique (G), and a mature silique (H) of 5-week-old plants. The arrowhead in (F) indicates the inflorescence meristem. Bars = 0.5 mm.
(I) and (J) Intracellular localization of GFP-fused GRF7 in root cells of 2-week-old transgenic plants.
(I) Schematic view of the 35Spro:GFP-GRF7 construct.
(J) Insets are enlargements of the boxed regions, showing from left to right: differential interference contrast (DIC) image, fluorescence from 4′,6-diamidino-2-phenylindole (DAPI) staining, GFP fluorescence, and the merged image of GFP and 4′,6-diamidino-2-phenylindole fields. Bars = 50 μm.
Subsequently, the expression profile of GRF members in response to various stimuli was analyzed with the help of the public microarray database Genevestigator (Hruz et al., 2008, 2011; https://www.genevestigator.com/). We found that the expression level of GRF7 is relatively low when compared with other GRF family members and representative stress-responsive transcription factors (see Supplemental Figure 3 online). In addition, the expression of GRF7 displays no obvious response to any plant hormones or abiotic stimuli (see Supplemental Figure 3 online). We evaluated GRF7 transcript levels under abscisic acid (ABA) or osmotic stress conditions, but no significant difference was detected in response to such treatments (see Supplemental Figure 3 online).
GRF7 Deficiency Elevates DREB2A Transcription under Nonstress Conditions
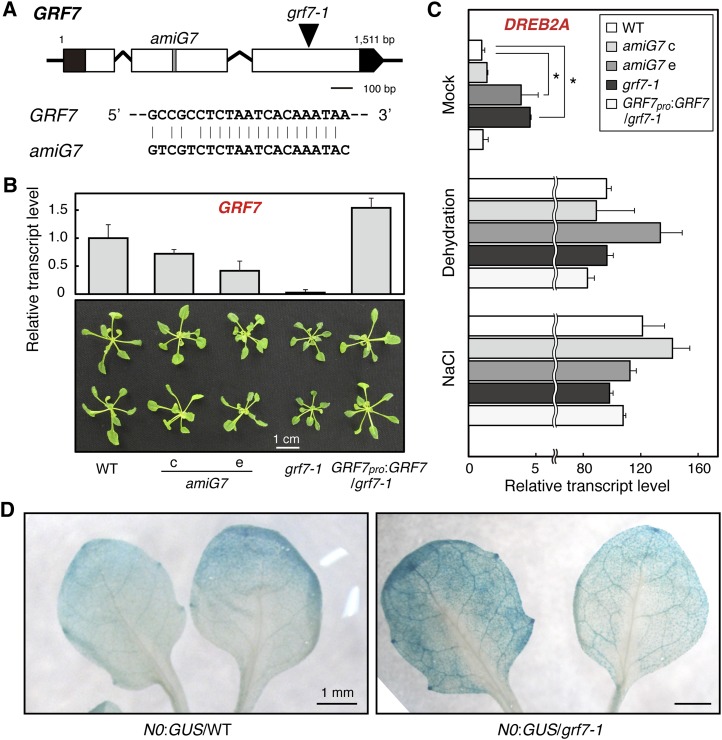
To analyze the function of GRF7 in planta, we obtained lines that were GRF7 deficient due to specific artificial miRNA expression (amiG7) or T-DNA insertion into GRF7 (grf7-1) (Figure 4A). Disturbed expression of GRF7 led to slightly smaller plant size than the wild type, and the size variation corresponded to the amount of GRF7 transcript remaining (Figure 4B; see Supplemental Figure 4 online). These lines were treated with dehydration or high salinity stress, and their DREB2A transcript levels were analyzed (Figure 4C). In a mock treatment, these GRF7-disrupted lines showed elevation of DREB2A transcript levels, which inversely corresponded with the GRF7 transcript levels (Figures 4B and 4C). The elevation of the DREB2A transcript levels and the reduced plant sizes in the grf7-1 mutant were rescued by introducing a construct expressing the coding region of GRF7 driven by its native promoter (GRF7pro:GRF7/grf7-1; Figures 4B and 4C; see Supplemental Figure 4 online). Under stress conditions, DREB2A transcript levels were less affected by these mutations (Figure 4C). These phenomena agreed with the results obtained from the truncation analyses of Region S in the DREB2A promoter (Figures 1 and 2).
Figure 4.
Effect of Reduced Expression of GRF7 on DREB2A Transcription.
(A) Schematic view of the GRF7 locus. The target site of amiG7 and the T-DNA insertion site of grf7-1 are indicated. Open boxes and angled lines indicate the coding sequence and introns, respectively. Closed boxes indicate untranslated regions. An alignment of amiG7 and the target sequence is shown.
(B) GRF7 transcript levels in two artificial miRNA lines (amiG7 c and e), a T-DNA insertion line (grf7-1), and a complementation line of grf7-1 (GRF7pro:GRF7/grf7-1). Data represent means of triplicate biological repeats, and the error bars indicate sd. Representative images of 3-week-old rosettes are shown for each line. WT, the wild type.
(C) DREB2A transcript levels after mock, dehydration, or high salinity stress treatments. Data represent means of triplicate biological repeats, and the error bars indicate sd. Asterisks denote significant differences (multiple t test with Bonferroni correction; *P < 0.01).
(D) Spatial expression pattern of DREB2A in the wild type and the grf7-1 mutant. The 1.8-kb DREB2A promoter-GUS construct (N0:GUS) was introduced into the wild type and grf7-1 mutant, and histochemical GUS assays were performed using 2-week-old seedlings of each plant. Representative images of GUS staining are shown.
Additionally, we introduced a construct in which GUS expression was driven by 1.8 kb of the DREB2A promoter (N0:GUS; Figure 1) into wild-type and grf7-1 backgrounds to evaluate the effect of GRF7 deficiency on the spatial expression pattern of DREB2A (Figure 4D). Compared with the GUS staining in the wild type, additional staining was observed in the grf7-1 plants at leaf veins, where clear staining was also observed in the GRF7pro:GUS plants (Figures 3E and 4D). This result indicates that GRF7 is a major negative regulator of DREB2A expression, at least in leaf veins. Furthermore, we used grf7-1 protoplasts for transient expression assays, as endogenous GRF7 (present in the wild type) could affect reporter activity even in the absence of the effector. The Region S–dependent repression of the reporter activity by coexpression of GRF7 was indeed more evident in grf7-1 protoplasts than in wild-type protoplasts (see Supplemental Figure 5 online; compare with Figure 2B). From these data, it is clear that the transcriptional repression of DREB2A by Region S is related to GRF7.
We also analyzed transgenic Arabidopsis plants expressing the 35Spro:GFP-GRF7 construct (see Supplemental Figures 2 and 6 online) and T-DNA insertion lines of GRF8, which is the most closely related family member to GRF7 (grf8-1 and grf8-2; see Supplemental Figures 1 and 7 online). No significant differences in the DREB2A transcript levels were found under nonstress or stress conditions in any of those plants (see Supplemental Figures 2, 4, 6, and 7 online). Along with the data from the DNA binding and transient expression assays (Figure 2), these results suggest that GRF7 is the main factor repressing the DREB2A promoter under nonstress conditions via interaction with Region S.
QLQ and WRC Domains Are Necessary for GRF7 Repressor Activity
Arabidopsis GRF family members were first identified as putative orthologs of rice (Oryza sativa) (Os-GRF1; van der Knaap et al., 2000). GRF family proteins contain two common domains in their N-terminal regions, QLQ and WRC. The QLQ domain has been suggested to have the ability to interact with other proteins (van der Knaap et al., 2000; Kim et al., 2003). The WRC domain is thought to function in DNA binding due to the presence of a conserved C3H-type zinc finger motif and a nuclear localization signal (van der Knaap et al., 2000; Kim et al., 2003). The C-terminal regions of the family members have limited similarity to each other although six members of the family, including GRF7, contain a conserved GGPL motif (see Supplemental Figure 1A online; Kim et al., 2003). To elucidate the function of each domain in GRF7, truncated derivatives of GRF7 were prepared, and their binding affinities for Region S were examined using Y1H analysis. As shown in Figure 5A, we obtained positive signals from full-length GRF7 (G7FL) and a QLQ-truncated form of GRF7 (G7ΔN), but not from a WRC-truncated form (G7ΔBD), suggesting an essential role for the WRC domain in binding to Region S. Nonetheless, the WRC domain alone (G7BD) did not show a positive signal. Next, we searched for sequences that allow binding to Region S in cooperation with the WRC domain using GRF7 derivatives containing the WRC domain and flanking sequences (Figure 5A; G7ΔC, G7ΔC2, and G7ΔC3). However, none of the tested derivatives gave rise to a binding signal.
Figure 5.
Truncation Analysis of GRF7.
(A) Y1H analysis using Region S as bait and various truncated forms of GRF7 as prey. The left panel shows schematic views of the truncated GRF7 derivatives. The right panel shows the representative growth status of yeast cells on SD/-UHL agar media with or without 3-AT from triplicate independent trials. Numbers on the top side of each photograph indicate relative densities of the cells. DPI, days postinoculation.
(B) Transient expression assays using Arabidopsis mesophyll protoplasts. Top and left panels show the schematic structures of the reporter and effectors, respectively. Right panel shows mean activity ratios of GUS and luciferase (LUC) from quadruplicate biological repeats, relative to the mean value obtained using the empty vector (VEC). The error bars indicate sd. The asterisk denotes a significant difference compared with VEC (multiple t test with Bonferroni correction; *P < 0.01). G, GGPL motif; Q, QLQ domain; W, WRC domain.
We analyzed the importance of the QLQ and WRC domains in GRF7 repressor activity using the transient expression system (Figure 5B). Full-length or truncated forms of GRF7 driven by the CaMV 35S promoter were used as effectors, and the DREB2A promoter-GUS construct (N0:GUS) was used as a reporter. Whereas G7FL repressed the promoter activity, neither G7ΔBD nor G7ΔN gave rise to such repression (Figure 5B). G7ΔBD did not show an interaction signal with Region S in Y1H, although G7ΔN did (Figure 5A). Taking into account these data, it is assumed that the transcriptional repression activity of GRF7 is based on a direct interaction with target DNA and might also require the function of the QLQ domain. The effect of ABA on the activity of GRF7 in Arabidopsis protoplasts was also examined, but GRF7 showed similar repression activity under control and ABA treatment conditions (see Supplemental Figure 8 online).
TGTCAGG Is a Target Sequence of GRF7
To date, target sequences of GRF family proteins have not been identified. We attempted to elucidate the target sequence of GRF7 in Region S by an electrophoretic mobility shift assay (EMSA) using glutathione S-transferase (GST)-tagged G7ΔN purified from Escherichia coli. First, Region S was divided into two overlapping fragments (Figure 6A, E1 and E2). The 32P-labeled E1 fragment showed delayed mobility due to specific interaction with the G7ΔN protein, whereas the 32P-labeled E2 fragment did not produce any shifted bands (Figure 6B). Next, to narrow down the GRF7 target sequence within E1, partially mutated E1 derivatives (Figure 6C, M1 to M5) were labeled with 32P and used for EMSA. The M4 and M5 oligomers lost the ability to bind the G7ΔN protein (Figure 6D). Finally, 13-bp sequences around the mutated region in M4 and M5 were screened with a different mutation series (Figure 6C, Ma to Mf). Using these mutated fragments as unlabeled competitors, we analyzed the affinities of these fragments for G7ΔN (Figure 6E). The unlabeled E1 fragment competed with the labeled E1 fragment, whereas the competition was not effective when unlabeled M4 and M5 fragments were tested (Figure 6E). Similarly, the competition was clear using the Ma and Mf fragments, whereas it was less evident with fragments Mb to Me (Figure 6E). From this series of trials, an essential 7-bp nucleotide (TGTCAGG) emerged as the target sequence of GRF7 in the DREB2A promoter (Figure 6C).
Figure 6.
EMSA for a GRF7 Target Sequence in the DREB2A Promoter.
(A) Relative locations and sequence information for the E1 and E2 fragments in Region S of the DREB2A promoter.
(B) Binding of the recombinant GRF7 protein (GST-G7ΔN) with Region S of the DREB2A promoter fragments in EMSA. Unlabeled probe DNA fragments were included as competitors in the indicated ratios. The shifted complexes indicate probes that exhibited delayed mobility due to interaction with recombinant GRF7. The results were reproduced in two independent experiments.
(C) Sequences of the E1 fragment and its derivatives used for EMSA. The gray block and underlined region (capitalized) indicate a putative target sequence of GRF7 deduced from the EMSA results.
(D) and (E) Rough (D) and fine (E) mapping of the binding sequence of GST-G7ΔN in the E1 fragment of the DREB2A promoter. Data are presented as in (B). The results were reproduced in two independent experiments.
To confirm the EMSA data, nucleotide substitutions were introduced into Region S in the N0 reporter construct and the effects were tested in Arabidopsis cells (Figure 7; see Supplemental Figure 9 online). The N0-gm1 reporter construct has a 4-bp mutation in the TGTCAGG sequence, and the N0-gm2 construct has a 4-bp mutation in nearby palindromic sequences (Figure 7A). In transient expression assays using Arabidopsis protoplasts, the N0-gm1 construct lost the repression of the promoter activity by G7FL, whereas N0-gm2 did not (Figure 7B). The transgenic plants harboring each reporter construct showed analogous results: The N0-gm1 construct showed significantly higher promoter activity than the N0 and N0-gm2 constructs under nonstress conditions, according to their GUS transcript levels (see Supplemental Figure 9 online). These data along with the EMSA results support the notion that the TGTCAGG sequence is a functional target sequence of GRF7 for repressing the DREB2A promoter activity through Region S.
Figure 7.
Effect of Nucleotide Substitutions in the Putative GRF7-Targeting Sequence.
(A) Schematic view and sequence information of the reporter and effector constructs.
(B) Transient expression assay using Arabidopsis mesophyll protoplasts. Values represent mean activity ratios of GUS and luciferase (LUC) from quadruplicate biological repeats. GUS/LUC ratios obtained using the empty effector plasmid (VEC) for each reporter construct were set to 1. The error bars indicate sd. Asterisks denote significant differences compared with VEC (multiple t test with Bonferroni correction; *P < 0.01). G, GGPL motif; Q, QLQ domain; S, Region S; VEC, empty effector vector; W, WRC domain.
GRF7 Represses the Expression of Osmotic Stress–Responsive Genes
To examine the role of GRF7 in the Arabidopsis transcriptional network, we compared the transcriptome of the grf7-1 mutant to that of the wild type under nonstress conditions using the Agilent Arabidopsis 3 Oligo Microarray system, which contains probes for >25,000 protein-coding loci. For these experiments, we used the aerial parts of the plants because GRF7 was predominantly expressed in these tissues. The grf7-1 mutant had 378 upregulated (Benjamini and Hochberg false discovery rate [FDR], P < 0.05; fold change, ≥4.00) and 31 downregulated genes (FDR, P < 0.05; fold change, ≤0.25) in comparison with the expression levels in the wild type (Figure 8A; see Supplemental Data Set 2 online). DREB2A was among the upregulated genes, with a 5.7-fold increase in its expression level (see Supplemental Data Set 2 online).
Figure 8.
Effect of the grf7-1 Mutation on the Transcriptome of Arabidopsis.
(A) Number of genes with altered expression in the grf7-1 mutant in comparison with the wild type. DN, downregulated gene group; UP, upregulated gene group.
(B) Heat maps indicating stimulus responsiveness of the top 100 upregulated (UP) and all downregulated (DN) genes in the grf7-1 mutant, as determined from Genevestigator stimulus data sets. For several genes, data were not available for analysis.
(C) Experimental validation by qRT-PCR analysis of several osmotic stress-inducible genes that were upregulated in grf7-1. The highest expression level for each gene was defined as 1.0. Data represent means of triplicate biological repeats, and the error bars indicate sd. WT, wild type.
(D) Effect of GRF7 deficiency on the high salinity tolerance of seedlings. The high salinity tolerance of GRF7-overexpressing (35Spro:GFP-GRF7 G; see Supplemental Figure 2 online) and -deficient (grf7-1 and amiG7 e) plants was evaluated. Top panel shows 13-d-old seedlings grown on a GM agar plate, and bottom panel shows 21-d-old seedlings grown for 4 d on GM agar plates, followed by 17 d on GM agar plates containing 150 mM NaCl. Survival rates (%; surviving individuals/tested individuals) on GM agar plates with 150 mM NaCl were calculated from the results of three independent experiments (n ≥ 15 for each experiment). The asterisks denote significant differences from the wild type (multiple t test with Bonferroni correction; *P < 0.05).
To characterize the genes with altered expression in grf7-1, gene ontological analysis was performed using the Gene Ontology Annotations database (Berardini et al., 2004; http://Arabidopsis.org/tools/bulk/go/). Compared with the Arabidopsis whole-genome categorization, the top 100 upregulated genes in grf7-1 showed higher ratios of gene groups termed “response to abiotic or biotic stimulus” (7.5% in the whole genome versus 37.2% in grf7-1) and “response to stress” (8.3% versus 35.1%) (see Supplemental Figure 10 online). Based on these data, we analyzed the expression patterns of genes that were up- or downregulated in grf7-1 with the Genevestigator database (Hruz et al., 2008). A high proportion of the top 100 upregulated genes were strongly responsive to ABA and osmotic stresses, and the downregulated genes were commonly repressed against such stimuli (Figure 8B). Analogous data were obtained by comparison analysis using microarray data from Arabidopsis plants after various abiotic stress treatments (E-MEXP-2116, Fujita et al., 2009; E-MEXP-2173, Maruyama et al., 2009; E-MEXP-2873, Yoshida et al., 2011). Among the 378 genes upregulated in grf7-1, 310 (82.0%) and 214 (56.6%) showed significantly increased expression in response to osmotic stress (i.e., dehydration or high salinity) and ABA treatment, respectively (see Supplemental Figure 11 and Supplemental Data Set 2 online). The responses of these genes to extreme temperatures were analyzed in the same way; however, relatively little overlap was detected in any case (see Supplemental Figure 11 and Supplemental Data Set 2 online). These data support the idea that GRF7 mainly represses the expression of osmotic stress– and/or ABA-responsive genes, including DREB2A. Subsequently, we confirmed the expression of several markedly upregulated genes in both amiG7 and grf7-1 mutant lines via qRT-PCR (Figure 8C). By analyzing upstream sequences, 103 of the 378 (27.3%) upregulated genes and six of the 31 (19.4%) downregulated genes were found to carry more than one putative GRF-targeting motif (TGTCAG or GTCAGG) in their 1-kb promoter regions (see Supplemental Data Set 2 online). In the upregulated genes, 92 of the 326 (28.2%) ABA- and/or osmotic stress–responsive genes harbor the putative GRF7-targeting motif in their promoter regions, a ratio higher than that of the other upregulated genes (11 of 52; 21.2%) (see Supplemental Data Set 2 online).
To evaluate the physiological consequences of such upregulation of osmotic-responsive genes by the grf7-1 mutation, we tested the drought and high salinity tolerance of GRF7-deficient or -overexpressing plant lines (Figure 8D; see Supplemental Figure 12 online). In high salinity conditions (150 mM NaCl), GRF7-deficient lines showed enhanced survival rates compared with the wild type (Figure 8D). Overexpression of GRF7 did not cause significant differences from the wild type (Figure 8D). An analogous result was obtained from a drought tolerance test; both GRF7-deficient lines presented higher survival rates than wild-type or GRF7-overexpressing plants (see Supplemental Figure 12 online).
DISCUSSION
DREB2A expression and activation promote plant survival in the face of severe environmental stress, including drought, high salinity, and high temperature, but also cause adverse effects on plant growth and reproduction (Sakuma et al., 2006a, 2006b). To mitigate these unfavorable effects, Arabidopsis is predicted to regulate the expression and/or activation of DREB2A tightly. In this report, we showed that Region S has repression activity within the DREB2A promoter under nonstress conditions (Figure 1). This represents one of many factors that regulate DREB2A expression in plant cells.
Since no cis-element was reliably predicted to occur in Region S, we conducted Y1H screening, and GRF7 was isolated as a candidate Region S–interacting protein (Figure 2; see Supplemental Table 1 online). Although Arabidopsis GRF family members have been expected to act as transcription factors, no specific evidence for this has been reported (Kim et al., 2003; Kim and Kende, 2004). In this study, we revealed that GRF7 can interact with the DREB2A promoter in vitro and in vivo and has Region S–dependent repressor activity (Figures 2, 6, and 7; see Supplemental Figures 2 and 9 online). The spatial expression pattern of DREB2A was extended in grf7-1 plants compared with the wild type (Figure 4D), and in the transient expression assay using protoplasts derived from grf7-1, GRF7 strongly repressed DREB2A promoter activity in a Region S–dependent manner (see Supplemental Figure 5 online). These results all support that GRF7 directly binds Region S of the DREB2A promoter and represses the expression of DREB2A in plant cells. Moreover, a GRF7 deficiency in plants leads to upregulation of many osmotic stress–responsive genes, including DREB2A, under normal growth conditions (Figure 8), which reflects the significance of GRF7 in the repression of a wide range of osmotic stress–responsive genes in Arabidopsis.
Additionally, a GTCA-containing sequence, TGTCAGG, was isolated from Region S as the target of GRF7 (Figures 6 and 7). Consistent with our results, a recently identified barley (Hordeum vulgare) GRF protein, BARLEY GROWTH REGULATING FACTOR1, binds to a region containing GTCA, although the target sequence was not specified (Osnato et al., 2010). Even though the TGTCAGG sequence shares the GTCA core sequence with W-boxes, nearby sequences are distinct from those of conventional W-boxes (A/GGTCAAA; Eulgem et al., 2000). Therefore, we define here the TGTCAGG sequence as a distinct cis-element, GTE (GRF7-targeting cis-element). GTE was enriched in the promoter regions of the genes upregulated in the grf7-1 mutant compared with those of the downregulated genes (see Supplemental Data Set 2 online). Based on our results, we propose a transcription pathway that comprises the newly identified transcriptional repressor GRF7 and the newly isolated cis-element GTE.
From analyses of a series of truncations of GRF7, we obtained several clues regarding how GRF7 represses promoter activity. Our Y1H results support the suggested functions of the QLQ and WRC domains as protein-interacting and DNA binding domains, respectively (van der Knaap et al., 2000; Kim et al., 2003) (Figure 5). GRF7 derivatives lacking the WRC domain failed to show a binding signal with Region S (Figure 5A). By contrast, GRF7 lacking the QLQ domain (G7ΔN) did show a Region S binding signal, which indicates that the QLQ domain is not necessary for the DNA binding ability of GRF7 (Figure 5A). In turn, G7ΔN could not repress the reporter activity in the transient expression system, which can be interpreted as indicating that the QLQ domain is important for the transcriptional repressor activity of GRF7 (Figure 5B). In addition, because the expression of GRF7 seems not to be affected by plant hormones or abiotic stresses (see Supplemental Figure 3 online), there could be a mechanism that regulates the activity of GRF7. Thus, we propose that an interaction with an unknown cofactor through the QLQ domain is important for transcriptional repression by GRF7. The GRF1-interating factor family proteins described in previous reports (Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009) are candidates for such cofactors.
Each member of the Arabidopsis GRF family contains a GGPL or TQL motif in its C-terminal region; however, the family members also show significant protein-specific differences in their primary amino acid sequences in these regions (Kim et al., 2003). Previous works suggest that several GRF proteins have transactivation activity in yeast, and the C-terminal region was suggested to be responsible for this activity (Choi et al., 2004; Kim and Kende, 2004). This is in contrast with our hypothesis that GRF7 can function as a transcriptional repressor. Since the GRF family is specific to plants, it is possible that yeast lack the QLQ-mediated repression system, which would explain the observed differences in GRF activity (van der Knaap et al., 2000). In addition, because the C-terminal sequence of GRF7 is distinct from those of other GRF proteins characterized to date (Kim et al., 2003), it is possible that the function of the C-terminal region in GRF7 is different from that of other GRF proteins. From the Y1H assays, we obtained a clue to the function of the C-terminal region of GRF7, which has a GGPL motif (Kim et al., 2003). GRF7 derivatives without GGPL and its flanking region (G7ΔC2 and G7ΔC3) lost the DNA binding signal in the Y1H analysis, suggesting the participation of the GGPL motif in the DNA binding activity of GRF7 (Figure 5A). Considering that other Arabidopsis GRF family members did not show the binding signal with Region S (Figure 2A), it is possible that the C terminus might affect the target sequence specificity of GRF7.
A GRF7 deficiency had less of an effect on DREB2A transcript levels under stress conditions, which suggests that GRF7 may not function under stress conditions (Figure 4). In another experiment, overexpression of GRF7 did not result in a significant difference in DREB2A expression under stress or nonstress conditions (see Supplemental Figures 2 and 6 online) and resulted in plants with rosettes of normal size and normal root elongation and water stress tolerance (Figure 8D; see Supplemental Figures 4, 6, and 12 online). One possible reason for the lack of effect of DREB2A overexpression is that GRF7 natively occupies the GTE regardless of whether or not the plants are stressed. Alternatively, it is possible that the binding state of GRF7 to the GTE is affected by the overexpression but that the DREB2A promoter activity in response to the stress signals is not. In either case, the stress response of the DREB2A promoter is expected to be unaffected by the binding of GRF7 to the GTE; therefore, at a minimum, the existence of an unknown mechanism to inactivate the GRF7 protein on the promoter is suggested. Based on the model of transcriptional repression proposed above, interaction with cofactors through the QLQ domain is a possible candidate for regulation of GRF7. Thus, characterization of the cofactors will be important to elucidate the regulation of transcriptional networks by GRF7.
Microarray analysis revealed roles of GRF7 in the Arabidopsis transcriptional network. Among the 409 genes that exhibited significantly altered expression in the nonstressed grf7-1 mutant plants, 378 (92.4%) were upregulated and 31 were downregulated (Figure 8A). This result is in agreement with our hypothesis that GRF7 functions as a transcriptional repressor in plant cells. In addition, more than half of the upregulated genes were osmotic stress– or ABA-inducible genes (Figure 8B; see Supplemental Figure 11 and Supplemental Data Set 2 online). These included genes for proteins involved in ABA metabolism and signaling, as well as stress-inducible transcription factors, protein kinases, and other proteins induced by osmotic stress (Figure 8C; see Supplemental Figure 11 online). Thus, GRF7 is expected to suppress basal levels of a wide range of osmotic stress responses by functioning as a transcriptional repressor of osmotic stress– and/or ABA-inducible genes under nonstress conditions.
In agreement with the transcriptomic change, GRF7 deficiency affected the osmotic stress tolerance of plants (Figure 8D; see Supplemental Figure 12 online). In both drought and high salinity conditions, grf7-1 and amiG7 plants showed clearly enhanced tolerance compared with the wild type. A related result was reported by other researchers (Liu et al., 2009), who found that the overexpression of miR396, which targets seven members of the GRF family, including GRF7, led to enhanced dehydration tolerance in Arabidopsis plants. They interpreted this phenomenon as originating from a decrease in the transpiration rate of leaves of the transgenic plants due to decreased stomatal density (Liu et al., 2009). However, the GRF7-deficient plants showed enhanced tolerance of high salinity conditions, which is not likely affected by transpiration (Figure 8D). Therefore, the increased expression of osmotic stress–inducible genes seems to be the factor responsible for the osmotic stress tolerance, although it is possible that regulation of morphogenesis by GRF7 affects osmotic stress tolerance through other pathways. The regulation of plant size by GRF7 could be another such pathway, since most studies of GRF family members have revealed that GRF-deficient plants are smaller than the wild type (Kim et al., 2003; Horiguchi et al., 2005; Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). These reports suggest that the size reduction is due to direct regulation of plant morphogenesis by GRF family proteins. In addition, we propose that another factor affecting plant size could be the released expression of osmotic stress–responsive genes, such as DREBs, STZ, and RD26, whose positive and negative effects on stress tolerance and plant growth, respectively, have been reported (Kasuga et al., 1999; Tran et al., 2004; Mittler et al., 2006; Sakuma et al., 2006a).
Data from this study and other groups are consistent in supporting that GRF7 has negative roles in osmotic stress responses but positive roles in plant growth and development (Figures 4 and 8; see Supplemental Figure 12 online) (Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). Under stress conditions, many stress-inducible genes are induced and function to improve stress tolerance. However, some of these genes inhibit plant growth. Under nonstress normal growth conditions, an individual plant can obtain better light conditions by growing a little larger than its neighbors. Therefore, it is important for plants to regulate stress responses strictly, and a suppressor of stress-responsive gene expression can be an effective method of maintaining plant growth. We propose that GRF7 plays such a role under nonstress conditions. GRF7-deficient lines (grf7-1 and amiG7) exhibited increased expression of stress-responsive genes, increased tolerance to osmotic stresses, and growth retardation (Figures 4 and 8; see Supplemental Figures 4 and 12 online), indicating that GRF7 minimizes the negative effect of stress-responsive genes on plant growth by transcriptionally regulating stress-responsive genes, including DREB2A (Figures 4 and 8).
In conclusion, we established a role for the transcriptional repressor GRF7 in the regulation of DREB2A expression. This protein can interact with DNA directly and repress the expression of a number of osmotic stress–responsive genes, including DREB2A, under nonstress conditions. A newly found cis-element, GTE, was identified as its target sequence. Through the GTE in the promoter region, GRF7 appears to minimize the adverse effects on plant growth by repressing the expression of stress-responsive genes under nonstress conditions. Complex positive and negative regulation in response to stress signals is expected to fine-tune the expression of stress-responsive genes, including DREB2A, to help maintain the ideal balance between plant growth and tolerance and thereby allow the plant to survive in rapidly changing environments.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used in this study. Seeds were sterilized and stratified at 4°C for 2 d in the dark, sown, and grown on germination media (GM) agar plates in a growth chamber with 16 h of fluorescent light at 40 ± 10 μmol m−2 s−1, 22°C, and 70% relative humidity (Kim et al., 2011). Arabidopsis transformation was achieved through the floral dip method (Clough, 2005). Harvested T1 candidate seeds were selected on GM agar plates including 50 mg L−1 kanamycin sulfate with 25 mg L−1 cefotaxime. To select homozygous lines, the same antibiotic selections were performed on the T2 and T3 generations. The verified T3 lines were used for further analyses.
For knocked-down expression of GRF7, a purchased artificial miRNA construct (catalog number AMR4844; Open Biosystems) was introduced into Arabidopsis plants. Transgenic candidates were selected on GM agar plates supplemented with 5 mg L−1 BASTA and 25 mg L−1 cefotaxime; subsequently, the transcript level of GRF7 was tested in each line by qRT-PCR. The T-DNA insertion lines for GRF7 and GRF8 were obtained from the ABRC (CS878963 for grf7-1, SALK_121691C for grf8-1, and SALK_001658 for grf8-2; Sessions et al., 2002; Alonso et al., 2003). The T-DNA insertion was confirmed by amplifying the LB-flanking region in genomic DNA of individual candidate lines. The primer sets used are presented in Supplemental Data Set 3 online.
Isolation of the DREB2A and GRF7 Promoter Fragments and Expression Vector Construction
The truncated DREB2A promoter and its derivatives were amplified from Arabidopsis genomic DNA. Nucleotide substitutions in the promoter construct were generated by mismatched amplification with each specific primer set (see Supplemental Data Set 3 online). The amplified promoter fragments were individually transferred into the BamHI restriction enzyme site of pGK-GUS (Qin et al., 2008). For the GRF7pro:GUS construct, the promoter region of 2980 bp from the transcription initiation site of GRF7 was amplified from Arabidopsis genomic DNA and transferred into the BamHI site of the pGK-GUS vector (Qin et al., 2008). For the GRF7pro:GRF7 construct, the GUS cassette of the GRF7pro:GUS construct was replaced by the GRF7 coding region between blunted XhoI and KpnI restriction enzyme sites. For the 35Spro:GFP-GRF7 construct, a clone containing the GRF7 coding region was obtained from the ABRC (clone No. U61879; Yamada et al., 2003). After confirmation of the sequence, the coding region of GRF7 was transferred into the EcoRV restriction enzyme site of the pGKX-NsGFP vector (Qin et al., 2008). The sequence of each construct was verified by comparison with The Arabidopsis Information Resource (TAIR; http://Arabidopsis.org/) database. Verified constructs were individually introduced into the Agrobacterium tumefaciens strain GV3101 (pMP90) by the freeze-thaw method and subsequently used for Arabidopsis transformation.
Computational cis-Element Prediction
PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002) and PLACE (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999) were used as databases for scanning of cis-elements in the DREB2A promoter sequence.
Phylogenetic Study
Sequence data for the GRF family members were obtained from the BioMart tool at the Phytozome database (version 7.0; http://www.phytozome.net/biomart/martview), where we retrieved all proteins containing both QLQ (Pfam, 08880; http://pfam.sanger.ac.uk/) and WRC (Pfam, 08879) domains. The phylogenic tree was constructed using the sequences of conserved regions from each member with the help of MEGA version 5.05 (http://www.megasoftware.net/), GeneDoc version 2.7.0000 (http://www.nrbsc.org/gfx/genedoc/), and ClustalX version 2.0.9 (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/). The distances between proteins were calculated by the neighbor-joining method with 1000 bootstrap trials, and Poisson correction was used to correct multiple substitutions at the same site. The nomenclature of the GRF family members from Arabidopsis and rice (Oryza sativa) is according to that used in previous reports (Kim et al., 2003; Choi et al., 2004).
qRT-PCR and Microarray Analysis
Total RNA was isolated from each plant sample using RNAiso plus (Takara Bio) according to the manufacturer’s instructions. Isolated RNA was employed for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen). qRT-PCR was performed using a 7500 real-time PCR system and 7500 system SDS software, version 1.4.0.25 (Applied Biosystems). SYBR Premix Ex Taq (Takara Bio) was used for amplification. Arabidopsis 18S rRNA was used as a quantitative control for the amount of template. The primer sets used are presented in Supplemental Data Set 3 online.
For genome-wide transcriptomic analysis, RNA samples from whole aerial parts of 2-week-old grf7-1 and wild-type seedlings grown on GM agar plates were used for microarray experiments using the Arabidopsis 3 Oligo Microarray Kit (4x44k; Agilent Technologies). For each of two biological replicates, the aerial parts from six plants were pooled to make a single RNA sample. The RNA samples were labeled with a Low RNA Input Linear Amplification/Labeling kit according to the manufacturer’s instructions (G4140-90050; Agilent Technologies). The cRNAs of grf7-1 and control samples were labeled with Cy5 and Cy3, or Cy3 and Cy5, respectively (dye swap experiment). Four microarrays were hybridized: two technical (dye swap) replicates for each of the two biological replicates. After hybridization, microarray slides were scanned (scanner model G2505C with scan control software, version A.8.5.1; Agilent Technologies), and the data were analyzed using Feature Extraction software, version 10.10.1.1 (Agilent Technologies), using the default settings. All microarray data analyses were performed according to Agilent methodology (G4460-90040; Agilent Technologies); signal intensities were normalized by the Lowess method, and significance was tested by an unpaired t test using GeneSpring GX software, version 11.5.1 (Agilent Technologies), where the Benjamini-Hochberg FDR method was used to obtain P values corrected for multiple testing. The average log2 ratio and sd of each probe were also calculated by GeneSpring GX. All microarray design information and data have been deposited to the ArrayExpress Archive (accession number E-MEXP-3559; http://www.ebi.ac.uk/arrayexpress/). To search for cis-elements in promoters, sequences 1 kb upstream of the transcription start sites were obtained from the TAIR database.
ChIP
ChIP assays were performed as described previously (Kim et al., 2011). Briefly, samples of 1 to 1.3 g were collected from whole 3-week-old Arabidopsis plants harboring the GFP-GRF7 protein-expressing construct (35Spro:GFP-GRF7) and subjected to ABA or dehydration treatment, as described previously (Kim et al., 2011). ChIP was performed using an EpiQuik Plant ChIP kit (Epigentek) according to the provided user guide. To immunoprecipitate GFP-GRF7, a GFP antibody (catalog No. 11814460001; Roche) was used. The primer sets used are presented in Supplemental Data Set 3 online.
Y1H Analysis
A modified MATCHMAKER One-Hybrid System (Clontech Laboratories) was used to perform the Y1H screening. Specifically, pHisi-1 and pLacZi vectors, both harboring quadruplicate tandem repeats of the Region S fragment in the EcoRI restriction enzyme site, were integrated into the genome of yeast strain YM4271 followed by selection on synthetic dextrose media agar plates without uracil and His (SD/-UH). We screened an Arabidopsis equalized full-length cDNA library (Mitsuda et al., 2010) on synthetic dextrose media without uracil, His, and Leu (SD/-UHL) supplemented with 30 mM 3-amino-1,2,4-triazole (3-AT). Candidate cDNA-harboring vectors were amplified and isolated via Escherichia coli. The sequence of each candidate was analyzed with the help of the TAIR database.
To verify the promoter–DNA interactions in yeast, the coding sequences of GRF family members and their derivatives were amplified from cDNA derived from Arabidopsis whole-plant RNA and introduced into the pGADT7 vector (Clontech Laboratories) individually using BamHI and XhoI restriction enzyme sites. The yeast transformation and the evaluation of interactions were performed as described previously (Kim et al., 2011). The primer sets used are presented in Supplemental Data Set 3 online.
Histochemical GUS Staining and Fluorescence Observation
Plants expressing the N0:GUS construct or the GRF7pro:GUS construct were collected at several growth stages and subjected to histochemical GUS assays as described previously (Kidokoro et al., 2009). Two-week-old transgenic plants harboring the 35Spro:GFP-GRF7 construct were collected and subjected to fluorescence signal observation as described previously (Kidokoro et al., 2009).
Stress and ABA Treatments
Stress and ABA treatments on Arabidopsis seedlings were performed as described previously (Kim et al., 2011). For high-salinity tolerance assays, 4-d-old seedlings on GM agar plates were transferred to GM agar plates with or without 150 mM NaCl. After 17 d of incubation under normal growth conditions, the survival rates were evaluated. Dehydration tolerance assays were performed as described in a previous report (Fujita et al., 2009).
Transient Expression Assays with Arabidopsis Mesophyll Protoplasts
Transient expression assays using Arabidopsis mesophyll protoplasts were performed as described by Kim et al. (2011).
Fusion Protein Preparation and EMSA
For GST-fused G7ΔN protein preparation, the coding sequence of G7ΔN was inserted into the BamHI and XhoI restriction enzyme sites of the pGEX-4T-1 vector (GE Healthcare Life Sciences) and expressed in the E. coli Rosetta (DE3) cell line (Merck KGaA). Expressed protein was extracted and purified using Glutathione Sepharose 4B (GE Healthcare Life Sciences) according to the manufacturer’s instructions. Subsequent probe labeling and EMSA steps were performed as described previously (Kidokoro et al., 2009). Sequence information for synthesized oligonucleotide probes is presented in Supplemental Data Set 3 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: DREB2A (AT5G05410), GRF7 (AT5G53660), GRF1 (AT2G22840), GRF2 (AT4G37740), GRF3 (AT2G36400), GRF4 (AT3G52910), GRF5 (AT3G13960), GRF6 (AT2G06200), GRF8 (AT4G24150), GRF9 (AT2G45480), RD26 (AT4G27410), STZ (AT1G27730), WRKY40 (AT1G80840), ABI1 (AT4G26080), AREB2/ABF4 (AT3G19290), NCED3 (AT3G14440), ATPK19 (AT3G08720), MPK11 (AT1G01560), and TSPO (AT2G47770).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of GROWTH-REGULATING FACTOR Family Members from Arabidopsis, Rice, S. moellendorffii, and P. patens.
Supplemental Figure 2. ChIP-PCR Assay in GFP-GRF7 Transgenic Plants.
Supplemental Figure 3. Expression Profile of GRF7.
Supplemental Figure 4. Rosette Radii of Genetically Modified Plants for GRF7 and GRF8.
Supplemental Figure 5. Transient Expression Assays of GRF7 in Protoplasts Derived from the grf7-1 Mutant.
Supplemental Figure 6. Gain-of-Function Analysis of GRF7.
Supplemental Figure 7. Loss-of-Function Analysis of GRF8.
Supplemental Figure 8. Effect of ABA on the Transcriptional Activity of GRF7 in Arabidopsis Mesophyll Protoplasts.
Supplemental Figure 9. Effect of Mutation at the GTE on the Transcriptional Activity of GRF7.
Supplemental Figure 10. Ontology of Genes Whose Expression Is Altered by GRF7 Deficiency According to the Microarray Data.
Supplemental Figure 11. Stress Responsiveness of Genes Whose Expression Is Altered by GRF7 Deficiency According to the Microarray Data.
Supplemental Figure 12. Dehydration Tolerance of GRF7-Deficient or -Overexpressing Plants.
Supplemental Table 1. List of Positive Clones Obtained from Yeast One-Hybrid Screening with Region S.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Supplemental Figure 1B.
Supplemental Data Set 2. Genes Regulated by GRF7, Identified by Microarray Analysis of the grf7-1 Mutant.
Supplemental Data Set 3. Oligonucleotides Used in This Study.
Supplementary Material
Acknowledgments
We thank Kaori Amano and Emiko Kishi for their excellent technical assistance, Kyoko Morimoto for protein work, Kyoko Yoshiwara for microarray analysis, and Masami Toyoshima for skillful editorial assistance. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (22119004) and Targeted Proteins Research Program (07088014) to K.Y.-S. and a Grant-in-Aid for Young Scientists (B) (21780314) to J.M. from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Science and Technology Research Partnership for Sustainable Development of the Japan Science and Technology Agency/Japan International Cooperation Agency, Genomics for Agricultural Innovation, Development of Abiotic Stress-Tolerant Crops by DREB Genes from the Ministry of Agriculture, Forestry, and Fisheries of Japan, and the Program for the Promotion of Basic Research Activities for Innovative Biosciences of Japan to K.Y.-S.
AUTHOR CONTRIBUTIONS
J.-S.K., J.M., K.S., and K.Y.-S. conceived and designed the research. J.-S.K. performed most of the experiments, and J.N. and K.N. helped him. S.K. performed the EMSA. K.M. performed the microarray experiment and analyzed the data. N.M., Y.T., M.O.-T., Y.K., T.Y., and M.M. contributed the Y1H screening. J.-S.K., J.M., and K.Y.-S. wrote the article.
Glossary
- ABRE
abscisic acid–responsive element
- GUS
β-glucuronidase
- Y1H
yeast one-hybrid
- qRT-PCR
quantitative RT-PCR
- ChIP
chromatin immunoprecipitation
- CaMV
cauliflower mosaic virus
- ABA
abscisic acid
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
- FDR
false discovery rate
- GM
germination media
- TAIR
The Arabidopsis Information Resource
- 3-AT
3-amino-1,2,4-triazole
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24: 23–58 [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg J., Aguilar X., Brännström K., Rautio L., Oloffson A., Wittung-Stafshede P., Björklund S. (2012). Interactions between DNA, transcriptional regulator Dreb2a and the Med25 mediator subunit from Arabidopsis thaliana involve conformational changes. Nucleic Acids Res. 40: 5938–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Kim J.H., Kende H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45: 897–904 [DOI] [PubMed] [Google Scholar]
- Clough S.J. (2005). Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., Nilsson A., Ulfstedt M., Ronne H., Wingsle G., Nilsson O., Björklund S. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., et al. (2011). RefGenes: Identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kidokoro S., Maruyama K., Nakashima K., Imura Y., Narusaka Y., Shinwari Z.K., Osakabe Y., Fujita Y., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Mizoi J., Yoshida T., Fujita Y., Nakajima J., Ohori T., Todaka D., Nakashima K., Hirayama T., Shinozaki K., Yamaguchi-Shinozaki K. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Ko J.H., Lee S., Lee Y., Pak J.H., Kim J.H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Liu H.C., Liao H.T., Charng Y.Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., Fujita M., Shinozaki K., Matsui M., Ohme-Takagi M. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51: 2145–2151 [DOI] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2010). Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M., et al. (2010). Cross talk between the KNOX and ethylene pathways is mediated by intron-binding transcription factors in barley. Plant Physiol. 154: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E., Kim J.H., Kende H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., Chang Z., Huang H., Cui X. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yoshida T., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.