Abstract
Background
Previous work showed that miRNAs play key roles in the regulation of metamorphosis in the hemimetabolan species Blattella germanica. To gain insight about which miRNAs might be important, we have constructed two miRNA libraries, one of the penultimate, pre-metamorphic nymphal instar (N5) and the other of the last, metamorphic nymphal instar (N6).
Results
High throughput sequencing gave 61 canonical miRNAs present in the N5 and N6 libraries, although at different proportions in each. Comparison of both libraries led to the identification of three and 37 miRNAs significantly more expressed in N5 and N6 respectively. Twelve of these 40 miRNAs were then investigated further by qRT-PCR and results indicated that miR-252-3p was well expressed in N5 but not in N6, whereas let-7-5p, miR-100-5p and miR-125-5p showed the reverse pattern. 20-Hydroxyecdysone (20E) tended to stimulate miRNA expression, whereas juvenile hormone (JH) inhibited the 20E stimulatory effect. Expression of let-7, miR-100 and miR-125 was increased by 20E, which has also been observed in D. melanogaster. The only miRNA that was inhibited by 20E was miR-252-3p. The involvement of let-7, miR-100 and miR-125 in metamorphosis has been demonstrated in other insects. Depletion of miR-252-3p caused growth and developmental delays, which suggests that this miRNA is involved in regulating these processes prior to metamorphosis.
Conclusions
The comparative analysis of miRNA libraries from pre-metamorphic (N5) and metamorphic stages (N6) of B. germanica proved to be a useful tool to identify miRNAs with roles in hemimetabolan metamorphosis. Three miRNAs emerged as important factors in the metamorphic stage (N6): let-7-5p, miR-100-5p and miR-125-5p, whereas miR-252-3p appears to be important in the pre-metamorphic stage (N5).
Keywords: MicroRNAs, Metamorphosis, Ecdysone, Juvenile hormone, Blattella, Cockroach, Drosophila
Background
Metamorphosis has been a key innovation in insect evolution, which had a decisive influence on the dramatic diversification of this animal class, with around one million presently described species [1]. There are two types of metamorphosis: hemimetaboly, or gradual metamorphosis, and holometaboly or abrupt metamorphosis [2], the latter being evolutionarily the most successful [1]. In all cases, insect metamorphosis is hormonally regulated, and the most important hormones are the ecdysteroid 20-hydroxyecdysone (20E), which determines the successive moults, and the terpenoid juvenile hormone (JH), which represses the expression of adult features. Each hormone exerts its respective action through a cascade of transcription factors that transduce the hormonal signal to the effector genes [3,4].
Recently, a number of reports have pointed to microRNAs (miRNAs) as important players in the metamorphic transition, in hemimetabolan as well as in holometabolan species [5]. miRNAs are small non-coding RNAs of about 21–22 nucleotides that modulate gene expression at a post-transcriptional level, often in the context of developmental and morphogenetic processes [6-8]. They undergo molecular processing before becoming mature miRNAs [9,10]. First, miRNAs are transcribed as part of a primary transcript (pri-miRNA), which contains one or more miRNA precursors (pre-miRNAs). In the nucleus, the pri-miRNAs are processed into hairpin-structured pre-miRNAs by the ribonuclease Drosha, and exported to the cytoplasm. Once in the cytoplasm, the pre-miRNAs are cleaved by the ribonuclease Dicer-1 into an imperfectly-paired duplex, whose 5’- and 3’-strands can either give two respective miRNAs or only one. Generally, only one of the strands give rise to a mature miRNA, whereas the other, called miRNA* or the star strand, is degraded [9]. However, given that both strands can give mature miRNAs in some cases [11], the miRNAs are presently labelled as miRNA-5p or miRNA-3p according to the position of the original strand in the pre-miRNA. This is the convention used in the present paper.
One of the more dramatic demonstrations that miRNAs are involved in insect metamorphosis was reported by Gomez-Orte & Belles [12], who silenced dicer-1 expression by RNAi in the last nymphal instar of the cockroach Blattella germanica, and obtained supernumerary nymphs after the following moult, instead of adults [12]. These results showed that Dicer-1 and miRNAs are important factors in the mechanisms regulating metamorphosis. However, which particular miRNAs were involved remained unclear.
B. germanica is a polyneopteran exopterigote insect that shows a gradual morphological transformation during the life cycle, and is a good representative of the less modified hemimetaboly. This, and the fact that it is especially sensitive to gene silencing by RNAi [13], have made B. germanica a favourite hemimetabolan model to study metamorphosis. However, the genome of B. germanica is yet to be sequenced. The first step to approach the study of miRNAs involved in metamorphosis was therefore to establish a baseline miRNA catalogue using high-throughput sequencing [14]. To gain more information about which miRNAs may be important for B. germanica metamorphosis, we constructed two miRNA libraries, one with RNA extracted around the peak of 20E of the penultimate nymphal instar (N5), and the other constructed equivalently but during the last instar (N6). The endocrine context differs between these two libraries with the presence of JH in N5 and its absence in N6. We therefore aimed to identify miRNAs differentially expressed in N5 and N6. Those that were identified with this approach were further studied by establishing the expression pattern in N5 and N6, and by investigating the effect of 20E alone and that of 20E plus JH in their expression. Finally, we selected miR-252-3p, the most interesting miRNA emerging from these studies, to carry out a functional analysis using a specific anti-miRNA.
Methods
Insects
Specimens of B. germanica were obtained from a colony reared in the dark at 30 ± 1°C and 60–70% relative humidity. Freshly ecdysed nymphs or adult females were selected and used at the appropriate ages. All dissections, treatments and tissue sampling were carried out on carbon dioxide-anaesthetized specimens. Samples for RNA extraction and quantification were frozen immediately after dissection, and stored at −80°C until use.
Samples to construct the miRNA libraries
To construct the two miRNA libraries, we carried out total RNA extraction from the whole body (excluding the head and the digestive tube, to avoid ocular pigments and intestine parasites) of B. germanica specimens, using the miRNeasy extraction kit (QIAGEN). Nine individuals taken in days 3, 4 and 5 (three each day) of fifth nymphal instar were pooled for the N5 library. The equivalent sampling was carried out on days 5, 6 and 7 of sixth (last) nymphal instar for the N6 library. The days chosen correspond to the 20E respective moulting peaks in N5 and N6 of B. germanica[15].
Analysis of miRNA libraries
Solexa technology (using an Illumina HiSeq 2000 sequencer) was used for small RNA sequencing in B. germanica. Sequences were received in FASTQ format, with the adaptor sequences trimmed. The first filter applied was based on quality values; considering that Illumina quality values range from 0 to 40 (using ASCII 64–104 in fastq) [16], we eliminated those reads having 80% of the sequence with values lower than 20. Then we applied a trimming process to delete regions with repetitive nucleotides or short motifs, and low quality regions, either at the beginning or at the end of the sequence. Sequences with the same length were grouped, then we obtained the counts collapsing the sequences using the suite of programs Fastx-toolkit (http://hannonlab.cshl.edu/fastx_toolkit), and those sequences with 10–28 nucleotides were kept. To eliminate artefacts, we mapped the sequences against Pfam database (excluding miRNAs), and against the genome sequences of Blattabacterium cuenoti, a bacteroid endosimbiont of B. germanica[17], B. germanica densovirus [18] and Escherichia coli. The remaining sequences were considered putative miRNAs. We identified the canonical miRNAs by Bowtie alignment [19] against mature miRNAs of the mirBASE database (November 2011 version) [20]. We followed the -3p or -5p nomenclature, instead of mature and star strand, assuming that the miRNA position in the precursor of B. germanica was conserved with respect to D. melanogaster. The seed was defined as the first 17 nucleotides and not mismatches were allowed [21]. Sequences shorter than 17 nucleotides were labelled as a given miRNA when they had 100% matching with the corresponding region of the miRNA seed. For expression analysis, different miRNAs length variants, or isoMIRs [22] of the same miRNA were consider together. Finally, the poorest represented miRNAs, i.e., the pool of those representing only 0.01% of the total reads, were eliminated from the differential expression analysis.
Comparison of miRNA libraries
In order to compare the N5 and N6 libraries, the counts were normalized by transforming them into RPKM (Reads Per Kilobase Mapped) [23] and were analysed by the NOISeq R package [24]. The counts of the different miRNA length variants were merged. The method of NOISeq applied was NOISeqsim, which assumes that there are not replicates for the experimental conditions and it simulates them. We used the default values to define those miRNAs differentially expressed (probability > 0.8). The evaluation of the results was based on the NOISeq scores: probability of being differentially expressed, M-value and D-value. The results were represented by a Volcano plot, as described by Cui and Churchill [25].
Expression patterns
miRNA expression patterns were established at 48 h intervals in female specimens of N5 and N6 and in freshly emerged adults. Three biological replicates of each age were studied. Whole body sampling and total RNA extraction was carried out as in the library construction. Quantification of miRNA levels was carried out by quantitative real-time PCR (qRT-PCR). An amount of 400 ng of total RNA was reverse transcribed with the NCode miRNA first-strand synthesis and qRT-PCR kit (Invitrogen), following the manufacturer protocol. Amplification reactions were carried out using IQTM SYBR Green Supermix (BioRad) and the following protocol: 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 30 s, in a MyIQ Real-Time PCR Detection System (BioRad). A dissociation curve was obtained to ensure that there was only one product amplified after the amplification phase. All reactions were run in triplicate. The endogenous reference gene used was U6, as in [26]; therefore, results are given as copies of RNA per 1000 copies of U6. Primer sequences for each miRNA are available on request.
Hormone treatments
In a first set of experiments, specimens were treated as freshly emerged sixth instar nymph (N6), by injecting 1 μL of 20E (Sigma) at a concentration of 1 μg/μL dissolved in water with 10% ethanol; controls where treated equivalently with 1 μL of water with 10% ethanol. Injection was performed through the membrane between fourth and fifth ventrites. In another experimental set, specimens were treated at the same stage, by applying topically 1 μL of JH III (Sigma) dissolved in acetone at a concentration of 2 μg/μL, and with 20E in the same conditions as in the previous set of experiments; controls were treated equivalently with 1 μL of acetone and 1 μL of water with 10% ethanol. JH III is the native JH of B. germanica[27], and given that the commercial JH III is a mixture of isomers containing ca. 50% of the biologically active (10R)-JH III, thus the active dose applied was around 1 μg per specimen. Total RNA extraction and quantification of dicer-1 expression and miRNAs were performed 24 h after the treatment following the procedures described above, and using three biological replicates. Statistical analysis of relative expression measurements was carried out with the REST software tool [28].
miRNA depletion
To deplete miR-252-3p expression we used miRCURY LNA™ microRNA Power Inhibitor (Exiqon). The sequence of LNA anti-miR-252-3p (GATAAGCACTTGAGCAGCAGG) is quite similar to the sequence of miR-252-5p (CTAAGTACTAGTGCCGCAGGA), which suggests that it might act as miR-252-5p mimic. However, the seed of LNA-antimiR-252-3p (ATAAGC) is very different to that of miR-252-5p (TAAGTA), which makes very improbable this possibility. A dose of 1 μL of LNA at 50 μM was injected in the abdominal cavity 24 h after the moult to the fifth instar nymph (N5), and a second injection at the same concentration was carried out 48 h later. Preliminary experiments administering a single dose 24 h after the moult to N5 depleted miR-252-3p less efficiently. Controls were injected with miRCURY LNA™ microRNA Inhibitor Negative Control A (Exiqon) at the same stages and concentrations. Specimens for miRNA quantification were collected 24 h after the second injection. RNA extraction, quantification of miRNA levels and statistical analysis were performed as described above. Specimens to study the phenotype were left alive until the adult moult.
Results
Comparison of libraries and miRNA selection for further studies
The sequencing of the N5 and N6 miRNA libraries of B. germanica resulted in 38,399,972 and 23,689,204 reads respectively, with sequence lengths between 10 and 40 nucleotides. From these, the application of the filters described in the Material and Methods section led to 2,260,210 and 2,174,640 reads in the N5 and N6 libraries respectively, corresponding to 61 canonical miRNAs. The conversion of the reads of each miRNA into RPKM in the N5 and N6 libraries gave an initial insight into which miRNAs were more and less expressed in each library (Table 1). Calculation of the probability of being differentially expressed, M-value and D-value led to the identification of those miRNAs that are differentially expressed in N5 and in N6. These calculations indicate that three miRNAs are more highly expressed in N5 (those with a positive M-value; Table 2), and 37 are more highly expressed in N6 (those with a negative M-value; Table 3). Results are summarized in a Volcano plot (Figure 1).
Table 1.
Counts and RPKM of the canonical miRNAs found in the N5 and N6 libraries
| miRNA |
N5 |
N6 |
||
|---|---|---|---|---|
| Counts | RPKM | Counts | RPKM | |
| bantam-3p |
4355 |
5939.042 |
10724 |
24195.476 |
| let-7-5p |
6238 |
8608.982 |
10157 |
22754.573 |
| miR-iab-4-5p |
618 |
856.282 |
559 |
1253.199 |
| miR-1-3p |
1162418 |
1676872.803 |
1101990 |
2579427.475 |
| miR-2-3p |
16839 |
30473.966 |
20102 |
44802.071 |
| miR-7-5p |
1115 |
1603.854 |
764 |
1782.859 |
| miR-8-3p |
129951 |
173738.073 |
185336 |
401628.852 |
| miR-8-5p |
36142 |
50136.290 |
37104 |
83764.303 |
| miR-9a-3p |
1316 |
1810.390 |
990 |
2209.068 |
| miR-9a-5p |
7345 |
10519.521 |
6705 |
15734.350 |
| miR-9b-3p |
444 |
610.332 |
504 |
1151.966 |
| miR-9c-5p |
2436 |
3731.535 |
3748 |
9306.244 |
| miR-10-3p |
2018 |
2857.459 |
1542 |
3543.522 |
| miR-10-5p |
10308 |
14188.293 |
9746 |
21761.685 |
| miR-12-5p |
607 |
811.513 |
714 |
1543.625 |
| miR-13a-3p |
923 |
1268.955 |
946 |
2090.234 |
| miR-13b-3p |
2436 |
3124.586 |
2431 |
5064.433 |
| miR-14-3p |
89736 |
124156.170 |
52803 |
118969.680 |
| miR-31-5p |
5091 |
7007.565 |
5468 |
12172.024 |
| miR-34-5p |
3124 |
4465.801 |
2343 |
5457.763 |
| miR-71-3p |
5006 |
6984.393 |
3798 |
8620.928 |
| miR-71-5p |
31574 |
43472.049 |
28524 |
63933.294 |
| miR-79-3p |
186 |
277.628 |
504 |
1151.966 |
| miR-87-3p |
4999 |
7205.065 |
7177 |
16792.743 |
| miR-92a-3p |
529 |
722.703 |
450 |
1001.511 |
| miR-92b-3p |
1915 |
2632.691 |
1375 |
3066.353 |
| miR-100-5p |
5943 |
8211.876 |
11666 |
26310.508 |
| miR-124-3p |
406 |
608.142 |
325 |
792.840 |
| miR-125-5p |
5131 |
7347.148 |
9416 |
21901.480 |
| miR-133-3p |
1951 |
2682.100 |
1404 |
3130.094 |
| miR-184-3p |
106965 |
167335.182 |
85068 |
200902.694 |
| miR-190-5p |
2223 |
2898.137 |
440 |
935.079 |
| miR-193-3p |
340 |
472.188 |
319 |
724.041 |
| miR-252-3p |
1587 |
2175.501 |
25 |
56.778 |
| miR-252-5p |
20378 |
29676.940 |
19167 |
45269.029 |
| miR-263a-5p |
30064 |
38608.305 |
34005 |
70511.600 |
| miR-263b-5p |
1470 |
1918.685 |
1669 |
3538.234 |
| miR-275-3p |
6538 |
19349.775 |
8708 |
19144.342 |
| miR-276-3p |
133114 |
192096.386 |
176820 |
394590.630 |
| miR-276-5p |
662 |
902.984 |
28 |
54.987 |
| miR-277-3p |
3881 |
5345.294 |
4880 |
10875.555 |
| miR-278-3p |
588 |
851.690 |
636 |
1492.383 |
| miR-279-3p |
33581 |
47196.919 |
43314 |
99043.811 |
| miR-281-3p |
346 |
479.946 |
442 |
1001.721 |
| miR-281-5p |
2412 |
3298.680 |
2637 |
5834.284 |
| miR-283-5p |
1684 |
2362.825 |
1891 |
4289.933 |
| miR-305-5p |
4620 |
6304.241 |
3952 |
8866.733 |
| miR-306-5p |
45159 |
61925.032 |
32982 |
73346.550 |
| miR-307-3p |
1829 |
2557.487 |
2247 |
5124.163 |
| miR-315-5p |
345 |
465.641 |
254 |
556.891 |
| miR-316-5p |
2143 |
2971.686 |
1923 |
4322.012 |
| miR-317-3p |
311250 |
429211.869 |
226595 |
509795.976 |
| miR-375-3p |
2744 |
3957.724 |
2693 |
6304.047 |
| miR-927-5p |
1167 |
1603.931 |
721 |
1608.971 |
| miR-965-3p |
420 |
554 |
381 |
816.976 |
| miR-993-3p |
371 |
531.079 |
170 |
398.526 |
| miR-998-3p |
242 |
328.263 |
221 |
484.999 |
| miR-2765-5p |
401 |
540.370 |
553 |
1210.667 |
| miR-2788-3p |
300 |
427.236 |
271 |
632.329 |
| miR-2796-3p |
903 |
1216.746 |
1172 |
2550.380 |
| miR-3770-5p | 1383 | 2086.531 | 1141 | 2800.141 |
Table 2.
miRNAs differentially expressed in N5 sorted by the absolute M-value
| miRNA | P | M-value | D-value |
|---|---|---|---|
| miR-252-3p |
0.917 |
5.260 |
2118.72 |
| miR-276-5p |
0.852 |
4.037 |
847.99 |
| miR-190-5p | 0.912 | 1.632 | 1963.06 |
The higher the M-value the more differentially expressed is the miRNA. P: Probability of being differentially expressed.
Table 3.
miRNAs differentially expressed in N6, sorted by the M-value
| miRNA | P | M-value | D-value |
|---|---|---|---|
| bantam-3p |
0.988 |
−2.026 |
18256.44 |
| miR-100-5p |
0.988 |
−1.680 |
18098.63 |
| miR-125-5p |
0.987 |
−1.576 |
14554.33 |
| let-7-5p |
0.986 |
−1.402 |
14145.59 |
| miR-9c-5p |
0.959 |
−1.318 |
5574.70 |
| miR-87-3p |
0.973 |
−1.221 |
9587.67 |
| miR-8-3p |
0.999 |
−1.209 |
227890.78 |
| miR-2765-5p |
0.830 |
−1.164 |
670.30 |
| miR-279-3p |
0.994 |
−1.069 |
51846.89 |
| miR-2796-3p |
0.884 |
−1.068 |
1333.63 |
| miR-276-3p |
0.998 |
−1.039 |
202494.24 |
| miR-277-3p |
0.956 |
−1.025 |
5530.26 |
| miR-307-3p |
0.923 |
−1.003 |
2566.67 |
| miR-12-5p |
0.837 |
−0.928 |
732.12 |
| miR-263b-5p |
0.895 |
−0.883 |
1619.55 |
| miR-263a-5p |
0.986 |
−0.869 |
31903.3 |
| miR-283-5p |
0.907 |
−0.860 |
1927.11 |
| miR-281-5p |
0.918 |
−0.823 |
2535.60 |
| miR-278-3p |
0.821 |
−0.809 |
640.69 |
| miR-31-5p |
0.951 |
−0.797 |
5164.45 |
| miR-8-5p |
0.984 |
−0.740 |
33628.01 |
| miR-13a-3p |
0.840 |
−0.720 |
821.27 |
| miR-13b-3p |
0.904 |
−0.697 |
1939.84 |
| miR-375-3p |
0.914 |
−0.672 |
2346.33 |
| miR-1-3p |
0.990 |
−0.622 |
902554.68 |
| miR-10-5p |
0.960 |
−0.617 |
7573.40 |
| miR-252-5p |
0.977 |
−0.609 |
15592.09 |
| miR-9a-5p |
0.947 |
−0.581 |
5214.83 |
| miR-71-5p |
0.977 |
−0.556 |
20461.24 |
| miR-2-3p |
0.974 |
−0.556 |
14328.10 |
| miR-316-5p |
0.878 |
−0.540 |
1350.32 |
| miR-305-5p |
0.910 |
−0.492 |
2562.49 |
| miR-3770-5p |
0.813 |
−0.424 |
713.61 |
| miR-71-3p |
0.8201 |
−0.304 |
1636.54 |
| miR-184-3p |
0.870 |
−0.264 |
33567.51 |
| miR-317-3p |
0.865 |
−0.248 |
80584.11 |
| miR-306-5p | 0.815 | −0.244 | 11421.52 |
The lower M-value, the more differentially expressed is the miRNA. P: Probability of being differentially expressed.
Figure 1.
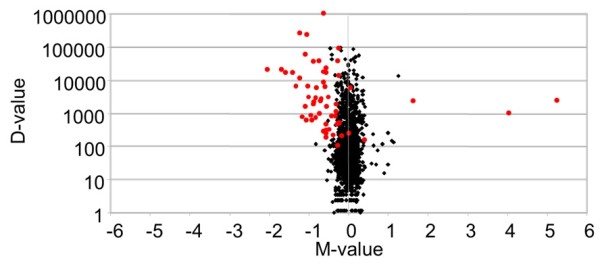
Volcano plot showing differentially expressed miRNAs in N5 and N6 libraries of Blattella germanica. M-D values in noise are represented by black dots, and differentially expressed miRNAs by red dots. M-value: log2(x1/x2). D-value: |x1-x2|. Where xi are the expression level in RPKMs in library i. The more external points represent the more differentially expressed values, and correspond to the miRNA analyzed.
Notably, both strands of the miRNA precursor miR-252 are well present in the two libraries, suggesting that both of them are biologically active miRNAs. The 5’ strand (miR-252-5p) is about 12-fold more abundant than the 3’ strand (miR-252-3p) in the N5 library and ca. 800-fold times more abundant in N6. Furthermore, from N5 to N6 there is an increase of miR-252-5p and a significant decrease of miR-252-3p (Table 1). miR-276 represents a similar case, with both strands represented in both libraries. In this case, however, the 3’ strand (miR-276-3p) is the predominant one in both libraries, being clearly more highly expressed in N6 with respect to N5, whereas the 5’ strand (miR-276-5p) is more highly expressed in the N5 with respect to N6.
For expression pattern studies and hormonal treatments, we selected the three miRNAs more highly expressed (M-value between 1.6 and 5.3) in the N5 library (miR-252-3p, miR-276-5p and miR-190-5p) and the four more differentially expressed (M-value between −1.4 and −2.0) in the N6 library (bantam-3p, miR-100-5p, miR-125-5p and let-7-5p). We also added three miRNAs that are similarly represented in N5 and N6: miR-252-5p and miR-276-3p, whose respective partner strands were chosen due to their higher expression in the N5 library, and miR-1-3p, which was the most abundant miRNA (accounting for more than 50% of total reads). Finally, we included two additional miRNAs (miR-14-3p and miR-34-5p) because of their potential interest in the context of moulting and metamorphosis. miR-14-3p has been described targeting the ecdysone receptor (EcR) in D. melanogaster[29], and miR-34-5p is inducible by the JH analogue methoprene in Drosophila S2 cells [30].
miRNA expression patterns
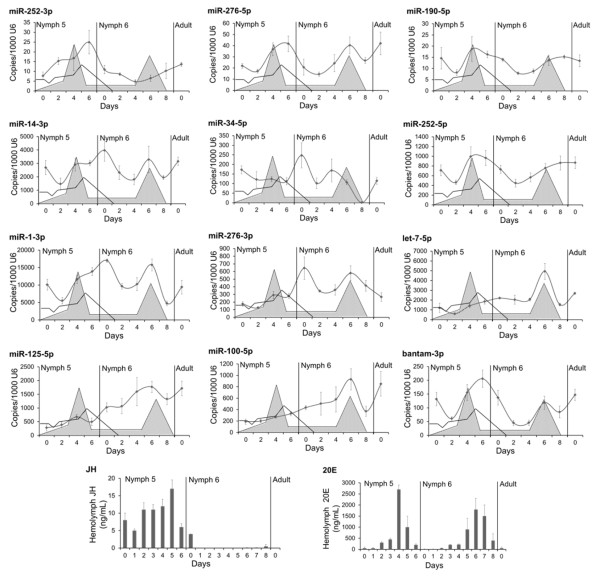
The miRNA expression patterns were determined in N5, and in N6, as well as in freshly emerged adults. One of the most peculiar patterns was that of miR-252-5p, which showed a burst of expression towards the end of N5, approximately one or two days after the peak of 20E, and then the expression steadily decreased during most of the N6, including the days of the peak of 20E of this stage (Figure 2). This pattern is consistent with counts data obtained from the bioinformatical analysis of N5 and N6. Also interesting are the patterns of let-7, miR-100 and miR-125, which show low expression in N5, which then increased in N6 reaching a burst around the days of the 20E peak (Figure 2); these patterns are also consistent with the sequencing data.
Figure 2.
Expression pattern of the 12 miRNA selected from the comparison of the N5 and N6 libraries in Blattella germanica. miRNA levels were measured on nymph 5, nymph 6 and in freshly emerged adults with qRT-PCR; data represent the mean ± SEM, and are indicated as copies of the respective miRNA per 1000 copies of U6; each point represents 3 biological replicates. Schematic patterns of juvenile hormone III (JH) and 20-hydroxyecdysone (20E) titers in nymph 5 and nymph 6 are superimposed in every graphic as empty and grey patterns, respectively. Actual values of JH and 20E titers are represented in the two bottom panels, according to the data of Treiblmayr et al. [41] (JH) and Romaña et al. [15] (20E).
The patterns of the remaining miRNAs examined show two bursts of expression. In the cases of miR-276-5p, miR-190-5p, miR-252-5p and bantam-3p, there were expression bursts seen both in N5 and in N6, approximately corresponding to the peaks of 20E (Figure 2). These data do not correlate with quantitative sequencing data, which predicted that miR-276-5p and miR-190-5p should have shown much higher expression levels around the 20E peak of N5, whereas those of miR-252-5p and bantam-3p should have been much lower at this stage. Two expression bursts of miR-1-3p, miR-34-5p and mir-276-3p occurred in N6, the first just after emergence and the other coinciding or close to the peak of 20E of this stage (Figure 2). Comparing the levels of expression around the peaks of 20E in N5 and N6, these approximately patterns correspond to what would be expected based on the sequencing data. Minor discrepancies, like in the case of miR-276-5p, miR-276-3p, miR-190-5p, can be due to individual variability, given that sequencing data come from a pool of nine specimens, whereas data of expression patterns is based on three particular specimens.
Effect of hormonal treatments
To mimic the endocrine situation of a nymph-adult transition at the peak of 20E (like in N6), freshly emerged last instar female nymphs were treated with 1 μg of 20E. The results (Figure 3A) show that, statistically, most miRNAs were unaffected by the treatment. However, the expression levels of miR-1-3p and miR-100-5p were significantly increased after treatment with 20E, whereas those of bantam-3p, miR-125-5p, miR-14-3p, miR-276-3p, miR-34-5p and let-7-5p showed a tendency to increase with respect to controls, although differences were not statistically significant. The expression of dicer-1 was not significantly affected by the treatment (Figure 3A).
Figure 3.
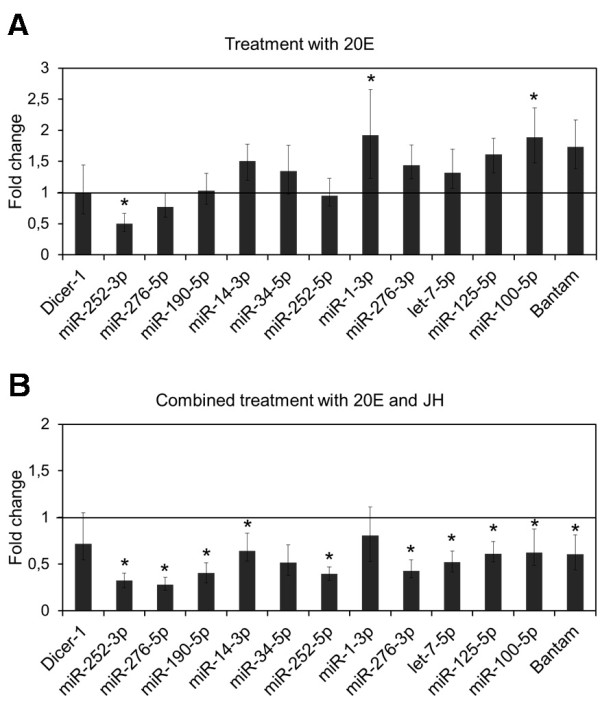
Effect of 20-hydroxyecdysone (20E) and 20E plus juvenile hormone III (JH) on miRNA expression in Blattella germanica. Specimens were treated with 1 μg of 20E (A) or with 1 μg of 20E plus 1 μg of JH (B) in freshly emerged last instar nymph and miRNAs were measured 24 h later. miRNA levels were measured by qRT-PCR, using U6 as a reference. Data represent 3 biological replicates (mean ± SEM) and are normalized against control females (reference value = 1); the asterisk indicates statistically significant differences with respect to controls (p < 0.05), according to the REST software tool [28].
On the other hand, the endocrine situation of a nymph-nymph transition at the peak of 20E (like in N5) was mimicked by treating equivalently staged specimens with 2 μg of JH and 1 μg of 20E. Results (Figure 3B) indicate that all the miRNAs studied, except miR-1-3p and miR-34-5p, had expression levels significantly lower than those of the controls, although the expression of the latter miRNA showed a tendency to be inhibited by the double hormonal treatment. The expression of dicer-1 was not significantly affected by the treatment (Figure 3B).
Functional analysis of miR-252-3p
Depletion of miR-252-3p was achieved with two doses of 1 μL of miR-252-3p LNA at 50 μM each injected in N5, one 24 h after the moult (N5D1) and another 48 h later (N5D3). On day 6 of the instar (N5D6), the levels of miR-252-3p had specifically decreased (Figure 4A), and had slightly recovered seven days later (N5D13), although they were still significantly lower than the levels of the controls (Figure 4B). Specimens treated with miR-252-3p LNA grew slower than controls, as observed on day 6 (N5D6; Figure 4C). On N5D6, when controls moulted to N6, the weight of treated specimens was significantly lower than that of controls (Figure 4D) and they did not moult until 4–7 days later (Figure 4E). Although they were observed to grow more slowly, treated specimens still moulted to the next (N6) nymphal instar with no apparent defects, and then developed normally in N6 and moulted to the adult stage within the developmental time schedule of controls and with no apparent morphological defects. According to these results, miR-252-3p seems not regulate metamorphosis, but may be rather related with growth and development in penultimate nymphal stage.
Figure 4.
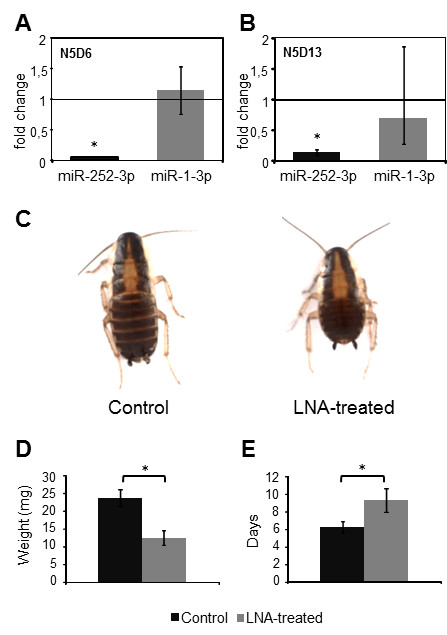
Effects of miR-252-3p depletion on nymphal development of Blattella germanica. Treated females received two injections of 50 μM of miR-252-3p LNA on day 1 and day 3 of fifth nymphal instar (N5D1 and N5D3, respectively); control females received an equivalent treatment with miRCURY LNA™ microRNA Inhibitor Negative Control A. A) Levels of miR-252-3p and miR-1-3p (used as negative control) on N5D6. B) Same data in N5D13. C) General aspect of a control specimen and a specimen treated with miR-252-3p LNA (LNA-treated) on N5D6. D) Weight of control and LNA-treated specimens in N5D6. E) Length (days) of N5 in control and LNA-treated specimens. qRT-PCR data in A and B represent 3 biological replicates and are normalized against the control females (reference value = 1); the asterisk indicates statistically significant differences with respect to controls (p < 0.05) according to the REST software tool [28]. Data in D and E represent 17 biological replicates; the asterisk indicates statistically significant differences with respect to controls (t-test, p < 0.01).
Discussion
High throughput sequencing of whole body miRNAs around the peak of 20E in the N5 and N6 libraries of B. germanica gave a total of 61 canonical miRNAs that were present in both libraries, although in different proportions in each. In a previous whole body miRNA library prepared during N6, but representing all eight days of the stage, we obtained a total of 49 canonical miRNAs [14]. Therefore, the number of miRNAs recovered in the present study, using animals collected around the peak of 20E, is higher. This suggests that most miRNAs might be more efficiently expressed around the peak of 20E, and that there was a dilution effect in the former library, which had all the days of the instar represented. Practically all canonical miRNAs reported by Cristino et al. [14] are present (except miR-1-5p and miR-184-5p), and are the most abundant in the present libraries. Some of the miRNAs (miR-8, miR-9a, miR-10, miR-71, miR-252, miR-276, miR-281) are represented by both strands, -3p and -5p, as was also observed in the previous N6 library [14].
The comparative analysis of N5 and N6 libraries allowed the identification of three and 37 miRNAs significantly more expressed in N5 and N6 respectively (Figure 1). Subsequently, 12 of these 40 miRNAs were investigated further by qRT-PCR to establish their expression patterns during N5 and N6. Results indicate that miR-252-3p is the only clear representative of miRNA that is well expressed in N5 and with decreased expression in N6, whereas let-7-5p, miR-100-5p and miR-125-5p are clear representatives of the reverse pattern (Figure 2). Whereas the expression pattern of miR-252-3p had yet to be studied in other insects, those patterns of let-7, miR-100 and miR-125 have been thoroughly examined. In D. melanogaster, these three miRNAs cluster in the same primary transcript, as first revealed by Bashirullah et al. [31] and Sempere et al. [30], and later confirmed in this fly and other insects [5,32]. Thus, the members of this miRNA cluster are expressed simultaneously, and studies in the holometabolan species, like D. melanogaster and B. mori have shown that expression concentrates in pre-metamorphic stages. In D. melanogaster, expression starts in late third (last) instar larvae, around the peak of 20E that triggers puparium formation, and continues until the imaginal moult [30,33]. In B. mori, let-7 expression starts in the moult leading to the penultimate larval instar, and continues until the emergence of the adult [34]. In the hemimetabolan B. germanica, the present study shows that expression of let-7, miR-100 and miR-125 concentrates in the last nymphal instar, having an expression burst around the peak of 20E that triggers metamorphosis (Figure 2).
Of note, among the miRNAs selected to study the expression pattern, we included miR-252 and miR-276, which were represented by both the -3p and -5p strands. In principle, the two respective strands originate from the same precursor and both should presumably be expressed at similar rates through parallel patterns. But this was not found to be the case. In the case of miR-252, the -3p strand is expressed at much lower levels than those of the -5p strand, whereas in the case of miR-276, the situation is the reverse; therefore, in each case the pattern of expression of one strand is clearly different to the expression of the other (Figure 2). These observations suggest that the expression is regulated differently in both strands, and that they play distinct miRNA functions [5]. In support of this notion, Liu et al. [35] reported that both strands of miR-276 are differentially expressed in different tissues and stages of B. mori, although miR-276-3p is always the most highly expressed strand.
Among the patterns examined, most of the expression peaks, either in N5 or N6, occur close to a peak of 20E (Figure 2), which suggests cause-effect relationships. This possible hormonal regulation of miRNA expression was therefore tested experimentally. In general, the experiments of hormonal treatments suggested that 20E alone tends to stimulate miRNA expression, whereas JH attenuates, or even inhibits the 20E stimulatory effect (Figure 3). Among those miRNAs that increased in expression after 20E treatment, we found let-7, miR-100 and miR-125. The increase in expression of miR-100-5p after 20E treatment is statistically significant, whereas let-7-5p and miR-125-5p had a tendency to increase (Figure 3A). This is consistent with previous studies in D. melanogaster that revealed that 20E increases the levels of these miRNAs [30,36] by inducing the expression of the polycistronic primary transcript that contains the let-7, miR-100 and miR-125 precursors [37]. In D. melanogaster, the burst of expression of let-7-5p in prepupae is important to ensure appropriate remodelling of the abdominal neuromusculature during metamorphosis [38]. Moreover, an independent work of Caygill and Johnston [39] showed that during D. melanogaster metamorphosis, the absence of let-7-5p and miR-125-5p results in temporal delays in the terminal cell-cycle exit in the wing, and in the maturation of neuromuscular junctions of imaginal abdominal muscles. The authors focused on the latter process by identifying the abrupt (ab) gene as a let-7-5p target, and by providing evidence that showed that let-7-5p regulates the maturation rate of abdominal neuromuscular junctions during metamorphosis by modulating ab expression [39].
Conversely, the significant stimulatory effect of 20E on miR-1-3p and the tendency of 20E to stimulate miR-34-5p that we observed in B. germanica contrasts with the results obtained in D. melanogaster, where 20E did not affect miR-1-3p and inhibited miR-34-5p [30]. We interpret these differences as reflections of different modes of regulation of these miRNAs in the two model species. The differences are also illustrated by the results obtained after JH treatment of B. germanica, which abolished the stimulatory effects of 20E on practically all miRNAs (Figure 3B), which was also the case for miR-34-5p. This contrasts again with the observations of Sempere et al. [30], which suggested that JH has a stimulatory effect on this miRNA in D. melanogaster.
Possibly the most interesting result of the experiments of hormonal treatments is that of miR-252-3p, which was the only miRNA that was inhibited by 20E alone (Figure 3A). Moreover, JH did not seem to counteract the 20E effect, as compared to other miRNAs (Figure 3B). Inhibition by 20E suggests that the decrease of miR-252-3p in N6, and the increase interrupted by a plateau around the 20E peak in N5 (Figure 2), might be due to the action of 20E. The other strand, classically considered the “mature” strand of miR-252, miR-252-5p, has been found in a number of insects [20], and its expression has been carefully studied in B. mori, where it shows a continuous high expression from the spinning larvae to pupal and adult stages [40]. Conversely, miR-252-3p has never been studied, nor has it been related with insect moulting and metamorphosis. It was for these reasons that we studied it functionally.
Depletion of miR-252-3p levels in the penultimate (N5) nymphal instar caused retarded growth and developmental delays within the instar (Figure 4). At the end, however, the treated insects were able to moult to the last (N6) nymphal instar and to the adult normally. Observed delays in N5 reached as long as 14 days in some cases, and levels of miR-252-3p measured in specimens on N5D13, which were presumably going to moult within the next one or two days, had somewhat recovered compared with levels measured on N5D6, although they were still significantly lower than levels of the miRNA of the controls (Figure 4). The data suggest that miR-252-3p regulates transcripts that are important for growth and development in N5, in the transition to the developmentally important N6 which precedes the imaginal moult. The fact that the treated specimens were finally able to moult to N6 and to the adult stage even with reduced levels of miR-252-3p, suggests that these levels, although still quite low, are above the operative threshold for this miRNA.
Conclusions
1. The comparative analysis of a miRNA library of the penultimate nymphal instar (N5) with an equivalent library from the last instar nymph (N6) of B. germanica revealed 40 canonical miRNAs that are differentially expressed in each library.
2. In terms of counts, three of these miRNAs were found to be significantly more expressed in N5, whereas 37 were found to be more expressed in N6. qRT-PCR studies confirmed that miR-252-3p is well expressed in N5 but not in N6, whereas let-7-5p, miR-100-5p and miR-125-5p showed the reverse pattern.
3. Hormonal treatments indicated that 20E increases the expression of let-7, miR-100 and miR-125, but inhibits that of miR-252-3p.
4. The involvement of let-7, miR-100 and miR-125 in metamorphosis has been shown in other insects. Depletion of miR-252-3p in B. germanica caused growth and developmental delays, which suggests that this miRNA is involved in regulating these processes prior to metamorphosis.
Competing interests
The authors wish to declare that they have no competing interests.
Author’s contributions
MR studied the miRNA expression patterns, the effect of hormonal treatments and the function of miR-252-3p, and drafted the manuscript. AdH carried out the comparison of the miRNA libraries. XB conceived and coordinated the study and wrote the final manuscript. All authors read and approved the final manuscript.
Contributor Information
Mercedes Rubio, Email: mercedes.rubio@ibe.upf-csic.es.
Anibal de Horna, Email: anibal.dehorna@ibe.upf-csic.es.
Xavier Belles, Email: xavier.belles@ibe.upf-csic.es.
Acknowledgements
Support for this research was provided by the Spanish MICINN (grant CGL2008-03517/BOS to X.B.) and by the CSIC (grant 2010TW0019, from the Formosa program, to X.B. and a predoctoral fellowship to M.R. from the JAE program) and from the Spanish National Institute for Bioinformatics (http://www.inab.org).
References
- Grimaldi D, Engel MS. Evolution of the insects. Cambridge University Press, Cambridge; 2005. [Google Scholar]
- Sehnal F, Svacha PJZ. In: Metamorphosis. Gilbert LI, Tata JR, Atkinson BG, editor. Academic Press, San Diego; 1996. Evolution of insect metamorphosis; pp. 3–58. [Google Scholar]
- Belles X. Encyclopedy of Life Sciences (ELS) John Wiley and Sons, Ltd, Chichester; 2010. Origin and Evolution of Insect Metamorphosis. [Google Scholar]
- Truman JW, Riddiford LM. Endocrine insights into the evolution of metamorphosis in insects. Annu Rev Entomol. 2002;47:467–500. doi: 10.1146/annurev.ento.47.091201.145230. [DOI] [PubMed] [Google Scholar]
- Belles X, Cristino AS, Tanaka ED, Rubio M, Piulachs MD. In: Insect Molecular Biology and Biochemistry. Gilbert LI, editor. Elsevier-Academic Press, Amsterdam; 2011. Insect MicroRNAs: From Molecular Mechanisms to Biological Roles; pp. 30–56. [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17(2):312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Orte E, Belles X. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc Natl Acad Sci USA. 2009;106(51):21678–21682. doi: 10.1073/pnas.0907391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X. Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae) PLoS One. 2011;6(4):e19350. doi: 10.1371/journal.pone.0019350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaña I, Pascual N, Belles X. The ovary is a source of circulating ecdysteroids in Blattella germanica (Dictyoptera, Blattellidae) Eur J of entomol. 1995;92(1):93–103. [Google Scholar]
- Horner DS, Pavesi G, Castrignano T, De Meo PD, Liuni S, Sammeth M, Picardi E, Pesole G. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief Bioinform. 2010;11(2):181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Neef A, Pereto J, Patino-Navarrete R, Pignatelli M, Latorre A, Moya A. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 2009;5(11):e1000721. doi: 10.1371/journal.pgen.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukha DV, Chumachenko AG, Dykstra MJ, Kurtti TJ, Schal C. Characterization of a new densovirus infecting the German cockroach, Blattella germanica. J Gen Virol. 2006;87(Pt 6):1567–1575. doi: 10.1099/vir.0.81638-0. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011;39(Web Server issue):W132–138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3' end processing of microRNAs in Drosophila. Curr Biol. 2011;21(22):1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21(12):2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Churchill GA. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4(4):210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Frentiu FD, Moreira LA, O'Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci USA. 2011;108(22):9250–9255. doi: 10.1073/pnas.1105469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps F, Casas J, Sánchez FJ, Messeguer A. Identification of juvenile hormone III in the hemolymph of Blattella germanica adult females by gas chromatography–mass spectrometry. Arch Insect Biochem Physiol. 1987;6:181–189. doi: 10.1002/arch.940060306. [DOI] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21(18):2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259(1):9–18. doi: 10.1016/S0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259(1):1–8. doi: 10.1016/S0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Liu S, Xia Q, Zhao P, Cheng T, Hong K, Xiang Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori) BMC Dev Biol. 2007;7:88. doi: 10.1186/1471-213X-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Gao S, Zhang D, Yin J, Xiang Z, Xia Q. MicroRNAs show diverse and dynamic expression patterns in multiple tissues of Bombyx mori. BMC Genomics. 2010;11:85. doi: 10.1186/1471-2164-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244(1):170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139(10):1788–1797. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22(12):1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Current biology : CB. 2008;18(13):943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang L, Li Q, Zhao P, Duan J, Cheng D, Xiang Z, Xia Q. MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori) BMC Genomics. 2009;10:455. doi: 10.1186/1471-2164-10-455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Treiblmayr K, Pascual N, Piulachs MD, Keller T, Belles X. Juvenile hormone titer versus juvenile hormone synthesis in female nymphs and adults of the German cockroach, Blattella germanica. J Insect Sci. 2006;6:1–7. doi: 10.1673/031.006.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]