Abstract
Catalyzed hairpin assembly (CHA) is a robust enzyme-free signal-amplification reaction that has a wide range of potential applications especially in biosensing. Although most studies on the analytical applications of CHA focus on the measurement of concentrations of biomolecules, we show here CHA can also be used to probe the spatial organization of biomolecules such as single-stranded DNA. The basis of such detection is the fact that a DNA structure that brings a toehold and a branch migration domain into close proximity can catalyze the CHA reaction. We quantitatively studied this phenomenon and applied it to the detection of domain reorganization that occurs during DNA self-assembly processes such as hybridization chain reaction (HCR). We also show CHA circuits can be designed to detect certain types of hybridization defects. This principle allowed us to develop a ‘signal-on’ assay that can simultaneously respond to multiple types of mutations in a DNA strand in one simple reaction, which is of great interest in genotyping and molecular diagnostics. These findings highlight the potential impacts of DNA circuitry on DNA nanotechnology and provide new tools for further development of these fields.
Keywords: Spatial organization, hairpin assembly circuits, enzyme-free
Although nucleic acids are best known as genetic materials, they have been engineered to perform a wide range of tasks. For example, DNAzymes and aptamers enable detection and manipulation of biomolecules,1 while more recently DNA tile assembly and DNA origami have been shown to be useful for the precise positioning of molecules with sub-nanometer precision.2 Enzyme-free DNA circuits have been shown to be capable computers.3
While the design of nucleic acid structures and circuits has proceeded quickly, the analytical methodologies capable of probing these DNA nanotechnologies have lagged. AFM can be used to look at the gross outline of DNA nanostructures,2c,4 and DNA circuits can be adapted to ensemble methods for detection, such as fluorescence and electrochemistry.5 However, the intimate probing of individual portions of the nanostructures themselves has generally been carried out with technically demanding methods, such as restriction digestion or chemical probing6.
In this manuscript, we both exploit the advances in DNA nanotechnology design and adapt these advances to much finer probing of structures and circuits by developing a new design principle, proximity detection within DNA nanostructures. We start with an enzyme-free signal-amplification circuit, catalyzed hairpin assembly (CHA)3d,5a, but make the execution of this circuit dependent upon the formation of a particular nanostructure, an assembled hybridization chain reaction (HCR)3a concatemer, whose molecular junctions are otherwise ‘unseeable’ with extant technologies (Figure 1). The sensitivity of this method is such that it can be used to probe defects in DNA nanomaterials caused by mismatches at the junctions of hybridized DNA strands. It has also proved to be a simple and effective analytical tool, allowing the direct readout of HCR in homogenous solution without the use of specialty fluorescent oligonucleotides as HCR substrates.
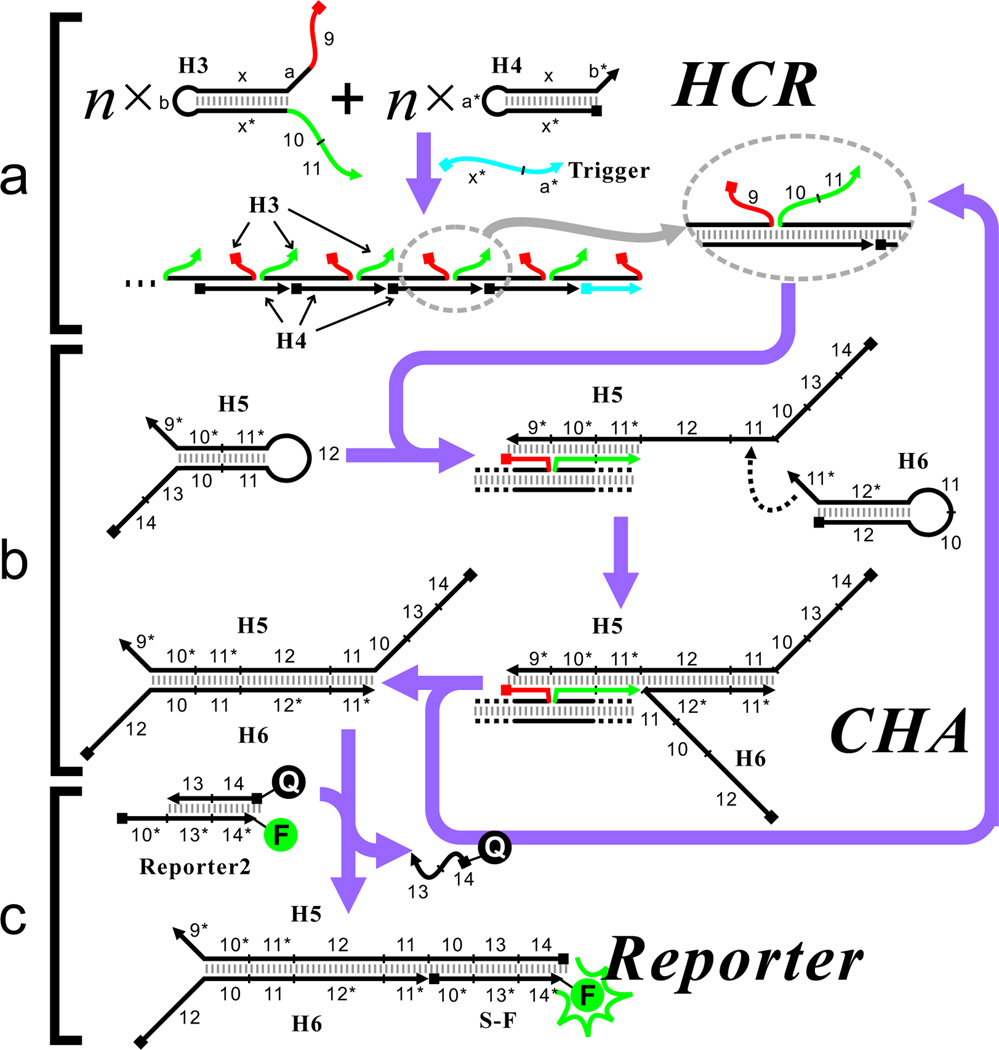
Figure 1.
Scheme of the CHA-based circuit that detects the degree of HCR assembly, (a) Scheme of HCR with extended domains. (b) CHA reaction catalyzed by the correctly formed HCR product. (c) Fluorescent reporter that detects the product of the CHA reaction.
The scheme for proximity detection is shown in Figure 1. A HCR reaction composed of two substrate hairpin DNAs H3 and H4 is initiated by the Trigger strand (Figure 1a). However, in contrast to traditional HCR,3a in our study hairpin H3 has been extended with a 10-nt segment ‘9’ (Figure 1, red) at its 5’ end and an 18-nt segment ‘10+11’ (Figure 1, green) at its 3’ end. Thus, when H3 is assembled into a concatenated HCR product, these two segments are co-localized through H4-mediated hybridization. In the attendant CHA reaction with H5 and H6, segments ‘9’ and ‘10+11’ act as a toehold and a branch-migration domain, respectively (Figure 1b). In short, the correct assembly of the H3:H4 concatemer via HCR is monitored via the H5:H6 CHA. The formation of the H5:H6 duplex can be easily detected using a fluorescent reporter named Reporter2, as shown in Figure 1c. As in all CHA-based circuits, the uncatalyzed hybridization of H5 and H6 should be extremely slow since interacting domains are sequestered in intramolecular secondary structures.
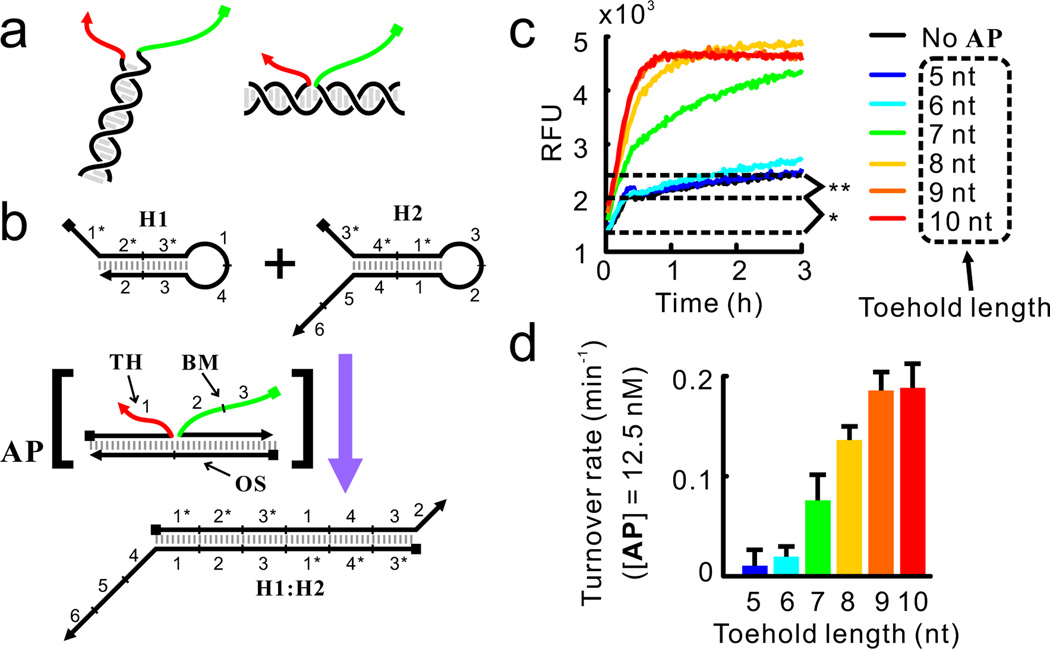
We have previously observed7 that co-localization of the two domains by direct hybridization (Figure 2a, left) leads to efficient strand displacement, and a simple reorganization of strand placement in the DNA structure shown in Figure 2a (right) ultimately led to the scheme shown in figure 1b. However, this latter design should allow any two oligonucleotide sequences to be brought together by an antisense oligonucleotide (for example, OS in Figure 2b) to function as a toehold (red in Figure 2a) + branch migration domain (green in Figure 2a). The expanded generality of this scheme is now shown in several examples below. We constructed a simple DNA structure named AP (standing for assembly product), in which strand TH (standing for toehold) and strand BM (standing for branch-migration) were hybridized to the organizer strand OS (Figure 2b). In essence, OS co-localized the toehold domain carried by TH and the branch-migration domain carried by BM.
Figure 2.
Feasibility of detecting HCR product using CHA-based circuits. (a) Toehold (red) and branch-migration domain (green) co-localized by direct (left) or indirect (right) hybridization. (b) Structure of the substrates (H1 and H2) and the catalyst (AP complex) of the CHA reaction. The toehold and branch-migration domain in the AP complex are shown in red and green, respectively. Detailed reaction pathway is shown in Figure S2. The fluorescent reporter for H1:H2, named Reporter, is similar to the Reporter2 complex shown in Figure 1c and is not shown here. (c) Kinetics of the CHA reaction catalyzed by the AP complex with different toehold length. In all reactions, [H1] = 75 nM, [H2] = [Reporter] = 50 nM, [TH] = [BM] = 15 nM, [OS] = 12.5 nM. Symbols * and ** denote two types of circuit leakage which are further discussed in Figure S1. (d) Turnover rates (rates of reaction divided by concentration of the catalyst) as function of toehold lengths.
As shown in Figure 2c, although uncatalyzed formation of H1:H2 was observable when 75 nM of H1 and 50 nM of H2 were mixed (black trace in Figure 2c, further dissected in Figure S1), the presence of 12.5 nM of AP substantially accelerated the reaction, presumably through the mechanism shown in Figure S2. Notably, catalysis is strictly dependent on the presence of the OS strand. In the absence of OS, the mixture of TH and BM did not yield any signal above background (Figures S1 and S3), highlighting the importance of the co-localization of the toehold and the branch-migration domain through correct DNA assembly. We have further studied how temperature and toehold length (Figure 2c–d) affect the rate of CHA. These data are described in Section 2 of Supporting Information and are largely in agreement with our understanding of the kinetics of CHA reactions.7–8
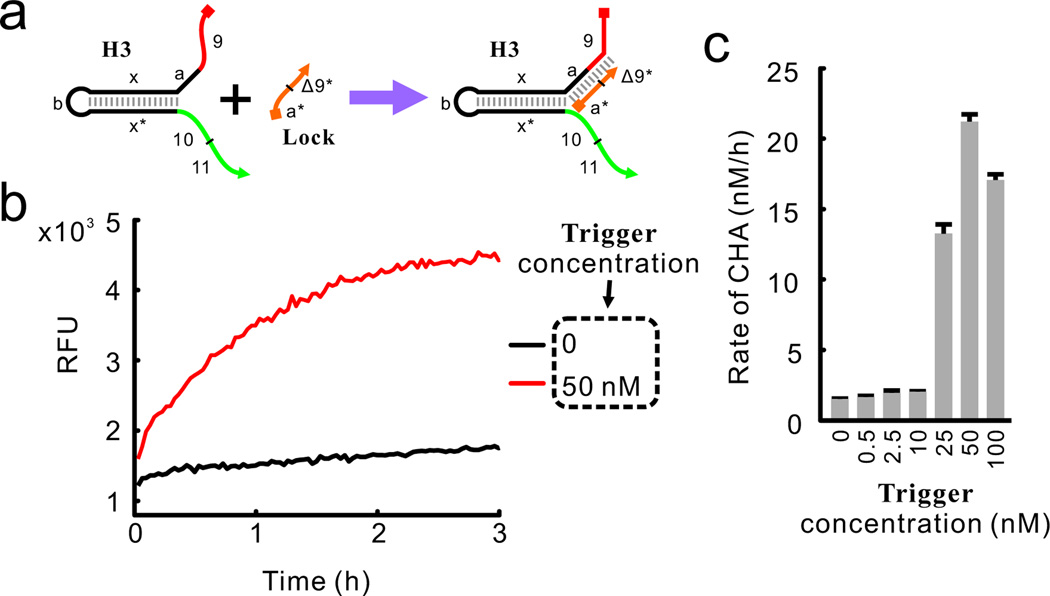
We next moved to determining whether we could detect multiple, different proximity junctions formed during HCR (Figure 1a). To make the detection of the HCR product more sensitive and specific, we introduced a 12-nt oligonucleotide named Lock that hybridized to the junction between segments ‘9’ and ‘a’ of H3 (Figure 3a). The function of Lock is two-fold: first, it blocks segment ‘a’ of H3, which allows timely termination of the HCR reaction (Figure S5, lane 8 and 16). Second, it abolishes the ability of monomeric H3 to initiate the CHA reaction by using segment ‘9’ as a ‘remote toehold’ (Figure S6).9 Some 200 nM of H3 and 200 nM of H4 were incubated in the presence or absence of 50 nM strand Trigger for 16 h before saturating concentrations of Lock were added. When the HCR product was analyzed by native electrophoresis, high molecular-weight HCR products were only found in the presence of Trigger, as expected (Figure S5). More importantly, when an aliquot of the HCR reaction was mixed with H5, H6, and Reporter2, the HCR reaction with Trigger led to ~10-fold faster fluorescent detection of H5:H6 than without Trigger (Figure 3b). While there was an initial burst in fluorescence (likely caused by stoichiometric opening of H5 by the HCR product, see Figure S7), when H6 was absent, the HCR product did not lead to a steady increase in fluorescent signal. This observation confirmed that the HCR product indeed initiates the CHA reaction cascade catalytically.
Figure 3.
Detection of HCR product using CHA circuits. (a) The Lock strand which can terminate HCR reaction and abolish the activity of monomeric H3 in catalyzing the CHA reaction. Segment Δ9* is complementary to the 6 nt to the 3’ terminus of segment 9. (b and c) Kinetics of the CHA reactions catalyzed by HCR products with different concentrations of the Trigger strand.
We then repeated this experiment using various concentrations of Trigger (Figure 3c). Interestingly, we consistently observed several non-linear relationships between the concentrations of Trigger and the rate of the CHA reaction (presumably reflecting the concentrations of correct assemblies formed by HCR). For example, the rate of the CHA reaction peaked when the Trigger concentration was roughly 1/8 to 1/4 of the concentration of H3 and H4 (the concentrations of H3 and H4 were held equal at 200 nM in all experiments). More Trigger actually led to a decreased rate of the CHA reaction (Figure 3c and data not shown). This effect is to be expected since saturation of the initial Trigger should lead to the formation of very short assemblies that have decreased numbers of active, proximity junctions. This phenomenon can also be observed via electrophoresis (Figure S5), where high concentrations of Trigger led to the formation of very short HCR chains.3a
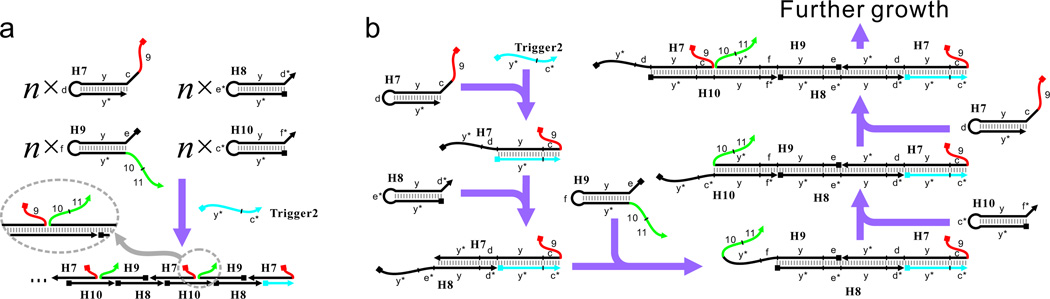
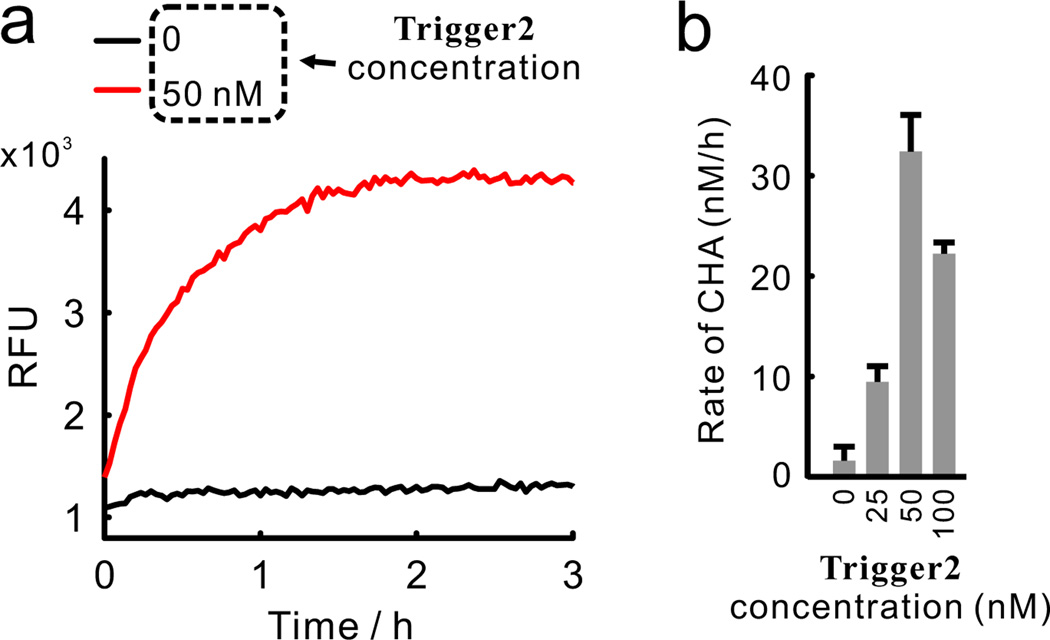
Having shown the ability to sense the triggered reorganization of two domains (toehold and branch-migration domain) on one molecule (H3), we next sought to test whether co-localization of such domains on separate molecules can also catalyze the downstream CHA reaction. To do this, we designed a new HCR system consisting of 4 hairpins: H7, H8, H9, and H10. The assembly of the HCR can be initiated by the Trigger2 strand (Figure 4). Since in this system the toehold and branch-migration domain are separated on two molecules (H7 and H9, respectively), no ‘Lock’ is needed to suppress the basal CHA reaction catalyzed by unassembled hairpins. As before, 200 nM of each of the four hairpins and 0 to 100 nM of Trigger2 were mixed for ~24 h, after which an aliquot of the HCR reaction was mixed with H5, H6, and Reporter2. As can be seen in Figure 5a, 50 nM Trigger2 led to a strong CHA signal whereas the absence of Trigger2 resulted in extremely low background. The rate of CHA reaction was dependent on the concentration of Trigger2 with a biphasic concentration dependence (Figure 5b) that was similar to that previously observed for a two-hairpin HCR (Figure 3c).
Figure 4.
Sensing the assembly of a 4-hairpin HCR using a CHA circuit. (a) Overall reaction of the 4-hairpin HCR reaction, where unlike H3 (see Figure 1a), the toehold (red) and branch-migration domain (green) locate on two separate molecules (H7 and H9, respectively), (b) Detailed reaction mechanism of the 4-hairpin HCR.
Figure 5.
Real-time kinetics (a) and rates (b) of the CHA reactions catalyzed by 4-hairpin HCR products with different concentrations of the Trigger2 strand (See Figure S8 for gel image).
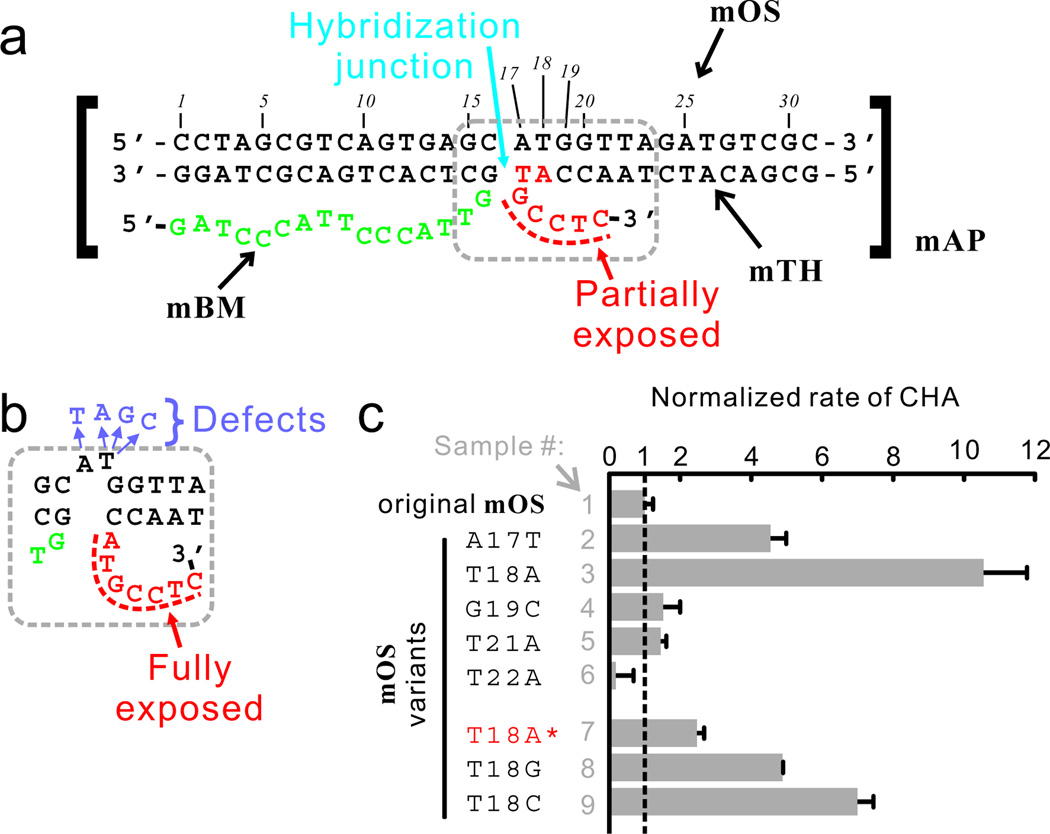
To further explore the utility of CHA circuits in structural probing, we asked whether DNA structures and CHA circuits could be designed so that small-scale structural difference (e.g. hybridization defects) could be sensed. To test this hypothesis, we designed a CHA reaction that could potentially be accelerated by hybridization defects near the hybridization junction of the two DNA strands (Figure 6). In this structure, named mAP (‘m’ standing for ‘modified’), the toehold and branch-migration domains (red and green in Figure 6a, carried by strands mTH and mBM, respectively) were co-localized via hybridization to the organizer strand mOS (Figure 6a). However, unlike the original AP, where the entire toehold for the CHA reaction (segment ‘9’, Figure 1a) was exposed, in mAP only 5 of 7 bases of the toehold (Figure 6a, red) were exposed and the other 2 bases were hybridized to mOS. While the mAP structure may be capable of triggering the CHA reaction H1 + H2 → H1:H2, catalysis is expected to be slow due to the partially sequestered toehold. By introducing hybridization defects between mTH and mOS near the junction the toehold region in mTH should be more readily liberated (Figure 6b), which may enhance the catalytic activity of mAP. Unpaired bases (in this case A17 and T18) should not abolish strand-displacement activity, and indeed we observe that adding one or two unpaired thymidines on the OS strand at the hybridization junction (Figure S9a) results in substantive (30%–50%) residual catalytic activity of the AP complex (Figure S9b–c).
Figure 6.
Detecting assembly defects using CHA circuits. In the perfectly formed mAP structure (a), the toehold (shown in red) is only partially exposed, whereas defects near the hybridization junction may fully expose the toehold (b). The hybridization defects can be mimicked by introducing point mutations on the mOS strand, which stimulate the activity of mAP to different extent (c). T18A* (Sample #7) denotes 1:5-diluted T18A variant of mOS.
In fact, single-point mutations in mOS near the hybridization junction (A17T, T18A, T18G, and T18C) significantly enhanced the catalytic activity of mAP (Figure 6c, sample #2, 3, 8, and 9), while mutations further away from the hybridization junction had little or no effect (Figure 6c, sample #4, 5, 6). Thus, we have designed a circuit that can sensitively detect hybridization defects at a particular site in a nanostructure. In addition, this system exemplifies the design of a ‘signal-on’ sensor that responds to mis-matches, rather than perfect matches. This advance, when coupled with appropriate pre-amplification (e.g. PCR) and concentration-normalization (e.g. a molecular beacon targeting an invariable region) techniques, may yield powerful genotyping tools which are of great interest in molecular diagnostics. First, ‘signal-on’ assays in general have much higher signal-to-background ratios and consequently much higher sensitivities than ‘signal-off’ assays. For example, even when the T18A variant of mOS is diluted by 5-fold it still led to roughly 150% higher signal over the background of undiluted, original (or in other words, ‘wildtype’) mOS (Figure 6c, compare sample #1 and #7). More importantly, in many cases multiple mutations in a small region of a gene can cause the amino-acid changes in the encoded protein that lead to disease or drug resistance.10 Having the ability to simultaneously respond to multiple mutations would dramatically reduce the number of genotyping tests required to identify such genes.
The findings presented in this work have implications in both nanotechnology and molecular diagnostics. First, with proper calibration and normalization, our circuit allows fast and more readily quantifiable observation of the quantity or quality of the assembly of DNA nanostructures. Second, our methods allow HCR and CHA, two enzyme-free signal-amplification processes that have been engineered to detect both nucleic acids and non-nucleic acid analytes3a,5,11, to be cascaded, potentially leading to greater amplification. Interestingly, although this signal amplification is larger, there are non-linearities in the response (compare output as a function of Trigger concentration between 10 nM and 25 nM in Figure 3d). This thresholding can potentially be explained by the presence of a small fraction of imperfectly formed, chain-terminating hairpins in the heterogeneous populations of H3 and/or H4. These ‘chain terminators’ are preferentially incorporated into the growing HCR chain, thus causing so-called ‘inhibitor ultra-sensitivity’12 (see Figure S10 for the detail of the proposed mechanism). This hypothesis is corroborated by our observation that the threshold is roughly proportional to the concentration of the hairpins used (data not shown). Interestingly, these insights provide another potential use for our methodology, the detection of defects in oligonucleotides that will be used for the construction of complex nanostructures. This is especially important as it is becoming increasingly clear that oligonucleotide purity is an issue in both circuit and structure assembly.13 This work, together with our earlier demonstrations,5a,7 strongly argues for the role of CHA circuits as a reliable and general-purpose signal amplifier, and suggests novel applications for assay and quality control of nanostructure assembly.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institute of Health (R01 AI092839-01, R01 GM094933-01), National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169), and Welch Foundation (F-1654). X.C. is partially supported by a postdoctoral trainee fellowship from Cancer Prevention Research Institute of Texas (CPRIT).
Footnotes
Supporting Information Available
Materials and methods. Effects of temperature and toehold length. Figure S1 to S10. Oligonucleotide sequence. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stojanovic MN, Landry DW. J. Am. Chem. Soc. 2002;124:9678–9679. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]; (c) Cho EJ, Yang LT, Levy M, Ellington AD. J. Am. Chem. Soc. 2005;127:2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]; (d) Lubin AA, Plaxco KW. Accounts Chem. Res. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Stojanovic MN, Stefanovic D. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]; (f) Zhang L, Zhu J, Li T, Wang E. Anal. Chem. 2011;83:8871–8876. doi: 10.1021/ac2006763. [DOI] [PubMed] [Google Scholar]
- 2.(a) Seeman NC. Nano. Lett. 2010;10:1971–1978. doi: 10.1021/nl101262u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wilner OI, Willner I. Chem. Rev. 2012;112:2528–2556. doi: 10.1021/cr200104q. [DOI] [PubMed] [Google Scholar]; (c) Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. Chem. Soc. Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mao CD, LaBean TH, Reif JH, Seeman NC. Nature. 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 3.(a) Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Qian L, Winfree E, Bruck J. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]; (c) Zhang DY. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]; (d) Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]; (e) Liu QH, Wang LM, Frutos AG, Condon AE, Corn RM, Smith LM. Nature. 2000;403:175–179. doi: 10.1038/35003155. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC, Lukeman PS. Rep. Prog. Phys. 2005;68:237–270. doi: 10.1088/0034-4885/68/1/R05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Li B, Ellington AD, Chen X. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin C, Rinker S, Wang X, Liu Y, Seeman NC, Yan H. Proc. Natl. Acad. Sci. USA. 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X. J. Am. Chem. Soc. 2012;134:263–271. doi: 10.1021/ja206690a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Zhang DY, Winfree E. J. Am. Chem Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]; (b) Soloveichik D, Seelig G, Winfree E. Proc. Natl. Acad. Sci. USA. 2010;107:5393–5398. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genot AJ, Zhang DY, Bath J, Turberfield AJ. J. Am. Chem. Soc. 2011;133:2177–2182. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- 10.(a) Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Pathol. Res. Pract. 2009;205:858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]; (b) Pozzi G, Meloni M, Iona E, Orru G, Thoresen OF, Ricci ML, Oggioni MR, Fattorini L, Orefici G. J. Clin. Microbiol. 1999;37:1197–1199. doi: 10.1128/jcm.37.4.1197-1199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Niu S, Jiang Y, Zhang S. Chem. Commun. 2010;46:3089–3091. doi: 10.1039/c000166j. [DOI] [PubMed] [Google Scholar]; (b) Shimron S, Wang F, Orbach R, Willner I. Anal. Chem. 2011;84:1042–1048. doi: 10.1021/ac202643y. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell JE. Trends Biochem. Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 13.(a) Bois JS, Venkataraman S, Choi HM, Spakowitz AJ, Wang ZG, Pierce NA. Nucleic Acids Res. 2005;33:4090–4095. doi: 10.1093/nar/gki721. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang DY, Winfree E. Nucleic Acids Res. 2010;38:4182–4197. doi: 10.1093/nar/gkq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.