This work characterizes the roles of two LysM-containing proteins, LYP4 and LYP6, in rice innate immunity. It finds that they act as unique dual-function pattern recognition receptors that can recognize microbe-associated molecular patterns from bacteria and fungi.
Abstract
Plant innate immunity relies on successful detection of microbe-associated molecular patterns (MAMPs) of invading microbes via pattern recognition receptors (PRRs) at the plant cell surface. Here, we report two homologous rice (Oryza sativa) lysin motif–containing proteins, LYP4 and LYP6, as dual functional PRRs sensing bacterial peptidoglycan (PGN) and fungal chitin. Live cell imaging and microsomal fractionation consistently revealed the plasma membrane localization of these proteins in rice cells. Transcription of these two genes could be induced rapidly upon exposure to bacterial pathogens or diverse MAMPs. Both proteins selectively bound PGN and chitin but not lipopolysaccharide (LPS) in vitro. Accordingly, silencing of either LYP specifically impaired PGN- or chitin- but not LPS-induced defense responses in rice, including reactive oxygen species generation, defense gene activation, and callose deposition, leading to compromised resistance against bacterial pathogen Xanthomonas oryzae and fungal pathogen Magnaporthe oryzae. Interestingly, pretreatment with excess PGN dramatically attenuated the alkalinization response of rice cells to chitin but not to flagellin; vice versa, pretreatment with chitin attenuated the response to PGN, suggesting that PGN and chitin engage overlapping perception components in rice. Collectively, our data support the notion that LYP4 and LYP6 are promiscuous PRRs for PGN and chitin in rice innate immunity.
INTRODUCTION
Plants, as sessile organisms, are constantly challenged by microbial pathogens from the environment. Due to lack of specialized immune cells, plants have evolved an elegant capability to trigger complex immune responses against invading microbes upon perception of microbe-associated molecular patterns (MAMPs), which are molecular signatures of whole classes of microbes (Ausubel, 2005). MAMP-triggered immunity (pattern-triggered immunity) plays a critical role in nonhost or basal resistance, which confers plants protection against a wide spectrum of potential pathogens (Nürnberger et al., 2004). During pattern-triggered immunity, recognition of different MAMPs by their cognate pattern recognition receptors (PRRs) at the plant cell surface and subsequent signal transduction across the plasma membrane are pivotal steps, which eventually lead to overlapping and specific defense responses to counteract invasion of different pathogens (Boller and Felix, 2009).
Most of the known MAMPs, regardless of their microbial origin, could be categorized into either polypeptide type or carbohydrate type. Well-characterized polypeptide MAMPs include the bacterial flagellin, elongation factor-Tu (EF-Tu), and Ax21 secreted protein. The bacterial flagellin and EF-Tu receptors in Arabidopsis thaliana have been identified as two leucine-rich repeat receptor-like kinases (LRR-RLKs), namely, FLS2 (for flagellin-sensitive 2) and EFR (for elongation factor EF-Tu receptor), respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). BAK1 (for brassinosteroid receptor BRI1-associated receptor kinase1), another Arabidopsis LRR-RLK, was found indispensable for intact flagellin signaling, presumably as a signal transducer or enhancer (Chinchilla et al., 2007; Heese et al., 2007). The rice (Oryza sativa) ortholog of Arabidopsis FLS2 was characterized as the flagellin receptor (Takai et al., 2008), and XA21 (resistance gene Xa21), another rice LRR-RLK, was identified as the receptor for the sulfated Ax21 protein (activator of XA21-mediated immunity) (Song et al., 1995; Lee et al., 2009). Interestingly, in addition to flagellin, Arabidopsis FLS2 can also recognize the Ax21 protein and the endogenous CLV3 peptide to trigger defense responses (Danna et al., 2011; Lee et al., 2011), suggesting that certain important PRRs may be assigned to multiple surveillance tasks in plant innate immunity.
Carbohydrate MAMPs, as represented by bacterial lipopolysaccharide (LPS) and peptidoglycan (PGN) and fungal chitin (Silipo et al., 2010), are mostly microbial cell envelop components. LPS is an outer membrane glycoconjugate from Gram-negative bacteria that is composed of a lipid A moiety, a covalently linked oligosaccharide core, and a polysaccharide moiety. It was found that the oligosaccharide core and the lipid A within LPS could induce distinct defense responses in plants, suggesting that plant cells likely sense sugar and lipid structures within LPS through different receptors (Silipo et al., 2005). Although the LPS receptors have been well characterized in animals (Ronald and Beutler, 2010), neither of these LPS receptors in plants has been identified so far.
Chitin, a homopolymer of β-(1,4) linked N-acetylglucosamine, is a common component of fungal cell walls. Its fragments, N-acetylchitooligosaccharides, which are released from chitin by plant chitinase digestion during plant–fungus interactions, can serve as elicitors in plant innate immunity (Silipo et al., 2010). Recent intensive efforts in investigating chitin recognition in plants have reaped progress. In rice, CEBiP (a chitin elicitor binding protein), a lysin motif (LysM)–containing plasma membrane protein, has been identified as the chitin receptor as it can physically bind to chitin and its silencing mutant exhibited significantly impaired chitin responses (Kaku et al., 2006). However, it is worth noting that Kaku et al. found that in their CEBiP-RNAi (for RNA interference) rice cells, where the CEBiP proteins became undetectable, ∼15% of the chitin-induced reactive oxygen species (ROS) generation was retained. More significantly, ∼29% (216 out of 746) of the genes induced by chitin in wild-type rice cells can be equally induced in the CEBiP-RNAi rice cells (Kaku et al., 2006). These data strongly suggested that rice hosts additional unknown receptors for chitin sensing (Kaku et al., 2006). Meanwhile, due to lack of an intracellular kinase domain, CEBiP was found to orchestrate with Os-CERK1 (for chitin elicitor receptor kinase 1), a LysM-containing RLK (LysM-RLK), for chitin signal transduction (Shimizu et al., 2010). Although Os-CERK1 has been suggested not to bind chitin directly (Shimizu et al., 2010), Arabidopsis CERK1 was repeatedly demonstrated to bind chitin on its own (Iizasa et al., 2010; Petutschnig et al., 2010; Liu et al., 2012), and its absence resulted in a complete loss of chitin responses in Arabidopsis (Miya et al., 2007; Wan et al., 2008). So far, the Arabidopsis counterpart of CEBiP has not been identified. It is also possible that such a component may not be necessary for chitin perception in Arabidopsis since CERK1 possesses the capabilities for both chitin binding and signal transduction (Liu et al., 2012).
PGN is an essential cell wall component in both Gram-positive and Gram-negative bacteria. This glycan is structurally closely related to chitin and consists of alternating residues of β-(1,4) linked N-acetylglucosamine and N-acetylmuramic acid where a short peptide chain is attached. The peptide chains from different PGN polymers can be cross-linked to form a three-dimensional mesh-like architecture, conferring structural strength and rigidity to the bacterial cell wall (Erbs and Newman, 2012). Since PGN is completely absent from eukaryotes, it is considered an ideal MAMP for the eukaryotic innate immune system (Dziarski and Gupta, 2005). Indeed, PGNs from Gram-positive and Gram-negative bacteria, despite with some biochemical variations, could both elicit defense responses in Arabidopsis (Gust et al., 2007; Erbs et al., 2008; Millet et al., 2010). Very recently, LYM1 and LYM3 have been identified as Arabidopsis PGN receptors (Willmann et al., 2011). Notably, LYM1 and LYM3 both possess LysM, which is also shared by all known chitin receptor components and the chitin-related rhizobial nodulation (Nod) factor receptors (Madsen et al., 2003; Radutoiu et al., 2003).
In this study, we initially aimed to pinpoint whether two LysM-containing proteins, LYP4 and LYP6, serve as rice PGN receptors. Surprisingly, in vitro ligand binding assays demonstrated that these proteins could not only bind to PGN but also to chitin. Silencing of either LYP gene significantly compromised PGN- and chitin-induced defense responses in rice, leading to increased susceptibility to both bacterial and fungal pathogens. Our findings suggested that LYP4 and LYP6 are dual function receptors sensing both bacterial PGN and fungal chitin in rice innate immunity.
RESULTS
Rice LYP4 and LYP6 Are LysM-Containing Proteins Localized at the Plasma Membrane
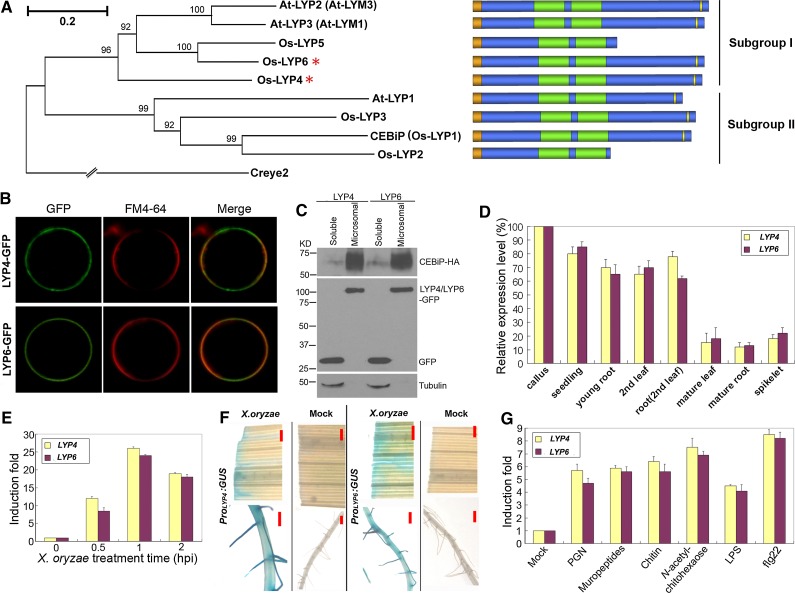
The initial goal of this study was to identify the PGN receptors in rice. As LysM was known as the binding motif for PGN in prokaryotes and the PGN-related chitin or Nod factor in plants (Bateman and Bycroft, 2000; Silipo et al., 2010), we postulated that the potential rice PGN receptor highly likely contains LysM. Phylogenetic analysis of the LysM-containing proteins (LYPs) from rice and Arabidopsis indicated that these LYPs could be categorized into two subgroups (Figure 1A). Os-LYP4 (Os09g27890), Os-LYP5 (Os02g53000), and Os-LYP6 (Os06g10660) belong to Subgroup I together with At-LYP2 (At1g77630) and At-LYP3 (At1g21880), while CEBiP (Os-LYP1) (Kaku et al., 2006), Os-LYP2 (Os11g34570), Os-LYP3 (Os09g37600), and At-LYP1 (At2g17120) are members of Subgroup II. Since CEBiP from subgroup II was previously characterized as the chitin receptor in rice (Kaku et al., 2006; Shimizu et al., 2010), we reasoned that the rice PGN receptor may be more likely to exist among LYP4, LYP5, and LYP6 from another branch (subgroup I) of the phylogenetic tree (Figure 1A). We therefore cloned these three rice genes, and their protein products consist of 401, 255, and 409 amino acids, respectively. Bioinformatic analysis predicted that LYP4 and LYP6 both have an N-terminal signal peptide, two characteristic LysMs, and a putative C-terminal glycosylphosphatidylinositol (GPI) anchor signal sequence (ω-site) (Figure 1A; see Supplemental Figure 1 online). This domain structure suggests that these proteins are localized at the plasma membrane through a lipid binding GPI anchor. Although LYP5 shows a substantial sequence similarity to the N terminus of LYP6, a long C-terminal portion (∼150 amino acids) including the GPI anchor signal sequence is absent from this protein (Figure 1A). Therefore, we focused only on LYP4 and LYP6, which have the GPI anchor for membrane attachment, in the subsequent study.
Figure 1.
LysM-Containing LYP4 and LYP6 Are Rice Plasma Membrane Proteins.
(A) Phylogenetic tree and domain structure diagram of LYPs in rice and Arabidopsis. The phylogenetic tree was generated using MEGA4. Full-length amino acids sequences of plant LYPs were selected for generating a bootstrap neighbor-joining phylogenetic tree. Creye2 from Chlamydomonas reinhardtii was used as an outgroup. Bootstrap probabilities were obtained from 1000 replicates. A scale bar is indicated. A text file of the alignment used to generate this tree is available as Supplemental Data Set 1 online. The N-terminal signal peptide, LysM, and the C-terminal GPI anchor signal (ω-site) of each LYP are colored in orange, green, and yellow, respectively.
(B) LYP4 and LYP6 localize at rice plasma membrane. Os-LYP4 and Os-LYP6 with GFP inserted behind the N-terminal signal peptide were individually expressed in rice protoplasts and visualized by confocal microscopy. FM4-64 dye was used to stain the plasma membrane.
(C) LYP4 and LYP6 localize in the microsomal fraction. LYP4-GFP and LYP6-GFP were individually coexpressed with CEBiP-HA and GFP in rice protoplasts. Microsomal and soluble fractions of protoplast lysates were separated through Suc gradient centrifugation. Distribution of individual proteins and the endogenous tubulin was analyzed through immunoblotting with anti-GFP, anti-HA, and antitubulin antibodies.
(D) Relative expression levels of LYP4 and LYP6 in different rice tissues and developmental stages. The expression levels of these genes were determined by qPCR, and the expression level of each gene in rice calli is set as 100%.
(E) Upregulation of LYP4 and LYP6 transcripts in rice seedlings by X. oryzae. Five-day-old rice seedlings were incubated with X. oryzae suspension (105 cells/mL) or mock treated (sterile water) for the indicated period. The expression levels of these genes were examined by qPCR, and the induction fold of each gene was calculated by the gene expression level in X. oryzae–treated seedlings relative to that in mock-treated seedlings at the same time point. hpi, h postinoculation.
(F) Induction of LYP4 and LYP6 expression in mature rice leaf and root by bacterial pathogen X. oryzae. The mature leaf (the fourth leaf) and the primary root from the indicated Promoter:GUS transgenic rice at the five-leaf stage were immersed into X. oryzae suspension (105 cells/mL) for 2 h before GUS staining. Bars = 1 mm.
(G) Induction of LYP4 and LYP6 expression by diverse MAMPs. Five-day-old rice seedlings were treated with 100 μg/mL of insoluble PGNXoo, soluble PGNXoo muropeptides, insoluble crab shell chitin, soluble chitin fragment N-acetylchitohexaose, soluble LPS, or 100 nM flg22 for 1 h, and the induction of each gene was examined by qPCR.
The experiments in (B) to (G) were repeated three times with similar results.
The data in (D), (E), and (G) represent the mean ± sd of nine samples from three independent tests.
All characterized homologs of LYP4 and LYP6 have been verified as plasma membrane proteins. These LYPs include rice CEBiP (Kaku et al., 2006), Arabidopsis LYM1 (At-LYP3), LYM2 (At-LYP1), and LYM3 (At-LYP2) (Borner et al., 2003; Willmann et al., 2011), and Medicago truncatula LYM1 and LYM2 (Fliegmann et al., 2011). To confirm the plasma membrane localization of Os-LYP4 and Os-LYP6, we inserted green fluorescent protein (GFP) behind the N-terminal signal peptide in both proteins. This fusion strategy was based on the fact that both the N-terminal signal peptide and the C-terminal GPI anchor signal sequence eventually would be removed from a mature GPI-anchored protein. Since plant protoplast systems have been used successfully to detect the cell surface localization of GPI-anchored proteins (Takos et al., 1997), we transiently expressed LYP4-GFP or LYP6-GFP using a monocot-specific constitutive Act1 promoter in rice green tissue protoplasts. In both cases, colocalization of GFP signal with the FM4-64–stained plasma membrane was readily detected under a confocal microscope (Figure 1B). To provide additional evidence for the plasma membrane localization of these proteins, we isolated the microsomal fraction from rice cells in which LYP4-GFP or LYP6-GFP was coexpressed with GFP and CEBiP-HA (HA tag behind the N-terminal signal peptide of CEBiP). These constructs were all transiently expressed in rice protoplasts under the control of the constitutive Act1 promoter. Successful preparation of rice microsomal fractions was verified in immunoblots showing that CEBiP (Kaku et al., 2006) was highly enriched in the microsomal fraction while GFP and endogenous tubulin proteins, as nonmembrane proteins, were exclusively found in the soluble fraction (Figure 1C). As expected, LYP4 and LYP6 were both visualized in the microsomal fraction through immunoblotting with anti-GFP antibodies (Figure 1C). Taken together, these data confirmed that LYP4 and LYP6 localize at the plasma membrane of rice cells.
LYP4 and LYP6 Expression Can Be Induced Quickly by Bacterial Pathogen Infection or Diverse MAMPs
To understand better the function of LYP4 and LYP6, we checked the expression profiles of these genes in different rice tissues and developmental stages by quantitative real-time PCR (qPCR). LYP4 and LYP6 were most abundantly expressed in rice callus cells, and both transcripts progressively decreased during maturation (Figure 1D). Furthermore, analysis of LYP4 and LYP6 expression patterns in Promoter:GUS (for β-glucuronidase) transgenic rice demonstrated strong GUS staining in young seedlings, particularly in the root meristem region and the lateral root primordium (see Supplemental Figure 2 online), resembling the expression patterns of their ortholog LYM1 in M. truncatula (Fliegmann et al., 2011). Interestingly, expression of LYP4 and LYP6 in rice seedlings, mature leaves, and roots could be quickly induced upon exposure to the rice bacterial pathogen Xanthomonas oryzae pv oryzae. In 5-d-old rice seedlings, 1 h X. oryzae treatment induced a 24- and 26-fold increase of endogenous LYP4 and LYP6 transcripts, respectively (Figure 1E). Moreover, incubation with X. oryzae suspension for 2 h rendered a strong GUS activity in mature leaves and roots of the Promoter:GUS transgenic rice, while incubation with sterile water had no effect on the GUS activity (Figure 1F). The possibility of false positive GUS staining due to pathogen contamination could be excluded as no GUS activity could be detected in the empty vector (pCAMBIA-1391Z containing an intact GUS gene without promoter) transgenic rice treated with X. oryzae (see Supplemental Figure 3 online). Moreover, we found that the transcripts of LYP4 and LYP6 could be promptly and significantly upregulated by treatment with diverse MAMPs, including PGN, chitin, LPS, and flg22 (Figure 1G). These findings hinted a potential involvement of LYP4 and LYP6 in rice innate immunity.
LYP4 and LYP6 Can Physically Bind PGN and Chitin
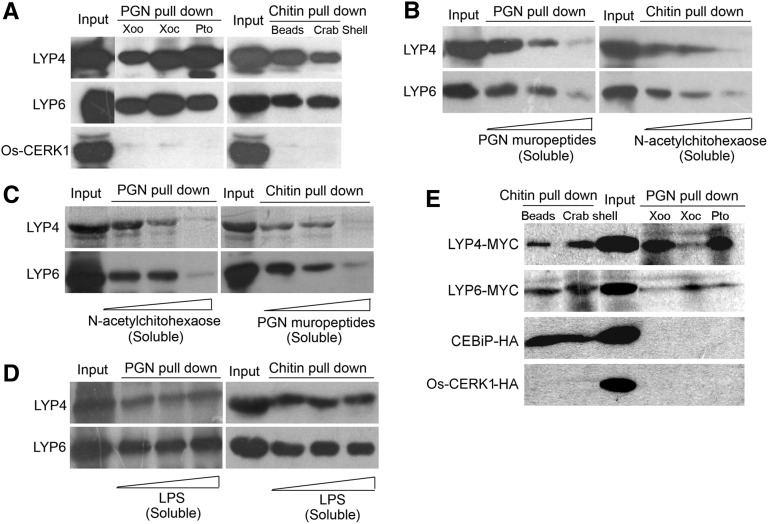
As we speculated that LYP4 and LYP6 are rice PGN receptors, we next addressed the question whether these proteins could physically bind PGN. Insoluble PGN purified from three different bacterial pathogens, X. oryzae pv oryzae (PGNXoo), X. oryzae pv oryzicola (PGNXoc), and Pseudomonas syringae pv tomato (Pto) DC3000 (PGNPto), was individually used to pull down the purified 6His-tagged recombinant LYP4 and LYP6 proteins in solution. Indeed, both LYP4 and LYP6 could coprecipitate with these PGNs (Figure 2A). We further found that the HPLC-purified soluble PGNXoo muropeptides, which are lysostaphin-digested products of PGNXoo, could compete with the insoluble PGNXoo for binding to these proteins, as the increase of muropeptides in solution was coupled with a decrease of PGNXoo-precipitated LYP4 and LYP6 (Figure 2B). Moreover, the analysis of PGN binding kinetics suggested that the association of PGNXoo to these proteins occurred as early as within 1 min and reached saturation in ∼30 min (see Supplemental Figure 4 online). In parallel, we conducted a PGN pull-down assay for the purified 6His-tagged recombinant Os-CERK1 (Os-LysM-RLK9) extracellular domain. In contrast with LYP4 and LYP6, CERK1 extracellular domain could be barely precipitated by any of these PGNs (Figure 2A). An effort to test CEBiP for PGN binding was hindered by the difficulty in producing 6His-tagged recombinant CEBiP proteins in Escherichia coli (data not shown). These data revealed a specific physical interaction between bacterial PGN and the two rice LYPs.
Figure 2.
LYP4 and LYP6 Selectively Bind PGN and Chitin but Not LPS.
(A) LYP4 and LYP6 coprecipitate with insoluble PGN or chitin. Sixty micrograms of PGN purified from bacterial pathogens X. oryzae (Xoo), X. oryzicola (Xoc), or P. syringae pv tomato DC3000 (Pto), commercial chitin beads (NEB), and crab shell chitin (Sigma-Aldrich) were individually used to pull down 1 μg purified 6His-tagged recombinant Os-LYP4, Os-LYP6, or Os-CERK1 in solution.
(B) Soluble PGN muropeptides and N-acetylchitohexaose compete with insoluble PGN and chitin, respectively, for LYP4 and LYP6 binding. Fifty micrograms of PGNXoo (left panel) or chitin beads (right panel) were used to pull down 1 μg of purified recombinant Os-LYP4 or Os-LYP6 in the presence of increasing amounts (0, 40, and 80 μg) of PGNXoo muropeptides (left panel) or N-acetylchitohexaose (right panel).
(C) Soluble PGN muropeptides and N-acetylchitohexaose cross-compete with insoluble chitin and PGN, respectively, for LYP4 and LYP6 binding. Fifty micrograms of PGNXoo (left panel) or chitin beads (right panel) were used to pull down 1 μg of purified recombinant Os-LYP4 or Os-LYP6 in the presence of increasing amounts (0, 40, and 80 μg) of N-acetylchitohexaose (left panel) or PGNXoo muropeptides (right panel).
(D) Soluble LPS shows no binding to LYP4 and LYP6. Fifty micrograms of PGNXoo (left panel) or chitin beads (right panel) was used to pull down 1 μg of purified recombinant Os-LYP4 or Os-LYP6 in the presence of increasing amounts (0, 40, and 80 μg) of LPS.
(E) LYP4 and LYP6 produced in planta coprecipitate with insoluble PGN or chitin. One hundred micrograms of PGN or chitin was individually used to pull down MYC-tagged LYP4 and LYP6 or HA-tagged CEBiP and CERK1, which was individually expressed in 2 mL of rice protoplasts. Triton X-100 (0.5%) was used to lyse protoplasts and solubilize membrane proteins.
One of the three biological repeats with similar results is shown, and results were obtained by immunoblotting using anti-His, anti-MYC, or anti-HA antibodies.
As PGN is structurally related to chitin (Silipo et al., 2010), we tested whether LYP4 and LYP6 could also physically bind to chitin. Chitin beads (NEB) and insoluble crab shell chitin (Sigma-Aldrich) were used to pull down the purified LYP4 and LYP6 proteins in solution. Surprisingly, these proteins were readily precipitated by either commercial chitin products (Figure 2A). Furthermore, when highly purified N-acetylchitohexaose (Isosep), a soluble hexamer of chitin oligosaccharide, was included in the pull-down assay to compete with the chitin beads, we observed a clear negative correlation between the amount of N-acetylchitohexaose added and the amounts of LYP4 and LYP6 precipitated by chitin beads (Figure 2B). Moreover, the assay of chitin binding kinetics suggested that the binding of chitin to these proteins occurred within 5 min and became saturated in ∼20 min (see Supplemental Figure 4 online). In parallel, we also performed the same chitin pull-down assay for the purified CERK1 extracellular domain. In agreement with the previous suggestion (Shimizu et al., 2010), we did not detect any coprecipitation of CERK1 with chitin beads or the crab shell chitin (Figure 2A). Our data suggested that LYP4 and LYP6 physically bind chitin in addition to PGN.
We next introduced cross-competition into the pull-down experiments and found that addition of excess soluble PGNXoo muropeptides disrupted the precipitation of these proteins by chitin beads (Figure 2C). Likewise, the presence of excess soluble N-acetylchitohexaose blocked the coprecipitation of these proteins with PGNXoo (Figure 2C). By contrast, addition of excess soluble LPS (Sigma-Aldrich) failed to affect the amounts of LYP4 and LYP6 precipitated by PGNXoo or chitin beads (Figure 2D). These data reinforced that LYP4 and LYP6 selectively bind PGN and chitin but not LPS.
Furthermore, LYP4 and LYP6 expressed in rice protoplasts also could be precipitated by different bacterial PGNs and commercial chitin products (Figure 2E), confirming the physical association of PGN and chitin to LYPs in rice. By contrast, the CEBiP proteins successfully expressed in rice protoplasts could be precipitated only by chitin but not PGN, while the full-length rice CERK1 proteins expressed in rice protoplasts could be pulled down by neither MAMP (Figure 2E).
Silencing of LYP4 or LYP6 Compromises Diverse PGN- and Chitin-Induced Defense Responses in Rice
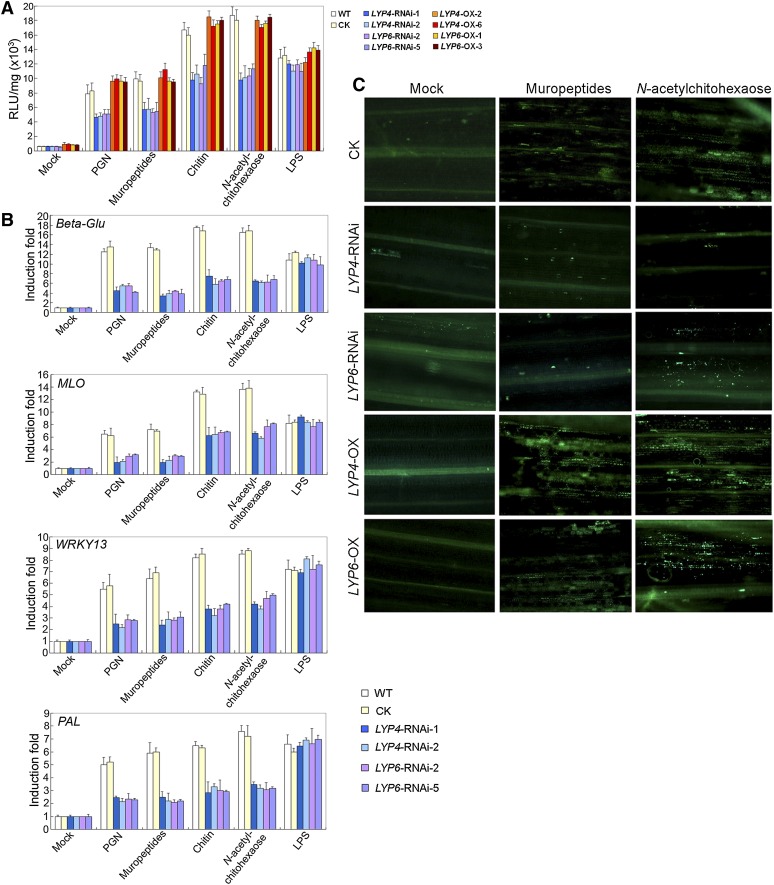
The findings regarding the plasma membrane localization of LYP4 and LYP6 and their physical interactions with both PGN and chitin pointed to a more exciting possibility that these proteins may be not only the PGN receptors but also previously unknown chitin receptors in rice. This motivated us to evaluate both PGN- and chitin-induced defense responses in LYP4 or LYP6 RNAi or overexpressing (OX) transgenic rice. Two representative lines of RNAi or OX transgenic rice were used for each gene in the subsequent analysis. The empty vector transgenic rice and the wild-type rice were used as controls. Although LYP4 and LYP6 genes share ∼80% identity, the LYP4 and LYP6 transcripts were specifically reduced by ∼80% by their cognate RNAi construct in the silencing lines (see Supplemental Figure 5A online). Notably, the expression of CEBiP and CERK1, the two known genes involved in rice chitin perception, was affected by neither RNAi construct (see Supplemental Figure 5B online). On the other hand, the expression of LYP4 in its OX lines was increased by 5.2- to 6.8-fold and that of LYP6 in its OX lines was increased by 14- to 15-fold (see Supplemental Figure 5C online). Using LYP RNAi or OX transgenic rice, we investigated three different cell responses occurring at different defense time points after PGN or chitin exposure. These defense responses included ROS generation (a very early response), defense gene activation (an early response), and callose deposition (a late response). LPS, as another glycoconjugate elicitor, was used as control MAMP during the examination of the two earlier defense responses.
Indeed, the amounts of ROS triggered by the purified PGNXoo or its HPLC-purified muropeptides declined by ∼40% in either LYP4 or LYP6 RNAi transgenic rice when compared with that produced in the control rice (Figure 3A). Similarly, the amounts of ROS induced by chitin or its soluble fragment N-acetylchitohexaose also decreased by 37 to 42% in LYP RNAi transgenic rice (Figure 3A). By contrast, LYP4 and LYP6 OX transgenic rice showed comparable ROS production after PGN or chitin treatment (Figure 3A). Notably, all transgenic rice lines and the wild-type rice treated with LPS exhibited no significant difference in ROS production (Figure 3A). Since ROS generation is one of the earliest defense responses (Boller and Felix, 2009), these results suggested that LYP4 and LYP6 function quite upstream within the PGN- and chitin-induced defense signaling pathways and strongly corroborated the notion that these proteins are potential receptors for the two MAMPs.
Figure 3.
RNAi Silencing or Overexpressing of LYP4 and LYP6 Specifically Modulate PGN- and Chitin- but Not LPS-Induced Defense Responses in Rice.
(A) ROS generation induced by PGN or chitin is significantly suppressed in LYP4 or LYP6 RNAi transgenic rice lines. Roots from 4-d-old wild-type or transgenic rice seedlings were incubated with 100 μg/mL PGNXoo, PGNXoo muropeptides, crab shell chitin, N-acetylchitohexaose, LPS, or sterile water (mock) for 30 min before ROS measurement. The data represent the mean ± sd of nine samples from three independent tests. CK, control (empty vector transgenic rice); RLU, relative light units; WT, the wild type.
(B) Defense gene activation induced by PGN or chitin is compromised in LYP4 or LYP6 RNAi transgenic rice lines. Callus cells from wild-type or RNAi transgenic rice lines were incubated with 100 μg/mL PGNXoo, PGNXoo muropeptides, crab shell chitin, N-acetylchitohexaose, LPS, or sterile water (mock) for 30 min, and the induction of four representative defense marker genes Beta-Glu, MLO, WRKY13, and PAL was determined by qPCR. The data represent the mean ± sd of nine samples from three independent tests.
(C) Callose deposition induced by PGN or chitin is substantially impaired in LYP4 and LYP6 RNAi transgenic rice but enhanced in LYP OX rice. The first leaf of 5-d-old wild-type, LYP4-RNAi-1, LYP6-RNAi-2, LYP4-OX-2, or LYP6-OX-3 rice seedling was immersed into solution containing 10 μg/mL PGNXoo muropeptides or N-acetylchitohexaose and vacuumed for 30 min. Callose staining was conducted 18 h later.
At least three biological repeats were conducted for individual experiments.
Moreover, the activation of four representative defense marker genes, namely, Beta-Glu, MLO, WRKY13, and PAL, by 30 min PGN/muropeptides or chitin/N-acetylchitohexaose treatment in wild-type rice was substantially suppressed in LYP4 or LYP6 RNAi transgenic rice (Figure 3B). However, these defense marker genes responded to 30-min LPS treatment equally well in both wild-type and LYP RNAi transgenic rice (Figure 3B). Furthermore, the callose staining spots on rice young leaves after PGNXoo muropeptides or N-acetylchitohexaose treatment were dramatically reduced in LYP4 or LYP6 RNAi transgenic rice when compared with those in the control rice (Figure 3C). By contrast, PGNXoo muropeptides or N-acetylchitohexaose treatment resulted in more callose deposition in LYP4 or LYP6 OX transgenic rice than in the control rice (Figure 3C). These data further strengthened the notion that LYP4 and LYP6 play crucial roles in PGN- and chitin-induced defense signaling in rice.
LYP4 and LYP6 Affect Rice Susceptibility to Both Bacterial and Fungal Pathogens
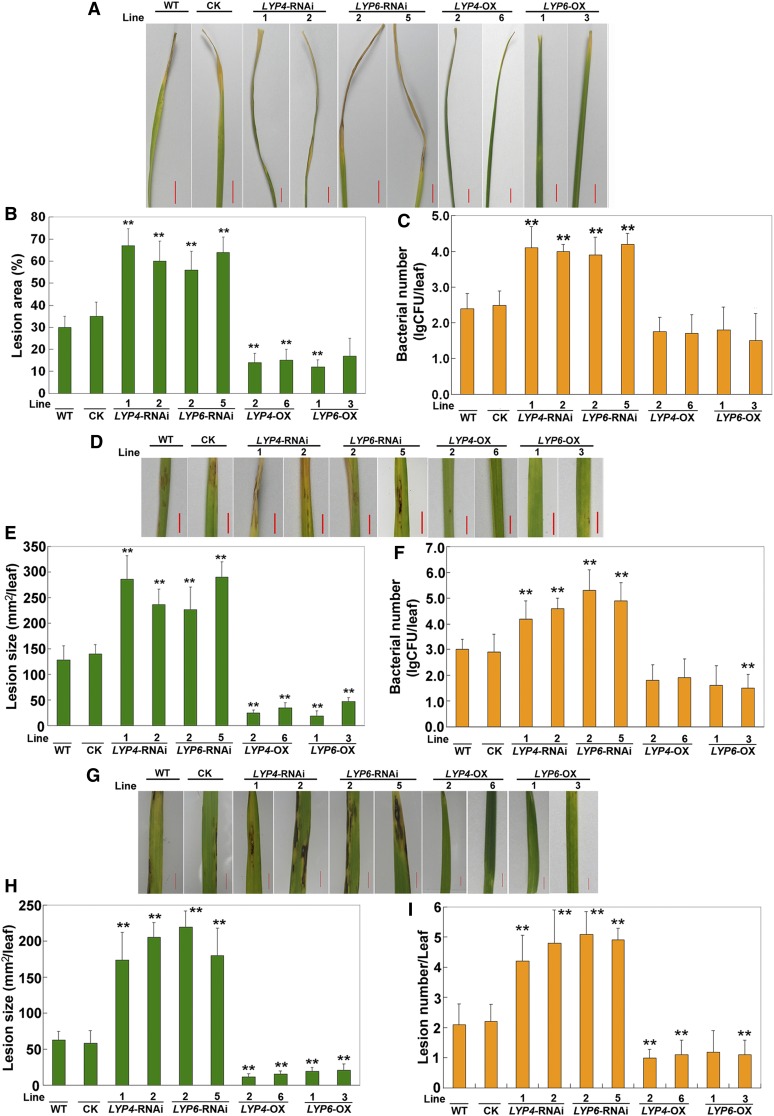
To validate the importance of LYP4 and LYP6 in rice innate immunity, we conducted pathogen growth assay in LYP RNAi or OX transgenic rice using the bacterial blight pathogen X. oryzae, the bacterial streak pathogen Xanthomonas oryzicola, and the fungal blast pathogen Magnaporthe oryzae. As expected, compared with the lesion area caused by X. oryzae infection in wild-type rice, the lesion areas in LYP4- or LYP6-silencing rice were significantly enlarged (Figures 4A and 4B). Accordingly, bacterial growth was increased by 25- to 50-fold in LYP silencing rice (Figure 4C). Similarly, the lesion area due to X. oryzicola infection also expanded significantly in LYP4- or LYP6-silencing rice relative to that in wild-type rice (Figures 4D and 4E). Consistent with this, more X. oryzicola growth was detected in LYP silencing rice (Figure 4F). In addition, the LYP silencing rice appeared to be more susceptible to fungal M. oryzae infection as the lesion size per leaf was considerably larger in LYP silencing rice than in wild-type rice (Figures 4G and 4H). Accordingly, the lesion number per leaf was increased by 0.9- to 1.4-fold in LYP silencing rice compared with that in wild-type rice (Figure 4I). These data suggested that knockdown of LYP4 and LYP6 expression in rice results in an increased susceptibility to both bacterial and fungal pathogens.
Figure 4.
RNAi Silencing or Overexpressing of LYP4 and LYP6 Affects Rice Innate Immunity against Bacterial and Fungal Pathogens.
(A) Representative phenotype of wild-type (WT) and LYP RNAi or OX transgenic rice infected by X. oryzae. Thirty leaves with equal size (the fourth leaf at the five-leaf stage) from each rice line were inoculated with X. oryzae (105 colony-forming units/mL), and phenotypes were recorded in 9 d after inoculation. CK, control (empty vector transgenic rice). Bars = 3 cm.
(B) The mean lesion area per leaf (lesion length/leaf length) of the 30 leaves used in (A).
(C) X. oryzae growth scored by lg colony-forming units (CFU)/leaf of the 30 leaves used in (A).
(D) Representative phenotype of wild-type and LYP RNAi or OX transgenic rice infected by X. oryzicola. Thirty leaves with equal size (the fourth leaf at the five-leaf stage) from each rice line were inoculated with X. oryzicola (105 colony-forming units/mL), and phenotypes were recorded in 9 d after inoculation. Bars = 3 cm.
(E) The mean lesion size per leaf of the 30 leaves used in (D).
(F) X. oryzicola growth scored by lg colony-forming units/leaf of the 30 leaves used in (D).
(G) Representative phenotype of wild-type and LYP RNAi or OX transgenic rice infected by M. oryzae. Fifteen leaves with equal size (the fourth leaf at the five-leaf stage) from each rice line were inoculated with M. oryzae, and phenotypes were recorded in 10 d after inoculation. Bars = 1 cm.
(H) The mean lesion size per leaf of the 15 leaves used in (G).
(I) The mean lesion number per leaf of the 15 leaves used in (G).
Three biological repeats were conducted, and similar results were observed. The data represent means ± sd (30 leaves in each transgenic line, three independent experiments). Statistical significance compared with the wild type is indicated by double asterisks (**P ≤ 0.01 and *P ≤ 0.05, Tukey’s test).
Conversely, the lesion area caused by X. oryzae infection decreased to 41 to 58% in LYP4 or LYP6 OX rice relative to that in wild-type rice (Figure 4B) and the X. oryzae growth in these LYP OX lines was reduced by more than 80% (Figure 4C). More significantly, the lesion size caused by X. oryzicola infection decreased to 15 to 38% in LYP OX rice relative to that in wild-type rice (Figure 4E), and the bacterial growth in these LYP OX rice dropped by more than 90% (Figure 4F). Moreover, the lesion area due to M. oryzae infection also shrank to 19 to 40% in LYP OX rice (Figure 4H), and the lesion number per leaf was reduced to 50 to 63% in comparison with that in wild-type rice (Figure 4I). These results indicated that upregulation of LYP4 and LYP6 expression in rice leads to an enhanced resistance against both bacterial and fungal infection.
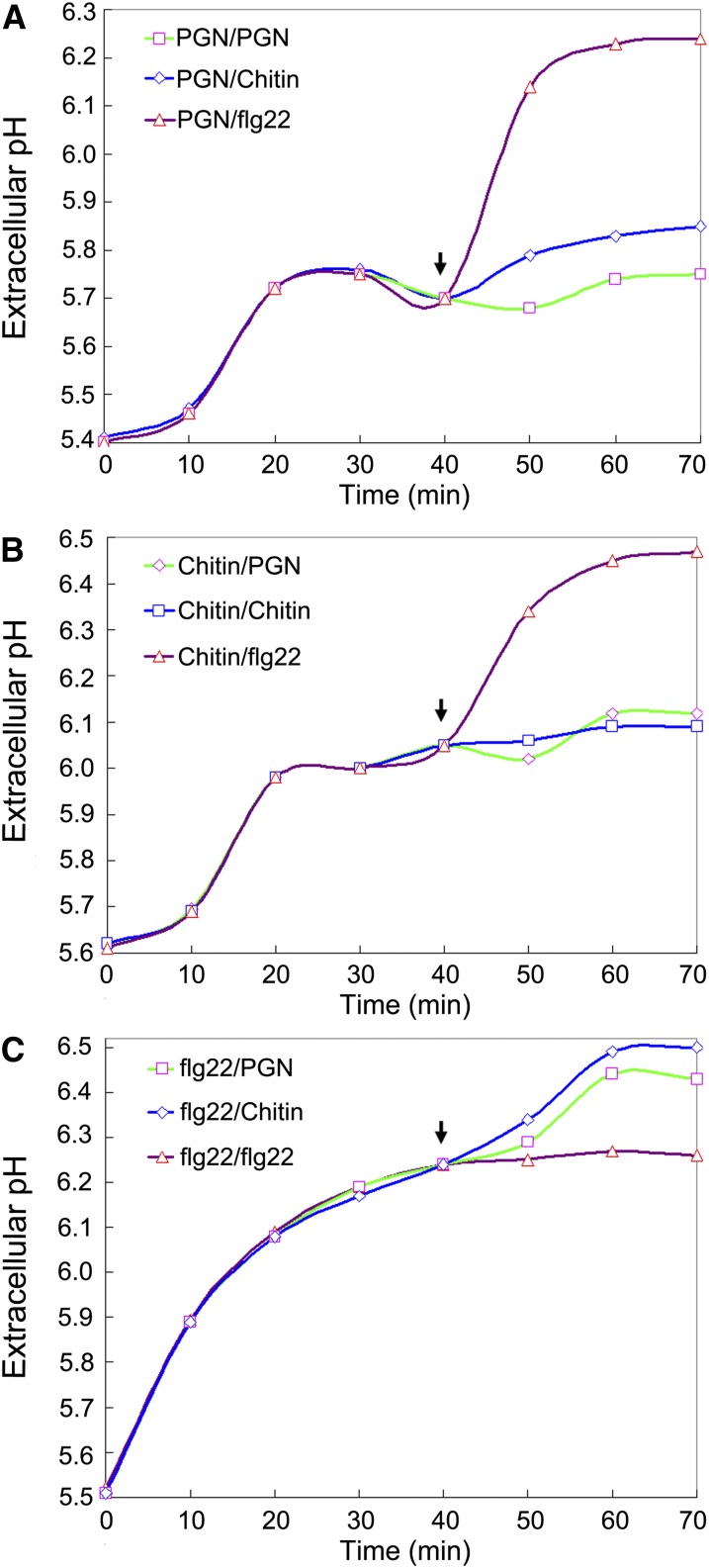
PGN and Chitin Engage Overlapping Perception Components in Rice
The aforementioned data suggested that LYP4 and LYP6 may be shared by both PGN and chitin perception systems in rice. If this were the case, the rice cells saturated with one of the two elicitors would become temporarily less responsive to another because the receptors, once occupied by the first elicitors, would not become immediately available for binding the second. To test this speculation, we set up three sets of tandem MAMP treatments for rice callus cells and closely followed the elevation of the medium pH that indicates the magnitude of alkalinization response of rice cells. In the first set of experiments, the rice cells were initially saturated with 600 μg/mL insoluble PGNXoo and then treated with 100 μg/mL PGNXoo muropeptides, 100 μg/mL N-acetylchitohexaose, or 10 nM flg22 (GenScript). It was obvious that the flg22 treatment after the saturating PGN treatment induced a second spike of medium alkalinization, while the N-acetylchitohexaose treatment led to only a slight increase of medium pH (Figure 5A). The alkalinization response to flg22 excluded the possibility that disappearance of the alkalinization response to chitin was due to depletion of certain components necessary for medium alkalinization by the saturating PGN treatment. Hence, these results suggested that chitin and PGN are perceived through overlapping receptor systems. In the second set of experiments, the rice cells were first saturated with 600 μg/mL crab shell chitin before the subsequent treatment with 100 μg/mL PGNXoo muropeptides, 100 μg/mL N-acetylchitohexaose, or 10 nM flg22. Likewise, although the flg22 treatment following the saturating chitin treatment could induce another dramatic elevation of medium pH, the PGNXoo muropeptides gave rise to only a negligible alkalinization response (Figure 5B). These results repeatedly suggested that chitin and PGN are sharing overlapping perception systems. In the third set of experiments, the rice cells were presaturated with 1 μM flg22 and then treated individually with 100 μg/mL PGNXoo muropeptides, 100 μg/mL N-acetylchitohexaose, and 10 nM flg22. In contrast with flg22, both PGNXoo muropeptides and N-acetylchitohexaose could provoke a further medium alkalinization (Figure 5C), verifying that PGNXoo muropeptides and N-acetylchitohexaose used in these experiments (Figures 5A to 5C) were active elicitors. Taken together, these data indirectly support the notion that LYP4 and LYP6 are dual function receptors for PGN and chitin in rice.
Figure 5.
PGN and Chitin Engage Overlapping Receptor Components in Rice.
(A) Rice cells pretreated with excess PGN have a dramatically attenuated alkalinization response to subsequent chitin treatment. Rice callus cells were first treated with 600 μg/mL insoluble PGNXoo for 40 min (endpoint marked by the arrow) and then treated with 100 μg/mL PGNXoo muropeptides (green curve), 100 μg/mL N-acetylchitohexaose (blue curve), or 10 nM flg22 (purple curve).
(B) Rice cells pretreated with excess chitin have a dramatically attenuated alkalinization response to subsequent PGN treatment. Rice callus cells were first treated with 600 μg/mL crab shell chitin for 40 min (endpoint marked by the arrow) and then treated with 100 μg/mL PGNXoo muropeptides (green curve), 100 μg/mL N-acetylchitohexaose (blue curve), or 10 nM flg22 (purple curve).
(C) PGNXoo muropeptides and N-acetylchitohexaose used in (A) and (B) are active. Rice callus cells were first treated with 1 μM flg22 for 40 min (endpoint marked by the arrow) and then treated with 100 μg/mL PGNXoo muropeptides (green curve), 100 μg/mL N-acetylchitohexaose (blue curve), or 10 nM flg22 (purple curve).
Three biological replicates were conducted for (A) to (C), and similar results were obtained.
DISCUSSION
Pattern-triggered immunity as the primary line of plant innate immunity solely relies on the prerequisite that the PRRs at plant cell surface successfully detect the approaching pathogens by recognizing their MAMPs. As such, identification of individual PRRs in plants, such as FLS2 for bacterial flagellin, EFR for bacterial EF-Tu protein, XA21 for bacterial Ax21 protein, and CEBiP and CERK1 for fungal chitin, have all become landmark achievements in plant innate immunity research. In this study, we added Os-LYP4 and Os-LYP6 to the inventory of plant PRRs, which, strikingly, recognize both bacterial PGN and fungal chitin elicitors. Our discovery of LYP4 and LYP6 as bacterial PGN receptors in rice resonated with a recent report that identified At-LYM1 and At-LYM3, orthologs of Os-LYP4 and Os-LYP6 (Figure 1A, Subgroup I), as PGN receptors in Arabidopsis. Meanwhile, our serendipitous finding that Os-LYP4 and Os-LYP6 also function as fungal chitin receptors in rice solved the puzzle raised by Kaku and colleagues. Dating back to the time when they discovered CEBiP as a rice PRR sensing fungal chitin, they had already realized that there must be additional chitin receptors present in rice (Kaku et al., 2006). In particular, in their CEBiP-RNAi rice cells where the CEBiP proteins were undetectable, ∼30% of the upregulated genes and 20% of the downregulated genes could still respond to chitin equally well as in wild-type rice cells (Kaku et al., 2006). Moreover, CEBiP-RNAi rice cells had reduced but noticeable chitin binding capacity (Kaku et al., 2006). Therefore, LYP4 and LYP6 uncovered in this study are very likely to be those additional chitin receptors in rice.
Intriguingly, in contrast with the dual specificities of Os-LYP4 and Os-LYP6 in binding PGN and chitin, At-LYM1 and At-LYM3, the orthologs of Os-LYP4 and Os-LYP6 in Arabidopsis, could bind only PGN but not chitin (Willmann et al., 2011). It has been recently reported that the same type of PRR from Arabidopsis and rice could behave differently in elicitor binding. For instance, Arabidopsis FLS2 could bind bacterial Ax21 peptides (Danna et al., 2011), whereas rice FLS2 could not (Lee et al., 2009). Also, Arabidopsis CERK1 could bind chitin (Petutschnig et al., 2010; Iizasa et al., 2010; Liu et al., 2012), whereas rice CERK1 could not (Figure 2; Shimizu et al., 2010). It is plausible that homologous PRRs from different plant species may have lost or acquired certain MAMP binding capability during evolution. In rice, it is tempting to assume that the loss of chitin binding capacity in CERK1 is compensated for by the gain of that ability in CEBiP and LYP4/6. Alternatively, in Arabidopsis, the chitin binding function of CERK1 may make unnecessary for the LYM1 and LYM3 proteins to have chitin binding capability.
The promiscuity of Os-LYP4 and Os-LYP6 in binding carbohydrate MAMPs PGN and chitin is reminiscent of At-FLS2 binding three different ligands, including flagellin and Ax21 as well as the endogenous CLV3 peptide (Danna et al., 2011; Lee et al., 2011). The promiscuity of PRRs in sensing multiple MAMPs provides a distinct physiological advantage to the host so that a limited number of PRRs would be able to perceive a maximum number of MAMPs. Considering plants in nature are often exposed concurrently to several groups of microbial pathogens, the advantage brought by LYP4 and LYP6 in rice is particularly spectacular in that these PRRs could detect PGN and chitin derived individually from the two major microbial groups, bacteria and fungi. As the expression of LYP4 and LYP6 genes could be rapidly upregulated upon recognition of either MAMP (Figure 1G), it seems that either type of microbial infection would quickly sensitize rice for further infection by both groups of microbes. Interestingly, although the transgenic rice overexpressing LYP4 or LYP6 indeed demonstrated an enhanced pathogen resistance (Figure 4), the PGN- or chitin-induced ROS production in these rice plants did not show significant difference compared that in wild-type rice (Figure 3A). Pathogen resistance is a complicated consequence of innate immunity, whereas ROS production is just one of the very early defense responses in plant innate immunity. The biological significance of ROS production in plant defense is not fully understood. It is very likely that the intensity of ROS generation is not linearly correlated with the magnitude of pathogen resistance, as the latter is shaped by complex interplay between multiple layers of defense responses that are induced by a cocktail of MAMPs from pathogens and occur with distinct dynamics.
The functions of other three evolutionarily related Os-LYPs (LYP2, LYP3, and LYP5) still remain enigmatic. LYP2 and LYP5 are presumably located in the apoplastic space rather than the plasma membrane due to lack of the GPI anchor (Figure 1A), whereas LYP3 likely resides at the plasma membrane like LYP4 and LYP6. LYP2, LYP3, and LYP5 all contain two intact LysMs (Figure 1A; see Supplemental Figure 1 online); thus, their binding capacity to PGN or chitin cannot be excluded at this moment. By inference, they may serve certain regulatory functions in PGN or chitin signaling in rice, similar to the case of Eix1 and Eix2 proteins in tomato. Both Sl-Eix1 and Sl-Eix2 were found to bind the fungal elicitor xylanase, where only the Eix2 receptor mediated defense signaling while Eix1 acted as a decoy receptor to attenuate the xylanase-induced defense signaling (Bar et al., 2010, 2011).
Both Os-LYP4 and Os-LYP6 are likely to be N-glycosylated when expressed in rice cells as the GFP hybrids of both proteins showed an actual molecular mass around 100 kD instead of the predicted molecular mass of 65 kD (Figure 1C), reminiscent of other LYP proteins expressed in planta (Kaku et al., 2006; Fliegmann et al., 2011; Willmann et al., 2011). Intriguingly, the N-glycosylation of these receptors appeared to be dispensable for ligand binding since the recombinant receptors expressed in E. coli were competent in PGN and chitin binding (Figure 2A). Similar findings were also obtained for their orthologs in Arabidopsis (Willmann et al., 2011). Recently, it has been revealed that the N-glycosylation of NFP, the LysM-RLK for Nod factor perception, was not essential for its biological activity including the ligand binding (Lefebvre et al., 2012). Our work and others thus suggest a role of N-glycosylation in regulating the protein trafficking of these receptors.
Although Os-LYP4 or Os-LYP6 are each able to bind PGN and chitin, our data suggest that they are not functionally redundant. This was because knockdown of single LYP gene expression in rice was sufficient to impair both PGN- and chitin-induced defense responses significantly (Figure 3) and to cause severe bacterial or fungal infection phenotypes (Figure 4). Similar observations recently have been made for Arabidopsis PGN receptors LYM1 and LYM3, where lym1 lym3 double mutant did not show increased susceptibility to bacterial pathogens when compared with lym single mutants (Willmann et al., 2011). Both studies favor a cooperative relationship between the pair of PRRs, suggesting that they may work in the same receptor complex.
The identification of LYP4 and LYP6 as additional chitin receptors in rice raises the question regarding their relationship with the previously identified rice chitin receptor CEBiP (Kaku et al., 2006). Kaku and coworkers found that 70% of the genes activated by chitin in wild-type rice cells lost responsiveness in CEBiP-RNAi rice cells, suggesting that CEBiP might play a major role in rice chitin perception. In line with this speculation, RNAi silencing of CEBiP diminished the chitin-induced ROS generation by 85% (Kaku et al., 2006), while silencing of LYP4/6 only reduced the chitin-induced ROS generation by ∼40% (Figure 3A). However, as 30% of the upregulated genes and 20% of the downregulated genes could respond to chitin equally well in wild-type and in CEBiP-RNAi rice cells, CEBiP and LYP4/6 proteins are very likely to work in different chitin receptor complexes. In support of this speculation, a major portion of CEBiP proteins were visualized as homodimers in blue native PAGE analysis (Shimizu et al., 2010). Nevertheless, a small fraction of CEBiP proteins did exist as larger-size oligomers (Shimizu et al., 2010), which makes it ambiguous whether some CEBiP proteins can be in the same complexes with LYP4/6. Further investigation of the composition and stoichiometry of rice chitin receptor complexes as well as chitin responses in CEBiP and LYP4/6 triple knockdown rice will be necessary to dissect fully their contributions in rice chitin perception and signaling.
In sum, we demonstrated that two LysM-containing proteins, LYP4 and LYP6, are dual function receptors for bacterial PGN and fungal chitin in rice. We provided three key lines of evidence pertaining to the function of these proteins. First, they are localized at plant cell surface. Second, they can specifically bind PGN and chitin. Third, knockdown of their expression perturbs the PGN- and chitin-induced defense responses in rice. LYP4 and LYP6 are unique among known PRRs in that they can recognize MAMPs across microbial groups. Future investigation of the LysMs in these PRRs will be meaningful not only for understanding the biochemical basis of LysM-PGN and LysM-chitin interactions, but also for guiding the engineering of promiscuous PRRs into other crop species to improve disease resistance. Moreover, comparison of the similarities and differences in PGN and chitin perception machineries between rice and Arabidopsis will provide valuable evolutionary insights for understanding critical mechanisms underlying innate immunity signaling in plants.
METHODS
Plant Growth
The rice (Oryza sativa) calli were cultivated at 25°C in the dark. Callus-regenerated T0 transgenic rice seedlings were first cultivated at 25°C under 16-h-light/8-h-dark conditions and then transferred into pots and grown under greenhouse conditions until setting seeds. Seeds were surface sterilized in sodium hypochlorite (10% [v/v]) for 15 min and germinated on half-strength Murashige and Skoog (MS) medium containing 50 μg/mL hygromycin at 25°C under 16-h-light/8-h-dark conditions for 4 d and subsequently transferred to Hoagland solution without hygromycin. After hydroponic culture for 2 weeks, the seedlings were moved into pots and grown under greenhouse condition.
Plasmid Construction
Routine molecular cloning techniques were followed to make the constructs. The primers used in this work are listed in Supplemental Table 1 online. The transient expression plasmids were built on the pUC19 vector containing the monocot-specific Act1 promoter, the coding region of each gene, and the NOS terminator. For GFP-, MYC-, or HA-tagged LYP proteins, the tag was inserted behind the N-terminal signal peptide of each protein, namely, behind Cys-30 in Os-LYP4, Gly-36 in Os-LYP6, and Cys-33 in CEBiP. The restriction site is indicated by the underlined nucleotides in Supplemental Table 1 online.
The recombinant binary plasmids used for RNAi-mediated silencing or overexpressing of Os-LYP4 and Os-LYP6 were designed as previously described (Zhong et al., 2007). In the RNAi constructs the sense and antisense fragments were separated by an intron element and expressed under the control of the Act1 promoter. The sense and antisense fragments were first cloned into an adaptor vector pRiAT, and then the entire expression cassette was extracted by KpnI and PmlI digestion and inserted into the identically digested binary vector pCAMBIA1301. The fragment of 1067 to 1498 bp within the Os-LYP4 coding sequence and that of 636 to 1246 bp within the Os-LYP6 coding sequence were selected as the RNAi target for each gene. Both strands of the RNAi targets were confirmed to have no contiguous 21-nucleotide identical hit in other rice genes when searched against the rice genome. Overexpression of Os-LYP4 or Os-LYP6 was also driven by the Act1 promoter. Similarly, the entire expression cassette was assembled in the adaptor vector pRiAT and then cloned into the binary plasmid pCAMBIA1301 with KpnI and PmlI digestion. The sequence-verified binary plasmids were transformed into Agrobacterium tumefaciens strain EHA105, which were used to transform the rice calli of Zhonghua11 explants (Zhou et al., 2005).
To generate the Promoter:GUS transgenic rice, the 1924-bp upstream sequence before the start codon of Os-LYP4 and the 1700-bp upstream sequence before the start codon of Os-LYP6 were amplified by genomic PCR from Zhonghua11 rice and then inserted into the promoter capture vector pCAMBIA-1391Z, which was transformed into rice via Agrobacterium.
The open reading frame encoding the predicted mature protein of Os-LYP4, Os-LYP6, or the extracellular domain of Os-CERK1 was PCR amplified and cloned into the pET-32a vector (Novagen). The resultant plasmids were respectively transformed into Escherichia coli BL21 (DE3) cells for recombinant protein expression.
Generation of Transgenic Rice Plants
The hygromycin-resistant clones of rice calli were selected using 50 mg/L hygromycin and differentiated into seedlings. The rice calli were cultured on NBD2 medium. The transgenic calli were regenerated on NB medium containing 5 mg/L 6-benzyl aminopurine, 1 mg/L naphthylacetic acid, 500 mg/L cephalothin, 50 mg/L hygromycin, and 3.0 g/L Phytagel. The regenerated shoots were transferred to half-strength MS medium with 0.5 mg/L naphthylacetic acid, 0.5 mg/L indole-3-acetic acid, and 3.0 g/L Phytagel to stimulate root differentiation. The seedlings were subsequently grown on half-strength MS medium for root elongation and better development. The expression levels of LYP4 and LYP6 in each transgenic rice line was determined by qPCR.
PGN and Muropeptide Purification
The total PGN of Xanthomonas oryzae, Xanthomonas oryzicola, or Pto DC3000 was purified as previously described (Gust et al., 2007; Erbs et al., 2008) with modifications. Briefly, the bacterial cells were boiled in 100 volumes of 5% SDS for 30 min. After cooling down to room temperature, the SDS-insoluble material was collected by centrifugation at 30,000g at 4°C for 30 min. The pellet was intensively washed with water until SDS became invisible and treated with 10% trichloroacetic acid at 45°C for 4 h followed by acetone wash and methanol wash to remove the lipid components. The pellet was then treated with trypsin and DNase I overnight to digest proteins and DNA. After washing with 8 M LiCl twice, the total PGN was resuspended in sterile water for storage. For muropeptide preparation, purified PGN (5 mg/mL) from X. oryzae was suspended in 100 mM sodium phosphate buffer, pH 6.8, and digested with 20 μg/mL lysostaphin (Sigma-Aldrich) at 37°C for 16 h. Digestion was terminated by boiling for 10 min followed by centrifugation. The supernatant was subjected to reverse phase HPLC using a Spherigel C-18 column (300A, 5 μm, 200 × 10 mm). Muropeptides were eluted with acetonitrile in a step gradient from 0 to 30% at a flow rate of 1 mL/min. Muropeptide peaks detected by the absorbance at 210 nm were pooled and concentrated in a rotary evaporator and then lyophilized.
Rice Protoplast Preparation, Transfection, and Microsomal Fractionation
The protoplast isolation from rice green tissues was performed as previously described (Zhang et al., 2011). Briefly, the 8-d-old rice seedlings were cut into ∼0.5-mm strips and incubated in enzyme solution (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES, pH 5.7, 10 mM CaCl2, and 0.1% BSA) for 4 to 5 h in the dark with gentle shaking (60 to 80 rpm). After digestion, the pellets were washed with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) and the protoplasts were collected by centrifugation at 1500g for 3 min. DNA (50 to 100 μg) was used to transfect every 1 mL (2 × 106 cells) of rice protoplasts. Transfected protoplasts were incubated in the dark at room temperature for 6 h for protein expression. Isolation of rice microsomal fraction through microcentrifugation was performed as described earlier (Abas and Luschnig, 2010) with modifications. Briefly, pelleted cells from 1 mL rice green tissue protoplasts expressing LYP4-GFP or LYP6-GFP were lysed in 200 μL extraction buffer (50 mM Tris-HCl, pH 7.5, 25% Suc, 5% glycerol, 10 mM EDTA, 1 mM DTT, and 2× Roche protease inhibitor cocktail) by vigorous vortexing. The lysate was centrifuged at 2000g for 3 min, and the supernatant was diluted by equal volume of sterile water and equally distributed into two 1.5-mL microcentrifuge tubes. The microsomal fraction was obtained as the pellet after a centrifugation in a benchtop microcentrifuge at 21,000g at 4°C for 3 h. The supernatant was kept as the soluble fraction and its proteins were concentrated by trichloroacetic acid precipitation before SDS-PAGE and immunoblot analysis. After washing with 200 μL washing buffer (20 mM Tris-HCl, pH 7.5, and 1 mM EDTA), the microsomal fraction was pelleted by centrifugation at 21,000g at 4°C for 1 h. The proteins were harvested from the microsomal fraction by boiling with SDS-PAGE loading buffer and analyzed by SDS-PAGE and immunoblotting with anti-HA (Roche), anti-GFP (Roche), or anti-tubulin (Sigma-Aldrich) antibodies.
For confocal laser scanning, the transfected protoplasts were incubated in the dark at room temperature for 6 h and stained with 8 μM FM4-64. The fluorescence was detected under a confocal laser scanning microscope (TSC-SP5 Leica).
Protein Expression in E. coli
The protein expression was induced at room temperature overnight by 0.2 mM isopropyl-β-d-thiogalactopyranoside. Bacteria were lysed by sonication and the 6His-tagged recombinant proteins were captured by the nickel-nitrilotriacetic acid-agarose (Novagen) affinity column and eluted with 60 mM imidazole. The Trx-tag fused with the purified protein products was removed by thrombin cleavage to avoid potential interference with target protein function and the 6His-tagged target protein was further purified with the thrombin cleavage capture kit (Novagen).
Ligand Binding Assay
The assay was performed according to the previously reported method (Tjoelker et al., 2000; Iizasa et al., 2010) with modifications. PGN or chitin, in some cases with soluble muropeptides or N-acetylchitohexaose as competitors, was coincubated with purified 6His-tagged recombinant proteins in binding buffer (50 mM Tris-HCl, pH 7.0, 100 mM NaCl, and 0.1 mM PMSF) at 4°C for 2 h with constant shaking. For binding assays of rice protoplast crude extracts, 0.5% Triton X-100 was added in the binding buffer. The glycan was spun down by centrifugation at 12,000g at 4°C for 10 min. The pellet was washed five times with the binding buffer and then boiled with SDS-PAGE loading buffer. The presence of target proteins in the glycan binding fraction was determined by SDS-PAGE and immunoblot analysis with anti-6His (Novagen), anti-MYC (Roche), or anti-HA (Roche) antibodies.
ROS Measurement
Hydrogen peroxide content was determined by the chemiluminescence method based on the Co(II) catalyzed oxidation of luminol as previously described (Pérez and Rubio, 2006). The polyvinylpolypyrrolidone (5%)–treated extracts after 1:100 dilution were measured by the Infinite M200 Monochromater (Tecan Trading) with the count time set as 10 s.
GUS Histochemical Assays
Histochemical GUS staining was performed using transgenic rice as previously described (Jefferson et al., 1987) with modifications. Briefly, the staining solution (100 mM phosphate buffer, pH 7.5, 0.5 mM potassium ferrocyanide, 0.5 potassium ferricyanide, 0.1% [v/v] Triton X-100, 10 mM EDTA, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) was added in a tightly capped tube to cover the tissues. After 24 h, the staining solution was removed and the sample was cleared with several changes of 70% (v/v) ethanol over 1 to 2 d at 37°C. GUS staining was observed using a stereomicroscope (Zeiss SteREO Lumar.V12).
Callose Staining
Rice seedlings treated with muropeptides or N-acetylchitohexaose were fixed in ethanol:acetic acid (3:1 [v/v]) solution for 5 h. The fixative was changed frequently to ensure thorough fixing and clearing of the tissues. Seedlings were then rehydrated successively in 70% ethanol for 2 h, in 50% ethanol for 2 h, and in water overnight. After being washed three times with water, seedlings were treated with 10% NaOH for 1 h to make the tissues transparent. After rinsing four times with water, seedlings were incubated in 150 mM K2HPO4, pH 9.5, containing 0.01% aniline blue (Sigma-Aldrich) for 4 h (Millet et al., 2010). The leaves were mounted on slide and callose spots were observed immediately under a Leica DM5000B microscope under UV channel (340 to 380 nm).
Bacterial and Fungal Pathogen Inoculation
Wild-type and transgenic rice plants at the five-leaf stage were inoculated with bacterial pathogens X. oryzae (strain GD4) using the leaf-clipping method (Chen et al., 2002) or X. oryzicola (strain GDx) using the spraying method (Chen et al., 2003). Rice plants at the same age were inoculated with fungal pathogen M. oryzae (strain Z-A) through the spraying method (Chen et al., 2003). All plants were cultured in a plant growth chamber with >90% humidity post inoculation.
Medium Alkalinization Measurement
Alkalinization response of rice callus cells was measured using semi-micro pH electrode (Mettler Toledo) as previously described (Kunze et al., 2004).
qPCR
The Actin1 gene was selected as the reference gene for qPCR, and the relative abundance of mRNA was analyzed using the SYBR PrimeScript RT-PCR kit (Takara) on an IQ5-Bio-Rad (Bio-Rad).
Bioinformatic Analysis
The phylogenetic analysis of rice and Arabidopsis thaliana LYPs were performed by the MEGA program (www.megasoftware.net). The phylogenetic tree was generated using MEGA 4 (Tamura et al., 2007). Full-length amino acid sequences of plant LYP proteins At-LYP1, At-LYP2, At-LYP3, Os-LYP1 (CEBiP), Os-LYP2, Os-LYP3, Os-LYP4, Os-LYP5, and Os-LYP6 were selected for generating a bootstrap neighbor-joining phylogenetic tree. Creye2 from Chlamydomonas reinhardtii was used as an outgroup. Bootstrap probabilities were obtained from 1000 replicates. The ClustalX alignment used to generate the tree is available as Supplemental Data Set 1 online. The GPI anchor signal sequence in Os-LYP4 and Os-LYP6 was predicted by the big-PI Plant Predictor (mendel.imp.ac.at/gpi/plant_server.html).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Os-LYP4, Os09g27890; Os-LYP6, Os06g10660; Os-CERK1, Os08g42580; Os-WRKY13, Os01g0750100; Beta-Glu, Os05g0495900, MLO, Os03g0129100; PAL, Os02g0627100; At-LYP1, At2g17120; At-LYP2, At1g77630; At-LYP3, At1g21880; Os-LYP1 (CEBiP), Os03g04110; Os-LYP2, Os11g34570; Os-LYP3, Os09g37600; Os-LYP5, Os02g53000; and Creye2, AAF43040, from C. reinhardtii.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of LYPs in Arabidopsis and Rice.
Supplemental Figure 2. LYP4 and LYP6 Are Strongly Expressed in Rice Young Tissues.
Supplemental Figure 3. No GUS Activity Is Induced by X. oryzae in pCAMBIA-1391Z Empty Vector Transgenic Rice.
Supplemental Figure 4. PGN and Chitin Binding Kinetic Analysis for LYP4 and LYP6.
Supplemental Figure 5. Relative Expression Levels of LYP4, LYP6, CEBiP, and CERK1 in LYP RNAi or Overexpressing Transgenic Rice.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1.
Supplementary Material
Acknowledgments
We thank Liexian Zeng (Plant Protection Research Institute, Guangdong Academy of Agricultural Science) for providing X. oryzae and X. oryzicola strains. We thank Xianzhang Bu (Sun Yat-sen University) for assistance in reverse phase HPLC. This research was supported by the National Natural Science Foundation of China (No. 30571069 and No. 31100874), the project of Science and Technology new star in Zhujiang city (2012-78), and the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20060558093).
AUTHOR CONTRIBUTIONS
H.-B.W., J.W., B.L., and J.-F.L. designed the research. B.L., J.Q., Y.A., J.S., J.-F.L., Y.Z., Z.L., J.L., D.F., K.Q., and Y.H. performed research. H.-B.W., J.-F.L., and B.L. analyzed data. J.-F.L., H.-B.W., and B.L. wrote the article.
Glossary
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- EF-Tu
elongation factor-Tu
- LRR-RLK
leucine-rich repeat receptor-like kinase
- LPS
lipopolysaccharide
- PGN
peptidoglycan
- LysM
lysin motif
- RNAi
RNA interference
- ROS
reactive oxygen species
- Nod
nodulation
- GPI
glycosylphosphatidylinositol
- GFP
green fluorescent protein
- qPCR
quantitative real-time PCR
- GUS
β-glucuronidase
- OX
overexpressing
- MS
Murashige and Skoog
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Bar M., Sharfman M., Avni A. (2011). LeEix1 functions as a decoy receptor to attenuate LeEix2 signaling. Plant Signal. Behav. 6: 455–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Sharfman M., Ron M., Avni A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63: 791–800 [DOI] [PubMed] [Google Scholar]
- Bateman A., Bycroft M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299: 1113–1119 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Borner G.H., Lilley K.S., Stevens T.J., Dupree P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang S., Xing Y., Xu C., Hayes P.M., Zhang Q. (2003). Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc. Natl. Acad. Sci. USA 100: 2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang S., Zhang Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology 92: 750–754 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Danna C.H., Millet Y.A., Koller T., Han S.W., Bent A.F., Ronald P.C., Ausubel F.M. (2011). The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. USA 108: 9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R., Gupta D. (2005). Peptidoglycan recognition in innate immunity. J. Endotoxin Res. 11: 304–310 [DOI] [PubMed] [Google Scholar]
- Erbs G., Newman M.A. (2012). The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol. Plant Pathol. 13: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G., Silipo A., Aslam S., De Castro C., Liparoti V., Flagiello A., Pucci P., Lanzetta R., Parrilli M., Molinaro A., Newman M.A., Cooper R.M. (2008). Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: Structure and activity. Chem. Biol. 15: 438–448 [DOI] [PubMed] [Google Scholar]
- Fliegmann J., Uhlenbroich S., Shinya T., Martinez Y., Lefebvre B., Shibuya N., Bono J.J. (2011). Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol. Biochem. 49: 709–720 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gust A.A., Biswas R., Lenz H.D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa E., Mitsutomi M., Nagano Y. (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Bevan M., Kavanagh T. (1987). The use of the Escherichia coli beta-glucuronidase as a gene fusion marker for studies of gene expression in higher plants. Biochem. Soc. Trans. 15: 17–18 [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chah O.K., Sheen J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Han S.W., Sririyanum M., Park C.J., Seo Y.S., Ronald P.C. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., Klaus-Heisen D., Pietraszewska-Bogiel A., Hervé C., Camut S., Auriac M.C., Gasciolli V., Nurisso A., Gadella T.W.J., Cullimore J. (2012). Role of N-glycosylation sites and CXC motifs in trafficking of Medicago truncatula Nod factor perception protein to plasma membrane. J. Biol. Chem. 287: 10812–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Z., Song C., Hu Y., Han Z., She J., Fan F., Wang J., Jin C., Chang J., Zhou J.M., Chai J. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164 [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Millet Y.A., Danna C.H., Clay N.K., Songnuan W., Simon M.D., Werck-Reichhart D., Ausubel F.M. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T., Brunner F., Kemmerling B., Piater L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198: 249–266 [DOI] [PubMed] [Google Scholar]
- Pérez F., Rubio S. (2006). An improved chemiluminescence method for hydrogen peroxide determination in plant tissues. Plant Growth Regul. 48: 89–95 [Google Scholar]
- Petutschnig E.K., Jones A.M., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Ronald P.C., Beutler B. (2010). Plant and animal sensors of conserved microbial signatures. Science 330: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A., Erbs G., Shinya T., Dow J.M., Parrilli M., Lanzetta R., Shibuya N., Newman M.A., Molinaro A. (2010). Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology 20: 406–419 [DOI] [PubMed] [Google Scholar]
- Silipo A., Molinaro A., Sturiale L., Dow J.M., Erbs G., Lanzetta R., Newman M.A., Parrilli M. (2005). The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J. Biol. Chem. 280: 33660–33668 [DOI] [PubMed] [Google Scholar]
- Song W.Y., Wang G.L., Chen L.L., Kim H.S., Pi L.Y., Holsten T., Gardner J., Wang B., Zhai W.X., Zhu L.H., Fauquet C., Ronald P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Takai R., Isogai A., Takayama S., Che F.S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Dry I.B., Soole K.L. (1997). Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 405: 1–4 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tjoelker L.W., Gosting L., Frey S., Hunter C.L., Trong H.L., Steiner B., Brammer H., Gray P.W. (2000). Structural and functional definition of the human chitinase chitin-binding domain. J. Biol. Chem. 275: 514–520 [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Wang H., Zhang D., Liu B., Wang J. (2007). Rice repetitive DNA sequence RRD3: A plant promoter and its application to RNA interference. J. Genet. Genomics 34: 258–266 [DOI] [PubMed] [Google Scholar]
- Zhou L.Y., Jiang D.G., Wu H., Zhuang C.X., Liu Y.G., Mei M.T. (2005). [Development of transformation system of rice based on transformation-competent artificial chromosome (TAC) vector]. Yi Chuan Xue Bao 32: 514–518 [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.