Summary
Ever since Hershey and Chase used phages to establish DNA as the carrier of genetic information in 1952, the precise mechanisms of phage DNA translocation have been a mystery [1]. Although bulk measurements have set a time-scale for in vivo DNA translocation during bacteriophage infection, measurements of DNA ejection by single bacteriophages have only been made in vitro. Here, we present direct visualization of single bacteriophages infecting individual Escherichia coli cells. For bacteriophage λ, we establish a mean ejection time of roughly 5 min with significant cell-to-cell variability, including pausing events. In contrast, corresponding in vitro single-molecule ejections are more uniform and finish within 10 s. Our data reveal that when plotted against the amount of DNA ejected, the velocity of ejection for two different genome lengths collapses onto a single curve. This suggests that in vivo ejections are controlled by the amount of DNA ejected. In contrast, in vitro DNA ejections are governed by the amount of DNA left inside the capsid. This analysis provides evidence against a purely intrastrand repulsion-based mechanism and suggests that cell-internal processes dominate. This provides a picture of the early stages of phage infection and sheds light on the problem of polymer translocation.
Results
A schematic of our experimental design is shown in Figure 1A. To visualize DNA translocation, we use a cyanine dye to stain the viral DNA while it still remains in the capsid [2]. Phages are first incubated in the appropriate DNA stain before dialyzing away excess dye. The stained phages are then briefly bound to bacterial cells, which are pipetted into a flow chamber. The sample is then washed with buffer and then imaged with time-lapse bright-field and fluorescence microscopy. The signature of an ejection event is a loss of fluorescence in the virus and a concomitant increase in the fluorescence within the bacterial cell (Figure 1B). We were encouraged that such an experiment was possible from previous studies that have shown that dye-stained phages can remain intact and can infect cells [2, 3]. Cyanine dyes have also been used to study the kinetics of viruses in living eukaryotic cells, demonstrating that some dyes have limited cytotoxicity [4]. A screen was performed and identified SYTOX Orange as a DNA stain with appropriate specificity/sensitivity (see Figure S1 available online) and physiological parameters (Figures S2–S4).
Figure 1.
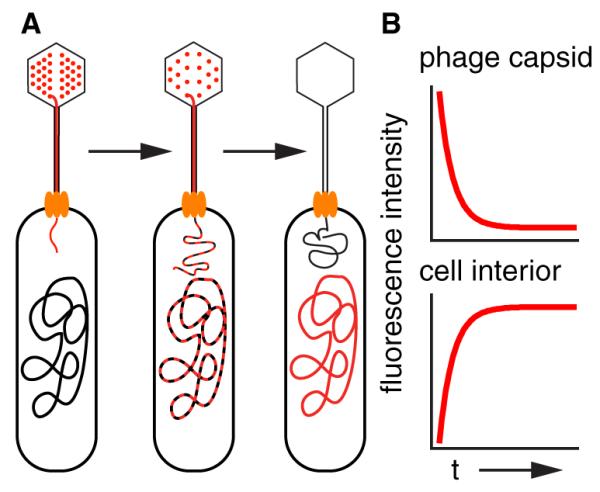
A Schematic for Monitoring DNA Translocation with Pre-Ejection Labeling
(A) The DNA is stained while still in the capsid. During ejection, the phage DNA carries its complement of cyanine dye with it, transferring fluorescence intensity from the virus to the cellular interior. Eventually, the dye falls off the phage DNA and rebinds to the bacterium’s genome.
(B) The timing of ejection is determined by measuring the loss of fluorescence intensity from the capsid; the concomitant increase in intensity in the cellular interior serves to verify that phage DNA has entered the cell. See also Figures S1–S3.
Single-Cell DNA Ejection Trajectories
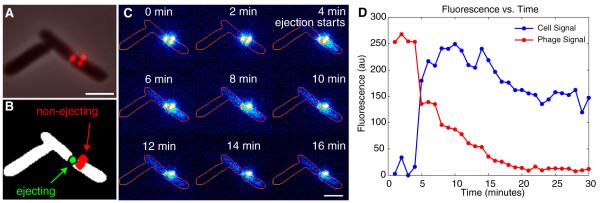
A typical in vivo ejection event for phage λcI60 (48.5 kbp) is shown in Figure 2. The attachment of the viruses to the host is revealed by diffraction-limited spots on the cell surface (Figure 2A). We identified pixels associated with either the virus or the cell (Figure 2B) and queried the fluorescence intensity as a function of time. As shown in Figure 2C, the ejection process is characterized by a loss of fluorescence intensity in the phage and a concomitant increase in fluorescence in the cellular interior (Figure 2D; representative Movies S1 and S2). The fluorescence inside the cell is diffuse: this reflects the dye molecules unbinding kinetics from the phage DNA (residence time ~1 s [5]) and redistributing themselves along the host genome, as verified in the Supplemental Experimental Procedures (Figures S3B and S3C). In the particular trajectory shown, the increase in cellular fluorescence is roughly equal to the decrease in phage fluorescence. The decrease in signal at the end of the trajectories in Figure 2D can be accounted for by photobleaching (Figure S3C).
Figure 2.
Dynamics of DNA Ejection
(A) Viruses attached to the cell surface in this fluorescence image merged with its bright-field counterpart.
(B) Segmentation masks of the cell (white), the phage that ejects its DNA (green), and the phages that do not eject their DNA (red).
(C) Time sequence of the fluorescence in the cell is shown. The edge of the cell is outlined for reference.
(D) Fluorescence intensity as a function of time is presented. The intensity of the phage segmented region and the cell segmented region are each plotted separately. The fluorescence intensity inside the nonejecting phage mask is stable; this is shown in Figure S4A. Note that in this ejection there appear to be steps and pauses.
The scale bars in (A) and (C) are 2 mm.
Single-Phage In Vivo Ejections Are Two Orders of Magnitude Slower Than In Vitro and Display Pausing
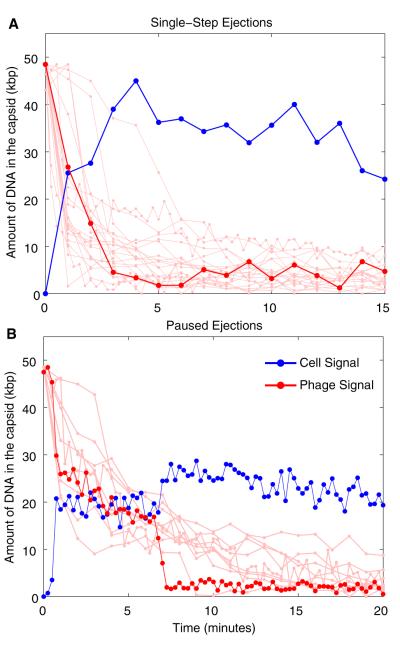
The results of a number of ejection events for λcI60 are shown in Figure 3. For the measurements shown here, the viralfluorescence decreases on a timescale of minutes, a factor of 10–100 times longer than the corresponding dynamics in vitro [6, 7]. Here, the ejection time is the time required for 80% of the fluorescence intensity to leave the viral capsid. The mean and SD for the ejection time for λcI60 was 5.2 ± 4.2 min (n = 45). In addition to the variability in ejection time, a number of ejections demonstrated pausing events, which we define as a nondecreasing fluorescence level greater than 2 min (our time resolution was typically 1 min); the mean pause time for λcI60 was 5.4 ± 4.1 min (n = 14). Both singlestep and paused ejections are shown in Figures 3A and 3B, respectively. A sample paused ejection is shown in Movie S3.
Figure 3.
Ejection Trajectories from Single-Cell Infections for λcI60
The red trajectories show the time history of the DNA intensity within the virus, and the blue trajectories show the concomitant increase in the fluorescence in the cellular interior. The solid-red color highlights a characteristic ejection, and the lighter-red color displays other ejection events for reference. The conversion between arbitrary units and kilobase pairs (kbp) was done by first subtracting each trace’s minimum observed fluorescence from itself. Each trace was then normalized by the maximum drop in the phage fluorescence and then multiplied by the genome length, which is 48.5 kbp for λcI60. Only two representative traces for the intensity within the cell are shown, with the remaining trajectories available online. Multiple lysate preparations from a single stock of CsCl-purified phages were used with similar results.
(A) Trajectories displaying a rapid and continuous ejection are illustrated.
(B) Trajectories that exhibit pausing events are demonstrated.
We next asked whether a reduction in driving force would produce significant differences in DNA translocation rates, an idea already used in our earlier in vitro measurements [7, 8]. Previously, the ejection of phage λcI60 (48.5 kbp) was compared to the ejection of phage strain λb221 (37.7 kbp). Through bulk and single-molecule in vitro experiments, it was shown that the amount of DNA inside the viral capsid was a control parameter for in vitro DNA ejection [7, 8]. Once λcI60 has ejected 10.8 kbp of DNA, the ejection forces and dynamics are equivalent to that of λb221. To explore the effect of genome length changes on DNA translocation rates in vivo, we performed our in vivo ejection assay for phage λb221. The mean time for ejection was 2.58 ± 2.34 min (n = 18). One paused ejection was also observed, with a pause time of 5 min. For λb221, we also observed a number of ejections (n = 10) that did not finish during the course of the movie, which we term a “stall.” Stalled ejections were not observed for λcI60. One possibility is that stalling events are related to λb221’s shorter genome and the consequent loss of a potential binding site that assists in entry. Stalled ejections were not included in the averages given earlier. The full set of trajectories for λcI60 and λb221 are online at http://www.rpgroup.caltech.edu/publications.
An Ensemble View of In Vivo Ejection Shows that the Amount of DNA in the Viral Capsid Is Not the Governing Control Parameter
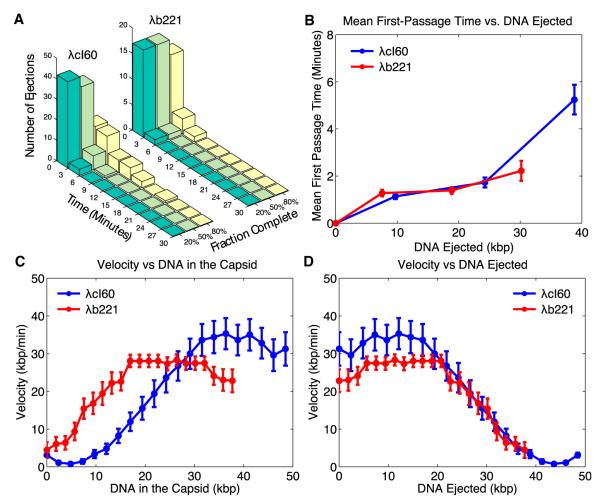
Measurements on both the wild-type and shortened genomes provide an opportunity to quantitatively examine the DNA translocation kinetics. One quantity of interest is the first-passage time for ejection: for each trajectory, we extracted the first-passage time for 20%, 50%, and 80% of completion, as determined by the decrease in the starting phage fluorescence. The first-passage time distributions for λcI60 and λb221 are shown in Figure 4A. By taking the mean of this distribution, we obtain the mean first-passage time (Figure 4B). We note that the mean first-passage time is a quantity that is amenable to theoretical calculations [9]. Another way to view the mean first-passage time is as an “average” ejection trajectory. When viewed in this way, one interpretation of Figure 4B is that, within the error of the measurement, the “average” trajectories for λcI60 and λb221 have considerable overlap. For both λcI60 and λb221, the velocities plateau after ~50% of the genome length (Figures 4C and 4D).
Figure 4.
An Ensemble View of Ejection Times and Dynamics for Phage λ
(A) Distributions of first-passage times for different fractions of completion of ejection are shown. The histograms are determined by setting ejection thresholds of 20%, 50%, and 80% complete as measured using the fluorescence. The distributions for both λcI60 (48.5 kbp) and λb221 (37.7 kbp) are shown.
(B) Mean first-passage times for λcI60 and λb221 are presented. The means of the distributions shown in (A) were used to calculate the mean first-passage time. There is little difference in the mean first-passage times of absolute amounts of DNA ejected between λcI60 and λb221.
(C) Velocity of ejecting DNA plotted as a function of the amount of DNA remaining in the capsid is illustrated. The initial portion of the ejection process is faster for λcI60 than λb221. There is no significant overlap between the two curves.
(D) Velocity of ejecting DNA plotted as a function of the amount of DNA ejected is demonstrated. There is a small difference between the two curves for the first 20 kbp and significant overlap after that.
Only trajectories without pauses were used to generate (C) and (D). Error bars for (B)–(D) represent SE (n = 45 for λcI60; n = 18 for λb221).
We also plotted the mean velocity at different amounts of DNA remaining in the capsid during an ejection (Figure 4C). There is little overlap between the two curves, and for lower amounts of DNA remaining in the capsid, the velocity for λb221 is higher than λcI60. This is to be contrasted with in vitro measurements where there is significant overlap between the two curves, with the “data collapse” in that case signifying that the dynamics are equivalent after λcI60 has ejected its first 10.8 kbp [7]. An alternative way to visualize these data is to plot the mean velocity versus the amount of DNA ejected into the cell (Figure 4D). When plotted in this fashion, there is considerable overlap between the two curves, with a small difference observed for the first 20 kbp of ejection. This analysis is consistent with the mean first-passage time analysis, which showed considerable overlap when the first-passage time was also plotted against the amount of DNA ejected.
Discussion
For bacterial viruses, genome delivery is at the heart of the viral life cycle, yet this critical process remains enigmatic as does the in vivo process of polymer translocation more generally. Beyond a purely intellectual understanding of this process, phage-mediated transfer of nucleic acids has medical and evolutionary implications [10–13]. Our objective was to design and perform an experiment with sufficient temporal resolution that would permit us to measure single-molecule DNA transfer in real time; we accomplished this for both WT (48.5 kbp) and mutant (37.7 kbp) λ phage using SYTOX Orange and fluorescence microscopy. These experiments reveal that the DNA translocation process is subject to strong cell-to-cell variability (1–20 min). A number of ejections also exhibited pauses and stalls. Our single-molecule measurements are consistent with earlier estimates of a minute timescale for in vivo genome delivery of phage λ from bulk experiments [14, 15].
A number of different hypotheses have been formulated for the actual translocation mechanism for phage λ. In addition to the driving force due to the packaged DNA, these models propose that thermal fluctuations, hydrodynamic drag, and active molecular motors might each play a role in bringing the viral DNA into the bacterial cell [9, 15–18]. Our results provide both surprises and useful insights that constrain the space of possible models and will guide future modeling efforts. One key result is that the length of DNA remaining inside the capsid is not the sole control parameter that governs the ejection dynamics, as it is in vitro. In the in vitro experiments, the approximate collapse of the data from the different genome lengths on a single curve revealed that the DNA within the capsid is driving the kinetics of ejection [7, 8]. By way of contrast, in the in vivo ejection experiments reported here, an approximate data collapse is only revealed when the velocity is plotted with respect to how much DNA is out of the capsid and in the cell rather than how much DNA remains within the capsid. Data collapse has been previously used to identify control parameters for in vitro DNA ejection as well as the lysis-lysogeny decision [7, 8, 19].
No collapse is seen when the velocity is plotted against the amount of DNA remaining inside the capsid. This has significant implications for the role energy stored in the compacted DNA plays during the in vivo ejection. If some significant portion of the ejection process were governed solely by the energy in the compacted DNA, then during that portion we would expect the dynamics of λcI60 and λb221 to be identical when the amount of DNA remaining in the capsid is identical. This is the in vitro case as studied in [7, 8]. Because the DNA-DNA repulsion inside the capsid is highest when the capsid contains more DNA, such a period would likely be at the beginning of the ejection process. As seen in Figure 4C, however, there exists no period of overlap between the velocity curves for the two phage strains and, hence, no period during the ejection process where the length of DNA in the capsid and, hence, intrastrand repulsion, is the sole control parameter. Two-step models in which the first half of the genome is delivered by the energy stored in the compacted DNA and the remainder is delivered by another mechanism are also not consistent with our data.
Another consequence of the data collapse (Figure 4D) is the possibility that the amount of DNA ejected (as opposed to the amount of DNA in the capsid) is a key control parameter for this system. This picture is consistent with models in which the mechanism is internal to the cell because the only information such a mechanism would utilize is the amount of DNA that has been brought inside the cell. One limitation to applying this argument is that only two genome lengths have been tested here. Such reasoning also does not exclude a mixed picture, as mentioned above.
The origin of the apparent pauses might provide information about the ejection mechanism, because DNA-based motors acting against a load have been observed to pause [20, 21]. However, the pauses observed here are much longer than the pauses observed for motors, and it is possible that they could simply be a reflection of the cell-to-cell variability in turgor pressure. Postpause resuming of DNA entry could thus indicate a secondary mechanism in conjunction with pressure, for high turgor cells. Another possibility is that the pauses observed here might also be related to mechanisms proposed for pauses observed in vitro for phage T5 [22, 23]. However, this is unlikely because pauses are not observed for phage λ in vitro [6, 7].
Our results are contrary to what was shown in T7, in which a constant DNA ejection rate was seen with bulk measurements [15]. T7 has a capsid similar in size to λ (60 versus 58 nm, respectively) with a 40 kbp genome; however, its tail is considerably shorter (23 versus 150 nm, respectively) [24, 25]. It has been suggested that a constant velocity is suggestive of a purely enzyme-driven model such as a molecular motor [15]. Such a feature is not seen in our data because Figure 4D shows that once ~20 kbp of DNA has been ejected, there is a marked decrease in the ejection velocity. However, the non-linear force-velocity relationship seen in vitro and the presence of pN level forces from the DNA-DNA repulsion inside the capsid make it unclear whether a constant ejection rate prediction would be true for λ.
The current data do not match previous calculations of ejection dynamics for mechanisms based on DNA binding proteins and thermal fluctuations [9]. Those calculations predict that after the first 10.8 kbp of DNA from λcI60 has been ejected, it should have the same dynamics as λb221, which is inconsistent with our data. Also perplexing is the timescale of ejection. The origin of the friction that sets the timescale for ejection is poorly understood, both in vitro and in vivo. A number of models assume a linear relationship between force and velocity, but it is now known that this assumption is not true in vitro [7, 9, 26]; we suspect that it is not true in vivo either.
In summary, we have examined the DNA ejection process for bacteriophage λ in vivo at the single-molecule level. We note that the techniques explored in this work may be generalizable to the study of other bacteriophages. It would be especially interesting to see a comparison between the bulk and single-molecule dynamics for bacteriophage T7 because bulk experiments have shown that the speed is constant throughout the ejection process in vivo [15, 24], as opposed to the variable rate reported here. We also note that the experimental platform presented here can be used to explore the effects of various genetic, chemical, and mechanical perturbations on the ejection process.
Experimental Procedures
Real-Time Imaging of DNA Ejection In Vivo
Glass coverslips were cleaned by sonication for 30 min in 1 M KOH followed by sonication in 100% ethanol with copious rinsing with purified water in between, and then dried on a hot plate. The coverslips were then briefly (5 s) immersed in a fresh solution of 1% polyethyleneimine, transferred into purified water, and finally dried with a stream of air. A microscope slide, double-sided tape, and the treated coverslip were then assembled into a flow chamber.
E. coli strain LE392 was grown up overnight in LB media at 37°C (see Supplemental Experimental Procedures for all buffer formulations). The saturated culture was then diluted 1:100 in M9 maltose-sup and grown for 3 hr at 37°C until the culture reached an optical density 600 of ~0.3. Plate lysate of the desired phage strain (see Supplemental Experimental Procedures) was centrifuged for 5 min at 13,000 × g to remove bacterial debris. The supernatant was recovered and then stained with SYTOX Orange at a final concentration of 500 nM for 3 hr at room temperature. Prior to binding stained phages to E. coli, free dye was removed by diluting 100 μl of the phage suspension and then centrifuging the sample across a 100 kDa (EMD Millipore,UFC910008) filter four times. Each round of centrifugation led to a 40-fold dilution of dye, reducing the final free concentration of dye to less than 200 pM. After the final round, the phages were brought up to the original volume of 100 μl with M9sup. Phages were then bound to cells by mixing ~50 μl of cells with phage at a multiplicity of infection (moi) of approximately seven for 1 min at room temperature. The cells were then flowed into the flow chamber and allowed to adhere to the surface for 2 min at room temperature. Occasionally, phages were bound to cells by mixing 10 μl of cells with phage at a moi of approximately one to five and incubating on ice for 30 min. Incubation of the cells in the flow chamber took place on ice as well. The initiation of phage ejection is slowed down considerably within this time period in either of these two conditions [19, 27, 28].
After the incubation, the flow chamber was washed with 200 μl of M9sup with 1% GODCAT mixture, 1% β-mercaptoethanol, and 0.5% glucose. The chamber was then sealed with valap and imaged on a Nikon Ti-E Perfect Focus microscope using a mercury lamp and TRITC filter (Semrock, LF561) set at 37°C. Snapshots of both the phase and fluorescence channels were taken either one or four times a minute, with a fluorescence excitation time of 500 or 300 ms, respectively. Images were collected using a Hamamatsu C8484 camera, a Photometrics CoolSNAP ES2 camera, or an Andor iXON EMCCD camera. We observed better conservation of fluorescence between the phage and the cell with the Hamamatsu and Photometrics cameras as opposed to the Andor camera. The electron multiplier gain of the Andor camera allowed for shorter exposure times and higher time resolution.
Supplementary Material
Acknowledgments
We are grateful to a number of people for help with experiments, advice, and critical commentary on the manuscript, including Heun Jin Lee, Maja Bialecka, Phillips laboratory, Talia Weiss, Vilawain Fernandes, Kari Barlan, Paul Grayson, Ido Golding, Lanying Zeng, Bill Gelbart, Chuck Knobler, Francois St. Pierre, and Drew Endy. We are also grateful to Ron Vale, Tim Mitchison, Dyche Mullins, and Clare Waterman as well as several generations of students from the MBL Physiology Course where this work has been developed over several summers. We also gratefully acknowledge financial support from several sources, including a National Institutes of Health (NIH) Medical Scientist Training Program Fellowship, a Yaser AbuMostafa Hertz Fellowship, and a NIH Director’s Pioneer Award. We also acknowledge the support of National Science Foundation grant number 0758343.
Footnotes
Supplemental Information Supplemental Information includes four figures, two tables, Supplemental Experimental Procedures, and three movies and can be found with this article online at doi:10.1016/j.cub.2012.05.023.
References
- 1.Hershey AD, Chase MM. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eriksson M, Härdelin M, Larsson A, Bergenholtz J, Akerman B. Binding of intercalating and groove-binding cyanine dyes to bacteriophage t5. J. Phys. Chem. B. 2007;111:1139–1148. doi: 10.1021/jp064322m. [DOI] [PubMed] [Google Scholar]
- 3.Mosier-Boss PA, Lieberman SH, Andrews JM, Rohwer FL, Wegley LE, Breitbart M. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl. Spectrosc. 2003;57:1138–1144. doi: 10.1366/00037020360696008. [DOI] [PubMed] [Google Scholar]
- 4.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan X, Habbersett RC, Yoshida TM, Nolan JP, Jett JH, Marrone BL. Probing the kinetics of SYTOX Orange stain binding to double-stranded DNA with implications for DNA analysis. Anal. Chem. 2005;77:3554–3562. doi: 10.1021/ac050306u. [DOI] [PubMed] [Google Scholar]
- 6.Wu D, Van Valen D, Hu Q, Phillips R. Ion-dependent dynamics of DNA ejections for bacteriophage lambda. Biophys. J. 2010;99:1101–1109. doi: 10.1016/j.bpj.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grayson P, Han L, Winther T, Phillips R. Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl. Acad. Sci. USA. 2007;104:14652–14657. doi: 10.1073/pnas.0703274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. The effect of genome length on ejection forces in bacteriophage lambda. Virology. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inamdar MM, Gelbart WM, Phillips R. Dynamics of DNA ejection from bacteriophage. Biophys. J. 2006;91:411–420. doi: 10.1529/biophysj.105.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 11.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467:982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One. 2011;6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 14.García LR, Molineux IJ. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 1995;177:4066–4076. doi: 10.1128/jb.177.14.4066-4076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 16.Grayson P, Molineux IJ. Is phage DNA ‘injected’ into cells—biologists and physicists can agree. Curr. Opin. Microbiol. 2007;10:401–409. doi: 10.1016/j.mib.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zandi R, Reguera D, Rudnick J, Gelbart WM. What drives the translocation of stiff chains? Proc. Natl. Acad. Sci. USA. 2003;100:8649–8653. doi: 10.1073/pnas.1533334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng L, Skinner SO, Zong C, Sippy J, Feiss M, Golding I. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 21.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc. Natl. Acad. Sci. USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangenot S, Hochrein M, Rädler J, Letellier L. Real-time imaging of DNA ejection from single phage particles. Curr. Biol. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 23.Chiaruttini N, de Frutos M, Augarde E, Boulanger P, Letellier L, Viasnoff V. Is the in vitro ejection of bacteriophage DNA quasistatic? A bulk to single virus study. Biophys. J. 2010;99:447–455. doi: 10.1016/j.bpj.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 25.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 26.Gopinathan A, Kim YW. Polymer translocation in crowded environments. Phys. Rev. Lett. 2007;99:228106. doi: 10.1103/PhysRevLett.99.228106. [DOI] [PubMed] [Google Scholar]
- 27.Mackay DJ, Bode VC. Events in lambda injection between phage adsorption and DNA entry. Virology. 1976;72:154–166. doi: 10.1016/0042-6822(76)90320-2. [DOI] [PubMed] [Google Scholar]
- 28.Moldovan R, Chapman-McQuiston E, Wu XL. On kinetics of phage adsorption. Biophys. J. 2007;93:303–315. doi: 10.1529/biophysj.106.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.