Abstract
The causative agent of malaria, Plasmodium, has to undergo complex developmental transitions and survive attacks from the mosquito's innate immune system to achieve transmission from one host to another through the vector. Here we discuss recent findings on the role of the mosquito's innate immune signaling pathways in preventing infection by the Plasmodium parasite, the identification and mechanistic description of novel anti-parasite molecules, the role that natural bacteria harbored in the mosquito midgut might play in this immune defense, and the crucial parasite and vector molecules that mediate midgut infection.
I. Introduction
Within the mosquito vector, malaria parasites must go through a series of complex developmental transitions before transmission to a human host occurs. After being ingested by a mosquito, male and female gametocytes fuse within the midgut lumen and will over a period of approximately 18 hours develop into a motile ookinete that will migrate to the midgut epithelium and invades a single epithelial cell. The ookinete must travel to the basal lamina before the infected cell is extruded from the epithelial layer. Once it arrives at the basal lamina, the parasite differentiates into an oocyst and then further develops over a period of about 10 days into thousands of sporozoites that are released into the mosquito hemolymph. Sporozoites migrate to and invade the salivary glands and can be transmitted when the mosquito takes another blood meal. A major bottleneck for Plasmodium development takes place during the ookinete invasion of the midgut epithelium. The majority of the parasite loss can be attributed to lumenal and epithelial immune responses mounted by the mosquito.
Insect (and especially dipteran) innate immunity has generally been resolved using the fruit fly Drosophila melanogaster as a model and bacteria or fungi as the challenging microbe. For a more thorough coverage of Drosophila immunity, the reader is referred to a review by Lemaitre and Hoffmann [1]. Cellular and humoral factors are major players in the response to microbial challenge, especially within the hemolymph (blood) of the insect. Hemocytes, the insect blood cells, are constantly circulating and can either engulf (by phagocytosis) or surround (encapsulation) a foreign invader as a defense mechanism. Humoral responses to pathogens involve melanization and antimicrobial effector molecules. During melanization, a serine protease cascade activates pro-phenoloxidases that, through a second catalytic cascade, generate the melanin and free radicals that are involved in killing microbes. Production of antimicrobial effector molecules are regulated by intracellular immune signaling pathways that are activated by pattern recognition receptors (PRRs) upon interaction with pathogen associated molecular patterns (PAMPs).
The intracellular immune signaling pathways have been extensively studied in Drosophila, with most information having been obtained by injection of bacteria or fungus directly into the fly hemolymph. Toll pathway activation occurs through pathogen detection by soluble peptidoglycan recognition proteins (PGRPs) that stimulate a serine protease cascade, culminating in the proteolytic activation of the extracellular ligand, Spätzle. Activation of a second pathway, the immune deficiency (IMD) pathway, occurs when a pathogen is detected by a membrane-bound class of PGRPs. From either pathway, extracellular signals initiate a series of intracellular reactions that lead to an increased expression of select immune-related genes, including antimicrobial peptides (Figure 1).
Figure 1. Toll and IMD immune signaling pathways involved in anti-Plasmodium defense.
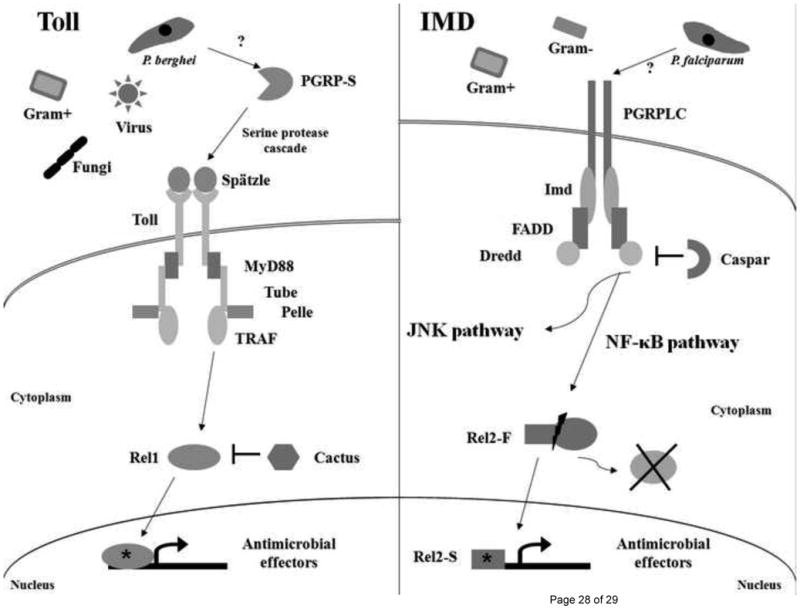
Following recognition of a microbe, or unknown Plasmodium ligand, by soluble PGRP molecules, the Toll pathway is stimulated by binding of the ligand Spätzle with the Toll transmembrane receptor. This triggers a series of molecular events that culminate in the activation (*) and translocation of Rel1 into the nucleus, up-regulating transcription of immune genes that are responsible for microbial killing. The IMD pathway is stimulated when the transmembrane PGRPLC receptor binds peptidoglycan or an unknown Plasmodium ligand that leads to the cleavage of Rel2-F and translocation of active Rel2-S (*) into the nucleus. A different set of anti-Plasmodium genes are up-regulated when the IMD pathway is stimulated. Branching of the IMD pathway is indicated, but the JNK pathway has not been extensively characterized in Anopheles mosquitoes.
Here we describe recent findings concerning the role of immune signaling pathways in preventing infection of the mosquito vector by the malaria parasite, the identification and mechanistic description of novel anti-parasite molecules, the role that natural bacteria harbored in the mosquito midgut might play in this overall immune response, and the crucial parasite and vector molecules that mediate midgut infection. The role of pattern recognition receptors in activating anti-Plasmodium defense will be discussed in the different sections.
II. Immune signaling pathways and Plasmodium infection
Immune signaling pathways, which direct insect immune responses to a variety of pathogens, have recently been shown to regulate anti-Plasmodium immunity in mosquitoes. The three major immune signaling pathways (Toll, IMD, and Jak/Stat) that were originally described in Drosophila or mammals have been identified through orthology in Anopheles gambiae [2]. A schematic representation of the Toll and IMD pathways is provided in Figure 1.
The Toll Pathway
The classical Toll pathway is activated upon infection with Gram-positive (G+) bacteria and fungi. It has also been implicated in the defense against viruses in fruit flies [3] and mosquitoes [4] and against the rodent malaria parasite P. berghei in Anopheles mosquitoes [5] (see below). PAMP recognition by Toll pathway PRRs is well documented, but the underlying mechanism is currently unresolved.
While Drosophila has two different transcription factors that separate the expression of Toll-mediated immune and developmental gene expression (Dif and Dorsal, respectively), Anopheles mosquitoes appear to express only an ortholog of Dorsal, the NF-kappaB-like REL1 (originally described as Gambif1) [2], [6], [7], [8], [9], [10]. Cactus, identified in Drosophila as a negative regulator of the pathway, sequesters REL1 to the cell cytoplasm [11], [12], [13], [14]. Directed degradation of cactus frees REL1 for nuclear translocation and subsequent transcription initiation at kappaB sites located upstream of canonical Toll pathway effector genes [6], [15], [16].
Using RNA interference (RNAi)-mediated gene silencing of cactus gene expression, Frolet and colleagues (2006) have shown that Toll pathway activation significantly decreases P. berghei parasite burden, while REL1 depletion increases infection levels in mosquito midguts. Importantly, they demonstrated that removal of a negative regulator could activate an immune response without pathogen challenge. They hypothesized that pre-formed effector molecules are able to immediately attack an invading pathogen and that early transcriptional activation is a means of replenishing molecules used in the directed immune response [5]. However, Toll pathway-mediated control of Plasmodium parasite infection may not be universal. Using multiple parasite-mosquito species combinations, Garver and coworkers (2009) showed for the first time that P. berghei infection of A. gambiae, A. stephensi, and A. albimanus is controlled through Toll pathway activation, while P. falciparum infection of the same mosquito species is independent of cactus depletion. Expression of numerous genes from diverse functional groups was regulated by cactus depletion, likely as a result of Toll pathway involvement in processes other than immunity (such as development); this widespread effect may contribute to the significant reduction in fitness seen in cactus-silenced mosquitoes [17].
In an Aedes aegypti-P. gallinaceum infection model, cactus silencing has been shown to protect mosquitoes from parasite infection via a mechanism that requires the expression of a second transcription factor, RUNT related transcription factor 4 (RUNX4) [18]. In this model, parasite melanization was mediated by prophenoloxidase gene expression, which was controlled cooperatively by REL1 and RUNX4. This dual-factor mechanism, or potentially another transcription factor such as REL2 (see below), might be used in mosquitoes to separate Toll pathway-mediated expression of different gene sets from disparate functional groups.
The IMD pathway
A second major immune signaling pathway, the IMD pathway, is likened to the tumor necrosis factor (TNF) signaling pathway in mammals [19], [20]. Microbe detection by PGRPs initiates intracellular signaling through the adaptor IMD protein and various caspase-like proteins and kinases, leading to a functional split in the pathway [21], [22], [23], [24], [25], [26], [27], [28]. One branch is similar to the c-Jun/JNK pathway of mammals and uses JNK to activate the transcription factor AP-1, while the other branch, an NF-kappaB activating branch, culminates in the processing of the transcription factor REL2 (Relish in Drosophila) [2], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38].
In the absence of immune stimulation, REL2 exists in two splice variants: a short form (REL2-S) lacking the inhibitory ankyrin domain that is constitutively active and responsible for basal immune gene expression, and a full-length form (REL2-F) that is inactive until immune stimulation occurs [29], [39]. IMD pathway activation stimulates cleavage of the carboxy-terminal end of REL2-F, exposing the nuclear localization signal for nuclear translocation and subsequent transcription initiation [30], [40]. Because REL2-S has no unique features when compared to REL2-F, dsRNA-mediated depletion of the short form alone is not possible. However, Meister et al. (2005) have suggested that REL2-F is essential for anti-Plasmodium activity because targeted depletion of the full-length form only or of both REL2 forms results in similar infection phenotypes in a P. berghei model. Their data also suggest that processing of REL2-F, carried out and regulated by components of the IMD pathway, is important for anti-Plasmodium gene transcription [29].
Depletion of caspar, a Fas-associating factor homolog that inhibits Relish activation in Drosophila [41] and is a putative IMD pathway inhibitor in Anopheles, produces a P. falciparum-refractory phenotype in A. gambiae females, indicating that the IMD pathway (at least downstream of caspar-mediated inhibition) controls transcription of genes involved in parasite elimination [17]. Caspar silencing is effective in limiting P. falciparum infection of three anopheline species but is less effective against P. berghei, indicating that IMD pathway-mediated control of parasite infection is mosquito species-independent and parasite species-dependent. It is interesting that the IMD and Toll pathways are both mosquito species-independent but limit infection for distinct parasite species. Depletion of caspar does not confer noticeable fitness effects in the laboratory environment, a finding that reflects the strictly immune-responsive nature of IMD [17].
The IMD pathway of A. gambiae may be more complex than has been reported for Drosophila. Preliminary data suggest that immune responses directed by components of the IMD pathway may be IMD protein-independent, and REL2 isoforms are regulated and utilized in a multi-faceted manner [29], [39], [42], [43].
The Jak/Stat Pathway
The third major immune signaling pathway, the Jak/Stat pathway, is named for the kinases (Jak) and transcription factors (STAT) that control its activation. Research on this pathway has intensified in Drosophila, where Jak/Stat has been shown to play an important role in the immune response against pathogenic bacterial infection in the gut [44], [45]. The pathway has also been associated with antiviral immunity in Drosophila [46] and Ae. aegypti [47] but until recently had been less extensively explored in the context of Anopheles-Plasmodium interactions.
Two STAT transcription factors (STAT-A and STAT-B) have been identified in A. gambiae, while only one STAT is present in Ae. aegypti and Drosophila. STAT-B apparently regulates the transcription of STAT-A, the ancestral transcription factor and predominant form expressed in adult mosquitoes. Translocation of STAT-A into the nucleus leads to up-regulation of anti-Plasmodium effector molecule expression. Recently, the pathway was shown to mediate the killing of P. falciparum and P. berghei parasites at a later infection stage, after midgut invasion. Depletion of STAT-A increased P. berghei oocyst intensity, while activation of the pathway (through depletion of the negative regulator SOCS) decreased infection levels [48].
Pathway divergence from Drosophila
Recent evidence suggests that the single pathogen-single pathway paradigm of immune activation in Drosophila may not be as clear-cut in mosquitoes. PGRP-LC, a receptor for IMD pathway activation, has been found to be essential for mosquito survival following Gram-negative (G-) and G+ bacterial challenge [42]. Also, the IMD pathway provides early protection from G+ bacterial infections and long-lasting protection against G- bacteria [43], in contrast to the pathogen specificity of the Toll (G+ bacteria and fungi) and IMD (G- bacteria) pathways that has been noted in Drosophila. Although Drosophila is a good model system for forward genetic screens and has been used in previous studies to identify anti-Plasmodium effectors, the data obtained correlate with mosquito immunity, especially with regard to Plasmodium infection, on a case-by-case basis [49].
Other pathways have been implicated in the defense against Plasmodium infection of the mosquito, including a kinase-kinase signaling cascade (MAPK-ERK) that is apparently regulated by mosquito ingestion of a human cytokine [50]. However, the specifics of how different microbes elicit immune responses, how the pathways discriminate among different methods of activation, how the alternative activation methods influence downstream gene transcription and microbe destruction, and why such branching is beneficial to the mosquito are all unclear at this point. When and where in the mosquito body these pathways are activated and on which parasite stage the response is targeted are areas of interest that are only beginning to be elucidated.
III. Effectors of the anti-Plasmodium response
The ultimate result of immune signaling pathway activation is an up-regulation of specific gene expression that is PAMP- and pathway-dependent. These immune effector genes form an important line of defense for the mosquito against invading pathogens. Microarray and RNAi-based studies have shown that effector molecules from diverse gene families play important roles in the killing of Plasmodium ookinetes and/or oocysts. The mechanisms employed by anti-parasite molecules are just beginning to be unraveled as new protein-protein interactions are discovered.
Leucine rich-repeat (LRR) domain-containing proteins
LRR domain-containing proteins play a key role in mediating anti-Plasmodium immunity in mosquitoes. This protein family encodes secreted, membrane-bound, and cytoplasmic proteins with numerous leucine-rich repeats (LRRs); these proteins are up-regulated in A. gambiae following infection with Plasmodium [2], [51], [52]. In this gene family, LRIM1 is a potent P. berghei antagonist, while LRRD19 (also known as APL1) and LRRD7 are involved in the defense against both human and rodent Plasmodium parasites [52], [53], [54].
Manual re-annotation has revealed that the LRRD19 (or APL1) locus encodes three distinct genes (APL1A, -B, and -C) that have arisen from recent duplications. Of the three, APL1A and -B have been proposed to have no effect on P. berghei infection while APL1C is suggested as the sole P. berghei antagonist [55]. LRIM1 and LRRD19 interact and play an important role in parasite melanization and killing during early P. berghei infection [53], [56], [57]. In A. gambiae, LRIM1 and APL1C form a disulfide-linked, high-molecular-weight complex that is secreted into the hemolymph. The heterodimeric complex interacts with the complement C3-like protein TEP1 and may be necessary to promote cleavage of TEP1 into an active form. The reactive TEP1 subsequently localizes to the surface of midgut-invading P. berghei, targeting the parasite for destruction. LRIM1 and APL1C are required for hemolymph circulation and binding of TEP1 to the parasite surface [43], [58]. These results reveal a role for LRR proteins as complement control factors that may function as part of a complement-like system in killing Plasmodium parasites.
LRIM1 does not interfere with natural P. falciparum infection but does control melanization in an anopheline species that is naturally refractory to P. berghei [56]. APL1A has however been suggested to be involved in P. falciparum resistance, both in laboratory colonies and field-collected mosquitoes [56], [54], [55], [59]. Other proteins such as fibrinogen immunolectin 9 (FBN9) [60] (see below) and/or other LRR proteins (such as LRRD7) may provide alternate mechanisms for TEP1-mediated parasite killing of human Plasmodium species.
More than 20 LRIM1 -related proteins have been identified in the genomes of vector mosquito species, but no orthologs have been found in other organisms, suggesting a mosquito-specific immune role for these genes [43]. APL1C and LRIM1 appear to be regulated by the Toll pathway [5], [58], [55]; however, the IMD pathway plays an essential role in mounting the anti-P. falciparum immune response through FBN9, LRRD7, and TEP1 in diverse anopheline mosquito species [17]. Expression of TEP1 from hemocytes circulating in the mosquito hemolymph may also be regulated by the Jak-Stat and Toll pathways, suggesting a multipartite regulation of this potent anti-Plasmodium effector [5], [48], [58].
The fibrinogen-related proteins (FREPs)
FREPs share a fibrinogen-like domain (FBG) that is evolutionarily conserved from invertebrates to mammals [61], [62], [63]. FREP gene families have been identified in mosquitoes and flies, with a significant expansion in the A. gambiae genome (58 genes) when compared to A. aegypti (37 genes) and D. melanogaster (14 genes) [60], [63], [64], [65]. A strong correlation between phylogeny, chromosomal location, and the expression pattern of FREP genes was identified in A. gambiae, implying conserved functions among the duplicated family members that apparently arose through tandem duplication and shuffling of FBG domains [60].
Transcriptomic and functional analyses have shown that FREP genes are involved in the mosquito's immune response to bacteria and Plasmodium parasites. RNAi-mediated gene silencing assays have implicated FBN8, FBN9, and FBN39 in the anti-Plasmodium defense, where FBN39 specifically protects the mosquito against P. falciparum [2], [51], [52], [66]. Functional studies of 38 members of this gene family in A. gambiae have revealed that FREP proteins have complementary and synergistic functions that are mediated by inter- and intra-molecular associations. Interestingly, in vitro bacterial binding assays show that FBN9 forms homodimers (and possibly heterodimers with other FREP proteins) that bind to bacterial surfaces with different affinities [60]. FREPs may use a mechanism similar to LRIM1/APL1C to form multimers as a means of increasing the mosquito's PRR repertoire and mediating anti-pathogen responses. However, the molecular basis for this mechanism remains to be elucidated.
C-type lectins (CTLs)
The C-type lectin (CTL) family is one of the largest and most diverse animal lectin families. CTLs bind carbohydrates in a Ca2+-dependent manner through the C-terminal carbohydrate recognition domain. These binding events mediate processes such as cell adhesion, cell/cell interactions, glycoprotein turnover, and pathogen recognition. In vertebrates, the collectins (collagenous lectins) serve as acute-phase proteins that mediate opsonization, clearance of microbial agents, and complement activation through the lectin pathway [67], [68], [69].
Twenty-three genes encoding C-type lectin domains have been identified in the A. gambiae genome. CTL4 and CTLMA2 have been identified as agonists of the rodent Plasmodium parasite, and silencing of either of these genes induces massive melanization of P. berghei ookinetes in the basal labyrinth of the midgut epithelium, blocking their development at the pre-oocyst stage [53]. However, CTLs do not appear to be involved in the defense against human Plasmodium parasites. A more recent examination has shown that CTL4 and CTLMA2 also significantly contribute to the mosquito's defense against G-, but not G+ bacteria. Like LRIM1/APLC, CTL4 and CTLMA2 each exist in the mosquito hemolymph as a disulfide-linked hetero-dimeric complex, a similarity that partially explains their similar roles in antibacterial defense and the melanization response to P. berghei [70]. However, whether a complement-like killing mechanism of action is utilized remains to be elucidated at the molecular level.
Other effector molecules
Other molecules have been implicated in killing Plasmodium parasites. Nitric oxide synthase (NOS), which is induced by the Jak-Stat or TGF-β1/MEK-ERK pathway, has potent anti-Plasmodium activity in the mosquito midgut and may be a late-stage line of defense against Plasmodium [48], [50]. NOS and antimicrobial proteins, such as gambicin and cecropin, were among the first anti-Plasmodium factors to be identified [71], [72]. Gambicin and a novel putative short secreted peptide, IRSP5, are more specific for defense against the rodent parasite P. berghei [52]. Among the putative pattern recognition receptors (PRRs) of A. gambiae, splice variants of the A. gambiae Down syndrome cell adhesion molecule gene (AgDscam) have been shown to protect mosquitoes against challenge with either P. berghei or P. falciparum [73] (Dong et al., unpublished data). An MD2-like receptor, AgMDL1, shows specificity in regulating resistance only to P. falciparum [52]. Among the 138 predicted immunoglobulin superfamily proteins of A. gambiae, IRID4 and IRID6 have been shown to be involved in limiting P. falciparum infection [74]. G- bacteria-binding proteins (GNBPs) are functionally diverse components of the A. gambiae innate immune system, within which GNBP4 acts as a key factor in the anti-Plasmodium responses [76], [75]. Figure 2 shows anti-Plasmodium effector molecules involved in parasite killing and at what stage of parasite development the killing may occur.
Figure 2. Parasite development and anti-Plasmodium effectors in the mosquito.
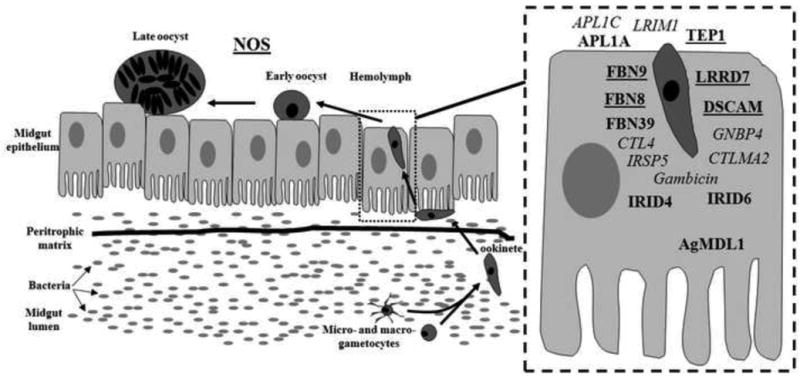
The Plasmodium parasite develops into the motile ookinete stage and will invade the midgut epithelium at approximately 18 hours after ingestion of an infected blood meal. At this stage, a number of effector molecules target the parasite for destruction (inset). NOS targets the parasite at a later developmental stage- the early/late oocyst transition. Effector molecules in underlined bold are involved in killing human and rodent Plasmodium species, those in bold italics have been shown to be effective against P. berghei only, and those in bold target P. falciparum.
IV. The influence of bacteria on the anti-Plasmodium response
When immune signaling pathways and anti-Plasmodium effectors are examined, a common theme emerges: The immune responses are active against both bacterial and plasmodial challenge. Therefore, it is not surprising that immune responses mounted against endogenous and exogenous bacteria are capable of modulating the infection by Plasmodium parasites, especially in the gut.
The process of blood feeding causes an extreme bloom in mosquito gut microbiota, presumably through an increase in available nutrients. Twenty-four hours after a blood meal, bacterial densities can reach 107 colony-forming units per milliliter and return to pre-blood meal concentrations in 3-5 days [77]. This dramatic increase in bacterial content loosely coincides with the invasion of the midgut by the parasite, and an immune response against bacteria at that time would be mounted when the parasite is most vulnerable.
There is considerable overlap between the anti-bacterial and anti-Plasmodium immune responses, with many molecules having activity against more than one type of microbe. Effector proteins including TEP1, FBN9, FBN39, LRRD7, and CTL4 have been identified through a microarray analysis of P. falciparum and P. berghei-infected mosquitoes and found to control resistance to bacterial and plasmodial challenge [52]. CTL4 and CTLMA2, first identified on the basis of their P. berghei melanization properties, are involved in anti-G- bacterial responses. Depletion of either molecule results in increased pathogenicity and decreased clearance of G- bacteria but has no effect on G+ bacteria [70]. GNBPs have been shown to regulate immune gene expression through the Toll or IMD pathway, and certain members are able to mediate Plasmodium oocyst intensities in A. gambiae [75].
Strong evidence for anti-bacterial/anti-Plasmodium immune synergy has recently been presented by Dong et al. (2009). Co-feeding live or heat-inactivated bacteria with parasites has been found to decrease the prevalence of P. falciparum oocysts after 8 days of incubation [78]. Experiments in which the bacterial populations in the midgut were lowered (through ingestion of antibiotic-containing sugar) have shown that mosquitoes without detectable bacteria in the midgut are more susceptible to P. falciparum infection [78], [79]. Differences in parasite infection have been observed when select immune genes are silenced in antibiotic-treated and untreated mosquitoes, allowing the identification of the natural gut microbiota as a major influence on Plasmodium development [78]. Basal immune gene expression is activated by gut bacteria and is responsible for controlling their proliferation, possibly acting through an IMD-mediated mechanism [42], [78]. This possibility is in line with the results of Drosophila studies suggesting that the gut epithelium-associated antibacterial immune responses that control proliferation of commensal microflora are dependent exclusively on the IMD pathway, while systemic responses are governed by both the IMD and Toll pathways [1].
Orthologs of Drosophila proteins that control commensal gut bacteria may play a role in early antiplasmodial immunity. The regulatory transcription factor Caudal inhibits IMD-dependent AMP over-expression in the absence of pathogenic challenge [80]. Removal of Caudal from Rel2 binding sites may be necessary for the expression of the IMD pathway components that are effective in killing Plasmodium in Anopheles midguts. The expression and activity of dual oxidase (DUOX), a membrane-associated protein involved in the regulation of reactive oxygen species (ROS), is dependent on the gut bacterial load [81]. PGRP-LC plays an important role in the DUOX-mediated response, but Relish (Rel2) is not involved in the regulation of ROS generation [82]. ROS are important for antibacterial and anti-P. berghei immunity in A. gambiae [83], suggesting that DUOX could play a role in parasite killing by controlling the production of free radicals.
If bacteria present in the mosquito midgut are capable of stimulating an immune response sufficient to interfere with Plasmodium development, then why are natural mosquito populations susceptible to parasite infection at all? Bacteria are not isolated from every field-collected mosquito, but this is most likely an artifact of the isolation processes that are culture-dependent and rely on the presence of bacteria capable of growth in the culture medium used. Also, the species composition of the midgut-associated bacteria may play a role: G- bacteria more robustly interfere with Plasmodium parasite infection than do G+ bacteria, and species-specific differences in G- inhibition are apparent [84], [85] (Cirimotich and Dimopoulos, unpublished results). Laboratory and field strains of G- bacteria have potent inhibitory effects on Plasmodium infection, significantly inhibiting parasite development when bacteria and parasite are introduced in the same blood meal [42], [77], [84], [85] (Cirimotich and Dimopoulos, unpublished results). However, no data have been collected on the co-prevalence of specific bacteria and Plasmodium infection in wild-caught mosquitoes. It would be interesting to look at the gut microbiome of mosquitoes from malaria-endemic and non-endemic areas to identify possible associations between bacteria species and natural parasite refractoriness. Increasing the prevalence of inhibitory bacteria, possibly through artificially baited sugar solutions, may create mosquito populations that are naturally refractory to parasite infection.
Current research efforts are devoted to teasing apart the tripartite interactions among bacteria, the parasite, and vector immunity. It is possible that the pathway-directed immune responses against parasites are elicited by the post-blood meal bacterial bloom, with an indirect effect on parasite survival. A second stimulus could be the introduction of bacteria into the hemolymph during the mechanical disruption of the midgut that occurs when the parasite invades the epithelial layer. However, the parasites themselves might be immunogenic, or a combination of anti-bacterial and anti-plasmodial mechanisms could be occurring. If mosquitoes indeed mount a Plasmodium-specific response, what are the parasite surface moieties that are detected by the mosquito's immune system, and which mosquito receptors are detecting them? Arrighi et al. (2008) have recently shown that the A. gambiae immune response can be stimulated through the introduction of parasite-derived glycosylphosphatidylinositols (GPI). Immune gene expression controlled by both the IMD and Jak-Stat pathways was increased soon after artificial bloodfeed; however, a more drastic increase in immune gene expression was observed when the mosquitoes were fed a GPI from Toxoplasma gondii, a response that potentially represents a less-conserved response to the unnatural pathogen [86]. In A. stephensi mosquitoes, NOS expression is induced by feeding with P. falciparum-derived GPI, and a second parasite-derived molecule, P. falciparum hemozoin, has been shown to trigger an immune response that may be mediated by the IMD pathway [87], [88]. The identification of the PRRs responsible for recognition of the parasite molecules will be crucial to determining the existence of a true Plasmodium-directed mosquito immune response.
V. Plasmodium–mosquito midgut interactions
In addition to the immune-mediated killing of Plasmodium parasites, physical barriers also present an obstacle to productive infection of the mosquito. Motile ookinetes in the midgut lumen must traverse the peritrophic matrix and invade the midgut epithelial cells. The ookinetes migrate through the epithelial cell to the intracellular space between the epithelial surface and basal lamina, where they form oocysts. Eventual maturation leads to rupture and release of sporozoites into the mosquito hemocoel. Crossing the peritrophic matrix and midgut epithelium are obligatory steps in the eventual transmission of the parasite by the mosquito vector.
Interactions with the peritrophic matrix
The peritrophic matrix (PM) is a thick chitin-containing layer that coats the lumenal side of the midgut epithelium after blood ingestion. The PM poses a major physical barrier to ookinete invasion because of its 1- to 20-μm thickness [89] and because the time of its maturation (24 hours after a blood meal) roughly coincides with the time of ookinete invasion of the midgut epithelium [90]. Although an artificially thickened PM in Ae. aegypti reduces P. gallinaceum oocyst formation [91], PM disruption by silencing chitin synthase results in a markedly lower oocyst count in the same vector-parasite system [92]. One possible explanation for these results is that the PM slows the diffusion of hydrolytic enzymes that may be detrimental to the parasite. However, the interaction of the parasite with the PM might be necessary for Plasmodium maturation. The A. gambiae PM proteome has recently been characterized by mass spectrometry [93]. A better understanding of the composition and structure of the PM may lead to modifications that can make it a complete barrier to Plasmodium infection.
Interactions with the midgut epithelium
Invasion of the midgut epithelium occurs once the ookinete has bound to an unknown receptor on the lumenal surface. The P. berghei membrane-attack ookinete protein (MAOP) is essential for midgut invasion and most likely acts by creating pores in target membranes. MAOP-disrupted ookinetes attach to the epithelium but are unable to enter the cytoplasm [94]. Other P. berghei ookinete surface proteins that are essential for invasion include the micronemal proteins circumsporozite and TRAP related protein (CTRP) and secreted ookinete adhesive protein (SOAP). Although the precise role of these molecules in midgut invasion has not been determined, CTRP (a member of the TRAP family of proteins, which are involved in motility and invasion in the Apicomplexa) plays a role in ookinete locomotion, while SOAP binds to laminin and may be required for adhesion to the basal lamina [95], [96].
Invasion may also require enzymatic processing of bound ligands for subsequent detachment and entry into the host cell. P. berghei Rhomboid 1 (PbROM1), a member of the rhomboid family of serine proteases, is able to cleave TRAP family members within their transmembrane domains [97]. PbROM1 gene disruptants are impaired in the ookinete-to-oocyst transition [98], indicating that proteolytic processing of invasion proteins may be required for efficient entry into midgut epithelial cells. Depletion of the Anopheles homologue of Croquemort, a Drosophila scavenger receptor expressed in the midgut in response to blood feeding, results in the inhibition of oocyst formation, suggesting an as-yet unknown role for this molecule in Plasmodium-midgut interactions [99].
Once inside the cell cytoplasm, the ookinete must quickly travel to the basal lamina before the cell undergoes apoptosis and is extruded from the epithelium; this process is known as the time-bomb model of ookinete invasion [90]. A key protein in this process is the cell-traversal protein for ookinetes and sporozoites (CelTOS). CelTOS-disrupted ookinetes are able to invade the midgut epithelium but fail to traverse the cell, indicating that the protein is needed for migration through the cytoplasm. The authors speculate that CelTOS may anchor the ookinete to molecules in the cytoplasm [100].
Cellular components also play a role in controlling ookinete invasion and oocyst formation. Decreasing actin polymerization increases P. berghei oocyst numbers, while depleting lipid transport activity reduces P. berghei oocyst formation [101], [102]. Although the role of lipid transport has not been fully determined, there appears to be a requirement for the host's cellular pathways in the parasite life cycle. There is apparent conservation involved here, since increasing actin polymerization or decreasing lipid transport components has similar effects on oocyst counts in P. falciparum field isolates [103].
There are other important interactions between the Plasmodium parasite and mosquito that must occur before transmission to a vertebrate host. Among the most important is parasite invasion of the salivary glands and movement into the salivary ducts. For further information on these interactions, a recent review by Ghosh and Jacobs-Lorena (2009) is recommended [104].
VI. Conclusions
The mosquito's immune response is paramount in limiting pathogen infection and, in the case of Plasmodium parasites, transmission. Using anti-pathogen molecules that are expressed as a result of immune signaling pathway activation, the mosquito is able to combat this infection process. These effector molecules are members of diverse protein families that in some instances appear to be mosquito-specific and may have evolved as anti-Plasmodium and anti-bacterial factors. Indeed, bacteria harbored in the mosquito midgut may play a significant role in modulating Plasmodium transmission through the stimulation of an immune response.
Recent efforts in the areas of vector biology and vector-borne diseases have focused on blocking infection of the insect as a means of disease control. A better understanding of the interactions between the parasite and the mosquito vector is crucial to achieving this goal. For instance, boosting the anti-Plasmodium immune response by temporally expressing Rel2 or a specific effector molecule in transgenic mosquitoes could create mosquito populations refractory to parasite infection. The introduction into and stable transmission of inhibitory bacteria in natural mosquito populations might also be a means of controlling Plasmodium transmission that does not require genetic modification or release of mosquitoes. Exploiting the anti-Plasmodium immune response of the mosquito is yet another potential mechanism for combating Plasmodium infections worldwide.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review of Immunology. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, et al. Immunity-related genes and gene families in Anopheles gambiae. Science (New York, NY) 2002;298(5591):159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 3.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(20):7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathogens. 2008;4(7):e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25(4):677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Barillas-Mury C, Charlesworth A, Gross I, Richman A, Hoffmann JA, Kafatos FC. Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. The EMBO Journal. 1996;15(17):4691–4701. [PMC free article] [PubMed] [Google Scholar]
- 7.Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. The Journal of Biological Chemistry. 1999;274(30):21355–21361. doi: 10.1074/jbc.274.30.21355. [DOI] [PubMed] [Google Scholar]
- 8.Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, González-Crespo S, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75(4):753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 9.Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. The EMBO Journal. 1999;18(12):3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes & Development. 1999;13(7):792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann A, Stein D, Geisler R, Hagenmaier S, Schmid B, Fernandez N, et al. A gradient of cytoplasmic Cactus degradation establishes the nuclear localization gradient of the dorsal morphogen in Drosophila. Mechanisms of Development. 1996;60(1):109–123. doi: 10.1016/s0925-4773(96)00607-7. [DOI] [PubMed] [Google Scholar]
- 12.Edwards DN, Towb P, Wasserman SA. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development (Cambridge, England) 1997;124(19):3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas E, Reichhart JM, Hoffmann JA, Lemaitre B. In vivo regulation of the IkappaB homologue cactus during the immune response of Drosophila. The Journal of Biological Chemistry. 1998;273(17):10463–10469. doi: 10.1074/jbc.273.17.10463. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Steward R. A multimeric complex and the nuclear targeting of the Drosophila Rel protein Dorsal. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14524–14529. doi: 10.1073/pnas.94.26.14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belvin MP, Jin Y, Anderson KV. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Genes & Development. 1995;9(7):783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- 16.Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392(6671):93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 17.Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathogens. 2009;5(3):e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou Z, Shin SW, Alvarez KS, Bian G, Kokoza V, Raikhel AS. Mosquito RUNX4 in the immune regulation of PPO gene expression and its effect on avian malaria parasite infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18454–18459. doi: 10.1073/pnas.0804658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal K, Silverman N. Positive and negative regulation of the Drosophila immune response. BMB Reports. 2008;41(4):267–277. doi: 10.5483/bmbrep.2008.41.4.267. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cellular Microbiology. 2005;7(4):461–469. doi: 10.1111/j.1462-5822.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- 21.Choe K, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science (New York, NY) 2002;296(5566):359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 22.Choe K, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(4):1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Developmental Cell. 2001;1(4):503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 24.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Current Biology: CB. 2002;12(12):996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- 25.Leulier F, Parquet C, Pili-Floury S, Ryu J, Caroff M, Lee W, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nature Immunology. 2003;4(5):478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 26.Kleino A, Valanne S, Ulvila J, Kallio J, Myllymäki H, Enwald H, et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. The EMBO Journal. 2005;24(19):3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nature Immunology. 2000;1(4):342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 28.Tanji T, Ip YT. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends in Immunology. 2005;26(4):193–198. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Meister S, Kanzok SM, Zheng X, Luna C, Li T, Hoa NT, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(32):11420–11425. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, et al. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Developmental Cell. 2002;3(5):711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 32.Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(19):10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Molecular Cell. 1999;4(5):827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 34.Hoa N, Zheng L. Functional characterization of the NF-κB transcription factor gene rel2 from Anopheles gambiae. Insect Science. 2007;14(3):175–184. [Google Scholar]
- 35.Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Rämet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes and Infection /Institut Pasteur. 2005;7(5-6):811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes & Development. 2000;14(19):2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sluss HK, Han Z, Barrett T, Goberdhan DC, Wilson C, Davis RJ, et al. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes & Development. 1996;10(21):2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 38.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Reports. 2000;1(4):347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luna C, Hoa NT, Lin H, Zhang L, Nguyen HLA, Kanzok SM, et al. Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Molecular Biology. 2006;15(6):721–729. doi: 10.1111/j.1365-2583.2006.00661.x. [DOI] [PubMed] [Google Scholar]
- 40.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Reports. 2000;1(4):353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(44):16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathogens. 2009;5(8):e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science (New York, NY) 2009;324(5924):258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host & Microbe. 2009;5(2):200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Cronin SJF, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science (New York, NY) 2009;325(5938):340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nature Immunology. 2005;6(9):946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 47.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, Zamora RE, et al. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host & Microbe. 2009;5(5):498–507. doi: 10.1016/j.chom.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics. 2008;180(3):1671–1678. doi: 10.1534/genetics.108.089748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surachetpong W, Singh N, Cheung KW, Luckhart S. MAPK ERK signaling regulates the TGF-beta1-dependent mosquito response to Plasmodium falciparum. PLoS Pathogens. 2009;5(4):e1000366. doi: 10.1371/journal.ppat.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, et al. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(13):8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathogens. 2006;2(6):e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science (New York, NY) 2004;303(5666):2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 54.Riehle MM, Markianos K, Niaré O, Xu J, Li J, Touré AM, et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science (New York, NY) 2006;312(5773):577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- 55.Riehle MM, Xu J, Lazzaro BP, Rottschaefer SM, Coulibaly B, Sacko M, et al. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PloS One. 2008;3(11):e3672. doi: 10.1371/journal.pone.0003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habtewold T, Povelones M, Blagborough AM, Christophides GK. Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species A. PLoS Pathogens. 2008;4(5):e1000070. doi: 10.1371/journal.ppat.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warr E, Lambrechts L, Koella JC, Bourgouin C, Dimopoulos G. Anopheles gambiae immune responses to Sephadex beads: involvement of anti-Plasmodium factors in regulating melanization. Insect Biochemistry and Molecular Biology. 2006;36(10):769–778. doi: 10.1016/j.ibmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Fraiture M, Baxter RHG, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, et al. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host & Microbe. 2009;5(3):273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Mitri C, Jacques J, Thiery I, Riehle MM, Xu J, Bischoff E, et al. Fine Pathogen Discrimination within the APL1 Gene Family Protects Anopheles gambiae against Human and Rodent Malaria Species. PLoS Pathog. 2009;5(9):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. The Journal of Biological Chemistry. 2009;284(15):9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujita T. Evolution of the lectin-complement pathway and its role in innate immunity. Nature Reviews Immunology. 2002;2(5):346–353. doi: 10.1038/nri800. [DOI] [PubMed] [Google Scholar]
- 62.Gokudan S, Muta T, Tsuda R, Koori K, Kawahara T, Seki N, et al. Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Zhao Q, Christensen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Middha S, Wang X. Evolution and potential function of fibrinogen-like domains across twelve Drosophila species. BMC Genomics. 9(2008):260. doi: 10.1186/1471-2164-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science (New York, NY) 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dimopoulos G, Casavant TL, Chang S, Scheetz T, Roberts C, Donohue M, et al. Anopheles gambiae pilot gene discovery project: identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annual Review of Immunology. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 68.Kuhlman M, Joiner K, Ezekowitz RA. The human mannose-binding protein functions as an opsonin. The Journal of Experimental Medicine. 1989;169(5):1733–1745. doi: 10.1084/jem.169.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsushita M, Fujita T. Activation of the classical complement pathway by mannose-binding protein in association with a novel C1s-like serine protease. The Journal of Experimental Medicine. 1992;176(6):1497–1502. doi: 10.1084/jem.176.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schnitger AKD, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. The Journal of Biological Chemistry. 2009;284(26):17616–17624. doi: 10.1074/jbc.M808298200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luckhart S, Li K, Dunton R, Lewis EE, Crampton AL, Ryan JR, et al. Anopheles gambiae immune gene variants associated with natural Plasmodium infection. Molecular and Biochemical Parasitology. 2003;128(1):83–86. doi: 10.1016/s0166-6851(03)00016-1. [DOI] [PubMed] [Google Scholar]
- 72.Vizioli J, Bulet P, Hoffmann JA, Kafatos FC, Müller HM, Dimopoulos G. Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(22):12630–12635. doi: 10.1073/pnas.221466798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biology. 2006;4(7):e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garver LS, Xi Z, Dimopoulos G. Immunoglobulin superfamily members play an important role in the mosquito immune system. Developmental and Comparative Immunology. 2008;32(5):519–531. doi: 10.1016/j.dci.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warr E, Das S, Dong Y, Dimopoulos G. The Gram-negative bacteria-binding protein gene family: its role in the innate immune system of anopheles gambiae and in anti-Plasmodium defence. Insect Molecular Biology. 2008;17(1):39–51. doi: 10.1111/j.1365-2583.2008.00778.x. [DOI] [PubMed] [Google Scholar]
- 76.Dimopoulos G, Richman A, Müller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(21):11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. The American Journal of Tropical Medicine and Hygiene. 1996;54(2):214–218. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- 78.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathogens. 2009;5(5):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beier MS, Pumpuni CB, Beier JC, Davis JR. Effects of para-aminobenzoic acid, insulin, and gentamicin on Plasmodium falciparum development in anopheline mosquitoes (Diptera: Culicidae) Journal of Medical Entomology. 1994;31(4):561–565. doi: 10.1093/jmedent/31.4.561. [DOI] [PubMed] [Google Scholar]
- 80.Ryu J, Kim S, Lee H, Bai JY, Nam Y, Bae J, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science (New York, NY) 2008;319(5864):777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 81.Ha E, Oh C, Bae YS, Lee W. A direct role for dual oxidase in Drosophila gut immunity. Science (New York, NY) 2005;310(5749):847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 82.Ha E, Lee K, Seo YY, Kim S, Lim J, Oh B, et al. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nature Immunology. 2009;10(9):949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 83.Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, et al. Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. The Journal of Biological Chemistry. 2008;283(6):3217–3223. doi: 10.1074/jbc.M705873200. [DOI] [PubMed] [Google Scholar]
- 84.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. Journal of Medical Entomology. 2003;40(3):371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 85.Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Plasmodium falciparum: inhibition of sporogonic development in Anopheles stephensi by gram-negative bacteria. Experimental Parasitology. 1993;77(2):195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 86.Arrighi RBG, Debierre-Grockiego F, Schwarz RT, Faye I. The immunogenic properties of protozoan glycosylphosphatidylinositols in the mosquito Anopheles gambiae. Developmental and Comparative Immunology. 2009;33(2):216–223. doi: 10.1016/j.dci.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 87.Akman-Anderson L, Olivier M, Luckhart S. Induction of nitric oxide synthase and activation of signaling proteins in Anopheles mosquitoes by the malaria pigment, hemozoin. Infection and Immunity. 2007;75(8):4012–4019. doi: 10.1128/IAI.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lim J, Gowda DC, Krishnegowda G, Luckhart S. Induction of nitric oxide synthase in Anopheles stephensi by Plasmodium falciparum: mechanism of signaling and the role of parasite glycosylphosphatidylinositols. Infection and Immunity. 2005;73(5):2778–2789. doi: 10.1128/IAI.73.5.2778-2789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abraham EG, Jacobs-Lorena M. Mosquito midgut barriers to malaria parasite development. Insect Biochemistry and Molecular Biology. 2004;34(7):667–671. doi: 10.1016/j.ibmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 90.Han YS, Thompson J, Kafatos FC, Barillas-Mury C. Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. The EMBO Journal. 2000;19(22):6030–6040. doi: 10.1093/emboj/19.22.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Billingsley PF, Rudin W. The role of the mosquito peritrophic membrane in bloodmeal digestion and infectivity of Plasmodium species. The Journal of Parasitology. 1992;78(3):430–440. [PubMed] [Google Scholar]
- 92.Kato N, Mueller CR, Fuchs JF, McElroy K, Wessely V, Higgs S, et al. Evaluation of the function of a type I peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aegypti. Vector Borne and Zoonotic Diseases (Larchmont, NY) 2008;8(5):701–712. doi: 10.1089/vbz.2007.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dinglasan RR, Devenport M, Florens L, Johnson JR, McHugh CA, Donnelly-Doman M, et al. The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochemistry and Molecular Biology. 2009;39(2):125–134. doi: 10.1016/j.ibmb.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(46):16310–16315. doi: 10.1073/pnas.0406187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, et al. CTRP is essential for mosquito infection by malaria ookinetes. The EMBO Journal. 1999;18(22):6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dessens JT, Sidén-Kiamos I, Mendoza J, Mahairaki V, Khater E, Vlachou D, et al. SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Molecular Microbiology. 2003;49(2):319–329. doi: 10.1046/j.1365-2958.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 97.Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathogens. 2006;2(10):e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srinivasan P, Coppens I, Jacobs-Lorena M. Distinct roles of Plasmodium rhomboid 1 in parasite development and malaria pathogenesis. PLoS Pathogens. 2009;5(1):e1000262. doi: 10.1371/journal.ppat.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.González-Lázaro M, Dinglasan RR, Hernández-Hernández FDLC, Rodríguez MH, Laclaustra M, Jacobs-Lorena M, et al. Anopheles gambiae Croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite Plasmodium berghei. Insect Biochemistry and Molecular Biology. 2009;39(5-6):395–402. doi: 10.1016/j.ibmb.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Molecular Microbiology. 2006;59(5):1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 101.Vlachou D, Zimmermann T, Cantera R, Janse CJ, Waters AP, Kafatos FC. Real-time, in vivo analysis of malaria ookinete locomotion and mosquito midgut invasion. Cellular Microbiology. 2004;6(7):671–685. doi: 10.1111/j.1462-5822.2004.00394.x. [DOI] [PubMed] [Google Scholar]
- 102.Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Current Biology: CB. 2005;15(13):1185–1195. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 103.Mendes AM, Schlegelmilch T, Cohuet A, Awono-Ambene P, De Iorio M, Fontenille D, et al. Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathogens. 2008;4(5):e1000069. doi: 10.1371/journal.ppat.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ghosh AK, Jacobs-Lorena M. Plasmodium sporozoite invasion of the mosquito salivary gland. Current Opinion in Microbiology. 2009;12(4):394–400. doi: 10.1016/j.mib.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]


