Abstract
Dengue fever (DF) is the most frequent arthropod-borne viral disease of humans, with almost half of the world's population at risk of infection1. The high prevalence, lack of an effective vaccine, and absence of specific treatment conspire to make DF a global public health threat1, 2. Given their compact genomes, dengue viruses (DENV 1-4) and other flaviviruses likely require an extensive number of host factors; however, only a limited number of human, and an even smaller number of insect host factors have been identified3-10. To discover insect host factors required for DENV-2 propagation, we carried out a genome-wide RNA interference screen in Drosophila melanogaster cells using a well-established 22,632 dsRNA library. This screen identified 116 candidate dengue virus host factors (DVHFs) (Supplementary Fig. 1). While some were previously associated with flaviviruses (e.g., V-ATPases and alpha-glucosidases)3-5, 7, 9, 10, most DVHFs were newly implicated in DENV propagation. The dipteran DVHFs had eighty-two readily recognizable human homologues and, using a targeted siRNA screen, we showed that forty-two of these are human DVHFs. This indicates remarkable conservation of required factors between dipteran and human hosts. This work suggests novel approaches to control infection in the insect vector and the mammalian host.
DENV 1-4 are transmitted from one human host to another by mosquitoes of the Aedes genus, principally Aedes aegypti and albopictus2. While there are important efforts to sequence and annotate the genomes of these vectors11-13, there is currently an unfortunate dearth of resources to carry out systematic functional genomics in Aedes. In contrast, there are robust materials and methods to do so in the related dipteran Drosophila melanogaster. In order to take advantage of these existing tools, DENV-2 New Guinea C (DEN2-NGC) was adapted by serial passage in D.Mel-2 cells over a period of four months (DEN2S2; Supplementary Fig. 2). To identify DVHFs required for efficient propagation of DENV2 in insect cells, we carried out a genome-wide RNA interference (RNAi) screen in Drosophila D.Mel-2 cells using the 22,632-dsRNA DRSC 2.0 library designed and provided by the Drosophila RNAi Screening Center (www.flyrnai.org)14. The screen was performed in duplicate, assaying 45,264 infections, excluding controls. If either duplicate resulted in fewer than 12,500 cells per well, the dsRNA was excluded from further analysis; a criterion that excluded 2,343 dsRNAs. The remaining 20,224 dsRNAs were scored by their effect on infection, which was determined by measuring expression of envelope protein (Supplementary Fig. 3). Each pair of duplicate dsRNA was assigned a Sum Rank score and those with scores expected with a frequency ≤ 0.065 by chance alone were selected for further analysis (Supplementary Fig. 4 and Methods). Of the 218 (1.1%) that met this criterion we were able to readily re-synthesize and re-screen 179 dsRNAs. We identified 118 dsRNAs, representing 116 unique DVHFs that inhibited infectivity by ≥ 1.5 fold with p < 0.05 (Supplementary Table 1).
The screen identified DVHFs previously known to be required for dengue and/or other flaviviral infection, such as an α-glucosidase and the V-ATPase proton pump (CG14476, VhaPP1 and Vha14 in Figs. 1b,c)3-5, 9, 10. The effect on V0 and V1 subunits of the V-ATPase provided strong evidence of a requirement for the holoenzyme. To obtain independent evidence for this, we tested the effect of bafilomycin, a specific V-ATPase inhibitor previously shown to inhibit flaviviruses3, 7, 9, on DENV infection of C6/36 Ae. albopictus cells. Bafilomycin treatment induced a dramatic inhibition of both DEN2-S2 and DEN2-NGC replication in these mosquito cells (Supplementary Fig. 5). These data demonstrated the validity of the screen since they generalize the findings to a well-studied DENV2 and cells of the natural vector Ae. albopictus.
Figure 1.
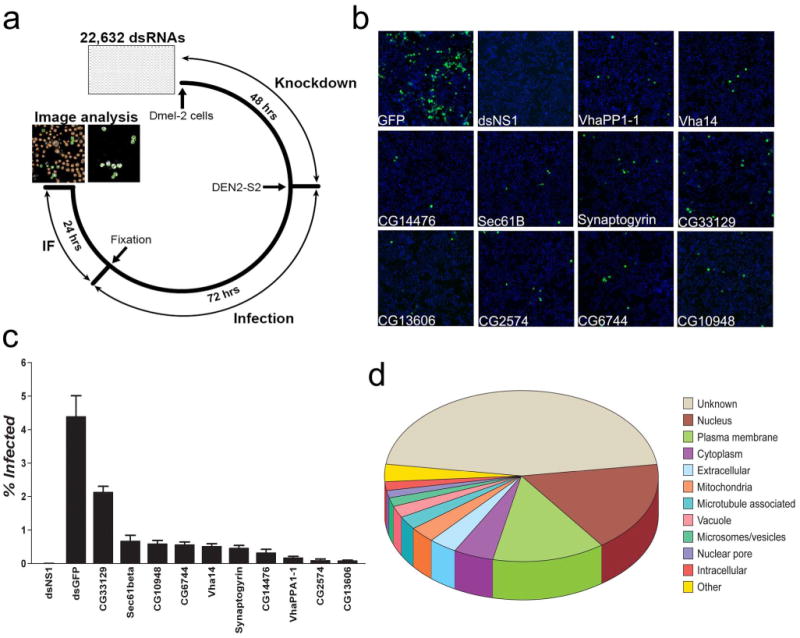
Genome-wide RNAi screen for dipteran DVHFs. 22,632 dsRNAs were assayed in duplicate for their effect on DEN2-S2 viral gene expression in D.Mel-2 cells. (a) Schematic of the experimental protocol used in the screen. (b) Representative images of dsRNA treated D.Mel-2 cells at 20× magnification with nuclei staining (blue) and dengue E protein staining (green). (c) The percentage of DEN2-S2 infected (% Infected) cells is indicated for controls and selected DVHFs. Error bars represent standard error of ≥ six independent observations. (d) Cellular localization of the 116 DVHFs identified in the Drosophila screen according to GO cellular component annotation (FlyBase: FB2008_05).
The overwhelming majority of DVHFs (111/116) had not been previously identified as such. Extant annotation (FlyBase: FB2008_05) predicted diverse cellular functions for DVHFs. DnaJ-1 and CG3061 are predicted to be involved in the unfolded protein response, which is activated upon DENV infection15, 16. α-Adaptin, cnir, lqf, synaptogyrin, Syx4, and Syx13, are all involved in vesicular transport and endocytosis17, which have been implicated in the entry and replication of a diverse group of viruses18, including DENV19. The novel DVHF lqf interacts with the Drosophila homologue of human EPS15, which is required for West Nile Virus (WNV) and DENV entry19-21. We posit that RNA binding proteins, such as bol, Unr and CG5205, and the 3′-5′exonuclease-like CG6744, assist in genome expression, replication and/or packaging (see below). Interestingly, the mosquito homologues of three DVHFs identified in our screen (pxb, H15 and Cyp6a19) were found to be differentially regulated after DENV infection in live mosquitoes22.
Gene Ontology (GO) annotation (GATHER, www.gather.genome.duke.edu/) of DVHFs indicated a surprisingly high number (22) of nuclear proteins. Although DENV gene products are known to transit through the nucleus during the course of infection23-26, it is also possible that DENV infection relocalizes many of these factors to the cytoplasm. Also remarkable was the large number of gene products predicted to be membrane-associated: 17 with the plasma membrane and 10 with intracellular membranes (ER, Golgi, vesicles and vacuole-like organelles) (Fig. 1d). The detection of many membrane bound gene products is fully consistent with the observations that viral infections cause remodeling of cellular membranes27.
In order to test whether DVHFs were required for propagation of DENV in the vector mosquito Ae. aegypti we tested the impact of depleting mosquito homologues of lola (AAEL009212), CG10320, a putative NADH dehydrogenase, (AAEL007054), and Cyp6a19 (AAEL009124) using an established method of RNAi-mediated gene silencing28. A dsRNA targeting AAEL009212 reduced the DEN2-NGC capacity to infect the midgut tissue at seven days after ingestion of infected blood (Fig. 2). The effect of DVHF gene silencing on DENV infection was likely underestimated because of an aberrantly low titer of one group of control GFP dsRNA injected mosquitoes (Fig. 2). A dsRNA targeting AAEL009124 did not affect infectivity. Where as inhibition of infectivity upon depletion of AAEL007054 was not statistically significant, exclusion of the point that appears to be an outlier leads to a reduction of infectivity that approaches significance and suggests that this gene product could be a DVHF in Ae. aegypti (Fig. 2). Given the complex spatio-temporal dynamics of DENV infection in the mosquito, the fact that the mosquitoes are genetically polymorphic, the inherent variability of blood meal infections29, and the uncertainty of achieving gene product depletion in the appropriate tissue and time after dsRNA injection, it was remarkable to obtain inhibition with these DVHFs. These data, together with those obtained above with Ae. albopictus cells, validate the use of the Drosophila screen to identify dipteran DVHFs.
Figure 2.
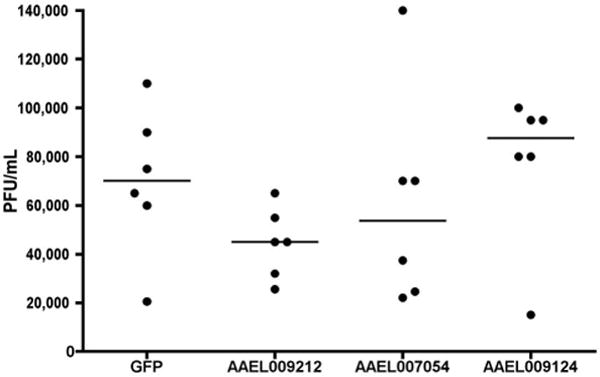
Injection of Ae. aegypti mosquitoes with dsRNA targeting a DVHF inhibits dengue virus propagation. Four-day old female mosquitoes were injected with dsRNAs targeting GFP, AAEL009212, AAEL007054, or AAEL009124. Three days after injection, mosquitoes were fed on a DEN2-NGC supplemented blood meal. Seven days later, 30 mosquitoes for each condition were randomly sorted into 6 groups of 5, their midguts removed, homogenized and titered. Data points indicate the combined titer of five dsRNA-treated mosquito midguts. Dashes indicate the median value of the six biological replicates. Significance at <0.05 level was determined using a one-sided student's T-test of viral titres (dsDVHF vs. dsGFP).
The 116 DVHFs had 82 readily identifiable human homologues, which we targeted with a library of siRNAs (Fig. 3a). We supplemented the library with siRNAs targeting gene products that were functionally associated with the V-ATPase but had not scored as DVHFs in the D.Mel-2 screen, for instance V-ATPase accessory proteins not found in insects. Of the 82 homologues of the dipteran DVHFs, 42 (51%) scored as human DVHFs (Fig. 3b, c; Supplementary Table 2). The remarkably high number of dipteran DVHFs that also were required for infection of human cells further validates the screen, and provides the first evidence for widespread conservation of flavivirus-host interactions between invertebrates and vertebrates (Supplementary Fig. 6 and Supplementary Table 3).
Figure 3.
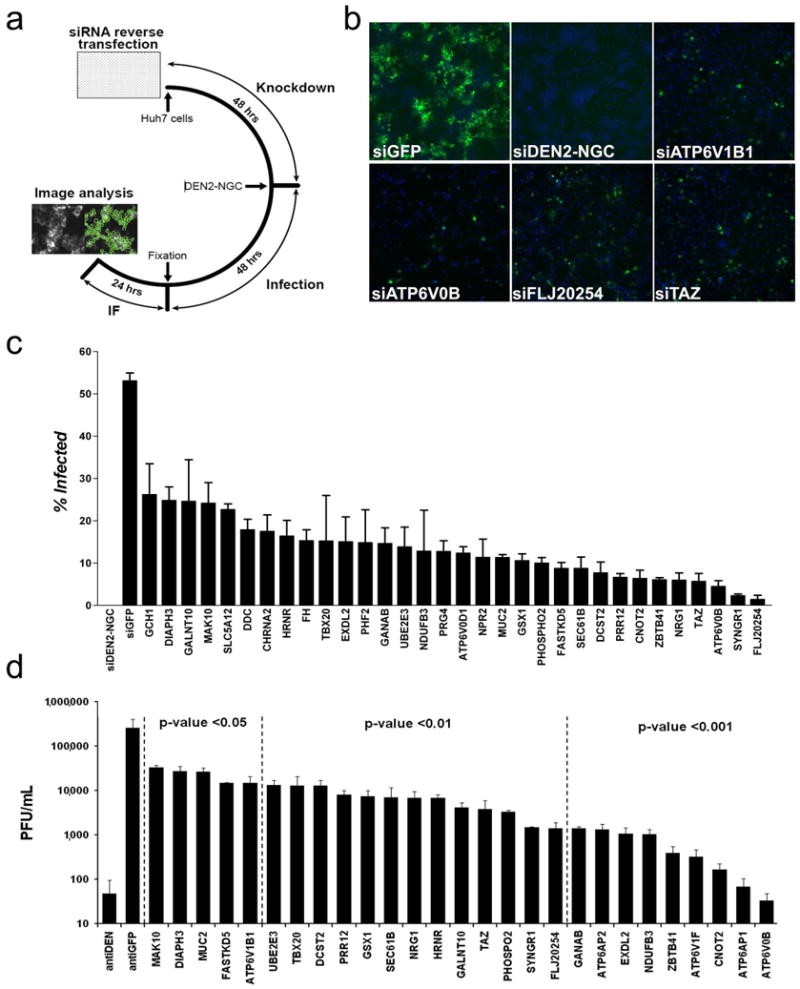
Screen for Human DVHFs. siRNAs targeting 82 human DVHF homologues were screened in HuH-7 cells for their ability to inhibit DEN2-NGC. (a) Schematic of the experimental protocol. (b) Representative images of siRNA treated HuH-7 cells at 1× magnification with nuclei staining (blue) and dengue E protein staining (green). (c) The percentage of DEN2-NGC infected cells is indicated for controls and selected DVHFs. (d) Viral propagation after treatment with control or DVHF siRNAs was measured 72 hours after DEN2NGC infection and plotted on a logarithmic scale. Error bars in (c) and (d) indicate standard error of three independent observations.
Knockdown of DVHFs in HuH-7 cells significantly reduced the formation of DEN2-NGC infectious particles (Fig. 3d). Knockdown of DVHFs predicted to be involved in entry, post-translational modifications and transcription accounted for the majority of those that resulted in greater than 10-fold inhibition of viral propagation. In order to ascertain whether or not the DVHFs were required for other viral infections we determined whether knockdown of DVHFs led to alterations in the gene expression for Yellow Fever virus (YFV) (17D vaccine strain), another flavivirus, and Coxsackie B3 (strain 20) (CB3), an enterovirus. Of six DVHFs that showed ≥ 2 fold inhibition of DEN2-NGC E protein expression by at least two independent siRNAs, only one (FLJ20254) scored by the same criteria as required for YFV and three (CNOT2, FLJ20254, TAZ) as required for CB3 gene expression (Supplementary Table 4). This suggests some shared host factors among these RNA viruses, but also points to the existence of dengue specific host factors.
To determine whether we had identified DVHFs that affected both early and late steps in the viral life cycle and to gain some early mechanistic insights we examined viral RNA accumulation in HuH-7 cells treated with siRNAs targeting NPR2, SEC61B, FLJ20254, TAZ, EXDL2, and CNOT2 transcripts. As expected, knockdown of these DVHFs reduced both the number of cells expressing DENV E protein and the titer of infectious virus recovered 48 hours post infection (pi) with DEN2-NGC (Fig. 4, a and b). Consistent with this, we noted a decrease in viral transcripts measured by qRT-PCR (Fig. 4e). Accumulation of viral transcripts at 18 and 24 hours pi was clearly decreased in cells depleted of FLJ20254, TAZ, EXDL2, and CNOT2 (c, d), indicating that these DVHFs act on steps required for the accumulation of RNA (e.g., early events). In contrast, knockdown of NPR2 did not result in lower RNA levels at early times pi, indicating that this DVHF acts at steps downstream of RNA accumulation. Given its location in the plasma membrane30, it is likely that NPR2 is involved in the assembly or exit of DENV.
Figure 4.
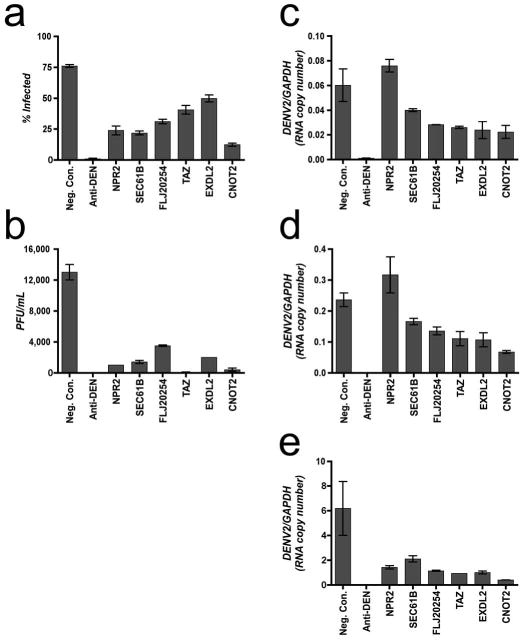
Analysis of DENV RNA accumulation after DVHF knockdown. (a) The percentage of DENV2-NGC infected (% Infected) cells 48 hours post infection (MOI ∼ 1.4) is indicated for controls and for six DVHFs. Error bars represent the standard deviation of six replicates. (b) Viral propagation 48 hours post infection was calculated for controls and six DVHFs. Error bars represent the range of duplicates. (c, d, e) Viral RNA accumulation was measured by RTqPCR at 18, 24, and 48 hours post infection respectively and normalized to GAPDH. Values represent the average of median qPCR measurements. Error bars represent the range of duplicates.
It is difficult to draw definitive conclusions for the 40 human homologues of dipteran DVHFs that did not recapitulate the effect on DENV infection (Supplementary Fig. 6). It is likely that some of these failed merely because their knockdown in human cells was ineffective. Nonetheless, these 40 genes that did not score in the human screen are not enriched in the aforementioned processes (e.g., endocytosis), but show highly significant enrichment for genes involved in immunity, suggesting that many represent dipteran-specific DVHFs.
The availability of resources to carry out en masse analysis in D. melanogaster and human cells permitted us to carry out a study that extended the known list of DVHFs by many fold. Given the likelihood of some false positives and the certainty of false negatives, this work represents an incomplete first version of what eventually will be a comprehensive DVHF list. Nonetheless, this study could lead to new targets for vector intervention. Furthermore, the information uncovered here should be used to explore the contribution of human DVHFs to disease severity and their potential in the treatment of DF, and related illnesses such as West Nile encephalitis/fever and yellow fever.
Methods Summary
The 22,632 dsRNA collection of the DRSC has been previously described 14. The DEN2-S2 virus used in the primary screen was isolated via serial passage of DEN2-NGC (a gift from Dr. A. de Silva – UNC-CH) in D.Mel-2 cells (Invitrogen). The experimental schedule for the primary and secondary screens is outlined in Fig. 1a. The human screen was done with HuH-7 cells treated independently with two siRNAs (Qiagen) for each gene product and DEN2-NGC according to the schedule outlined in Fig. 2a. Infected cells were labeled with the anti-E, 4G2 primary antibody (isolated from the DI-4G2-4-15 hybridoma (ATCC#HB-112)) and Alexa-488 anti-mouse secondary (Invitrogen), and counterstained with Hoescht 33342 (Sigma). Imaging and analysis of infection was done with a Cellomics ArrayScan Vti HCS machine.
Selection of candidates from the primary screen was done using a nonparametric approach, the Sum Rank algorithm, in order to produce an appropriate summary statistic of each dsRNA tested in duplicate. Briefly, within each plate, wells were ranked by the percentage of infected cells, with the well with the lowest percentage infected cells given rank = 1. For each dsRNA tested in duplicate, we calculated a Sum Rank (SR) statistic using the formula: SR = Rank on plate #1 + Rank on plate #2. SRs at either extreme are less likely to be observed by random chance. The number of times a given SR is expected to occur near the lower extreme (SR = 2) for a single pair of duplicate plates is given by: E[SR] = (SR - 1) / (# Valid Wells). SRs were calculated for every dsRNA in the Drosophila genome, with E[SR] scores below 0.065 used to select potential targets (218 dsRNAs) for further analysis (See Supplementary Fig. 4).
Standard experimental procedures were used for all other experiments. Please see Supplementary Information for more detail.
Supplementary Material
Acknowledgments
We thank A. de Silva, C. Lambeth, E. Wagner (UNC-Chapel Hill), F. Scholle and P. Florez de Sessions (North Carolina State University), J. Umbach, S. Braderick, B. Cullen, J. Nevins, M. Marengo, and M. Gromeier (Duke University), R. Wharton (Ohio State University), D. Gubler, E. E. Ooi and S. Vasudevan (Duke-NUS Graduate Medical School, Singapore), and members of the Triangle Flavivirology Group for reagents, time, and insights. MGB thanks the late Dr. Robert Shope for his inspiration and mentorship. We thank the DRSC (Harvard Medical School), which is funded by NIH grant RO1 GM067761, for expert assistance. We give special thanks to B. Mathey-Prevot (previously at the DSRC and now at Duke University) for his support, advice and expertise, and for pointing out the important overlap of our screen with others previously published. We acknowledge funding from NIH (R21-AI64925 and 5U54-AI057157-05S to MGB, AI076442 to PLY, and 1R01AI061576-01 to GD); ASM (to JL. Ramirez); Johns Hopkins Malaria Research Institute (to JA. Souza-Neto). MAR is a Karnovsky Fellow. PLY acknowledges an award from the Giovanni Armenise –Harvard Foundation. We also acknowledge funding from NCBC (to J. Nevins and MGB), NIH (1SA0RR024572-1 to MGB), Duke Center for RNA Biology, Duke University School of Medicine, Institute of Genome Sciences and Policy, and the Duke Comprehensive Cancer Center (5P30-CA14236) in support of the RNAi Facility.
Footnotes
Author Contributions: All authors contributed to the strategy and implementation of the work.
The authors declare no competing financial interests.
Author Information: Complete list of hits and dsRNA sequence information are available at the DRSC website (http://www.flyrnai.org).
Supplementary Information accompanies the paper on www.nature.com/nature
References
- 1.Kyle JL, Harris E. Global Spread and Persistence of Dengue. Annu Rev Microbiol. 2008 doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 2.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 3.Andoh T, et al. Effect of bafilomycin A1 on the growth of Japanese encephalitis virus in Vero cells. J Neurovirol. 1998;4:627–31. [PubMed] [Google Scholar]
- 4.Chapel C, et al. Antiviral effect of alpha-glucosidase inhibitors on viral morphogenesis and binding properties of hepatitis C virus-like particles. J Gen Virol. 2006;87:861–71. doi: 10.1099/vir.0.81503-0. [DOI] [PubMed] [Google Scholar]
- 5.Courageot MP, Frenkiel MP, Dos Santos CD, Deubel V, Despres P. Alpha-glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J Virol. 2000;74:564–72. doi: 10.1128/jvi.74.1.564-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–6. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinz FX, et al. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–17. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 8.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–5. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nawa M. Effects of bafilomycin A1 on Japanese encephalitis virus in C6/36 mosquito cells. Arch Virol. 1998;143:1555–68. doi: 10.1007/s007050050398. [DOI] [PubMed] [Google Scholar]
- 10.Whitby K, et al. Castanospermine, a potent inhibitor of dengue virus infection in vitro and in vivo. J Virol. 2005;79:8698–706. doi: 10.1128/JVI.79.14.8698-8706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–7. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson D, et al. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–5. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–23. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–5. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 15.Umareddy I, et al. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol J. 2007;4:91. doi: 10.1186/1743-422X-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–80. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littleton JT. A genomic analysis of membrane trafficking and neurotransmitter release in Drosophila. J Cell Biol. 2000;150:F77–82. doi: 10.1083/jcb.150.2.f77. [DOI] [PubMed] [Google Scholar]
- 18.Pelchen-Matthews A, Raposo G, Marsh M. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 2004;12:310–6. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Krishnan MN, et al. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–5. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu JJ, Ng ML. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol. 2004;78:10543–55. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosso C, Galvan-Mendoza IJ, Ludert JE, del Angel RM. Endocytic pathway followed by dengue virus to infect the mosquito cell line C6/36 HT. Virology. 2008;378:193–9. doi: 10.1016/j.virol.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor M, et al. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–6. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 24.Pryor MJ, et al. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8:795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 25.Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J Virol. 2006;80:5451–64. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang SH, Syu WJ, Hu ST. Identification of the homotypic interaction domain of the core protein of dengue virus type 2. J Gen Virol. 2004;85:2307–14. doi: 10.1099/vir.0.80067-0. [DOI] [PubMed] [Google Scholar]
- 27.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006;4:371–82. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y, et al. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franz AW, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103:4198–203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


