Abstract
Experimental evidence suggests that microfilaments and microtubules play contrasting roles in regulating the balance between motility and stability in neuronal structures. Actin-containing microfilaments are associated with structural plasticity, both during development when their dynamic activity drives the exploratory activity of growth cones and after circuit formation when the actin-rich dendritic spines of excitatory synapses retain a capacity for rapid changes in morphology. By contrast, microtubules predominate in axonal and dendritic processes, which appear to be morphologically relatively more stable. To compare the cytoplasmic distributions and dynamics of microfilaments and microtubules we made time-lapse recordings of actin or the microtubule-associated protein 2 tagged with green fluorescent protein in neurons growing in dispersed culture or in tissue slices from transgenic mice. The results complement existing evidence indicating that the high concentrations of actin present in dendritic spines is a specialization for morphological plasticity. By contrast, microtubule-associated protein 2 is limited to the shafts of dendrites where time-lapse recordings show little evidence for dynamic activity. A parallel exists between the partitioning of microfilaments and microtubules in motile and stable domains of growing processes during development and between dendrite shafts and spines at excitatory synapses in established neuronal circuits. These data thus suggest a mechanism, conserved through development and adulthood, in which the differential dynamics of actin and microtubules determine the plasticity of neuronal structures.
Neuronal circuits need to maintain a delicate balance between stability and plasticity. On the one hand, the synaptic connections they make must be stable enough to support reliable signal transmission, while on the other, they must be sufficiently plastic to accommodate changes in connectivity that are necessary for the long-duration adaptation of behavior to sensory experience. How is neuronal structure organized and regulated to accommodate these diverse needs? Increasingly, experimental evidence implicates the neuronal cytoskeleton in regulating morphological plasticity in adult as well as developing tissue. More than any other cell type, neurons depend for their distinctive morphology on the cytoskeleton whose protein components are organized in a set of microdifferentiated compartments that mirror the polarized form of the cell and play a significant role in determining its development (1–3). Microfilaments and microtubules act together to guide and support the growth and differentiation of axons and dendrites. Whereas dynamic actin filaments drive the exploratory activity of growth cones as they respond to external guidance cues, microtubules stabilize the structure of the newly established process (4–10).
Recent results suggest that a similar “division of labor” between the two cytoskeletal filament systems may persist in dendrites beyond the developmental period. In adult brain the highest concentrations of actin are associated with dendritic spines that form the postsynaptic component of most excitatory synapses (11–13). This dendritic spine actin retains a capacity for dynamic activity and can drive rapid changes in their shape (14–18). By contrast, the highest concentrations of the microtubule components, including tubulin and the microtubule-associated proteins (MAPs), occur in the shafts of dendrites (19–23). This is consistent with ultrastructural studies showing microtubule bundles as the predominant cytoskeletal components of dendrite shafts whereas the cytoplasm of spines is characterized by a meshwork of fine filaments consistent with the predominance of actin-containing microfilaments (24–26).
Despite these indications for separation between the two filament systems, the nature of the interface between the microtubule and microfilament domains has remained uncertain because of immunohistochemical data suggesting that MAP2, the major dendritic MAP, is present at postsynaptic sites and in dendritic spines (19, 27, 28). MAP2 can bind to actin in vitro (29, 30) so if it is present in spines this might suggest that it can act as a bridge between actin and microtubules at spine synapses. Studies using transfected fibroblastic cells have yielded diverse results regarding potential interactions between MAP2 and the actin cytoskeleton (31–35). Because cytoskeletal components are likely to be important in determining the locus of anatomical plasticity in dendrites (16, 18, 36–39) we have re-examined the distribution of actin and MAP2 in primary neurons by using fluorescent protein tags that allow the both the location and the dynamics of cytoskeletal proteins to be determined in living cell (40). The results show a striking compartmentalization of the cytoskeleton in dendrites with microtubule proteins limited to the dendritic shaft whereas actin is overwhelmingly concentrated in spines. This distribution is accompanied by a differentiation of dendrite structure into highly motile postsynaptic elements, the spines, and morphologically more stable elements, the dendrite shafts.
Methods
Eukaryotic expression constructs containing actin and MAP2c and MAP2b tagged with green fluorescent protein (GFP) under control of chicken β-actin sequences and techniques for preparing time-lapse recordings from transfected hippocampal neurons were as described (14, 33). The topaz spectral variant of GFP (41), here referred to as YFP (yellow fluorescent protein), was obtained from Packard Bioscience and was used to replace GFP in existing vectors by standard techniques. Transgenic mice expressing actin-GFP have been described (42). Transgenic mice expressing MAP2-GFP were generated by cloning a fragment containing the MAP2-GFP coding sequence (33) into the pTSC vector containing a modified Thy-1 promoter (43). A 9.5-kb EcoRv/PvuI fragment was injected into oocytes of B6CF1 strain mice, and transgenic lines were established by standard techniques. Positive progeny were identified by PCR using GFP-specific primers and by Southern blot analysis.
Organotypic slice cultures from transgenic mice were established as described by Gahwiler et al. (44). After at least 4 weeks in culture individual slices were mounted in purpose-built observation chambers (Life Imaging Services, Olten, Switzerland) and perfused with artificial cerebrospinal fluid or Tyrode's solution. No difference was apparent between buffers. Time-lapse recordings were made by using a Leica IRBE inverted microscope equipped with a Nipkow disk-microlens confocal system (Life Science Resources, Cambridge, U.K.). To display time-dependent changes in printed figures, a subtraction protocol was used to sum differences between images in time-lapse recordings by using metamorph software (Universal Imaging, West Chester, PA). The results were displayed by using a pseudocolor look-up table with dark blue indicating lack of change and red to yellow increasing amounts of motility (42).
Results
Comparison of Actin and MAP2 Distributions Using Spectral Variants of GFP.
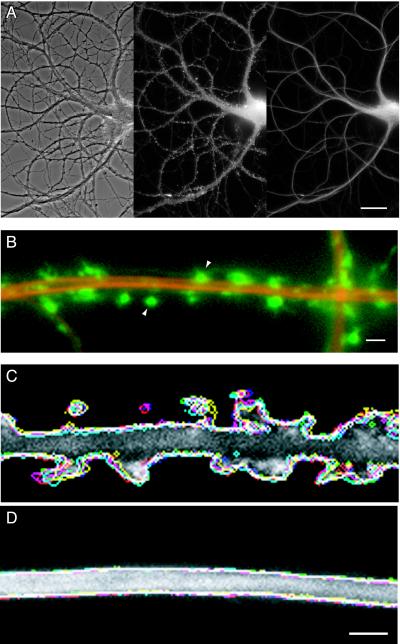
To asses the distribution and dynamics of microfilaments and microtubules in dendrites, we prepared eukaryotic expression vectors containing actin labeled with GFP and MAP2c labeled with YFP. To compare their properties within the same dendrite, hippocampal neurons from 18-day rat embryos were simultaneously transfected with actin-GFP and MAP2c-YFP and maintained in dispersed cell culture for at least 3 weeks. By this time most excitatory synapses are made onto dendritic spines of mature appearance that are contacted by presynaptic terminals whereas earlier immature lateral filopodia are abundant (45–49). Fig. 1A shows a living cell in such a culture visualized by phase-contrast microscopy (Left) and with filter sets selective for GFP (Center) or YFP (Right). Even at the low magnification shown in Fig. 1A, punctate labeling along dendrites, indicative of actin-GFP accumulation in dendritic spines, was evident (Center). By contrast the same dendrites visualized by MAP2-YFP were smooth in appearance (Right), indicating the absence of MAP2 from dendritic spines.
Figure 1.
Actin and MAP2 differ in both distribution and dynamics in living hippocampal neurons. (A) Distribution of actin and MAP2 in a transfected hippocampal neuron in cell culture for 24 days, simultaneously expressing actin-GFP and MAP2c-YFP. The phase-contrast image (Left) shows the arrangement of the cell body and processes of the transfected cell interspersed with the network of axonal processes of untransfected cells. The original gray-scale images for actin-GFP (Center) and MAP2c-YFP (Right) images were prepared by using appropriate selective filter sets. (Bar = 20 μm.) (B) Comparative distribution of actin and MAP2 in a dendrite segment produced by overlaying pseudocolored images for actin-GFP (green) and MAP2c-YFP (red). The high concentration of actin in dendritic spines (arrowheads) contrasts with the confinement of MAP2 to dendrite shafts. (Bar = 2 μm.) (C and D) Time-dependent changes in the configuration of actin and MAP2 in dendrites. Six frames from a single time-lapse recording for actin-GFP (C) and MAP2-YFP (D) images, recorded alternately 30 s apart, were converted into profile outlines. Each outline was assigned a different color and overlaid onto a single gray-scale image from the same recorded sequence. Variations between the different color outlines indicate regions of morphological change that are evident in the actin images of dendritic spines (C) but are absent from the MAP2 images of the dendrite shaft (D). (Bar = 2 μm.) Refer to supplemental Movie 1 for the original time-lapse sequence.
To study these distributions in more detail, actin-GFP and MAP2c-YFP images were taken at higher magnification and compared by assigning them contrasting colors (actin, green; MAP2c, red) and overlaying the images. Fig. 1B shows the results of this procedure for a segment of dendrite from a doubly transfected cell in which the strong targeting of actin into spines and the contrasting restriction of MAP2 to the dendrite shaft is evident. The data shown in Fig. 1B were taken from a time-lapse recording in which successive images were captured alternately by using the GFP or YFP filter sets. Such recordings show the same rapid dynamics of actin in dendritic spines described in previous studies (14, 42). By contrast, MAP2 showed no detectable dynamic activity over the 15 min of recording (see Movie 1, which is available as supplemental data on the PNAS web site, www.pnas.org). To represent this result in still images, six frames of actin-GFP and six frames of MAP2c-YFP, recorded alternately 30 s apart, were converted into profile outlines by using a computer routine. Each was assigned a different color and all six then were overlaid onto a single gray-scale fluorescence image from the same time-lapse series. Changes in the shape of dendritic spines then are revealed by the separately colored outlines representing the successively recorded images (Fig. 1C). By contrast, the same procedure applied to images of MAP2c shows no detectable change during the period of recording (Fig. 1D).
This tight localization of MAP2 to dendritic microtubules was not only seen for the juvenile MAP2c splice variant but also for the high molecular weight MAP2b form that is expressed in the adult brain (50). Fig. 2 shows results for hippocampal neurons transfected with MAP2b-GFP. Like the embryonic MAP2c form, adult MAP2b is localized in dendrites but not within axons (arrow in Fig. 2 A and B). Both here and in higher magnification images MAP2b was bound to microtubules in dendrite shafts and did not enter dendritic spines (Fig. 2 C and D).
Figure 2.
MAP2 is absent from dendritic spines. (A and B) Like the embryonic low-molecular weight variant MAP2c, high-molecular weight MAP2b is confined to the somatodendritic domain of hippocampal neurons and is absent from the axon (arrow). Shown are both a phase (A) and a fluorescence (B) image of a live neuron transfected with GFP-tagged MAP2b and kept in culture for 14 days. (Bar = 25 μm.) (C and D) This neuron transfected with MAP2b-GFP was maintained in culture for 4 weeks, by which time cells carry many dendritic spines. In the enlarged image (D) of the area outlined in C, the restriction of MAP2b-GFP fluorescence to microtubule bundles in the dendritic shaft is obvious. No fluorescence is detected in spine protuberances from the dendrite. (Bars: C = 15 μm; D = 2 μm.)
Time-Lapse Recording of MAP2-GFP in Tissue Slices from Transgenic Mice.
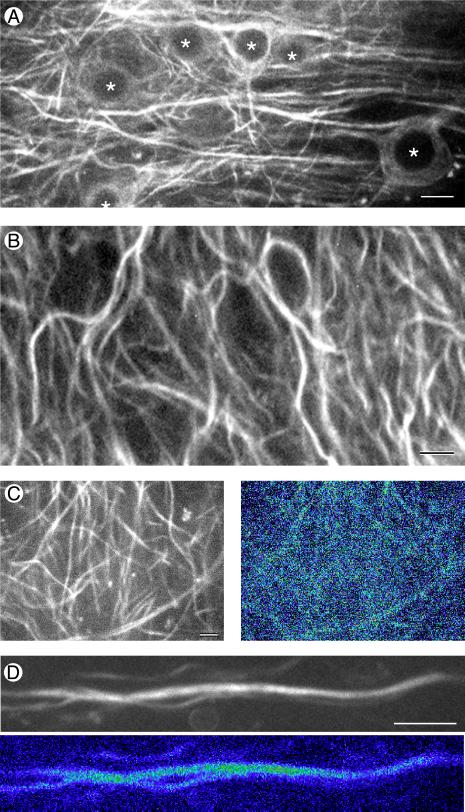
Time-lapse recordings of GFP-labeled MAP2c in dendrites of dispersed cells consistently failed to show dynamic activity of the microtubule cytoskeleton over periods of up to 30 min. However, it remained possible that changes might occur over longer periods, particularly because transfection experiments using fibroblastic cells indicate that, although MAP2 slows microtubule dynamics, it does not inhibit them entirely (33). Indeed, substantial changes in configuration of the microtubule cytoskeleton are visible when time-lapse recordings are made from MAP2c-GFP transfected cells over periods of several hours (33). To address the question of whether comparable changes occur in dendrites we raised transgenic mice expressing MAP2c-GFP in central nervous system neurons (Fig. 3). Like the actin-GFP expressing animals we have previously described (42), MAP2c-GFP mice are indistinguishable from nontransgenic litter mates in morphology, fertility, and lifespan and show no obvious behavioral abnormalities nor deficits in the Morris water maze (H.P., P. Kelly, and A.M., unpublished data). This lack of overt effects of expressing exogenous MAP2 is consistent with results we previously obtained for transgenic mice expressing high levels of epitope-tagged MAP2c (51). In organotypic hippocampal slice cultures established from these animals MAP2c-GFP is readily detectable in dendrites with weaker expression occurring in cell bodies (Fig. 3A). In more than 50 independently established cultures MAP2 was always limited to the shafts of dendrites (see, for example, Fig. 3B). In several hundred cells examined within these cultures we have failed to find any evidence for the presence of MAP2c-GFP in dendritic spines.
Figure 3.
Time-lapse recording of MAP2 in hippocampal tissue slices from transgenic mice stably expressing MAP2c-GFP. (A) Confocal GFP fluorescence image taken near the cell body layer of area CA1 in an organotypic slice culture established from an 11-day-old transgenic mouse and maintained in culture for 25 days. Nuclei in cell bodies are marked with *. (Bar = 10 μm.) (B) MAP2 localization in hippocampal neurons is limited to the shafts of dendrites. Single frame taken from a time-lapse recording of MAP2c-GFP fluorescence in the CA1 neuropil of a hippocampal slice maintained in culture for 4 weeks. (Bar = 5 μm.) (C and D) Short-term time-lapse assay for MAP2 dynamics. (C Left) A single confocal gray-scale image of MAP2-GFP fluorescence in area CA1 of a 5-week-old hippocampal slice culture. (C Right) A pseudocolor “difference image” produced by summing gray-scale differences between images taken 30 sec apart over 10 min of time-lapse recording. Compare the overall lack of change in the MAP2 image (dark blue color) during the recording period to the high degree of change (green, yellow, and red) in actin images of similar configuration (Fig. 4A Right). (Bar = 5 μm.) (D) Long-term timelapse assay for MAP2 dynamics. Original gray-scale fluorescence image (Upper) and pseudocolor difference image (Lower) of a dendrite segment followed over a 3-h time period. The dark blue color again indicates an overall lack of change in MAP2 distribution during this longer recording period. (Bar = 5 μm.)
Confocal time-lapse recordings of MAP2c-GFP in hippocampal slices from transgenic mice showed a surprising lack of motility. Fig. 3C shows data from a 4-week-old culture where the general distribution of MAP2c-GFP is shown by the single frame of original fluorescence data (Left). Fig. 3C Right shows a “difference image” prepared by subtracting gray scale values for pixels in 30 successive frames and then summing the differences (see ref. 42). The values are displayed on a pseudocolor scale in which areas where there was little change are colored blue while those where large changes occurred appear red and white. As the overall blue color of Fig. 3C Right shows, there was little change in the MAP2c-GFP image during the 10 min of recording. Similar time-lapse recordings of MAP2c-GFP in hippocampal slices were made for periods of up to 3 h (n = 12). Fig. 3D shows an example focused on a single dendrite recorded continuously for 3 h in which the blue coloration of the MAP2c-containing dendrite indicates the lack of change in the image (compare this to the actin-GFP pseudocolor image of dendritic spines shown below in Fig. 4B). We considered the possibility that microtubules in dendrites might not show dynamic activity except under conditions of enhanced stimulation. Because both long-term potentiation of synaptic responses and stimulation induced increases in dendritic spine numbers are associated with activation of N-methyl-d-aspartate (NMDA) receptors (52–56) we made time-lapse recordings of MAP2c-GFP in slices during exposure to NMDA or to the NMDA receptor antagonist MK-801. In neither case was any change in MAP2c-GFP images detectable in recordings of up to 2 h duration.
Figure 4.
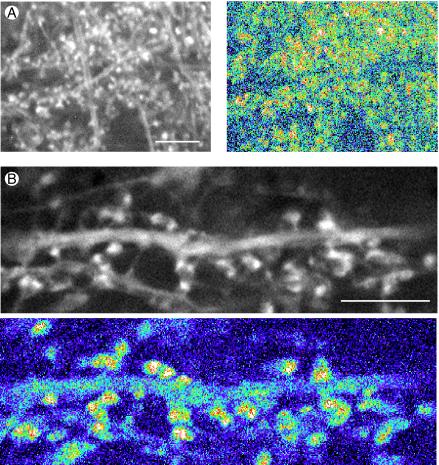
Time-lapse recording of actin dynamics in dendrite spines of hippocampal tissue slices from transgenic mice expressing actin-GFP. (A) (Left) An original fluorescence image in a single frame from a time-lapse recording in which frames were collected 30 sec apart. (Right) Changes in actin distribution over 10 min displayed by difference imaging using a pseudocolor scale (see text for details). Red and yellow patches indicate areas of high motility associated with dendritic spines. (Bar = 10 μm.) (B) Single gray-scale frame (Upper) and pseudocolor difference image at higher magnification. Shape changes are associated with dendritic spines (red and yellow patches) whereas the dendrite shaft shows little dynamic activity (Lower). (Bar = 10 μm.)
Actin-GFP Shows High Motility in Dendritic Spines of Transgenic Mice.
Because MAP2-GFP in dendrites showed so little dynamic activity, we made comparable time-lapse recordings of actin-GFP in hippocampal slice cultures from transgenic mice. As previously reported (42), actin-GFP was concentrated in heads of dendritic spines (Fig. 4 A Left and B Upper). Time-lapse recordings made from these cultures confirmed that this spine actin is rapidly dynamic. This is shown by the pseudocolored difference images in Fig. 4 in which areas where there were large changes in the image during the 10 min of recording are colored red and yellow and areas where little change occurred appear blue.
Discussion
Our data indicate that the cytoskeleton in neuronal dendrites is partitioned into distinct microtubule and microfilament domains associated with dendrite shafts and spines, respectively. This finding is in contrast to earlier immunocytochemical studies, which reported the presence of MAP2 in dendritic spines (19, 27, 28). A possible reason for this discrepancy is that the reaction product of immunoperoxidase staining used to detect MAP2 in the earlier studies spread from its origin at microtubules in the dendrite shaft into dendritic spines. Electron microscope studies generally confirm the results of our live cell imaging observations by showing that while microtubules are abundant in dendrites they are absent from spines that instead contain a meshwork of microfilaments consistent with the presence of high actin concentrations (11, 24–26). An exception is the presence of microtubules in large, branched spines in area CA3 of the hippocampus but these spines also contain ribosomes, multivesicular bodies, and mitochondria (57) emphasizing the special status conferred by their large size. Microtubules are not found in other CA3 spine types of more usual size, supporting the conclusion that microtubules generally do not extend into the spine cytoplasm.
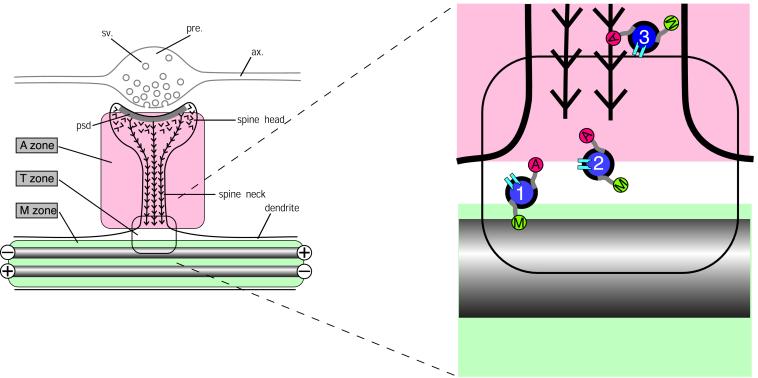
Based on the partitioning of microfilaments and microtubules between shaft and spine the dendrite cytoplasm can be considered, from the perspective of plasticity, as divided into separate microtubule (M) and actin (A) zones (Fig. 5). Interesting questions arise concerning events at the transition zone (T, Fig. 5). Because most neurons are postmitotic, their molecular components must be continuously replaced. For some proteins, including MAP2 (58), this is achieved by export of mRNA into dendrites, presumably followed by local synthesis of the protein product (59, 60). However, for most neuronal proteins, synthesis occurs in the cell body followed by transport into axons and dendrites. This process is best understood for membrane proteins that are transported on vesicles. These are conveyed along microtubules by motor molecules of the kinesin and dynein families, which can confer directional specificity toward axon or dendrite (61–64). The recent identification of a dendrite-specific kinesin, KIF17, bound to NMDA-2B glutamate receptor subunits as part of a vesicular complex (65) confirms the existence of a mechanism for transporting functional components of spine synapses along dendritic microtubules. The ultimate destination of the NMDA receptor subunits is the postsynaptic membrane, raising the question of how the vesicle that contains them travels from the microtubule transport system of the dendrite shaft to the postsynaptic membrane at the tip of a dendritic spine. Evidence for interactions between microtubule- and actin-based transport mechanisms near the cell surface (66, 67), together with the demonstration of a direct interaction between microtubule and actin transport motors (68), suggest that this transition may be accomplished by transfer of vesicles possessing motors for both systems from microtubules to microfilaments. A hypothetical scheme for this transfer is indicated diagrammatically in Fig. 5 where vesicles move from microtubule to microfilament transport systems at the base of the spine. The management of this putative transition remains to be determined because a thin cortical layer of actin filaments is also present within dendrite shafts. The mechanisms responsible for delivering materials via the spine cytoplasm to sites in the postsynaptic junction have significant implications for synaptic plasticity in view of growing evidence for physical exchange of receptor molecules in the postsynaptic membrane of glutamatergic synapses (69–72).
Figure 5.
Hypothetical scheme for the partitioning of cytoskeletal microdomains between shaft and spine in dendrites. (Left) Part of dendrite in the region of a spine synapse. The axonal component (ax.), with its swollen presynaptic (pre.) bouton containing synaptic vesicles (sv.) is outlined in gray. It forms a synapse at the tip of a dendritic spine head. Inside the spine head the junctional region is marked by the postsynaptic density (psd.), a complex of scaffolding proteins that acts as the platform for assembling functional molecules such as neurotransmitter receptors and ion channels. The cytoskeleton of the dendritic spine is composed of actin filaments (barbed lines) that are inserted into the psd. The cytoskeleton of the underlying dendrite consists predominantly of microtubules (gray rods), which in dendrites are bidirectionally oriented so that some have the plus ends distally and others the minus end distally as indicated. This distribution of cytoskeletal filaments demarcates three cytoplasmic zones, an M zone in the dendrite shaft, where microtubules predominate, an A zone in the dendritic spine, where actin filaments predominate, and a T, or transition, zone. (Right) The expanded diagram shows the relationship of these zones to the delivery of materials to the synaptic domain as suggested by current evidence. Transport vesicles (blue filled circles) carry cargoes of functional molecules, such as NMDA receptors (pale blue symbols), bound for the postsynaptic membrane. These vesicles bear both microtubule-dependent (M, kinesin and dynein) and actin-dependent (A, myosin) motor molecules. Transitory detachment of kinesin and dynein from microtubule tracks provides the opportunity for the myosin motors of transport vesicles to engage with the actin filaments of dendritic spines along which they travel to the synaptic domain. Single chevrons in the vicinity of the postsynaptic membrane represent the presence of labile actin filaments in this zone.
The necessity of special mechanisms for transferring materials from shaft to spine raises the question of why such a partitioning of dendrite structure should exist at all. One possibility, suggested by the results of the present study, is that this separation is a specialization for regulating anatomical plasticity. As our time-lapse recordings show, the actin and microtubule domains are associated with distinct rates of plasticity. Whereas actin in dendritic spine defines a region of rapid morphological change occurring over seconds and minutes (14, 15, 17), time-lapse imaging of MAP2 suggests that microtubules in the dendrite shaft undergo little change in periods of up to 3 h. This does not exclude that dynamic changes in dendritic microtubules may occur over longer periods. Indeed time-lapse imaging of MAP2-containing microtubule bundles in transfected epithelial cells shows that gradual alterations in the configuration of the microtubule cytoskeleton can occur over periods of several hours (33). This finding suggests that MAP2-containing neuronal microtubules may have a capacity for morphological plasticity although at a rate intrinsically slower than that of actin filament arrays, which appear constantly motile in comparable recordings (14). That gradual changes in the extent and branching of dendrites can occur has been demonstrated by repetitive imaging of dendrites in superior cervical ganglia of adult rats where substantial changes in dendritic arbors have been documented over periods of weeks and months (73, 74). However, other studies support the idea that dendritic spines are the predominant site of activity-dependent morphological plasticity in the brain in vivo (for example, refs. 17 and 75–78).
Taken together these observations suggest that microdifferentiation of the dendritic cytoskeleton in mature neurons may be a cellular specialization for dividing the structural support of dendrites into two levels of stability. One of these, involving microtubules, appears to respond slowly, providing morphological stability to dendrite arbors while still allowing for long-term flexibility, whereas the other, involving motile actin filaments, allows for rapid, activity-dependent changes in synaptic structure.
Supplementary Material
Acknowledgments
We thank Thierry Doll and Jean-Francois Spetz for assistance in preparing transgenic animals.
Abbreviations
- MAP
microtubule-associated protein
- GFP
green fluorescent protein
- YFP
yellow fluorescent protein
- NMDA
N-methyl-d-aspartate
Footnotes
This paper was presented at the National Academy of Sciences colloquium, “Molecular Kinesis in Cellular Function and Plasticity,” held December 7–9, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Matus A, Huber G, Bernhardt R. Cold Spring Harbor Symp Quant Biol. 1983;48:775–782. doi: 10.1101/sqb.1983.048.01.079. [DOI] [PubMed] [Google Scholar]
- 2.Craig A M, Banker G. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- 3.Bradke F, Dotti C G. Science. 1999;283:1931–1934. doi: 10.1126/science.283.5409.1931. [DOI] [PubMed] [Google Scholar]
- 4.Mitchison T, Kirschner M. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 5.Smith S J. Science. 1988;242:708–715. doi: 10.1126/science.3055292. [DOI] [PubMed] [Google Scholar]
- 6.Gordon-Weeks P R. BioEssays. 1991;13:235–239. doi: 10.1002/bies.950130506. [DOI] [PubMed] [Google Scholar]
- 7.Avila J, Dominguez J, Diaz-Nido J. Int J Dev Biol. 1994;38:13–25. [PubMed] [Google Scholar]
- 8.Tanaka E, Sabry J. Cell. 1995;83:171–176. doi: 10.1016/0092-8674(95)90158-2. [DOI] [PubMed] [Google Scholar]
- 9.Heidemann S R. Int Rev Cytol. 1996;165:235–296. doi: 10.1016/s0074-7696(08)62224-x. [DOI] [PubMed] [Google Scholar]
- 10.Letourneau P C. Perspect Dev Neurobiol. 1996;4:111–123. [PubMed] [Google Scholar]
- 11.Matus A, Ackermann M, Pehling G, Byers H R, Fujiwara K. Proc Natl Acad Sci USA. 1982;79:7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fifkova E. Brain Res. 1985;356:187–215. [PubMed] [Google Scholar]
- 13.Kaech S, Fisher M, Doll T, Matus A. J Neurosci. 1997;17:9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 15.Dunaevsky A, Tashiro A, Majewska A, Mason C, Yuste R. Proc Natl Acad Sci USA. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halpain S. Trends Neurosci. 2000;23:141–146. doi: 10.1016/s0166-2236(00)01576-9. [DOI] [PubMed] [Google Scholar]
- 17.Lendvai B, Stern E A, Chen B, Svoboda K. Nature (London) 2000;404:876–881. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- 18.Matus A. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 19.Matus A, Bernhardt R, Hugh-Jones T. Proc Natl Acad Sci USA. 1981;78:3010–3014. doi: 10.1073/pnas.78.5.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhardt R, Matus A. J Cell Biol. 1982;92:589–593. doi: 10.1083/jcb.92.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgoyne R D, Cumming R. Neuroscience. 1984;11:156–167. doi: 10.1016/0306-4522(84)90220-3. [DOI] [PubMed] [Google Scholar]
- 22.De Camilli P, Miller P E, Navone F, Theurkauf W E, Vallee R B. Neuroscience. 1984;11:817–846. [PubMed] [Google Scholar]
- 23.Huber G, Matus A. J Cell Biol. 1984;98:777–781. doi: 10.1083/jcb.98.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters A, Palay S L, Webster H d F. The Fine Structure of the Nervous System. Philadelphia: Saunders; 1976. [Google Scholar]
- 25.Landis D M, Reese T S. J Cell Biol. 1983;97:1169–1178. doi: 10.1083/jcb.97.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen R S, Chung S K, Pfaff D W. Cell Mol Neurobiol. 1985;5:271–284. doi: 10.1007/BF00711012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caceres A, Banker G, Steward O, Binder L, Payne M. Brain Res. 1984;315:314–318. doi: 10.1016/0165-3806(84)90167-6. [DOI] [PubMed] [Google Scholar]
- 28.Morales M, Fifkova E. Cell Tissue Res. 1989;256:447–456. doi: 10.1007/BF00225592. [DOI] [PubMed] [Google Scholar]
- 29.Sattilaro R F. Biochemistry. 1986;25:2003–2009. doi: 10.1021/bi00356a025. [DOI] [PubMed] [Google Scholar]
- 30.Selden S C, Pollard T D. Ann NY Acad Sci. 1986;466:803–812. doi: 10.1111/j.1749-6632.1986.tb38464.x. [DOI] [PubMed] [Google Scholar]
- 31.Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan N J, Hirokawa N. J Cell Sci. 1992;103:953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- 32.Weisshaar B, Doll T, Matus A. Development (Cambridge, UK) 1992;116:1151–1161. doi: 10.1242/dev.116.4.1151. [DOI] [PubMed] [Google Scholar]
- 33.Kaech S, Ludin B, Matus A. Neuron. 1996;17:1189–1199. doi: 10.1016/s0896-6273(00)80249-4. [DOI] [PubMed] [Google Scholar]
- 34.Ozer R S, Halpain S. Mol Biol Cell. 2000;11:3573–3587. doi: 10.1091/mbc.11.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colella R, Lu C, Hodges B, Wilkey D W, Roisen F J. Brain Res Dev Brain Res. 2000;121:1–9. doi: 10.1016/s0165-3806(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 36.van Rossum D, Hanisch U K. Trends Neurosci. 1999;22:290–295. doi: 10.1016/s0166-2236(99)01404-6. [DOI] [PubMed] [Google Scholar]
- 37.Harris K M. Curr Opin Neurol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- 38.Jontes J D, Smith S J. Neuron. 2000;27:11–14. doi: 10.1016/s0896-6273(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 39.Parnass Z, Tashiro A, Yuste R. Hippocampus. 2000;10:561–568. doi: 10.1002/1098-1063(2000)10:5<561::AID-HIPO6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 40.Ludin B, Matus A. Trends Cell Biol. 1998;8:72–77. [PubMed] [Google Scholar]
- 41.Ormö M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 42.Fischer M, Kaech S, Wagner U, Brinkhaus H, Matus A. Nat Neurosci. 2000;3:887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- 43.Botteri F M, van der Putten H, Wong D F, Sauvage C A, Evans R M. Mol Cell Biol. 1987;7:3178–3184. doi: 10.1128/mcb.7.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gahwiler B H, Thompson S M, Audinat E, Robertson R T. In: Culturing Nerve Cells. Banker G, Goslin K, editors. Cambridge, MA: MIT Press; 1991. pp. 379–411. [Google Scholar]
- 45.Bartlett W P, Banker G A. J Neurosci. 1984;4:1954–1965. doi: 10.1523/JNEUROSCI.04-08-01954.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papa M, Bundman M C, Greenberger V, Segal M. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziv N E, Smith S J. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 48.Boyer C, Schikorski T, Stevens C F. J Neurosci. 1998;18:5294–5300. doi: 10.1523/JNEUROSCI.18-14-05294.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiala J C, Feinberg M, Popov V, Harris K M. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matus A. Annu Rev Neurosci. 1988;11:29–44. doi: 10.1146/annurev.ne.11.030188.000333. [DOI] [PubMed] [Google Scholar]
- 51.Marsden K M, Doll T, Ferralli J, Botteri F, Matus A. J Neurosci. 1996;16:3265–3273. doi: 10.1523/JNEUROSCI.16-10-03265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bliss T V, Collingridge G L. Nature (London) 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 53.Andersen P, Moser E, Moser M B, Trommald M. J Physiol (Paris) 1996;90:349. doi: 10.1016/s0928-4257(97)87917-x. [DOI] [PubMed] [Google Scholar]
- 54.Engert F, Bonhoeffer T. Nature (London) 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 55.Maletic-Savatic M, Malinow R, Svoboda K. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 56.Toni N, Buchs P A, Nikonenko I, Bron C R, Muller D. Nature (London) 1999;402:421–425. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- 57.Chicurel M E, Harris K M. J Comp Neurol. 1992;325:169–182. doi: 10.1002/cne.903250204. [DOI] [PubMed] [Google Scholar]
- 58.Garner C C, Tucker R P, Matus A. Nature (London) 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 59.Steward O, Banker G A. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 60.Schuman E M. Neuron. 1999;23:645–648. doi: 10.1016/s0896-6273(01)80023-4. [DOI] [PubMed] [Google Scholar]
- 61.Brady S T. Trends Cell Biol. 1995;5:159–164. doi: 10.1016/s0962-8924(00)88980-1. [DOI] [PubMed] [Google Scholar]
- 62.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 63.Burack M A, Silverman M A, Banker G. Neuron. 2000;26:465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- 64.Terada S, Hirokawa N. Curr Opin Neurobiol. 2000;10:566–573. doi: 10.1016/s0959-4388(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 65.Setou M, Nakagawa T, Seog D H, Hirokawa N. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 66.Gavin R H. Int Rev Cytol. 1997;173:207–242. doi: 10.1016/s0074-7696(08)62478-x. [DOI] [PubMed] [Google Scholar]
- 67.Goode B L, Drubin D G, Barnes G. Curr Opin Cell Biol. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 68.Huang J D, Brady S T, Richards B W, Stenolen D, Resau J H, Copeland N G, Jenkins N A. Nature (London) 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 69.Nishimune A, Isaac J T, Molnar E, Noel J, Nash S R, Tagaya M, Collingridge G L, Nakanishi S, Henley J M. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- 70.Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir R L. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 71.Shi S H, Hayashi Y, Petralia R S, Zaman S H, Wenthold R J, Svoboda K, Malinow R. Science. 1999;284:1811–1816. doi: 10.1126/science.284.5421.1811. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J J, Esteban J A, Hayashi Y, Malinow R. Nat Neurosci. 2000;3:1098–1106. doi: 10.1038/80614. [DOI] [PubMed] [Google Scholar]
- 73.Purves D, Hadley R D. Nature (London) 1985;315:404–406. doi: 10.1038/315404a0. [DOI] [PubMed] [Google Scholar]
- 74.Purves D, Hadley R D, Voyvodic J T. J Neurosci. 1986;6:1051–1060. doi: 10.1523/JNEUROSCI.06-04-01051.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valverde F. Exp Brain Res. 1967;3:337–352. doi: 10.1007/BF00237559. [DOI] [PubMed] [Google Scholar]
- 76.Coss R G, Brandon J G, Globus A. Brain Res. 1980;192:49–59. doi: 10.1016/0006-8993(80)91007-0. [DOI] [PubMed] [Google Scholar]
- 77.Woolley C S, McEwen B S. J Comp Neurol. 1993;336:293–306. doi: 10.1002/cne.903360210. [DOI] [PubMed] [Google Scholar]
- 78.Moser M B, Trommald M, Egeland T, Andersen P. J Comp Neurol. 1997;380:373–381. doi: 10.1002/(sici)1096-9861(19970414)380:3<373::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.