Abstract
BACKGROUND
Examination of spontaneously occurring phasic muscle activity from the human polysomnogram may have considerable clinical importance for patient care, yet most attempts to quantify the detection of such activity have relied upon laborious and intensive visual analyses. We describe in this study innovative signal processing approaches to this issue.
METHODS
We examined multiple features of surface electromyographic signals based on 16,200 individual 1-second intervals of low impedance sleep recordings. We validated which of those features most closely mirrored the careful judgments of trained human observers in making discriminations of the presence of short-lived (100-500 msec) phasic activity, and also examined which features provided maximal differences across 1-second intervals and which features were least susceptible to residual levels of amplifier noise.
RESULTS
Our data suggested particularly promising and novel features (e.g., Non-linear energy, 95th percentile of Spectral Edge Frequency) for developing automated systems for quantifying muscle activity during human sleep.
CONCLUSIONS
The EMG signals recorded from surface electrodes during sleep can be processed with techniques that reflect the visually based analyses of the human scorer but also offer potential for discerning far more subtle effects, Future studies will explore both the clinical utility of these techniques and their relative susceptibility to and/or independence from signal artifacts.
Keywords: Electromyography, Sleep, Muscle Activity, Phasic Activity, Polysomnography, Computer Detection
1.1 INTRODUCTION
Polysomnography (PSG), the recording of multiple physiologic signals during sleep, generates huge amounts (typically 500 megabytes or more for an overnight sleep period) of electrophysiologic data that are typically acquired digitally but remain analyzed visually by human expert scorers [1]. Of PSG parameters typically recorded, the electroencephalogram (EEG), by far, has been the most extensively examined using advanced signal processing techniques for data reduction, including approaches such as spectral analysis using the Fast Fourier Transform (FFT) [2], period amplitude analysis [3], and non-linear dynamics employing estimates of dimensional complexity [4]. Although such approaches have stimulated many exciting studies of the functional significance of the sleep EEG [5, 6], other components of the human PSG has received far less attention from the signal processing perspective. Some work, for example, has attempted to automate identification of the eye movements of rapid eye movement sleep (REM) [7], whereas other studies have attempted to quantify muscle activity from surface electromyography (EMG) during sleep [8-11]. In recent years, greater attention has focused on the analysis of EMG signals derived from sleep, because of known aberrations of such activity during REM as a component of some neurodegenerative conditions [12-16]. We describe here the initial derivation of an unsupervised, computer-based approach for quantifying surface EMG during sleep that is both compatible with discriminations provided by the trained human scorer, as well as potentially extending such quantification to incorporate unique signal processing features.
2.0 METHODS
2.1. Overall Approach
To establish a viable digital approach of the human PSG for the quantification of EMG activity, we initially relied upon judgments of trained visual scorers. Five different individuals (A,B,C,D,E), each representing different levels of experience, independently evaluated surface EMG signals from a human PSG recording, as recorded with a low impedance (< 10,000 ohms), bipolar derivation placed directly over a limb muscle group frequently associated with discharge in sleep (anterior tibialis). Our general strategy was to use a consensus of human scorers’ determinations of the presence or absence of phasic activity for each 1-second interval. Although EMG signals during sleep are frequently recorded from surface sensors located above other muscle groups as well (e.g., mentalis), such locations can be problematic for introducing certain kinds of artifacts (e.g., snoring) and would represent a more complex issue for human scorer discrimination, at least at this early stage of developing computer-based learning approaches. Scorers varied in experience level from an individual with over 30 years of experience in examining PSG to a lab assistant with very modest (< 6 months) experience. Our rationale for inclusion of such a wide array of scorer expertise was to enhance generalizeability of any algorithm that could result from our analyses.
Sleep (PSG) data were collected on an Embla (MedCare, Bloomfield, CO) Model N7000 data acquisition unit using the sleep data collection software program Somnologica ® 2.0. PSG data were converted from Somnologica .edf format, for EMG feature processing, via the computer software program MATLAB (version 7.8 R2009a) using the biosig toolbox version 2.54 (Schloegl A-Graz University of Technology, Graz, Austria) and were digitized at a sampling rate of 200 Hz.
Visual analyses occurred on 17″ monitors. Each of the 5 scorers was asked to evaluate a total of 16,200 seconds (4.5 hours) of bilateral EMG signals distributed approximately equally from REM and non-REM sleep, marking each second (via drag-and-click function) for the presence or absence of a phasic muscle discharge of 4 times the surrounding background activity and lasting between 100 and 500 msec. This definition resembles what we have used previously for visual analyses of the phasic EMG during sleep [17]. Screen display was held constant at 10 seconds per viewing window. Because we relied on a 1-second epoch to define presence or absence of phasic activity, scorers were instructed to score an event as present or absent for the 1-second interval, rather than attempt to quantify the number of phasic events within the given interval (see Figure 1), thus allowing for repetitive returns to baseline during that interval. Each scorer was provided an unmarked recording and had no access to the judgments made by any of the other visual scorers during the entire process.
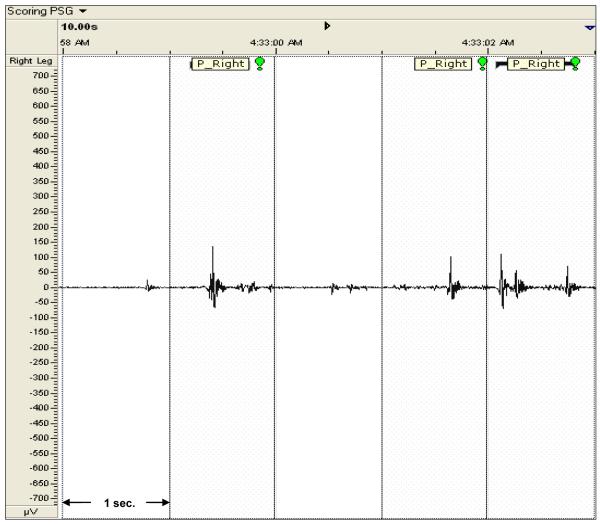
Figure 1.
Somnologica ® 2.0 RemLogic plot of a 5-Second segment of Right Leg EMG data with phasic activity labeling from Scorer B. Phasic activity is demarcated with shaded dotted boxes and “P_RIGHT” labels.
2.2. Computer-based Feature Extraction from EMG Recordings
Feature extraction was used to translate EMG signal information into a compact quantitative format and characterize signal information for the identification of seconds with and without phasic muscle events. Exhaustive review of prior EMG sleep signal analysis, bio-signal feature extraction techniques, and statistical analysis methods were used to compile a library of 15 features, listed below. Formulae associated with the calculations are numbered and shown in bolded parentheses. The final data matrix was obtained by evaluating feature values for each data channel in one second epochs using eight equal non-overlapping moving windows.
1. Relative EMG Frequency Power (Prel): a frequency domain feature that provides a sub-band analysis of the high frequency EMG signal components (frequency band [12.5;32 Hz ]) [18] (sampled at 200Hz) with the power spectra (P(f)) extracted using the Fast Fourier Transform (FFT) [19, 20]
| (1) |
2. Spectral Edge Frequency 95th percentile (SEF 95): the frequency up to which 95 percent of the total signal power is accumulated [21].
| (2) |
where fs is the sampling frequency
3. Skewness (Skew): a time domain feature that measures the asymmetry of the probability distribution of the EMG signal amplitude [22].
| (3) |
with M representing the number of data samples of the EMG signal in an 1-second epoch interval and symbolizing the sample mean within that interval,
4. Variance (S2): the distance, spread, between a data sample with respect to the mean [22].
| (4) |
5. Kurtosis (Kurt): a measure of the peakedness or flatness of the probability distribution of the signal amplitude [23].
| (5) |
6. Entropy (Ent): an information domain feature that calculates the amount of uncertainty or unpredictability of the EMG signal amplitude.
| (6) |
with M symbolizing the length of the data signal, n representing the number of bins, the optimal bins being obtained from the Freedman-Diaconis rule [24], to estimate the histogram of the data signal with binj indicating the number of data samples from x(i) in the jth histogram bin [25].
7. Mobility (Mobi): a time domain feature that measures the relative average slope of the EMG signal. It is expressed as the standard deviation (std) of the slope (signal’s first derivative dx/dt) with reference to the standard deviation of the signal amplitude [25].
| (7) |
where for the discrete EMG signal x(i), and , using a first order approximation of the derivative.
8. 75th Amplitude Percentile (75_Amp): the amplitude value below which 75% of the total EMG signal amplitude resides [25]. So, the value separates lowest 75% and highest 25% of the data. It is also called upper quartile or third quartile.
| (8) |
where M is the number of samples x(i) of the EMG signal in one epoch and card represents the number of elements within the sample set (set’s cardinality).
9. Complexity (Comp): the ratio of the mobility (Mobi) of the first derivate of the signal to the mobility of the signal amplitude. Complexity expresses the average EMG wave-shape in relation to a pure sine wave [25].
| (9) |
10. Mean Absolute Amplitude (MAA): a time domain feature that measures the absolute value of the mean EMG amplitude [26].
| (10) |
11. Curve Length (L): the sum of the value of the first order differences of the EMG signal amplitude values [27].
| (11) |
12. Mean Energy (MnE): a time domain feature that measures the squared EMG signal amplitude [27].
| (12) |
13. Zero Crossings (ZC): defined as the number of crossings of the EMG signal over the ordinate, where the axis equals zero [27].
| (13) |
14. Average Nonlinear Energy (NE): a non-linear feature that is sensitive to signal fluctuations in the time and frequency domain, with respect to the following non-linear operator (NLO):
the NLO is weighted with a Hanning window and then the mean/Average Nonlinear Energy (NE) is calculated as follows:
| (14) |
where, NLOw is the Hamming windowed version of the non-liner operator, NLO, with N being the data epoch [27].
15. Spectral Entropy (SE): defined as the amount of uncertainty or unpredictability of the EMG signal in the frequency domain [27].
| (15) |
2.3 Performance Models Comparing Features
We evaluated the relative performance of each feature using three analytic approaches. First, we examined the relative accuracy afforded by each feature by examining percentage agreement for each feature by use of an unsupervised, probabilistic model of likelihood of class assignment in relation to human scorer consensus. Accuracy was defined as the sum of the percentage of correctly (i.e., consensually) identified seconds with phasic activity (true positive) plus the percentage of correctly (i.e., consensually) identified seconds without phasic activity (true negative). Consensus was defined as unanimous agreement among all 5 human scorers. These calculations were made separately for left and right leg channels to provide internal replication of the performance of various features.
Unsupervised classification of 1-second epochs with phasic versus non-phasic activity were conducted using Gaussian Mixture Modeling (GMM) [28]. GMMs are a member of the probabilistic model based clustering framework. GMMs assume that the data come from a multivariate finite mixture model of the form
where K is the number of components , πk are the mixing proportions (weights) and Nk (x;μk, ∑k) are multivariate normal distributions of the form
where n is the dimension of the feature vector x , μk is the mean vector and ∑k is the covariance matrix.
Estimates for the model parameters were obtained using the expectation maximization (EM) algorithm, a common approach in GMM parameter selection, which is an iterative algorithm used to determine the maximum likelihood (other criteria can be used but the maximum likelihood is the most common) of the data given the parametric model [29]. Once the parameters are determined each object is assigned to one the K clusters (components representing phasic or non-phasic activity) on the basis of estimated posterior probabilities of cluster membership. In other words the object x , feature value of each 1-second epoch, was assigned to cluster i if
A second approach to evaluating the relative utility among features was to compare parametric values across consensually agreed upon seconds containing phasic EMG activity. Because visual analyses were limited to a simple binary classification, we reasoned that the additional information incorporated by some features would result in higher coefficients of variation (COVs) for those features relative to other features. Higher values would be suggestive of potentially greater sensitivity to fluctuations in occurrence (either as density within the time domain and/or amplitude within the voltage domain) of muscle activity that would be difficult to detect visually. For example, Figure 1 demonstrates three distinct seconds as identified as demonstrating phasic activity, but the visual appearance of these recording segments varied widely. Ideally, any computer-derived feature would be sensitive to such variation across discrete 1-second intervals and demonstrate relatively larger, rather than smaller, COVs.
A final approach for comparison was to determine the relative success of each feature to deal with the low amplitude, high frequency, residual noise present within the N7000 recording system. Even within the context of low impedance recordings and the high performance amplification with a substantial common mode rejection ratio (CMRR) > 90dB at 60 Hz for the Embla N7000 system, some non-physiologic, low voltage (< 1 uV) signals are always detectable. The CMRR measures the tendency of the amplifier to reject input signals that are common to both EMG electrodes and a high CMRR (>90dB) is assumed to suppress signals common to both electrodes sites, such as from power sources, electromagnetic devices and muscles distant from the electrode radius [30]. Assuming the latter, we reasoned that, for seconds with consensus agreement on absence of phasic activity, the ratio of such so-called “residual noise” across left and right leg EMG channels would approach unity. Features that showed ratios considerably higher or lower than 1.0 would be considered to have greater susceptibility to spurious signals, regardless of their origin.
3.0 RESULTS
3.1 Visual Analyses
Approximately 10% of all seconds evaluated contained phasic muscle activity. The percentages of 1-second intervals detected for each of the 5 scorers were: 7.67, 7.90, 8.80, 9.44 and 12.70. The overall consensus (i.e., the proportion of 1-second intervals in which all 5 scorers agreed on the presence or absence of phasic muscle activity was very high (93.52%; Kappa = .818, p < .0001). Because we were able to establish high reliability among scorers with regards to the presence and absence of phasic activity, we limited further our machine-learning analysis to only those 1-second intervals with consensus agreement on presence (N = 1,090) or absence (N = 14,060) of EMG phasic activity.
3.2 Relative Classification Performance of Computer-derived Features from Unsupervised Learning Models
Tables 1 and 2 present results evaluating the performance of each of the 15 features for the unsupervised models derived from the EMG signals using the GMM algorithm. With few exceptions, most showed substantial differentiation between seconds with and without consensually agreed phasic muscle activity. Nine of the features demonstrated accuracy above 90% and of these, only Comp (complexity) showed substantial discrepancy between seconds identified accurately with phasic activity (true positives) versus seconds identified accurately as not containing phasic activity (true negatives).
Table 1.
Accuracy (% agreement) for Consensus Visual Scoring (left leg)
| Feature Name (Number)a | Accuracyb | TPc | TNd |
|---|---|---|---|
| Relative High Frequency Power (1) | 63.530 | 13.849 | 66.917 |
| Spectral Edge Frequency 95th Percentile (2) | 91.838 | 95.723 | 91.573 |
| Skewness (3) | 89.862 | 68.839 | 91.295 |
| Variance (4) | 90.265 | 97.352 | 89.782 |
| Kurtosis (5) | 88.316 | 83.707 | 88.629 |
| Entropy (6) | 45.646 | 50.917 | 45.287 |
| Mobility (7) | 89.537 | 45.214 | 92.559 |
| Amplitude 75th Percentile (8) | 96.829 | 91.853 | 97.168 |
| Complexity (9) | 91.786 | 20.774 | 96.626 |
| Mean Absolute Amplitude (10) | 92.228 | 96.945 | 91.906 |
| Curve Length (11) | 92.488 | 97.759 | 92.128 |
| Energy (12) | 90.265 | 97.352 | 89.782 |
| Zero Crossing (13) | 73.447 | 56.823 | 74.580 |
| Non-Linear Energy (14) | 92.995 | 97.556 | 92.684 |
| Spectral Entropy (15) | 94.957 | 86.354 | 95.544 |
Relevant formulae are shown in parentheses (see text)
Accuracy = % of correct phasic and non-phasic GMM predictions compared to consensus visual scoring
TP = true positives (% of seconds with phasic activity assessed visually that were correctly classified by the GMM for that feature)
TN = true negatives (% of seconds without phasic activity assessed visually that were correctly classified by the GMM for that feature).
Table 2.
Accuracy (% agreement) for Consensus Visual Scoring (right leg)
| Feature Name (Number)a | Accuracyb | TPc | TNd |
|---|---|---|---|
| Relative High Frequency Power (1) | 64.713 | 17.028 | 68.879 |
| Spectral Edge Frequency 95th Percentile (2) | 90.410 | 97.496 | 89.791 |
| Skewness (3) | 89.820 | 66.611 | 91.848 |
| Variance (4) | 88.747 | 98.164 | 87.925 |
| Kurtosis (5) | 86.347 | 84.641 | 86.496 |
| Entropy (6) | 59.134 | 96.350 | 13.419 |
| Mobility (7) | 89.311 | 33.222 | 94.210 |
| Amplitude 75th Percentile (8) | 95.789 | 94.825 | 95.873 |
| Complexity (9) | 90.585 | 20.868 | 96.675 |
| Mean Absolute Amplitude (10) | 90.370 | 97.997 | 89.704 |
| Curve Length (11) | 90.813 | 98.498 | 90.141 |
| Energy (12) | 88.774 | 98.164 | 87.954 |
| Zero Crossing (13) | 71.473 | 54.090 | 72.991 |
| Non-Linear Energy (14) | 92.073 | 97.997 | 91.556 |
| Spectral Entropy (15) | 97.425 | 84.641 | 98.542 |
Relevant formulae are shown in parentheses (see text)
Accuracy = % of correct phasic and non-phasic GMM predictions compared to consensus visual scoring
TP = true positives (% of seconds with phasic activity assessed visually that were correctly classified by the GMM for that feature)
TN = true negatives (% of seconds without phasic activity assessed visually that were correctly classified by the GMM for that feature).
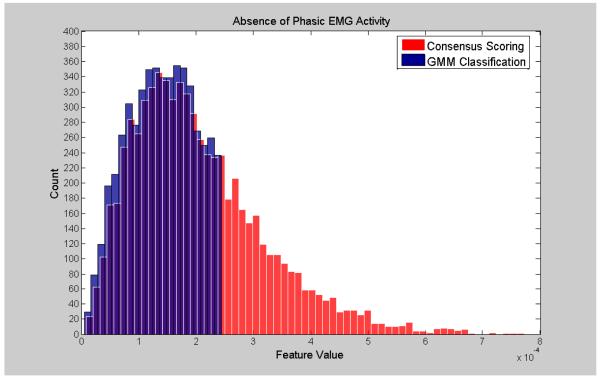
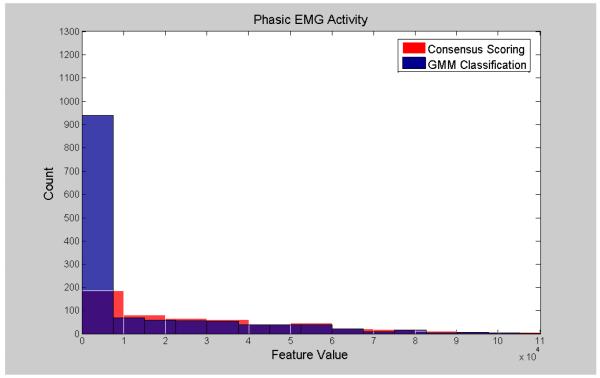
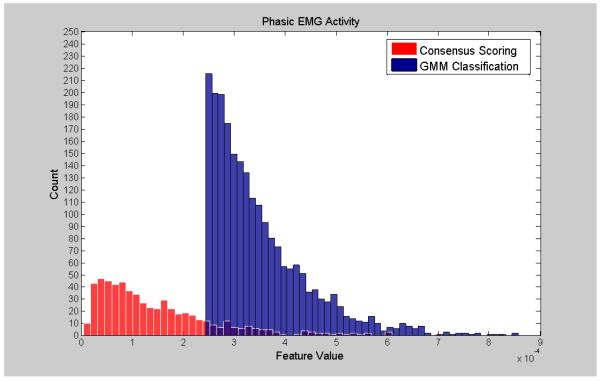
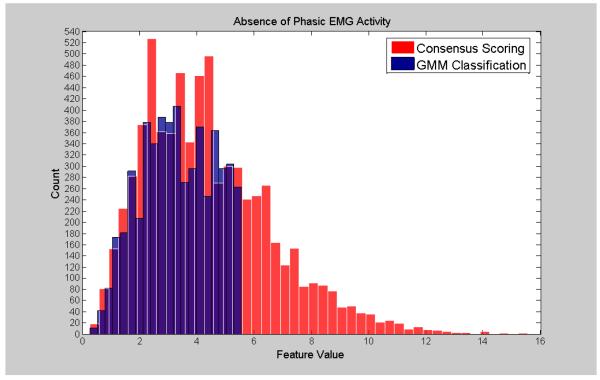
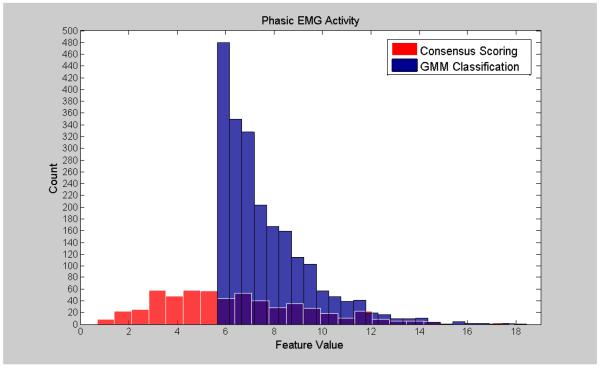
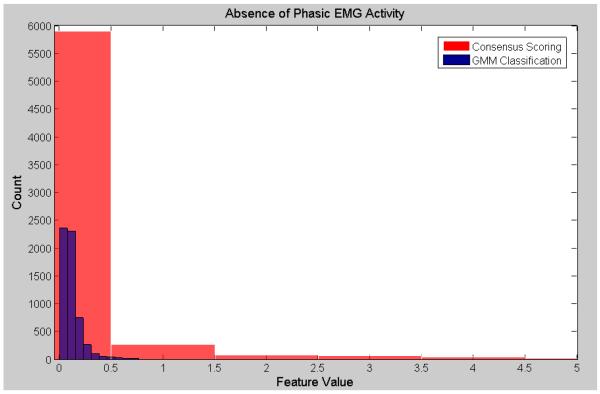
Visualization of the agreement/overlap between consensus scoring and GMM classifications were displayed using histogram plotting for three selected features (Figures 2-7; right leg data shown; results were similar using left leg). This information follows directly from Table 2 and illustrates the accuracy of the unsupervised GMM algorithm in classifying absence of phasic and phasic EMG activity with respect to consensus scoring. Histograms for a poorly performing feature, Relative High Frequency Power (formula 1), are displayed in Figures 2 and 3, with overlap values of 68.9% and 17.0% for absence of phasic and presence of phasic EMG activity, respectively. Results for a mid-level performing feature, Zero-Crossing (formula 13) are displayed in Figures 4 and 5 with overlap values of 73.0% and 54.1% for absence of phasic and presence of phasic EMG activity, respectively. Lastly, histograms for an optimally performing feature, Variance (formula 4), are displayed in Figures 6 and 7 with overlap values of 87.9% and 98.2% for absence of phasic and presence of phasic EMG activity, respectively.
Figure 2.
Histogram plots of the absence of phasic activity for consensus visual scoring and the GMM algorithm for a poorly performing feature: Relative High Frequency Power (1) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
Figure 7.
Histogram plots of the presence of phasic activity from consensus visual scoring and GMM for an optimally performing feature: Variance (4) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
Figure 3.
Histogram plots of the presence of phasic activity agreement labeling from consensus visual scoring and the GMM algorithm for a poorly performing feature: Relative High Frequency Power (1) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
Figure 4.
Histogram plots of the absence of phasic activity for consensus visual scoring and the GMM algorithm for a mid-level performing feature: Zero Crossing (13) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
Figure 5.
Histogram plots of the presence of phasic activity agreement labeling of the consensus visual scoring and GMM algorithm for a mid-level performing feature: Zero Crossing (13) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
Figure 6.
Histogram plots of the absence of phasic activity for consensus visual scoring and the GMM algorithm for an optimally performing feature: Variance (4) (Right Leg). Purple color indicates overlapping sections of the consensus scoring and GMM classification histograms.
3.3 Coefficients of Variation (COVs) across Features
Table 3 presents COVs established for left and right leg separately for various EMG features for consensually agreed upon seconds with phasic activity. Although some differences occurred for particular features in left and right leg recordings, features (2) (SEF, Spectral Edge), (4) (S2, Variance), (12) (MnE, Energy), (14) (NE, non-linear energy) and (15) (SE, Spectral Entropy) showed consistently higher COVs.
Table 3.
Coefficients of Variation (COV) across 1-second Intervals for Consensually Agreed Seconds with Phasic Activity
| Feature Name (Number)a | COV (Left) |
COV (Right) |
|---|---|---|
| Relative High Frequency Power (1) | 77.3 | 80.5 |
| Spectral Edge Frequency 95th Percentile (2) | 711.5 | 916.0 |
| Skewness (3) | 75.5 | 74.1 |
| Variance (4) | 880.2 | 807.9 |
| Kurtosis (5) | 129.1 | 118.6 |
| Entropy (6) | 58.8 | 45.0 |
| Mobility (7) | 60.8 | 63.7 |
| Amplitude 75th Percentile (8) | 292.7 | 329.3 |
| Complexity (9) | 204.1 | 222.1 |
| Mean Absolute Amplitude (10) | 188.9 | 183.8 |
| Curve Length (11) | 200.1 | 200.1 |
| Energy (12) | 912.1 | 809.3 |
| Zero Crossing (13) | 51.1 | 49.1 |
| Non-Linear Energy (14) | 613.5 | 1308.4 |
| Spectral Entropy (15) | 1602.3 | 1343.5 |
Relevant formulae are shown in parentheses (see text)
3.4 “Residual Noise” across Features
Table 4 presents the ratio between left and right consensually agreed upon seconds without phasic activity. Ratios approximating 1.0 suggested features that were relatively robust to residual noise within the recording system. Selected features (1, 3, 5, 6, 7, 13) all approximated unity. Several features with promising discrimination in the unsupervised models (Tables 1 and 2) and/or in the COV analysis (Table 3), e.g., SEF, Spectral Edge (2); S2, Variance (4); SE, Spectral Entropy (15), showed substantial divergence, suggesting a higher susceptibility to artifact.
Table 4.
Ratio of Left-to-Right Feature Activity for Consensually Agreed Seconds without Phasic Activity
| Feature Name (Number)a | Ratio (L/R) |
|---|---|
| Relative High Frequency Power (1) | 0.82 |
| Spectral Edge Frequency 95th Percentile (2) | 0.13 |
| Skewness (3) | 0.98 |
| Variance (4) | 0.12 |
| Kurtosis (5) | 1.04 |
| Entropy (6) | 0.95 |
| Mobility (7) | 0.97 |
| Amplitude 75th Percentile (8) | 0.68 |
| Complexity (9) | 0.81 |
| Mean Absolute Amplitude (10) | 0.62 |
| Curve Length (11) | 0.70 |
| Energy (12) | 0.12 |
| Zero Crossing (13) | 0.98 |
| Non-Linear Energy (14) | 0.46 |
| Spectral Entropy (15) | 0.20 |
Relevant formulae are shown in parentheses (see text)
4.0 DISCUSSION
When based solely on accuracy in relation to visually analyzed EMG recordings, a number of different computer-derived features (SEF, S2, Kurt, 75_Amp, MAA, L, MnE, NLE, SE) appeared promising approaches to quantifying the human sleep EMG signal. However, when examined from standpoint of maximizing differences among consensually agreed upon seconds with phasic activity (Table 3), only 5 of these features (SEF, S2, MnE, NLE, SE) retained highest variability, although these also appeared to be more susceptible to residual noise artifact (Table 4). Features promising lower influence of residual noise (Prel, Skew, Kurt, Ent, Mobi, ZC) showed reduced variability across consensually agreed upon seconds with phasic activity and/or lower accuracy in relation to consensually agreed upon seconds with and without phasic activity. These data suggest that further studies applying computer-derived metrics derived from digitally processed muscle activity during sleep should examine multiple features simultaneously to determine whether susceptibility to residual noise bears upon feature discrimination.
Recent interest in quantification of the EMG during sleep has been prompted, at least in part, by the recognition that a particular class of neurodegenerative disease (i.e., the synucleinopathies, including Parkinson’s disease, Dementia with Lewy Bodies, Multi-system Atrophy, idiopathic Rapid Eye Movement Sleep Behavior Disorder) are characterized by elevated muscle activity in sleep, particularly in REM, but often in Non-REM sleep as well [17, 31, 32]. Numerous investigators, including ourselves, have quantified muscle activity visually during sleep, typically following a variant of the manual (c.f., computer detected) approach outlined in the original study of Lapierre and Montplaisir [33]. Studies such as these have shown robust differentiation from controls [34, 35], progression of severity [36] or development of Parkinson’s disease [37, 38] over time, short-term stability across consecutive nights [39, 40], validation with video monitoring of motor behavior [41, 42] and spouse or caregiver questionnaires [43], and high synchronization of firing across muscle groups during REM [44]. Taken together, these diverse findings indicate that careful evaluation of muscle activity during sleep may hold important diagnostic and prognostic significance for this class of neurodegenerative diseases. However, given the tedious, time-consuming, and labor-intensive nature of such visual analysis, the foregoing findings also suggest the necessity of an automated approach to this process.
Our approach to quantification differs from those used by other investigators of the surface EMG in sleep in several important respects. First, although there is a long history of attempts to quantify muscle activity during sleep, our study is the initial attempt to extend automated approaches to include a complete library of techniques that incorporate several novel measures for EMG quantification, more typically applied for the sleep EEG but not for sleep-related muscle activity (e.g., non-linear dynamics, entropy). Early studies quantifying muscle activity of sleep employed cumbersome analog-to-digital hardware, which did little more than summate voltage repetitively to a pre-established criterion level [8]. Later work using 100-128 Hz digitization of EMG signals, focused largely on reducing muscle activity to a binary measure predicting desynchronization in the EEG [45] or examining activation levels over intervals of 20 seconds to examine associations between muscle tone and REM versus –Non-REM sleep [9]. The latter study’s reliance on variance measures anticipated to some extent the later variance-based approach of Burns et al. [10] who, using 3-second windows and a 200Hz sampling rate, defined high levels of mentalis activity as transient occurrence in the 5th percentile of the variance distribution across all such intervals during the entire night of sleep. In another recent study, Ferri et al. [11] provided a detailed analysis of amplitudes of surface mentalis EMG signals within 1-second intervals and demonstrated differences in the voltage and time domain as a function of REM versus Non-REM sleep in healthy young controls, differences between younger controls and aged controls in REM sleep, and (in REM) differences between aged controls and patients with varied synucleinopathies. The approaches used by Burns et al. [10] and Ferri et al. [11] resembled several features examined here in our study in quantifying variance (formula 4) and amplitude (formulae 1, 8 and 10). By contrast, Mayer et al. [46] also employed an amplitude-derived smoothing function to determine a moving baseline, examined both voltage and time domains, but reported that slightly longer duration (0.5-10.0 sec; most less than 2.0 sec) elevations in muscle activity provided the clearest, clinically relevant differences.
A second way that our approach differs from at least some of the recent studies examining the sleep EMG is that investigators often have attempted to distinguish between short-duration (< 100 msec) activity representing motor unit activity and relatively longer duration (up to 5 or, in some cases, 30 seconds) tonic activity, presumably representing the recruitment of additional motor units within a muscle group [47]. Although the latter situation occurs, albeit variably, within the sleep EMG, the ability of human scorers to make this distinction visually (c.f., Figure 1B, pg 447 in [48] vs Figure 3, pg 2048 in [34]) can be difficult. Some automated digitization approaches [10, 11] simply allow the time domain to vary without arbitrary restrictions on duration of activity, whereas others make a binary differentiation of short versus long durations (e.g., [46]). Because phasic activity in a surface muscle recording most closely represents the fundamental activity of a firing motor unit, our validation has focused on phasic events (< 500 msec duration) occurring in the context of low impedance, high quality surface EMG recording free from artifacts and gross movement. Longer durations of motor activation [46], some resulting in behaviorally observable movement in which engagement of multiple motor units and involvement of multiple muscle groups are likely [47], might be expected to present a more complex challenge for analysis. Recordings with spurious signals, such as balistocardiographic artifact, also will represent an analytic challenge. Recordings from other muscle groups, such as the mentalis, which might be subject to different kinds of artifacts (e.g., inspiratory and/or expiratory vibratory noise), would represent yet another technical consideration. Our future work using the features analyses described in this study will pursue such issues.
Finally, our approach to automated signal processing of the human EMG signal subscribes to the concept that before any utilization of any system can be implemented at the level of the individual patient (i.e., by characterizing an entire overnight PSG), the most fundamental validation should mirror accurately the operation of a human visual scorer on a second-to-second, rather than a whole-night, basis. Ultimately, such an elemental step is essential before any attempt is made to apply complex signal processing to the vagaries clinical diagnosis of disease, an exceedingly complex issue in its own right. In the case of synucleinopathies, definitive disease definition of requires neuropathologic verification [49], although clinical criteria have been agreed upon consensually and are widely used [50]. Perhaps more relevant to the current effort is that for relatively new application of a potentially diagnostic tool like phasic sleep EMG, epidemiologic considerations involving sensitivity, specificity, positive predictive value and negative predictive value must all be examined on a population-wide basis. Clinical studies of the sleep EMG like those cited above vastly overselect for likelihood of synucleinopathic conditions and might be expected to be biased in favor of more accurate discrimination on a case-by-case (c.f., a second-by-second) basis. By way of example, although the prevalence of conditions like rapid eye movement sleep behavior disorder have been estimated at 1.6 cases per 100 normals in the general population [51] gross oversampling of such cases in clinical studies of the sleep EMG (e.g., 48 patients vs 25 controls [46]; 17 patients to 6 controls [43]; 80 patients to 80 controls [34]; 54 patients to 35 controls [35]) have the potential to create highly biased upwards estimates of diagnostic accuracy. It remains uncertain how well phasic EMG activity could discriminate incipient synucleinopathies in samples mirroring true prevalence of patterns of muscle activity during sleep observed in the general population. Initial validation of signal-processing methodology for phasic EMG should take place independently from such issues of case identification.
5.0 CONCLUSIONS
In summary, we have described in this study an approach for computerized detection of phasic EMG activity from surface electrodes during prolonged intervals of human PSGs. Several of the novel signal processing features tested here appeared to be promising for second-to-second validation with the trained human visual scorer, yet were not unduly influenced by system noise and retained the ability to detect a broad range of activity not otherwise detectable by the human scorer. Future studies will examine the relative resilience of such features to recording segments that will incorporate more diverse patterns of motor activation during human sleep.
HIGHLIGHTS.
Surface recordings of phasic EMG in sleep may be prognostic for neurodegeneration
Most attempts to quantify such activity have relied upon visual judgments
We describe computer-derived, digitally based features for quantifying EMG in sleep
Features reflect human scorers but discriminate beyond visually based approaches
Several unique signal processing features are promising for future studies
ACKNOWLEDGMENTS
This work was supported by 1 R01 NS-050595; 1 R01 NS-055015; 1 F32 NS-070572 and the “Action support post-doctoral fellows of the Operational Programme Education and Lifelong Learning” of the Greek Ministry of Education, Lifelong Learning and Religious Affairs, co-financed by the European Union.
We gratefully acknowledge the following individuals for their assistance in conducting this study: Sophia Greer, Shannon Hollars, Dr. David Rye, Dr. Lynn Marie Trotti, Ray Williams, and Anthony Wilson.
Abbreviations
- PSG
Polysomnography
- EMG
Electromyography
- EEG
Electroencephalography
- REM
Rapid Eye Movement
- COV
Coefficient of Variation
- GMM
Gaussian Mixture Modeling
- EM
Expectation Maximization
- Prel
Relative High Frequency Power
- SEF
Spectral Edge Frequency
- S2
Variance
- Kurt
Kurtosis
- Ent
Entropy
- Mobi
Mobility
- 75_Amp
75th Amplitude Percentile
- Comp
Complexity
- MAA
Mean Absolute Amplitude
- L
Curve Length
- MnE
Mean Energy
- ZC
Zero Crossings
- NE
Nonlinear Energy
- SE
Spectral Entropy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Redline S, Sanders MH, Lind BK, Quan SF, Iber C, Gottlieb DJ, et al. Methods for obtaining and analyzing polysomnography data for a multicenter study. Sleep Heart Health Research Group. Sleep. 1998;21:759–67. [PubMed] [Google Scholar]
- 2.Borbely AA, Baumann F, Brandeis D, Strauch I, Lehmann D. Sleep deprivation: effects on sleep stages and EEG power density in man. Electroencephalogr Clin Neurophysiol. 1981;51:483–95. doi: 10.1016/0013-4694(81)90225-x. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg I, March JD, Fein G, Floyd TC, Walker JM, Price L. Period and amplitude analysis of 0.5-3 c/sec activity in NREM sleep of young adults. Electroencephalogr Clin Neurophysiol. 1978;44:202–13. doi: 10.1016/0013-4694(78)90266-3. [DOI] [PubMed] [Google Scholar]
- 4.Fell J, Roschke J, Mann K, Schaffner C. Discrimination of sleep stages: a comparison between spectral and nonlinear EEG measures. Electroencephalogr Clin Neurophysiol. 1996;98:401–10. doi: 10.1016/0013-4694(96)95636-9. [DOI] [PubMed] [Google Scholar]
- 5.Massimini M, Huber R, Ferrarelli F, Hill S, Tononi G. The sleep slow oscillation as a traveling wave. J Neurosci. 2004;24:6862–70. doi: 10.1523/JNEUROSCI.1318-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Mamelak A, Hobson JA. Nightcap: a home-based sleep monitoring system. Sleep. 1989;12:157–66. doi: 10.1093/sleep/12.2.157. [DOI] [PubMed] [Google Scholar]
- 8.Bliwise D, Coleman R, Bergmann B, Wincor MA, Pivik RT, Rechtschaffen A. Facial muscle tonus during REM and NREM sleep. Psychophysiology. 1974;11:447–508. doi: 10.1111/j.1469-8986.1974.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 9.Brunner DP, Dijk DJ, Borbely AA. A quantitative analysis of phasic and tonic submental EMG activity in human sleep. Physiol Behav. 1990;48:741–8. doi: 10.1016/0031-9384(90)90219-t. [DOI] [PubMed] [Google Scholar]
- 10.Burns JW, Consens FB, Little RJ, Angell KJ, Gilman S, Chervin RD. EMG variance during polysomnography as an assessment for REM behavior disorder. Sleep. 2007;30:1771–8. doi: 10.1093/sleep/30.12.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferri R, Manconi M, Plazzi G, Bruni O, Vandi S, Montagna P, et al. A quantitative statistical analysis of the submentalis muscle EMG amplitude during sleep in normal controls and patients with REM sleep behavior disorder. J Sleep Res. 2008;17:89–100. doi: 10.1111/j.1365-2869.2008.00631.x. [DOI] [PubMed] [Google Scholar]
- 12.Rye DB, Bliwise DL. Movement disorders specific to sleep and the nocturnal manifestations of waking movement disorders. In: Watts RL, Koller WC, editors. Movement Disorders: Neurologic Principles and Practice. 2nd Edition McGraw Hill; New York: 2004. pp. 855–890. [Google Scholar]
- 13.Boeve BF, Silber MH, Ferman TJ, Lucas JA, Parisi JE. Association of REM sleep behavior disorder and neurodegenerative disease may reflect an underlying synucleinopathy. Mov Disord. 2001;16:622–30. doi: 10.1002/mds.1120. [DOI] [PubMed] [Google Scholar]
- 14.Iranzo A, Santamaria J, Tolosa E. The clinical and pathophysiological relevance of REM sleep behavior disorder in neurodegenerative diseases. Sleep Med Rev. 2009;13:385–401. doi: 10.1016/j.smrv.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Mahowald M, Bornermann MAC, Schenck CH. When and where do the synculeinopathies begin? Neurology. 2010;75:488–9. doi: 10.1212/WNL.0b013e3181ec804a. [DOI] [PubMed] [Google Scholar]
- 16.Kempfner J, Sorensen G, Zoetmulder M, Jennum P, Sorensen HB. REM behavior disorder detection associated with neurodegenerative diseases. Conf Proc IEEE Eng Med Biol Soc. 2010:5093–6. doi: 10.1109/IEMBS.2010.5626212. [DOI] [PubMed] [Google Scholar]
- 17.Bliwise DL, He L, Ansari FP, Rye DB. Quantification of electromyographic Activity during sleep: a phasic electromyographic metric (PEM) J Clin Neurophysiol. 2006;23:59–67. doi: 10.1097/01.wnp.0000192303.14946.fc. [DOI] [PubMed] [Google Scholar]
- 18.Zoubek L, Charbonier S, Lesecq S, Buguet A, Chapotot F. Feature selection for sleep/wake stages classification using data driven methods. Biomed Signal Process Control. 2007;2:171–9. [Google Scholar]
- 19.Hayes MH. Statistical Digital Signal Processing and Modeling. John Wiley & Sons, Inc.; New York: 1996. The DFT and FFT, The Periodogram. [Google Scholar]
- 20.Proakis JG, Manolakis DG. Digital signal processing: principles, algorithms and applications. 4th Edition Prentice Hall; New York: 2006. [Google Scholar]
- 21.Long CW, Shah NK, Loughlin C, Spydell J, Bedford RF. A comparison of EEG determinants of near-awakening from isoflurane and fentanyl anesthesia. Spectral edge, median power frequency, and delta ratio. Anesth and Analg. 1989;69:169–73. [PubMed] [Google Scholar]
- 22.Wasserman L. All of Statistics. A concise course in statistical inference. Springer; New York: 2004. Statistical functionals. [Google Scholar]
- 23.Filliben J. NIST/SEMATECH e-Handbook of Statistical Methods. U.S. Department of Commerce; Washington DC: 2003. Measures of skewness and kurtosis. Quantitative Techniques; p. 1.3.5. [Google Scholar]
- 24.Freedman D, Diaconis P. On the Histogram as a Density Estimator - L2 Theory. Zeitschrift Fur Wahrscheinlichkeitstheorie Und Verwandte Gebiete. 1981;57:453–76. [Google Scholar]
- 25.Zoubek L. Automatic Classification of Human Sleep Recordings Combining Artifact Identification and Relevant Features Selection (Ph.D. Dissertation) University of Ostrava; Grenoble: 2008. [Google Scholar]
- 26.Agarwal R, Gotman J, Flanagan D, Rosenblatt B. Automatic EEG analysis during long-term monitoring in the ICU. Electroencephalogr Clin Neurophysiol. 1998;107:44–58. doi: 10.1016/s0013-4694(98)00009-1. [DOI] [PubMed] [Google Scholar]
- 27.D’Alessandro MM. The Utility of Intracranial EEG Feature and Channel Synergy for Evaluating the Spatial and Temporal Behavior of Seizure Precursors, (Ph.D. Dissertation) Georgia Institute of Technology; Atlanta: 2001. [Google Scholar]
- 28.Bishop C. Pattern recognition and machine learning. Springer; New York: 2006. Mixtures of Gaussians; pp. 430–43. [Google Scholar]
- 29.Hand D, Mannila H, Smyth P. Principles of Data Mining. MIT Press; Cambridge: 2001. [Google Scholar]
- 30.Fridlund AJ, Cacioppo JT. Guidelines for human electromyographic research. Psychophysiology. 1986;23:567–89. doi: 10.1111/j.1469-8986.1986.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 31.Bliwise DL, Rye DB. Elevated PEM (phasic electromyographic metric) rates in rapid eye movement behavior disorder patients on nights without behavioral abnormalities. Sleep. 2008;31:853–7. doi: 10.1093/sleep/31.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bliwise DL, Trotti LM, Greer SA, Juncos JJ, Rye DB. Phasic muscle activity in sleep and clinical features of Parkinson’s Disease. Ann Neurol. 2010;68:353–9. doi: 10.1002/ana.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapierre O, Montplaisir J. Polysomnographic features of REM sleep behavior disorder: development of a scoring method. Neurology. 1992;42:1371–4. doi: 10.1212/wnl.42.7.1371. [DOI] [PubMed] [Google Scholar]
- 34.Montplaisir J, Gagnon J-F, Fantini ML, Postuma RB, Dauvilliers Y, Desautels A, et al. Polysomnographic diagnosis of idiopathic REM sleep behavior disorder. Mov Disord. 2010;25:2044–51. doi: 10.1002/mds.23257. [DOI] [PubMed] [Google Scholar]
- 35.Ferri R, Rundo F, Manconi M, Plazzi G, Bruni O, Oldani A, et al. Improved computation of the atonia index in normal controls and patients with REM sleep behavior disorder. Sleep Med. 2010;11:947–9. doi: 10.1016/j.sleep.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Iranzo A, Tarri PL, Casanova-Molla J, Serradell M, Vilaseca I, Santamaria J. Excessive muscle activity increases over time in idiopathic REM sleep behavior disorder. Sleep. 2009;32:1149–53. doi: 10.1093/sleep/32.9.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postuma RB, Gagnon JF, Rompre S, Montplaisir JY. Severity of REM atonia loss in idiopathic REM sleep behavior disorder predicts Parkinson disease. Neurology. 2010;74:239–44. doi: 10.1212/WNL.0b013e3181ca0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iranzo A, Molinuevo JL, Santamaria J, Serradell M, Marti MJ, Valldeoriola F, et al. Rapid-eye-movement sleep behavior disorder as an early marker for a neurodegenerative disorder: a descriptive study. Lancet Neurol. 2006;5:572–7. doi: 10.1016/S1474-4422(06)70476-8. [DOI] [PubMed] [Google Scholar]
- 39.Cygan F, Oudiette D, Leclair-Visonneau L, Leu-Semenescu S, Arnulf I. Night-to-night variability of muscle tone, movements and vocalizations in patients with REM sleep behavior disorder. J Clin Sleep Med. 2010;6:551–5. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang JH, Lam SP, Ho CKW, Li AM, Tsoh J, Mok V, et al. Diagnosis of REM sleep behavior disorder by video-polysomnographic study: is one night enough? Sleep. 2008;31:1179–85. [PMC free article] [PubMed] [Google Scholar]
- 41.Manni R, Terzaghi M, Glorioso M. Motor-behavioral episodes in REM sleep behavior disorder and phasic events during REM sleep. Sleep. 2009;32:241–5. doi: 10.1093/sleep/32.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frauscher B, Gschliesser V, Brandauer E, Ulmer H, Peralta CM, Muller J, et al. Video analysis of motor events in REM sleep behavior disorder. Mov Disord. 2007;10:1464–70. doi: 10.1002/mds.21561. [DOI] [PubMed] [Google Scholar]
- 43.Consens FB, Chervin RD, Koeppe RA, Little R, Liu S, Junck L, et al. Validation of a polysomnographic score for REM sleep behavior disorder. Sleep. 2005;28:993–7. doi: 10.1093/sleep/28.8.993. [DOI] [PubMed] [Google Scholar]
- 44.Frauscher B, Iranzo A, Hogl B, Cassanova-Molla J, Salamero M, Gschliesser V, et al. Quantification of electromyographic activity during REM sleep in multiple muscles in REM sleep behavior disorder. Sleep. 2008;31:724–31. doi: 10.1093/sleep/31.5.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilcher JJ, Schulz H. The interaction between EEG and transient muscle activity during sleep in humans. Hum Neurobiol. 1987;6:45–9. [PubMed] [Google Scholar]
- 46.Mayer G, Kesper K, Ploch T, Cainsius S, Penzel T, Oertel W, et al. Quantification of tonic and phasic muscle activity in REM sleep behavior disorder. J Clin Neurophysiol. 2008;25:48–55. doi: 10.1097/WNP.0b013e318162acd7. [DOI] [PubMed] [Google Scholar]
- 47.Hallett M. Clinical neurophysiology of movement disorders. In: Chokroverty S, Hening WA, Walters AS, editors. Sleep and Movement Disorders. Butterworth Heinemann; Philadelphia: 2003. pp. 153–64. [Google Scholar]
- 48.Boeve BF, Molano JR, Ferman TJ, Smith GE, Lin SC, Bieniek K, et al. Validation of the Mayo Sleep Questionnaire to screen for REM sleep behavior disorder in an aging and dementia cohort. Sleep Med. 2011;12:445–53. doi: 10.1016/j.sleep.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braak H, Del Tredici K, Rub U, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 50.McKeith IG, Dickson DW, Lowe J, et al. Diagnosis and management of dementia with Lewy bodies: third report of the DLB consortium. Neurology. 2005;65:1863–72. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 51.Ohayon M, Schenck C. Violent behavior in sleep: prevalence, comorbidity and consequences. Sleep Med. 2010;11:941–6. doi: 10.1016/j.sleep.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]