Grain texture is a key agronomic trait for cereal crops. The positional cloning of maize classical seed mutant opaque1 revealed a molecular mechanism for seed hardness determination. The findings highlight that maize myosin XI-I plays an important role in protein body biogenesis by affecting endoplasmic reticulum morphology and motility.
Abstract
Myosins are encoded by multigene families and are involved in many basic biological processes. However, their functions in plants remain poorly understood. Here, we report the functional characterization of maize (Zea mays) opaque1 (o1), which encodes a myosin XI protein. o1 is a classic maize seed mutant with an opaque endosperm phenotype but a normal zein protein content. Compared with the wild type, o1 endosperm cells display dilated endoplasmic reticulum (ER) structures and an increased number of smaller, misshapen protein bodies. The O1 gene was isolated by map-based cloning and was shown to encode a member of the plant myosin XI family (myosin XI-I). In endosperm cells, the O1 protein is associated with rough ER and protein bodies. Overexpression of the O1 tail domain (the C-terminal 644 amino acids) significantly inhibited ER streaming in tobacco (Nicotiana benthamiana) cells. Yeast two-hybrid analysis suggested an association between O1 and the ER through a heat shock protein 70–interacting protein. In summary, this study indicated that O1 influences protein body biogenesis by affecting ER morphology and motility, ultimately affecting endosperm texture.
INTRODUCTION
In cereals, the endosperm stores carbohydrates and proteins, which provide nutrients during embryo development and germination as well as food for humans and raw materials for industry (Wang et al., 2012a). The endosperm contains four major cell types: transfer cells, aleurone cells, starchy endosperm cells, and a region of embryo-surrounding cells (Sabelli and Larkins, 2009). The starchy endosperm contributes 80 to 90% of the grain weight and plays a determinant role in seed texture and nutritional quality. In maize (Zea mays), it typically consists of two distinct regions: the vitreous endosperm and the starchy endosperm. Kernel hardness, a key agronomic trait in maize, is influenced by the relative amounts of vitreous and opaque portions. It has been hypothesized that there is a positive correlation between zein proteins and kernel vitreousness (Moose et al., 2004). Therefore, either quantitative or qualitative reductions in zein proteins may result in starchy or opaque endosperm phenotypes.
Dozens of soft and starchy endosperm maize mutants have been identified (Neuffer et al., 1997). At least 18 of these have been positioned on the genetic map (Gibbon and Larkins, 2005). So far, only seven genes corresponding to these mutants have been cloned, including opaque2 (o2), o5, o7, floury1 (fl1), fl2, Mucronate (Mc), and Defective endosperm B30 (De-B30) (Schmidt et al., 1990; Coleman et al., 1997; Kim et al., 2004, 2006; Holding et al., 2007; Miclaus et al., 2011; Myers et al., 2011; Wang et al., 2011b). The majority of these mutants result from defects in zein protein synthesis. O2 encodes a bZIP transcription factor that regulates expression of 22-kD α-zeins (Schmidt et al., 1990). O7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis, particularly the 19- and 22-kD α-zeins (Wang et al., 2011b). Other mutations directly affect zein genes themselves: fl2 encodes a defective 22-kD α-zein (Coleman et al., 1997); Mc encodes a defective 16-kD γ-zein (Kim et al., 2006), and De-B30 encodes a defective 19-kD α-zein (Kim et al., 2004). The protein bodies in these mutants are small and misshapen.
Correspondingly, direct disruption of zein genes expression by RNA interference phenocopies the opaque phenotype (Segal et al., 2003; Wu and Messing, 2010). Thus, it is likely that the soft, opaque phenotype of the mutant endosperm is related to altered zein protein accumulation and packing. However, some opaque/floury mutants, such as o1, o5, and fl1, show no notable alterations in zein proteins (Hunter et al., 2002). FL1 encodes an endoplasmic reticulum (ER) membrane protein possibly involved in targeting 22-kD α-zeins to the interior of protein bodies (Holding et al., 2007). O5 encodes a monogalactosyldiacylglycerol synthase (MGD1) that affects amyloplast membranes surrounding starch granules (Myers et al., 2011). Transcript profiling showed that the unfolded protein response (UPR) is stimulated in many opaque mutants, including o1, o2, fl2, Mc, and De-B30 (Hunter et al., 2002). Although the defect of several opaque/floury mutants has been determined at the molecular level, the mechanism underlying endosperm opacity remains puzzling and elusive.
o1 is a classical recessive opaque mutant (Emerson et al., 1935). It was shown that, in the W64A background, the amount of protein (zein and nonzein) and their amino acid compositions in o1 endosperm are nearly identical to the wild type (Nelson et al., 1965; Hunter et al., 2002). Although o1 does not have a higher Lys content than the wild type, the molecular nature of its action could provide invaluable insight into factors that influence seed hardness.
Here, we report the map-based cloning of o1, which encodes a plant-specific myosin XI protein. The actin-based actomyosin system plays an essential role in the organization and dynamics of the plant cell interior, including the endomembrane system and trafficking network (Sparkes, 2011). Plant genomes contain two classes of myosins, VIII and XI; the latter is closely related to the metazoan and fungal class V myosins (Avisar et al., 2008). Metazoan and fungal myosin V proteins are processive molecular motors involved in organelle and vesicle transport and partitioning during cell division, mitotic spindle positioning, mRNA localization, and the establishment of cell polarity (Li and Nebenführ, 2008; Hammer and Sellers, 2012). By contrast, very little is known about the functions of plant myosins and most of the data have been obtained from immunological, physiological, and biochemical studies. Myosin VIII proteins are associated with endocytosis, cytokinesis, and plasmodesmal function (Reichelt et al., 1999; Golomb et al., 2008; Wu et al., 2011), whereas class XI myosins are involved in cytoplasmic streaming and organelle motility and structure (Li and Nebenführ, 2007; Avisar et al., 2008, 2009; Sparkes et al., 2009a; Ueda et al., 2010; Yokota et al., 2009, 2011).
Functional characterization of plant myosin XI has also been retarded due to the functional redundancy of different isoforms present in genome. For example, among all 13 class XI myosins of Arabidopsis thaliana, only XIK and MYA2 resulted in detectable phenotypes under normal growth conditions (Peremyslov et al., 2008). Single mutants of either gene exhibited shorter root hairs. The double mutant xik mya2 displayed reduced fecundity and was stunted, suggesting that XIK and MYA2 have overlapping and additive effects on the root hair elongation (Prokhnevsky et al., 2008). Moreover, triple and quadruple myosin mutation analysis indicated that myosin is required for both polarized elongation and diffuse growth of several plant cell types (Peremyslov et al., 2010). In the moss Physcomitrella patens, simultaneous silencing of myosin XIA and XIB resulted in severely stunted plants, indicating that myosin is essential for tip growth (Vidali et al., 2010).
We used a forward genetics strategy to elucidate the function of myosin XI/O1 in maize endosperm development. The mutations in O1 cause dilated ER; small, misshapen protein bodies; and an opaque endosperm phenotype. Subcellular fractionation assays demonstrated that O1 is associated with the ER and protein bodies. Dominant-negative study indicated that O1 is responsible for ER motility. We provide evidence for the function of class XI myosins in the organization and movement of the ER network and the biogenesis of protein bodies from ER.
RESULTS
The Maize o1 Mutant Produces Dilated ER and Small, Misshapen Protein Bodies
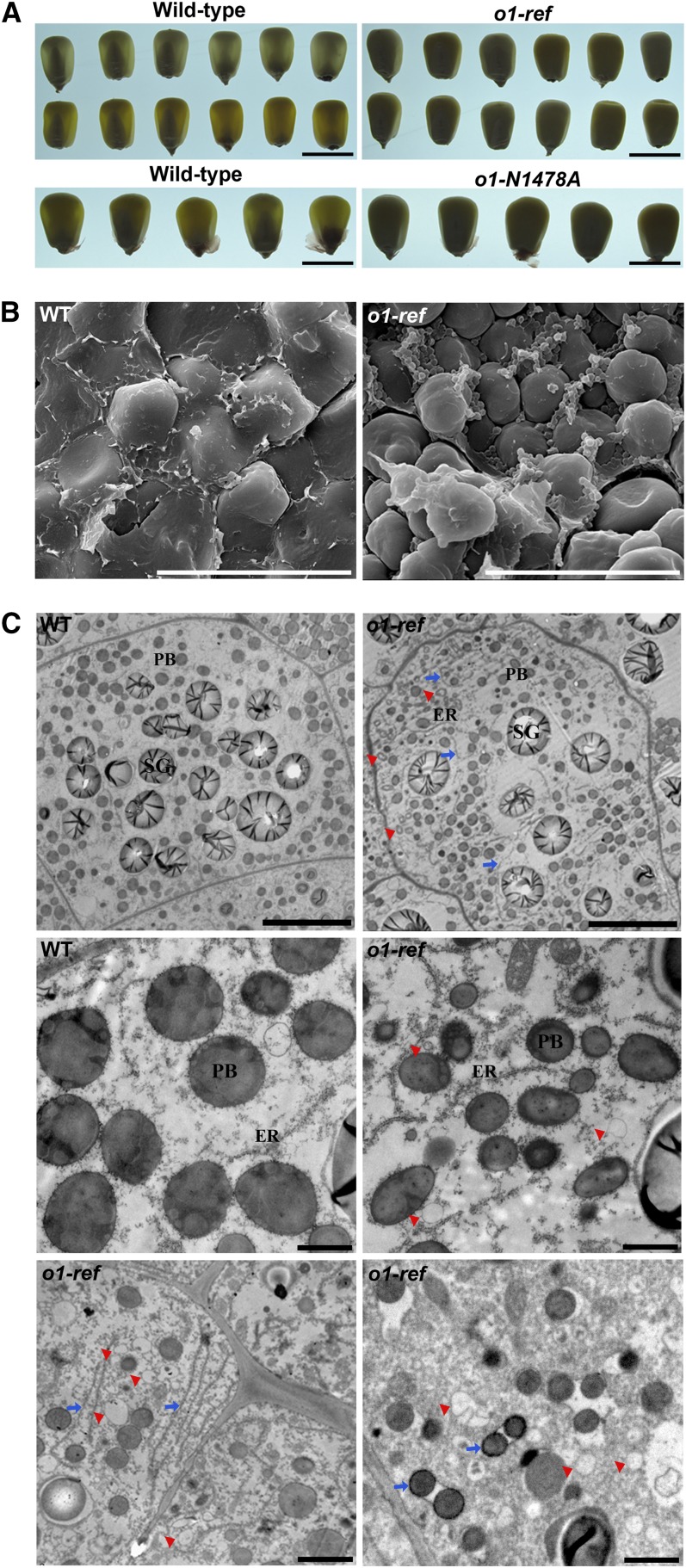
The o1-ref mutant obtained from the Maize Genetics Cooperation stock center was introgressed into the W22 background until the backcross (BC) 4 generation. The mutant kernels have an obvious opaque appearance at maturity (Figure 1A). To investigate the mature endosperm architecture, kernels of o1-ref and the wild type were analyzed by scanning electron microscopy. Compared with the wild type, o1-ref had observable alterations at the peripheral part of endosperm. In the normally vitreous region, the starch granules in o1-ref were loosely packed. There were no prominent contacts between starch granules and protein bodies (right panel, Figure 1B). Starch granules in the wild type were embedded in a matrix that completely filled the spaces between them (left panel, Figure 1B). We also compared the ultrastructure of endosperm cells at 25 d after pollination (DAP) using transmission electron microscopy. In wild-type endosperm cells, protein bodies (1 to 2 μm in diameter) were spherical and surrounded by rough ER (left panel, Figure 1C). In o1-ref endosperm, protein bodies were smaller (0.5 to 1 μm in diameter) and were irregularly shaped (right panel, Figure 1C). The cisternal ER was often dilated in the o1-ref endosperm (Figure 1C, blue arrow). Numerous small polygonal ring or vesicle-like structures accumulated peripheral to the cell walls in o1-ref (Figures 1C, red arrowhead). These structures seem to originate from the endomembrane system.
Figure 1.
Phenotypic Features of Maize o1 Mutants.
(A) Light transmission by mature kernels. Wild-type and mutant kernels were randomly selected from BC ears of o1-ref and o1-N1478A and viewed on a light box. Bars = 1 cm.
(B) Scanning electron microscopy analysis of the peripheral regions of mature wild-type (WT) and o1 endosperm. Bars = 30 μm.
(C) Ultrastructure of developing endosperms of the wild type (WT) and o1. Top (25 DAP): Low-magnification images of endosperm cells in the wild type and o1. Bars = 10 μm. Middle (25 DAP): Smaller and misshapen protein bodies in o1. Bars = 1 μm. Bottom (20 DAP): Dilated ER (blue arrows) and polygonal ring structures (red arrowheads) in o1. Bars = 2 μm. PB, protein body; SG, starch granules.
Mutations of O1 Do Not Impair Zein Biosynthesis
Zeins overwhelmingly accumulate in protein bodies and appear to make an essential contribution to the kernel vitreousness. We measured the protein content of the endosperm from o1-ref to determine if it caused large qualitative or quantitative differences in zein and nonzein proteins composition compared with the wild type. The analysis indicated that the amount of total proteins, zeins, and nonzeins in o1 mutants were similar to the wild type (Table 1). This result was also validated by SDS-PAGE, which indicated no obvious difference in the major zein components between different o1 alleles and their corresponding wild-type genotypes (see Supplemental Figure 1 online).
Table 1. Protein Contents of o1-ref and Wild-Type Endosperm.
| Genotype | Total Protein | Zeins | Nonzeins |
|---|---|---|---|
| Wild type | 91.00 ± 1.18 | 71.92 ± 0.90 | 22.92 ± 0.16 |
| o1-ref | 91.25 ± 1.08 | 72.67 ± 0.54 | 22.58 ± 0.49 |
| P value | 0.881 | 0.501 | 0.544 |
Values shown are mg of protein per g of endosperm dry weight ± se. P values are from Student’s t tests, compared to the wild type.
Noticeably, the zein content was not altered, although the protein bodies are smaller and misshapen in o1 mutants compared with the wild type. To explain this, we quantified the number and size of protein bodies in the fourth and fifth endosperm cell layers from aleurone at 20 and 25 DAP (Table 2). At 25 DAP, in o1-ref endosperm, the average protein body area was 0.79 μm2, or ∼60% of that in the wild type (1.40 μm2). However, the number of protein bodies in o1-ref was increased ∼1.3-fold compared with the wild type. Thus, it appears that protein bodies in o1-ref are smaller but their larger numbers compensate to maintain total zein content equivalent to wild-type.
Table 2. Size and Number of Protein Bodies in the Fourth and Fifth Cell Layers from Aleurone of o1-ref and Wild-Type Endosperm at 20 and 25 DAP.
| Genotype | Stage (DAP) | Mean Area (μm2) | se | P Value | Number (per 100 μm2) | se | P Value |
|---|---|---|---|---|---|---|---|
| Wild type | 20 | 1.35 | 0.80 (n = 230) | 23.00 | 5.57 (n = 15) | ||
| o1-ref | 20 | 0.74 | 0.55 (n = 382) | <0.01 | 31.83 | 6.34 (n = 19) | <0.01 |
| Wild type | 25 | 1.40 | 0.50 (n = 381) | 22.80 | 4.34 (n = 16) | ||
| o1-ref | 25 | 0.79 | 0.42 (n = 564) | <0.01 | 28.73 | 6.77 (n = 22) | <0.01 |
p values are from Student’s t tests.
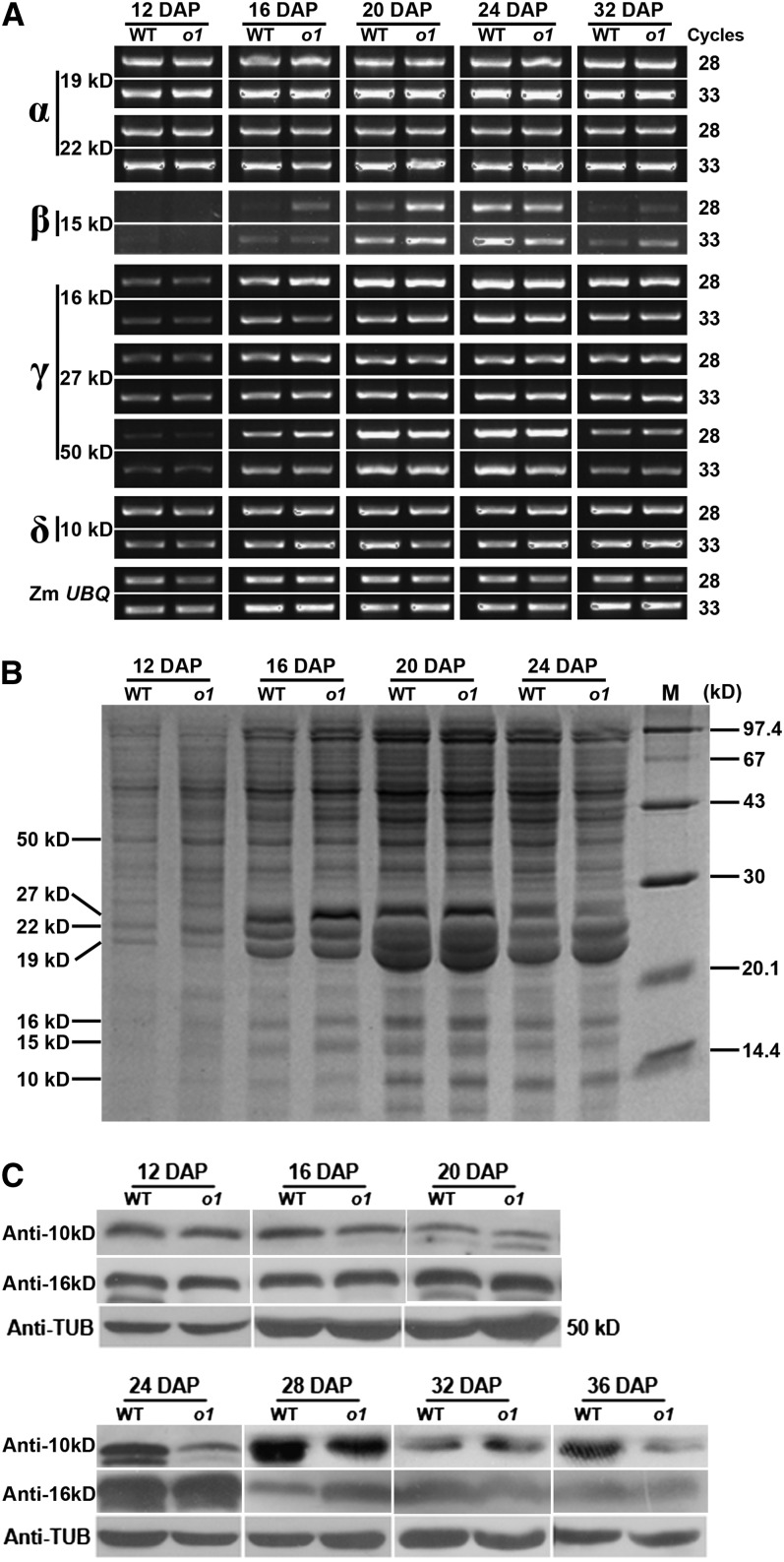
To ascertain whether zein synthesis contributes to misshapen protein bodies in the o1-ref mutant, zein gene expression was examined temporally during maize kernel development at the transcriptional and translational levels. The results indicated that the expression of members in α-, β-, γ-, and δ-zein classes was similar between o1-ref mutant and its wild-type counterpart (Figure 2).
Figure 2.
Comparison of Zein Biosynthesis and Accumulation in Wild-Type and o1-ref during Kernel Development.
(A) RNA expression profiles of representative zein genes. WT, the wild type.
(B) SDS-PAGE analysis of total protein extracted from endosperm.
(C) Immunoblot comparing 16- and 10-kD zein accumulation in wild-type and o1-ref developing endosperms.
Positional Cloning of O1
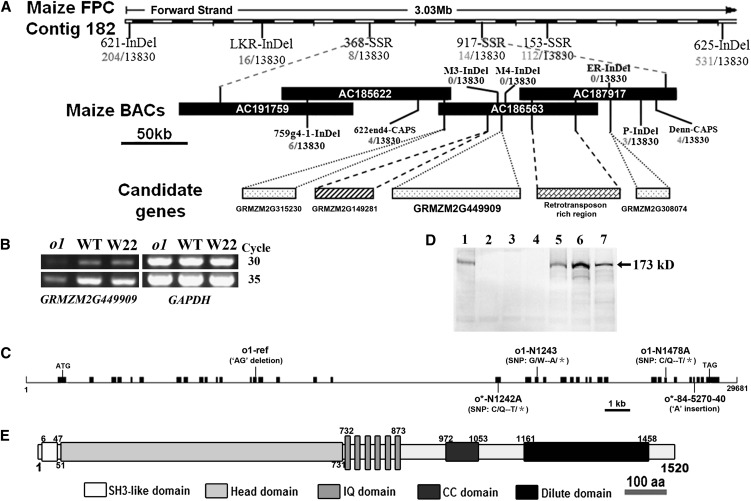
We used a map-based cloning approach to isolate the O1 gene. We first crossed the o1-ref mutant into the W22 inbred line and then backcrossed with o1-ref to generate a BC mapping population. The o1 locus was initially placed between the simple sequence repeat markers umc2187 and bnlg2291 in Bin 4.07. By screening a mapping population of nearly 14,000 individual BC lines, o1 was mapped between markers 662end4-CAPS (four recombinants) and P-InDel (three recombinants) (Figure 3A). This interval is ∼250 kb in length and contains one complete (AC186563) and two partial (AC185622 and AC187917) BAC clones, using the molecular markers listed in Supplemental Table 1 online.
Figure 3.
Map-Based Cloning and Identification of O1.
(A) The o1 locus was mapped to a 250-kb region on chromosome 4 with four candidate genes. See Supplemental Table 1 online for detailed information.
(B) The candidate gene GRMZM2G449909 is downregulated in o1. WT, the wild type.
(C) Structure and mutation sites of the O1 gene. Lines represent introns, and black boxes represent exons.
(D) Immunoblot comparing accumulation of O1 protein in endosperms of the wild type, o1 alleles, and other opaque mutants at 20 DAP. Lanes: 1, the wild type; 2, o1-ref; 3, o1-N1478A; 4, o1-N1243; 5, o5; 6, o7; 7, W22.
(E) Schematic diagram of maize O1 protein structure. aa, amino acids.
Annotation analysis of this 250-kb DNA segment identified four putative open reading frames and a 90-kb retrotransposon-rich region (Figure 3A; see Supplemental Table 2 online). Expression analysis revealed that one of the open reading frames, GRMZM2G449909 (collinear to rice [Oryza sativa] Os02g0777700), was downregulated in o1-ref (Figure 3B). Genomic DNA sequences of the four candidate genes revealed a 2-bp deletion (of AG) in GRMZM2G449909 exon 14 of o1-ref, causing a frameshift and a premature stop codon (Figure 3C). The GRMZM2G449909 gene region was examined in four additional o1 allelic mutants (Figure 3C). In o1-N1243, a G/A transition at exon 21 would change a Trp to a stop codon. A C/T transition at exon 32 would also cause a truncated protein, from Gln to stop in o1-N1478A. In two newly identified o1 alleles, o*-84-5270-40 has a 1-bp insertion (of A) at exon 35 would result in a premature stop codon, and o*-N1242A has a C/T transition at exon 19 would change a Gln to a stop codon. The corresponding cDNAs for these five o1 alleles as well as for wild-type inbreds W22 and B73 were amplified by RT-PCR and sequenced, and the results confirmed the genomic sequences from the mutants and inbred lines.
Using affinity-purified antibody against the protein encoded by GRMZM2G449909, we determined that a protein with an expected molecular mass of 173 kD is present in developing wild-type kernels, whereas this protein was not detectable in the three analyzed o1 mutants (Figures 3D). Immunoblot analysis revealed that this protein is present in developing kernels of other opaque mutants, such as o5 and o7. Together, these results are consistent with GRMZM2G449909 encoding the O1 protein.
O1 Encodes a Plant-Specific Myosin XI Motor Protein
Analysis of genomic DNA data (http://www.maizeGDB.org) revealed that the O1 (GRMZM2G449909) gene spans ∼30 kb and contains 38 exons and 37 introns (Figure 3C). The longest identified mature transcript (full length) is a transcript 5131 bp in length with a transcript (open reading frame) having 4563 bp of coding sequence and noncoding regions of 205 and 360 bp at the 5′- and 3′-ends, respectively. It is predicted to encode a 173-kD protein of 1520 amino acids (Figure 3D).
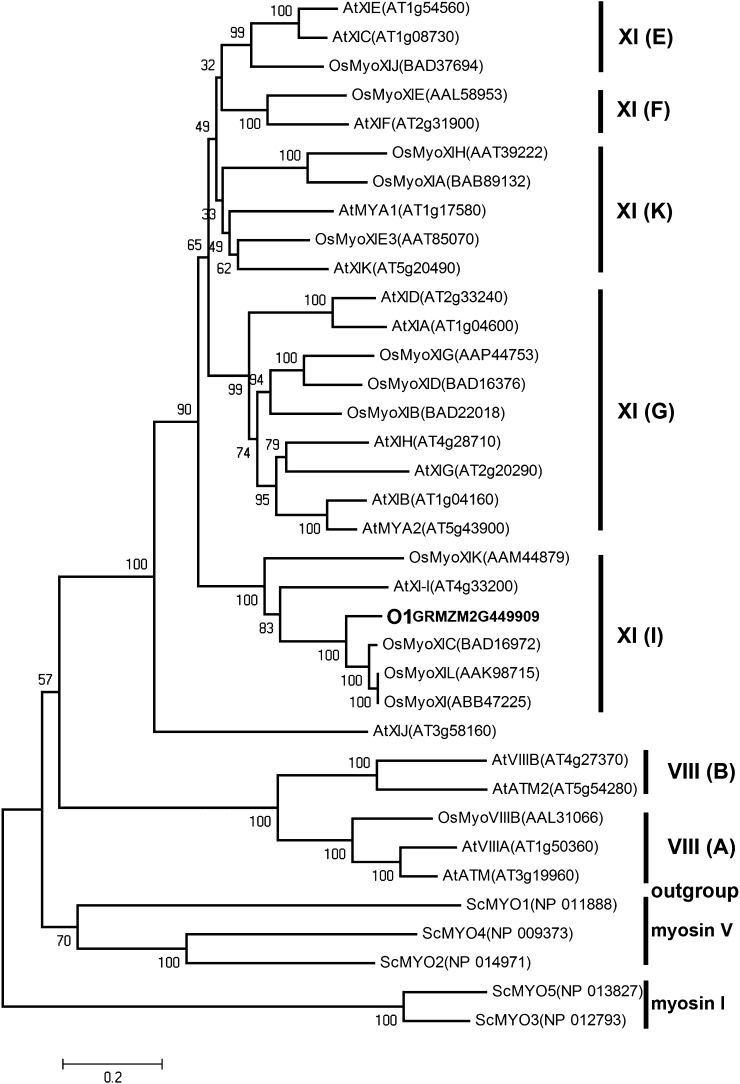
This gene encodes a putative plant-specific myosin XI motor protein with several conserved domains (Figure 3E; see Supplemental Figure 2 and Supplemental Table 3 online). The N-terminal SH3-like domain is 41 amino acids long and occurs in many other myosins with unknown function. The myosin head motor domain is composed of 681 amino acids, with predicted function as actin binding, ATP binding and hydrolysis, and force generation. The neck domain contains six IQ motifs and serves as binding sites for myosin light chains and calmodulin. The coiled-coil domain consists of ∼82 amino acids and is used for homodimerization. In addition, the protein has a DIL (dilute; Mercer et al., 1991) domain that anchors itself to cargoes via attachments to organelle-specific receptors at the C terminus. We used the sequences of the full-length myosin proteins to make a phylogenetic tree with members from the genomes of Arabidopsis, rice, and yeast (Saccharomyces cerevisiae) (Figure 4). The resulting topology is consistent with a hypothesis of two myosin ancestors (class VIII and class XI) that existed in land plants before lineage-specific expansions (Peremyslov et al., 2011). O1 was placed in a well-supported clade XI (I) composed of Arabidopsis XI-I and four rice myosins, suggesting a burst of gene duplication in the rice genome. Unlike in rice, O1 appeared to be the only member belonging to this clade in the maize genome. Unexpectedly, Arabidopsis XI-C, E, and J were not clustered into subgroup XI (J) (Peremyslov et al., 2011) but were in a highly credible clade that we named XI (E).
Figure 4.
Phylogenetic Analysis of Plant Myosins.
Maize O1 and currently identified myosins in rice (monocot) and Arabidopsis (dicot) plants were aligned by ClustalW. The phylogenetic tree was constructed using MEGA 5.0, and yeast class I and V myosins were used as outgroups (see Methods). At, Arabidopsis thaliana; Os, Oryza sativa; Sc, Saccharomyces cerevisiae. See Supplemental Data Set 1 online for the sequences and alignment used.
O1 Is Constitutively Expressed at a High Level in Developing Kernel
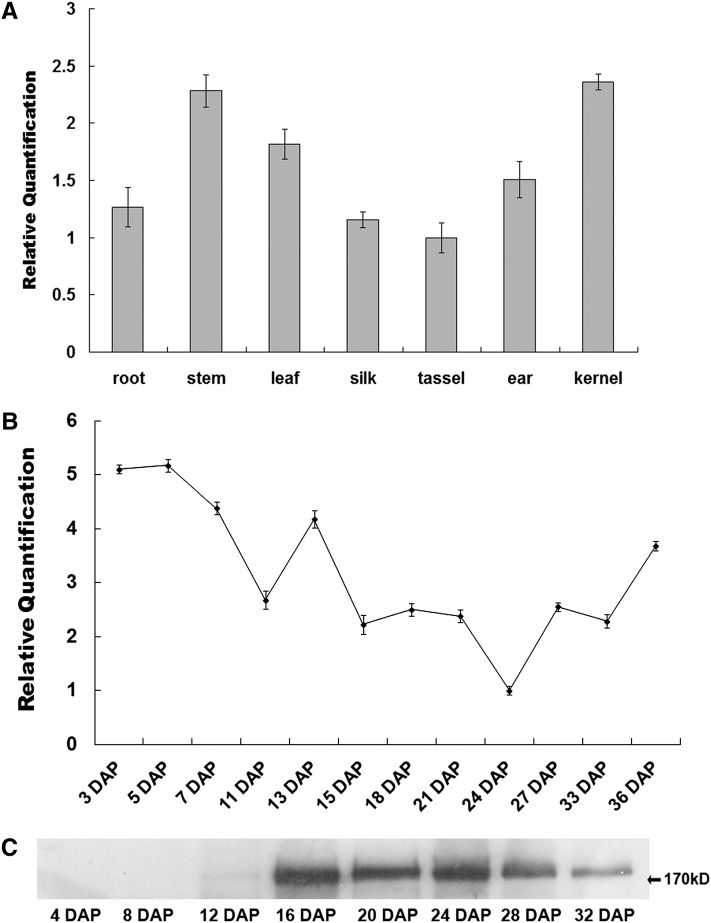
The spatial expression of O1 was examined by real-time quantitative RT-PCR. O1 is constitutively expressed in all tested tissues (Figure 5A). The transcript levels of O1 are high in kernel and stem, intermediate in ear and leaf, and low in root, silk, and tassel. During kernel development, the expression of O1 was high at an early stage, then decreased and fluctuated at a mid stage, and finally slightly increased at a late stage (Figure 5B). The O1 protein was first detectable at 12 DAP and accumulated in abundance, peaking between 16 and 28 DAP (Figure 5C). The inconsistency between RNA level and protein accumulation suggests that O1 may be posttranscriptionally regulated.
Figure 5.
Expression Pattern of O1.
(A) RNA expression level of O1 in various tissues. Ubiquitin was used as an internal control. For each RNA sample, three technical replicates were performed. Representative results from two biological replicates are shown. Error bars represent sd. Transcript abundance is indicated relative to tassel.
(B) Expression profiles of O1 during maize kernel development. Ubiquitin was used an internal control. For each RNA sample, three technical replicates were performed. Representative results from two biological replicates are shown. Error bars represent sd. Transcript abundance is indicated relative to 24-DAP kernels.
(C) Immunoblot analysis of O1 protein accumulation during kernel development.
O1 Is Predominantly Associated with the ER Membrane
To determine the subcellular localization of the O1 protein, we first attempted to use an eYFP-O1t reporter, which harbored a globular tail domain fused with enhanced yellow fluorescent protein (eYFP) at the N terminus (see Supplemental Figure 3A online). However, the subcellular location of the O1 tail fusion varied, from the cytosol and punctuate structures to the nuclear envelope (see Supplemental Figures 3B to 3D online). Consequently, we used a fractionation approach to detect the presence of O1 in the subcellular fractions using the O1-specific antibody.
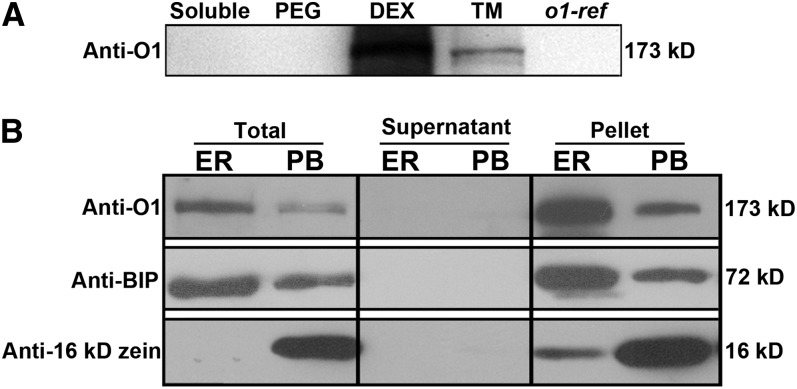
Total proteins extracted from 20-DAP kernels were separated into soluble, total membrane, plasma membrane (polyethylene glycol), and endomembrane (dextran T500) fractions. O1 was most abundant in the endomembrane fraction and was barely detectable in the soluble protein or plasma membrane fractions (Figure 6A). Discontinuous Suc gradient centrifugation of a microsomal fraction was performed to separate cisternal ER and protein body membranes, two core organelles essential for storage protein accumulation (Habben et al., 1993). The O1 protein was abundantly present in the ER membrane fraction and also existed in protein body fraction (Figure 6B). These data indicated that O1 protein is likely associated with the intracellular membrane system, particularly the ER membrane.
Figure 6.
O1 Is Associated with the ER Membrane.
(A) Immunoblotting of O1 in fractionated plant proteins with anti-O1 antibody.
(B) Immunoblots showing that O1 is preferentially associated with the ER membrane fraction. Fractions were subjected to immunoblot analysis with antibodies against O1, BIP (ER marker), or 16-kD zein (protein body [PB] marker). DEX, the dextran T500 phase containing the endomembrane fraction; PEG, the polyethylene glycol 3350 phase containing the plasma membrane fraction; TM, total membranes.
O1 Does Not Directly Interact with Some Intrinsic ER or Protein Body Proteins
In yeast and plants, it has been shown that myosin plays an important role in forming and extending ER tubules (Estrada et al., 2003; Yokota et al., 2011). Several integral membrane proteins, including the reticulon family protein Rtn4a/NogoA (Voeltz et al., 2006) and its interacting protein DP1/Yop1p (Shibata et al., 2008), and a dynamin family GTPase atlastin Sey1p (Hu et al., 2009), are indispensable in the generation of the tubular ER network. To ascertain whether myosin is required for the formation and regulation of tubular ER structures in vivo, putative maize orthologs were isolated and tested for interaction with O1 using the yeast two-hybrid assay. As shown in Supplemental Figure 4A online, none of the proteins tested directly interacted with the O1 cargo binding domain. Similarly, neither zeins nor FL1, intrinsic proteins of protein bodies, interacted with O1 in the yeast two-hybrid assay (see Supplemental Figure 4B online).
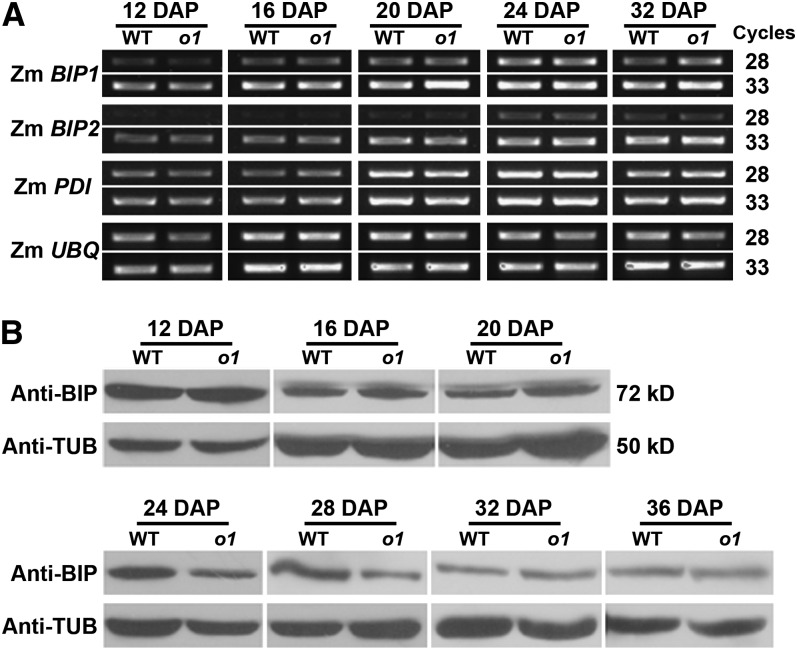
A common feature of previously characterized opaque mutants is elevated expression of molecular chaperones, such as heat shock protein 70 (HSP70) and PDI, suggesting that a stimulated UPR is the possible cause of kernel opacity (Hunter et al., 2002). To investigate this possibility for o1, we examined the expression of chaperone genes. Transcript levels of maize BIP1, BIP2, and PDI did not differ between o1-ref and the wild type in developing seeds (Figure 7A). The level of ER lumen binding protein (BIP) was also not markedly affected in o1-ref during seed development (Figure 7B). Therefore, a mechanism other than UPR appears to be responsible for the opaque phenotype of o1.
Figure 7.
Stimulated UPR Was Not Observed in the o1 Mutant.
(A) RT-PCR analysis of ER stress sensors Zm BIP1, Zm BIP2, and Zm PDI transcript in the wild type (WT) and o1 during kernel development. Ubiquitin was used as an internal control.
(B) Immunoblot comparing BIP accumulation in wild-type and o1 developing kernels. Anti-TUB was used as a sample loading control.
Overexpression of the O1 Tail Domain Significantly Inhibits ER Motility in Tobacco
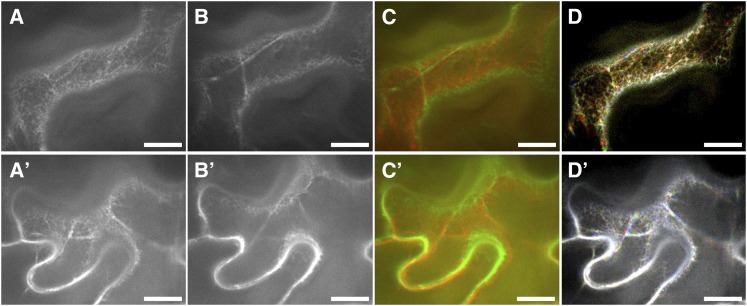
To further explore the functional relationship between O1 and the ER, a dominant-negative strategy was employed to investigate the role of O1 in movement of the ER network. We transiently coexpressed the O1 tail domain fused downstream to an eYFP construct with an ER marker (mCherry-HDEL) in tobacco (Nicotiana benthamiana) leaves using agroinfiltration. ER dynamics were analyzed for at least 30 40- and 80-s movies with 2- and 1.6-s intervals, respectively, in four independent experiments. The streaming patterns of ER in control cells solely expressing mCherry-HDEL are shown in Figures 8A to 8D, which includes the first and last frames of a 50-frame movie, the overlapped image of the first and last frames, and the merged image of three successive frames with red, green, and blue, respectively (see Supplemental Movies 1 and 2 online). As illustrated in Figure 8D (the brighter the more static), the ER movement velocity was very fast, comparable to that previously reported in tobacco and Arabidopsis (Sparkes et al., 2009a; Ueda et al., 2010). Compared with controls, ER motility was dramatically suppressed when the O1 tail domain was coexpressed (Figures 8D and 8D’; see Supplemental Movies 3 and 4 online). The ER morphology was generally similar between O1 truncation and control cells (Figures 8A, 8A’, 8B, and 8B’). The interior region of the ER network is more dynamic whereas the peripheral ER is more static. These results indicate that O1 is associated with ER streaming.
Figure 8.
ER Streaming Is Suppressed in the Presence of a Dominant-Negative O1 Tail Construct Fused to eYFP.
(A) to (D) N. benthamiana leaves were infiltrated with the ER marker, mCherry-HDEL, alone.
(A) and (B) The first (A) and last frame (B) of a 50-frame 80-s movie (see Supplemental Movie 2 online).
(C) Merger of the first (colored red) and last frame (colored green) showing the total motility pattern.
(D) Three random, successive frames are colored green, red, and blue, respectively, and then overlapped together to generate an integrity image.
(A’) to (D’) N. benthamiana leaves were infiltrated with the ER marker, mCherry-HDEL, with eYFP-O1t. One image was acquired including the two channels to ensure that both reporters were present in the same cell, and a time-lapse movie was acquired using only the mCherry channel.
(A’) and (B’) The first (A’) and last frame (B’) of a 50-frame 80-s movie (see Supplemental Movie 4 online).
(C’) Merger of the first (colored red) and last frame (colored green) showing total motility pattern.
(D’) Three random, successive frames are colored green, red, and blue, respectively, and then overlapped together to generate an integrity image.
The brighter color indicates the slower movement. Bars = 10 μm.
Association between O1 and the ER Might Depend on an HSP70-Interacting Protein
To identify the adaptor responsible for O1 and ER association, a yeast two-hybrid assay was performed to search for O1 interacting proteins. The cargo binding tail domain of O1 was fused in frame with the GAL4 DNA binding protein. This bait, without self-activation, allowed us to screen a developing maize kernel cDNA library for interacting proteins (Wang et al., 2010). After screening 8 × 106 clones, we identified seven putative O1-interacting proteins, which included some membrane-related proteins (Table 3).
Table 3. Summary of Putative Proteins Interacting with O1 Using a Yeast Two-Hybrid Assay.
| Contiga | BLASTNb | Identityc | Predicted Functiond |
|---|---|---|---|
| 1 | NM_001150131.1 | 99 | Pleckstrin homology domain–containing protein |
| 2 | XM_002468636.1 | 93 | Putative kinesin-related protein |
| 3 | NM_001112518.1 | 98 | Putative splicing factor |
| 4 | XM_002443897.1 | 91 | HIP |
| 5 | NM_001196447.1 | 97 | Pro-rich receptor-like protein kinase |
| 6 | NM_001154497.1 | 98 | Chloroplast outer envelope protein |
| 7 | NM_001175737.1 | 99 | β-Coat protein |
Assembly of independent positive clones after DNA sequencing.
GenBank accession numbers with the highest nucleotide sequence similarity to the Y2H clone shown in the left column.
The percentage of nucleotide sequence identity based on BLASTN analysis.
Based on the BLASTX result for the TopBLASTN sequences.
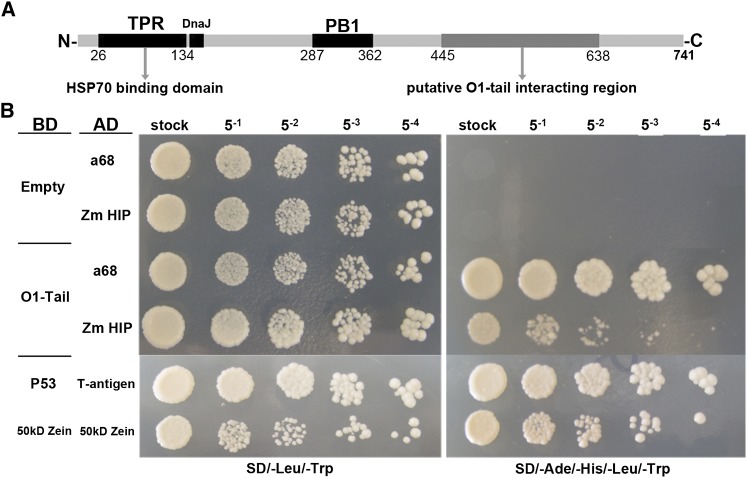
Considering that O1 is associated with the ER, we focused primarily on the interaction of O1 with a HSP70-interacting protein (HIP), which is a tetratricopeptide repeat protein (TPR) that interacts with the ATPase domain of chaperons, such as HSP70 and HSP90 (Prapapanich et al., 1996). The HIP gene (GRMZM2G023275) encodes a protein of 741 amino acids with a molecular mass of 85 kD and includes N-terminal TPR and DnaJ domains and a middle PB1 domain (Figure 9A). All the identified HIP independent clones contain a common fragment, which is probably the putative O1 tail interacting region (Figure 9A). We further tested the interaction between O1 and the full-length HIP protein. As shown in Figure 9B, we found an identified clone a68 containing only the HIP C terminus (from Glu-445 to the stop codon) that showed a stronger interaction than the full-length HIP with the O1 tail domain. Based on these data, we propose HIP acts as an adaptor, which binds O1 at its C terminus and binds the HSP70 and HSP90 proteins by the N-terminal TPR domain (Prapapanich et al., 1996).
Figure 9.
Interaction between O1 and HSP70-Interacting Protein, HIP.
(A) Schematic diagram of maize HIP protein structure.
(B) Yeast two-hybrid interactions between the O1 tail domain and HIP. a68 is one of the identified clones only containing HIP C terminus (from Glu-445 to stop codon). Interaction between T-antigen and P53 and 50-kD zein itself were used as positive control. AD, activating domain; BD, binding domain.
[See online article for color version of this figure.]
DISCUSSION
O1 Encodes a Plant-Specific Myosin XI Motor Protein in Maize
Using a positional cloning approach, we identified O1 as a plant-specific myosin XI gene (GRMZM2G449909) and four additional independent o1 alleles confirmed its identity (Figure 3). Mutations in all five independent alleles cause premature stop codons, generating truncated proteins. Immunoblotting assays failed to detect the expected protein in the three analyzed o1 allelic mutants, suggesting they are also null mutations. Taken together, these data suggest that the identified myosin defects cause the o1 mutant phenotype.
Myosins constitute a superfamily of actin-based motor proteins that generate motion through an ATP hydrolysis cycle. They are involved in various cellular activities within plant cells, such as organelle trafficking (Avisar et al., 2009), ER movement and remodeling (Sparkes et al., 2009a; Ueda et al., 2010), and organelle inheritance (Sheahan et al., 2004). O1 encodes a plant-specific myosin XI protein with several conserved domains, including an N-terminal SH3-like domain, a myosin head, six IQs, a coiled-coil region, and a DIL tail. O1 is an ortholog of Arabidopsis myosin XI-I. However, the function of XI-I is unknown because its T-DNA insertional lines produce no discernible phenotypes during vegetative growth (Peremyslov et al., 2008). The five o1 alleles produce truncated myosin proteins missing the cargo binding tail domain. Interestingly, the o1-N1478A and o*-84-5270-40 alleles lack only a part of the dilute domain but still have a strong mutational phenotype. This suggests that the dilute motif plays an essential role in myosin function. Analogous to yeast myosin myo2p, organelle targeting of plant myosin XI is mediated by the interaction of the two globular tail subdomains GT1 and GT2 (Li and Nebenführ, 2007). Recently, it was proposed that two conserved Arg residues in the dilute domain regulate the cytoplasm dynamics of myosin XIK (see Supplemental Figure 2 online; Avisar et al., 2012).
O1 Affects the Morphology and Motility of the ER Network
The ER is the entry port for secretory proteins of the endomembrane system. It is typically composed of three structurally and functionally distinct subdomains: tubules, cisternae, and the nuclear envelope (Sparkes et al., 2009b). We observed that the ER lumen is often dilated and numerous polygonal ring structures accumulate in o1-ref mutant (Figure 1C). In maize root apical cells, the cortical ER network becomes dilated and vesiculated following treatment with the myosin inhibitor drug 2,3-butanedione-2-monoxime (Samaj et al., 2000). After transient overexpression of the Arabidopsis myosin XIK tail domain in tobacco epidermal cells, persistent ER tubules were longer and more abundant, and they also included polygonal rings (Sparkes et al., 2009a). Furthermore, the xik mya1 mya2 triple knockout mutant had unevenly distributed thick, sheet-like structures of the ER network, large ER aggregates, and abnormally elongated ER bodies (Ueda et al., 2010). Thus, these results indicate that myosin XI is involved in determining proper ER organization, probably by affecting the endomembrane system.
However, colocalization was not found for O1 and the ER. This may be caused by the strong fluorescence of the cytosolic pool of eYFP-O1t, which is similar to that previously reported for myosin XI (Sparkes et al., 2008; Avisar et al., 2009). Nevertheless, our subcellular fractionation assay indicated that O1 is associated with the ER membrane (Figure 6). In Arabidopsis, XIK did not colocalize with ER, but it did physically associate with the ER (Ueda et al., 2010). An antibody against a 175-kD tobacco myosin XI appeared to label the ER in BY-2 suspension cell culture cells and it cosedimented with the ER fraction (Yokota et al., 2009). Notably, it has been shown that myosin plays an essential role in ER tubule formation in yeast and plants (Estrada et al., 2003; Yokota et al., 2011). We failed to detect an interaction between O1 and several putative key components involved in this process (see Supplemental Figure 3A online). One possibility is that we did not yet isolate all putative interacting candidates. For example, reticulon-like proteins belong to large gene families, with 21 isoforms in Arabidopsis (Nziengui et al., 2007). Only five members share ER location, topology, and membrane-shaping properties (Sparkes et al., 2010). Therefore, the question of whether myosin is directly involved in forming ER tubules or with novel participating proteins needs further study.
Because the attachment of cargo, such as organelles, is mediated by myosin tails, overexpression of headless tails is expected to either bind to cargo or titrate out accessory factors required for endogenous myosin recruitment, and these may interfere with the tail binding capacity of organelles and inhibiting their transport. We used this dominant-negative strategy to investigate whether O1 is responsible for ER movement. A live-cell assay indicated that overexpression of the O1 tail domain significantly inhibited ER motility (Figure 8; see Supplemental Movies 1 to 4 online). When using the actin-disrupting drug latrunculin b and overexpression of the Arabidopsis XIK tail domain in tobacco, it was obvious that both actin and myosin XIK are required to maintain ER movement and remodeling (Sparkes et al., 2009a). In parallel, ER dynamics were characterized in the cotyledonary petioles of Arabidopsis single, double, and triple T- DNA insertional myosin mutants (Ueda et al., 2010). The results also indicated that myosin XIK plays a role in ER streaming. Together, these data suggest that the actomyosin system is required for ER tubule growth and shrinkage through actin polymerization and myosin processivity (Sparkes, 2011). A model for how the actomyosin system regulates ER movement and remodeling has been proposed, in which the movement effectively pulls out excess membrane from cisternal regions to form the tubular network and sliding of tubules forms closed polygonal rings (Sparkes et al., 2009b).
O1 Affects Zein Protein Body Biogenesis
In contrast with other opaque mutants, such as o2, o7, fl2, and DeB30, o1 has no apparent effect on zeins biosynthesis but nevertheless creates a soft, starchy endosperm (Table 1, Figure 1). This suggests that o1 has a different mechanism for influencing kernel texture. It is believed that zein protein bodies play an important role in the vitreous endosperm phenotype at kernel maturity. Several maize opaque mutants, such as o2, o7, fl2, De-B30, and Mc (Schmidt et al., 1990; Gillikin et al., 1997; Kim et al. 2004, 2006; Wang et al., 2011b), result in quantitative or qualitative alternations in zein accumulation. Consistent with a previous report (Hunter et al., 2002), we found that all the o1 alleles result in no significant changes in zein or nonzein proteins at maturity (Table 1; see Supplemental Figure 1 online). Therefore, it seems there is no causative relationship between o1 opaque phenotype and zein proteins. However, ultrastructural observation did demonstrate that o1-ref has small, misshapen protein bodies (Figure 1), similar to what is observed in some other opaque mutants (Schmidt et al., 1990; Coleman et al., 1997; Kim et al., 2004, 2006; Wang et al., 2011b). Could this change be caused by reduced synthesis of zein proteins in the o1 mutant? Expression profiles of representative zein genes were similar between o1-ref and the wild type during kernel development (Figure 2). Thus, we believe that O1 has no effect on zein proteins synthesis and accumulation. This conclusion is supported by the increased number of protein bodies in o1-ref (Table 2).
Considering the morphological alterations of protein bodies in o1, we propose that O1 has an effect on zein protein body assembly. When protein bodies were gently isolated from maize endosperm in vitro, they appeared to be enmeshed within a cytoskeleton network of fine actin filaments and myosins (Abe et al., 1991). In rice endosperm cells, the transport of prolamin mRNAs to the surface of the prolamin protein bodies is likely dependent on intact actin filaments and myosins (Hamada et al., 2003), and this conserved mechanism has also been proposed for maize zein mRNAs (Washida et al., 2004, 2009). Our data does not support a potential role of O1 in zein mRNA transport during protein body biogenesis. First, none of the zein proteins’ accumulation, downstream from the RNA transport, was reduced or affected in the o1-ref mutant. If mRNA transport was defective, the translational efficiency would be impaired (Kindler et al., 2005). Second, the overall distribution of protein bodies within endosperm cells appears to be unaffected in o1 mutant alleles.
A question to be answered is whether O1 directly affects protein body assembly. None of the putative O1-interacting proteins from yeast two-hybrid assay is predicted to be associated with protein bodies. On the other hand, the direct interaction tests between O1 and protein body intrinsic proteins indicated that neither zein proteins nor FL1 could interact with O1 (see Supplemental Figure 3B online). The protein body is an ER-localized organelle and mainly includes zein proteins surrounded by a domain of the ER membrane (Lending and Larkins, 1989). Recently, a proteomic analysis of protein bodies in tobacco leaves by transient transformation with a Zera sequence (the first 112 amino acids in N-terminal Pro-rich domain of 27-kD γ-zein) characterized many ER resident and secretory proteins (Joseph et al., 2012). These data suggest that the ER plays an important role in protein body formation. Given that O1 has an effect on ER morphology and motility, it could indirectly affect protein body assembly, possibly by regulating ER network.
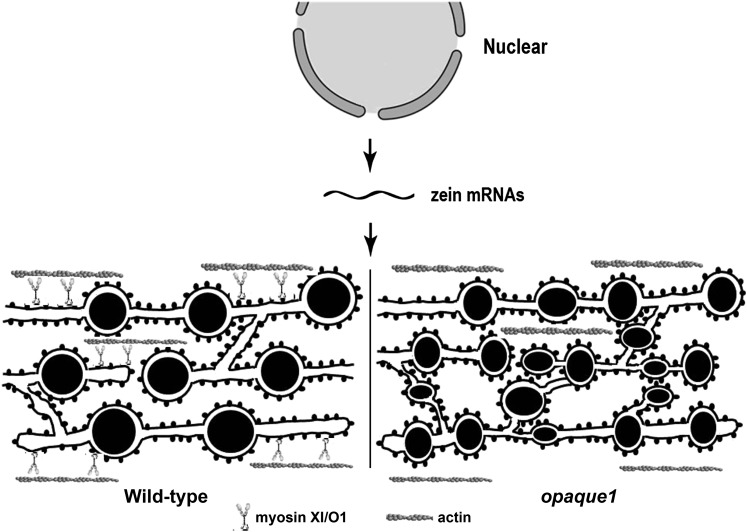
Based on the results, we propose a model for how the actomyosin system affects zein protein body biogenesis by regulating ER network in maize endosperm cell (Figure 10). In plants, myosin XI is the major protein responsible for ER streaming (Sparkes et al., 2009a; Ueda et al., 2010). When myosin was suppressed or disrupted by overexpressing the XIK tail domain or by using inhibitor 2,3-butanedione-2-monoxime in plant cells, the ER was dilated and more abundant, and also formed polygonal rings (Samaj et al., 2000; Sparkes et al., 2009a). We observed similar ER morphological changes in the o1 mutant. Due to the defective O1 function, ER movement is reduced and the ER structure is dilated. Zein mRNAs were found to be symmetrically distributed on ER membranes based on in situ hybridization (Kim et al., 2002). The increased ER membrane in o1 could provide more initiation loci for protein body biogenesis at early stage. However, because the zein biosynthesis is not affected, there are simply more but smaller protein bodies in o1. Opacity of the endosperm at the mature seed stage could reflect the corresponding change in the proteinaceous matrix surrounding starch granules that is mainly made by these small and misshapen protein bodies.
Figure 10.
Model Depicting the Function of Myosin in Protein Body Formation.
In the wild type, zeins are synthesized and stored in the ER as protein bodies, accompanying normal ER morphology and streaming mainly driven by a myosin motor. In the o1 mutant, ER dynamics were suppressed due to the O1 (myosin) defect, giving rise to the enlarged ER lumen, in which the zein protein bodies assemble and are stored. Therefore, there are more loci for zein aggregation in o1. Given that the total zein protein synthesis is not altered, the protein bodies reasonably become smaller. Moreover, because the ER motor system is out of homeostasis, this could affect the normal zein assembly, generating the misshapen protein bodies.
METHODS
Plant Materials
The lines containing the mutations o1-ref, o1-N1243, o1-N1478A, o*-84-5270-40, and o*-N1242A were obtained from the Maize Genetics Cooperation stock center. The mutations were crossed into the W22 genetic background for at least three generations, and o1-ref was backcrossed into the heterozygote for four generations to generate a BC mapping population. Kernels were collected from self-pollinated o1-ref/+ and o1-N1243/+ ear in the W22 genetic background segregating 25% homozygous mutant kernels and 75% wild-type kernels (o1/+ or +/+). All plants were cultivated in the field at the campus of Shanghai University, Shanghai, China. Root, stem, the third leaf, silk, tassel, and ear tissues were collected from at least three W22 plants at the V12 stage (Wang et al., 2010). Immature seeds were harvested at 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 24, 27, 30, 33, and 36 DAP. Nicotiana benthamiana plants were grown and transformed according to Sparkes et al. (2006).
Scanning Electron Microscopy and Transmission Electron Microscopy
For scanning electron microscopy, o1 and wild-type mature kernels harvested from the same self-pollinated ear were analyzed according to the method described previously (Wang et al., 2011a). For transmission electron microscopy, developing kernels (20 and 25 DAP) of o1 and the wild type were treated as described (Lending and Larkins, 1989; Wang et al., 2011b). Protein body analysis was performed using Image-Pro Plus 6.0 software (MediaCybernetics). All measurements were subjected to statistical analysis using the Student’s t test program using SigmaPlot 9.0 (Systat Software).
Protein Quantification
Endosperm of o1 and wild-type mature kernels was separated from the embryo and pericarp by dissection after soaking in water. The samples were dried to constant weight, pulverized with a mortar and pestle in liquid N2, and then measured according to a previously described protocol (Wang et al., 2011b). All measurements were replicated at least three times. Total protein and zein and nonzein proteins were extracted from 50 mg of each pooled endosperm flour sample according to the method of Wallace et al. (1990). A bicinchoninic acid protein assay kit (Pierce) was used for measurement of the extracted proteins, according to the manufacturer’s instructions.
Map-Based Cloning
The o1 locus was mapped and cloned using 13,830 plants from a BC population of o1-ref and W22. Molecular markers distributed throughout maize (Zea mays) chromosome 4 were used for preliminary mapping. Molecular markers for fine mapping (see Supplemental Table 1 online) were developed to localize the mutated locus to a 250-kb region. The corresponding DNA fragments were amplified from five o1 alleles and wild-type plants using KOD Plus DNA polymerase (Toyobo) and sequenced using a MegaBACE 4500 DNA analysis system (Amersham Biosciences).
Phylogenetic Analysis
Amino acid sequences deduced from cDNA nucleotide sequences were aligned with ClustalW program at the BCMsearch launcher (Baylor College of Medicine, Houston, TX) using their default settings (pairwise alignment options: gap opening penalty 10, gap extension penalty 0.1; multiple alignment options: gap opening penalty 10, gap extension penalty 0.2, gap distance 5, no end gaps, protein weight matrix using Gonnet) for protein multiple alignment. A rooted phylogenetic tree of plant myosin genes was constructed by the neighbor-joining method using the MEGA5.0 software package. The evolutionary distances were computed using the Poisson correction analysis. A bootstrap method with heuristic search was used with the following options: 10,000 random seeds, simple stepwise addition, and yeast class I and V myosins as reference sequences.
RNA Extraction and RT-PCR Analysis
Developing kernel RNA was extracted as described by Wang et al. (2012b), and the remaining RNA samples were isolated by TRIzol (Tiangen). The residual contaminating DNA was removed by an RNase-free DNase I (Takara) treatment. RNA was reverse transcribed to cDNA using ReverTra Ace reverse transcriptase (Toyobo).
Quantitative real-time RT-PCR was performed using a Mastercycler ep realplex 2 (Eppendorf) with SYBR Green Real-Time PCR Master Mix (Toyobo) according to the manufacturer’s protocol. Specific primers for O1 and Ubiquitin were designed by Primer Express Software V3.0 (Applied Biosystems) (see Supplemental Table 1 online). The real-time RT-PCR experiments were performed with two independent sets of RNA samples. Each RNA sample was extracted from a pool of tissues/kernels collected from at least three individual W22 plants. For each RNA sample, three technical replicates were performed in a final volume of 20 μL containing 10 μL 2× SYBR Green PCR buffer, 1 μL reverse transcribed cDNA (1 to 100 ng), and 1.8 μL 10 μM/L forward and reverse primers. Representative results from two biological replicates are shown. The specificity of the PCR amplification procedures was checked with a heat dissociation curve protocol. Quantification of the relative changes in gene expression was performed using the 2−ΔΔcycle threshold method as described previously (Livak and Schmittgen, 2001), and Ubiquitin was used as a reference gene.
Polyclonal Antibodies
For anti-O1 antibody production, a specific cDNA fragment of O1 (2770 to 3399 sequence site, representing amino acids 924 to 1133) was cloned into pGEX-4T-1 (Amersham Biosciences), and the glutathione S-transferase–tagged O1 fusion protein was purified using the AKTA protein purification system (GE Healthcare) with a GSTrap FF column. For 16- and 10-kD zein antibody production, zein proteins were separated by SDS-PAGE, the corresponding bands were cut out, and the proteins were recovered by preparative electrophoresis. Antibodies were produced in rabbits using standard protocols at Shanghai ImmunoGen Biological Technology.
Subcellular Fractionation and Immunoblot Analysis
Developing kernels were ground in liquid N2 and homogenized in a twofold excess of extraction buffer (60 mM Tris-Cl, pH 6.8, 1.5% SDS, 2 mM DTT, 15 mM β-mercaptoethanol, 10% Suc, and proteinase inhibitor cocktail [Sigma-Aldrich]) for direct immunoblotting. For subcellular fractionation, the powder of 20-DAP kernels was dissolved in resuspension buffer (25 mM Tris-HCl, pH 7.5, 0.25 M Suc, 2 mM EDTA, 2 mM DTT, 15 mM β-mercaptoethanol, 10% glycerol, and proteinase inhibitor cocktail). After removing cell debris by centrifuging at 500g (for ER and protein body separation) or 10,000g (for aqueous polymer two-phase partitioning) for 20 min at 4°C, the supernatant was treated as follows. For aqueous two-phase separation of the plasma membrane and endomembrane fractions, the sample was treated as described before with a few modifications (Xiong et al., 2010). Briefly, the supernatant was ultracentrifuged at 100,000g for 1 h at 4°C to obtain a microsomal pellet. Then, the microsomal proteins was redissolved in 10 mL suspension phase buffer (5 mm K2HPO4-KH2PO4, pH 7.8, 250 mM Suc, and 2 mM DTT) using a glass homogenizer, and then diluted in the two-phase system (6.2%, the final concentrations [w/w] of Dextran T500 and polyethylene glycol 3350, respectively), which consists of 12.4 g 20% (w/w) Dextran T500, 6.2 g 40% (w/w) polyethylene glycol 3350, 7.5 mL 1 M Suc in 20 mM K2HPO4-KH2PO4, pH 7.8, and 1 mL 0.8 M NaCl, and finally was added with water to 40 g total weight. The sample was thoroughly mixed by inversions and centrifuged at 8,000g for 10 min at 4°C. The same mixtures without cell membranes sample were centrifuged in parallel and used for the following washing. The upper (PEG, polyethylene glycol 3350, the plasma membrane fraction) and the lower (DEX, Dextran T500, the endomembrane fraction) phases were separately collected and washed with fresh lower and upper phase solution of fractionation buffer without samples, respectively. Both purified phases were diluted 10-fold with phase buffer and centrifuged at 100,000g for 1 h at 4°C and dissolved in a proper volume of suspension buffer. For separating ER and protein bodies, the supernatant was applied to discontinuous Suc gradients according to a pervious method (Habben et al., 1993).
Proteins were separated by SDS-PAGE and transferred electrophoretically to polyvinylidene difluoride membrane (0.45 μm; Millipore). The membrane was then incubated with antibodies and visualized using the Super Signal West Pico chemiluminescent substrate kit (Pierce) according to the manufacturer’s instructions. The purified anti-O1 and 16- and 10-kD zein antibodies were used at 1/500, while the α-tubulin antibody (Sigma-Aldrich) and BIP (at-95) antibody (Santa Cruz Biotechnology) were used at 1/1000.
ER Movement Assay
The cDNA sequence corresponding to the tail domain of O1 was amplified by PCR using KOD plus polymerase (Toyobo) with primers listed in Supplemental Table 1 online. Amplified fragments were subcloned into pENTR/D-TOPO using the Gateway TOPO cloning kit (Invitrogen) and sequenced. To express eYFP-O1t fusion protein, the right entry clone was introduced into pB7WGY2 plant expression vector by LR reaction of the Gateway system (Invitrogen) (Karimi et al., 2005). The well-established fluorescent protein marker mCherry-HDEL was used for labeling ER (Nelson et al., 2007). The binary expression vectors were transformed into Agrobacterium tumefaciens strain GV3101.
The ER in the adaxial side of leaf epidermal pavement cells was imaged. Dual imaging of YFP and mCherry was done using multitracking in-line switching mode on a spinning-disk confocal microscope (Andor). eYFP was excited with a 514-nm argon laser and mCherry with a 587-nm laser, and their emissions were detected using a 514/587 dichroic mirror and 530- to 580-nm and 590- to 630-nm band-pass filters. All imaging was performed using a 100 × 1.45–numerical aperture oil immersion objective and 512 × 512 pixel resolution. The time-lapse images of mCherry-HDEL were captured by two methods: 50 frames per 80 s and 20 frames per 40 s. For samples where cells were coexpressing a fluorescent myosin tail domain and mCherry-HDEL, coexpression was verified before time-lapse imaging of the mCherry-HDEL alone was performed. All images were analyzed using the Image J software (National Institutes of Health).
Yeast Two-Hybrid Assay
The O1 protein tail domain was generated by PCR and ligated into the pMD18-T vector (Takara). After sequence confirmation, it was fused downstream to GAL4 BD domain in pGBKT7 at the EcoRI and PstI sites. We used this bait to screen a maize developing kernel cDNA library fused downstream to GAL4 AD domain in pGADT7 (Wang et al., 2010). Zeins, FL1, and other putative genes were cloned from maize and inserted in frame into pGADT7. Yeast transformation and screening procedures were performed according to the manufacturer’s instructions (Clontech). Yeast strain Y187 was cotransformed with pGBKT7-O1t and pGADT7-Preys. The human P53 and maize 50-kD zein were used as positive control. The putative positive clones were further spotted with a series of dilution on SD/-Ade/-His/-Leu/-Trp medium. The resulting clones were transformed into Escherichia coli TOP10 for DNA sequencing.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: 10 kD δ-Zein, AF371266 (GRMZM2G100018); 15 kD β-Zein, M12147 (GRMZM2G086294); 16 Kd γ-Zein, AF371262 (GRMZM2G060429); 19 kD α-Zein, M12146 (AF546188.1); 22 kD α- Zein, NM_001112529 (GRMZM2G044625); 27 kD γ-Zein, AF371261 (GRMZM2G138727); 50 kD γ-Zein, BT062750 (GRMZM2G138689); FL1, NM_001112594 (GRMZM2G094532); Zm GAPDH, NM_001111943 (GRMZM2G046804); Zm HIP, GRMZM2G023275; Zm HVA22, NM_001153512 (GRMZM2G092925); Zm Reticulon, NM_001136619 (GRMZM2G142409); Zm Sey, NP_001151750; Zm Rab5a, EU966300 (GRMZM2G015159); Zm BIP1, NM_001112423 (GRMZM2G114793); Zm BIP2, NM_001112424 (GRMZM2G415007); Zm PDI, NM_001112284 (GRMZM2G091481); and Zm UBQ, BT018032.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SDS-PAGE Analysis of the Zein Fraction from Mature Endosperm of Wild-Type and o1 Alleles.
Supplemental Figure 2. Multiple Alignment of Amino Acid Sequences Encoded by Maize Opaque1 and Its Homologs.
Supplemental Figure 3. Subcellular Localization of O1.
Supplemental Figure 4. Yeast Two-Hybrid Interactions between the O1 Tail Domain and Other Proteins.
Supplemental Table 1. A List of Primers Used in This Study.
Supplemental Table 2. A Summary of Candidate Gene of O1.
Supplemental Table 3. Major Domains Represented in the Maize Opaque 1 Gene Product.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 4.
Supplemental Movie 1. ER Streaming (mCherry-HDEL) in a Control Tobacco Epidermal Cell (20 Frames with 2-s Interval).
Supplemental Movie 2. ER Streaming (mCherry-HDEL) in a Control Tobacco Epidermal Cell (50 Frames with 1.6-s Interval).
Supplemental Movie 3. ER Streaming in a Tobacco Epidermal Cell Coexpression mCherry-HDEL and the eYFP-O1 Tail Domain (20 Frames with 2-s Interval).
Supplemental Movie 4. ER Streaming in a Tobacco Epidermal Cell Coexpression mCherry-HDEL and the eYFP-O1 Tail Domain (50 Frames with 1.6-s Interval).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (31000747 and 31171559), the Ministry of Science and Technology of China (2012AA10A305 and 2009CB118400), and the Science and Technology Commission of Shanghai Municipality (11DZ2272100). We thank Brian A. Larkins (University of Arizona) for critical reading the article, Ming Yuan and Tonglin Mao (China Agricultural University) for ER movement assay, Yulong Ren (Nanjing Agricultural University) for providing the ER marker, and our colleague Yihan Chen for genetic characterization of the o1 mutant allele, o*-N1242A.
AUTHOR CONTRIBUTIONS
Guifeng W. and R.S. designed the experiment. Guifang W., Fang W., Gang W., Fei W., X.Z., M.Z., J.Z., and D.L. performed the experiments. Guifeng W., Gang W., Fang W., Y.T., Z.X., and R.S. analyzed the data. Guifeng W. and R.S. wrote the article.
Glossary
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- DAP
days after pollination
- BC
to be defined
- eYFP
enhanced yellow fluorescent protein
- TPR
tetratricopeptide repeat protein
References
- Abe S., You W., Davies E. (1991). Protein bodies in corn endosperm are enclosed by and enmeshed in F-actin. Protoplasma 165: 139–149 [Google Scholar]
- Avisar D., Abu-Abied M., Belausov E., Sadot E. (2012). Myosin XIK is a major player in cytoplasm dynamics and is regulated by two amino acids in its tail. J. Exp. Bot. 63: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Abu-Abied M., Belausov E., Sadot E., Hawes C., Sparkes I.A. (2009). A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150: 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A.I., Makarova K.S., Koonin E.V., Dolja V.V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Clore A.M., Ranch J.P., Higgins R., Lopes M.A., Larkins B.A. (1997). Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R.A., Beadle G.W., Fraser A.C. (1935). A summary of linkage studies in maize. Cornell Univ. Agric. Exp. Stn. Memoir 180: 1–83 [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., Ferro-Novick S. (2003). Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163: 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Larkins B.A. (2005). Molecular genetic approaches to developing quality protein maize. Trends Genet. 21: 227–233 [DOI] [PubMed] [Google Scholar]
- Gillikin J.W., Zhang F., Coleman C.E., Bass H.W., Larkins B.A., Boston R.S. (1997). A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol. 114: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L., Abu-Abied M., Belausov E., Sadot E. (2008). Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben J.E., Kirleis A.W., Larkins B.A. (1993). The origin of lysine-containing proteins in opaque-2 maize endosperm. Plant Mol. Biol. 23: 825–838 [DOI] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Choi S.B., Wang C., Singh S., Kawai N., Franceschi V.R., Okita T.W. (2003). The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J.A., III, Sellers J.R. (2012). Walking to work: Roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13: 13–26 [DOI] [PubMed] [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., Blackstone C. (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter B.G., Beatty M.K., Singletary G.W., Hamaker B.R., Dilkes B.P., Larkins B.A., Jung R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14: 2591–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph M., Ludevid M.D., Torrent M., Rofidal V., Tauzin M., Rossignol M., Peltier J.B. (2012). Proteomic characterisation of endoplasmic reticulum-derived protein bodies in tobacco leaves. BMC Plant Biol. 12: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kim C.S., Gibbon B.C., Gillikin J.W., Larkins B.A., Boston R.S., Jung R. (2006). The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. Plant J. 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Kim C.S., Hunter B.G., Kraft J., Boston R.S., Yans S., Jung R., Larkins B.A. (2004). A defective signal peptide in a 19-kD alpha-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol. 134: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.S., Woo Ym Y.M., Clore A.M., Burnett R.J., Carneiro N.P., Larkins B.A. (2002). Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell 14: 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D., Tiedge H. (2005). RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21: 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending C.R., Larkins B.A. (1989). Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.F., Nebenführ A. (2007). Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. J. Biol. Chem. 282: 20593–20602 [DOI] [PubMed] [Google Scholar]
- Li J.F., Nebenführ A. (2008). The tail that wags the dog: The globular tail domain defines the function of myosin V/XI. Traffic 9: 290–298 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mercer J.A., Seperack P.K., Strobel M.C., Copeland N.G., Jenkins N.A. (1991). Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature 349: 709–713 [DOI] [PubMed] [Google Scholar]
- Miclaus M., Wu Y., Xu J.H., Dooner H.K., Messing J. (2011). The maize high-lysine mutant opaque7 is defective in an acyl-CoA synthetase-like protein. Genetics 189: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose S.P., Dudley J.W., Rocheford T.R. (2004). Maize selection passes the century mark: A unique resource for 21st century genomics. Trends Plant Sci. 9: 358–364 [DOI] [PubMed] [Google Scholar]
- Myers A.M., James M.G., Lin Q., Yi G., Stinard P.S., Hennen-Bierwagen T.A., Becraft P.W. (2011). Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nelson O.E., Mertz E.T., Bates L.S. (1965). Second mutant gene affecting the amino acid pattern of maize endosperm proteins. Science 150: 1469–1470 [DOI] [PubMed] [Google Scholar]
- Neuffer M.G., Coe E.H., Wessler S.R. (1997). Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press)
- Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. (2007). Reticulon-like proteins in Arabidopsis thaliana: Structural organization and ER localization. FEBS Lett. 581: 3356–3362 [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Mockler T.C., Filichkin S.A., Fox S.E., Jaiswal P., Makarova K.S., Koonin E.V., Dolja V.V. (2011). Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol. 155: 1191–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Avisar D., Dolja V.V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prapapanich V., Chen S., Toran E.J., Rimerman R.A., Smith D.F. (1996). Mutational analysis of the hsp70-interacting protein Hip. Mol. Cell. Biol. 16: 6200–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky A.I., Peremyslov V.V., Dolja V.V. (2008). Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt S., Knight A.E., Hodge T.P., Baluska F., Samaj J., Volkmann D., Kendrick-Jones J. (1999). Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 19: 555–567 [DOI] [PubMed] [Google Scholar]
- Sabelli P.A., Larkins B.A. (2009). The development of endosperm in grasses. Plant Physiol. 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J., Peters M., Volkmann D., Baluska F. (2000). Effects of myosin ATPase inhibitor 2,3-butanedione 2-monoxime on distributions of myosins, F-actin, microtubules, and cortical endoplasmic reticulum in maize root apices. Plant Cell Physiol. 41: 571–582 [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Burr F.A., Aukerman M.J., Burr B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G., Song R., Messing J. (2003). A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M.B., Rose R.J., McCurdy D.W. (2004). Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 37: 379–390 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. (2011). Recent advances in understanding plant myosin function: Life in the fast lane. Mol. Plant 4: 805–812 [DOI] [PubMed] [Google Scholar]
- Sparkes I., Runions J., Hawes C., Griffing L. (2009a). Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. (2009b). The plant endoplasmic reticulum: A cell-wide web. Biochem. J. 423: 145–155 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Teanby N.A., Hawes C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: A genetic tool for the next generation. J. Exp. Bot. 59: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V.V., Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wallace J.C., Lopes M.A., Paiva E., Larkins B.A. (1990). New methods for extraction and quantitation of zeins reveal a high content of gamma-zein in modified opaque-2 maize. Plant Physiol. 92: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Gao Y., Wang J., Yang L., Song R., Li X., Shi J. (2011a). Overexpression of two cambium-abundant Chinese fir (Cunninghamia lanceolata) α-expansin genes ClEXPA1 and ClEXPA2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol. J. 9: 486–502 [DOI] [PubMed] [Google Scholar]
- Wang G., Sun X., Wang G., Wang F., Gao Q., Sun X., Tang Y., Chang C., Lai J., Zhu L., Xu Z., Song R. (2011b). Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G., Wang G., Wang F., Song R. (2012a). A transcriptional roadmap for seed development in maize. In Seed Development: Omics Technologies toward Improvement of Seed Quality and Crop Yield, G.K. Agrawal and R. Rakwal, eds (Dordrecht, The Netherlands: Springer), in press.
- Wang G., Wang G., Zhang X., Wang F., Song R. (2012b). Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 23: 159–163 [DOI] [PubMed] [Google Scholar]
- Wang G., Wang H., Zhu J., Zhang J., Zhang X., Wang F., Tang Y., Mei B., Xu Z., Song R. (2010). An expression analysis of 57 transcription factors derived from ESTs of developing seeds in maize (Zea mays). Plant Cell Rep. 29: 545–559 [DOI] [PubMed] [Google Scholar]
- Washida H., et al. (2009). Identification of cis-localization elements of the maize 10-kDa delta-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J. 60: 146–155 [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Messing J., Esen A., Okita T.W. (2004). Asymmetric localization of seed storage protein RNAs to distinct subdomains of the endoplasmic reticulum in developing maize endosperm cells. Plant Cell Physiol. 45: 1830–1837 [DOI] [PubMed] [Google Scholar]
- Wu S.Z., Ritchie J.A., Pan A.H., Quatrano R.S., Bezanilla M. (2011). Myosin VIII regulates protonemal patterning and developmental timing in the moss Physcomitrella patens. Mol. Plant 4: 909–921 [DOI] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2010). RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 153: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Li R., Qian Q., Song X., Liu X., Yu Y., Zeng D., Wan J., Li J., Zhou Y. (2010). The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J. 64: 56–70 [DOI] [PubMed] [Google Scholar]
- Yokota E., Ueda H., Hashimoto K., Orii H., Shimada T., Hara-Nishimura I., Shimmen T. (2011). Myosin XI-dependent formation of tubular structures from endoplasmic reticulum isolated from tobacco cultured BY-2 cells. Plant Physiol. 156: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Ueda S., Tamura K., Orii H., Uchi S., Sonobe S., Hara-Nishimura I., Shimmen T. (2009). An isoform of myosin XI is responsible for the translocation of endoplasmic reticulum in tobacco cultured BY-2 cells. J. Exp. Bot. 60: 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.