Abstract
Summary: Gibberellic acids (GAs) are key plant hormones, regulating various aspects of growth and development, which have been at the center of the ‘green revolution’. GRAS family proteins, the primary players in GA signaling pathways, remain poorly understood. Using sequence-profile searches, structural comparisons and phylogenetic analysis, we establish that the GRAS family first emerged in bacteria and belongs to the Rossmann fold methyltransferase superfamily. All bacterial and a subset of plant GRAS proteins are likely to function as small-molecule methylases. The remaining plant versions have lost one or more AdoMet (SAM)-binding residues while preserving their substrate-binding residues. We predict that GRAS proteins might either modify or bind small molecules such as GAs or their derivatives.
Contact: aravind@ncbi.nlm.nih.gov
Supplementary Information: Supplementary Material for this article is available at Bioinformatics online.
1 INTRODUCTION
Gibberellic acids (GAs) are a large family of diterpene molecules that are currently known to be synthesized by plants, fungi and bacteria (Yamaguchi, 2008). In plants they are key hormones that regulate various aspects of development such as seed germination, stem elongation, leaf expansion, flower development and asymmetric cell division in roots. The major plant players implicated in GA response are GRAS family proteins, which include diverse members such as GA insensitive (GAI) (Koornneef et al., 1985; Peng et al., 1997), Repressor of GA1-3 (RGA1/DELLA) (Silverstone et al., 1997), Short-root (Benfey et al., 1993), Scarecrow (SCR) (Di Laurenzio et al., 1996) and Nodulation Signal Pathway 1 and 2 (NSP1 and NSP2) (Kalo et al., 2005; Smit et al., 2005). While the GRAS family has been studied intensely for more than a decade as the so-called ‘green revolution’ genes (Peng et al., 1999), there is little clarity about their actual mode of action in GA pathways. Genetic and molecular studies have suggested that at least 23 members of the GRAS family have functions related to GA signaling in Arabidopsis (Lee et al., 2008). Of these, GAI and RGA1 help in establishing repressive chromatin as the default state at promoters of genes that are regulated by GA (Peng et al., 1997; Silverstone et al., 1997). The presence of GA results in recruitment of GAI1 to the GA receptor, a protein of the α/β hydrolase superfamily, which in turn results in the GAI1 being targeted for proteasomal destruction via ubiquitination by a F-Box-Skp complex (Murase et al., 2008). Thus, GA-sensitive genes are derepressed in the presence of a GA signal. In contrast, several GRAS members such as SCL3 appear to act as positive regulators downstream of GA by removing the block imposed by the negative regulators such as GAI and RGA1 (Heo et al., 2011). Consistent with their opposing roles in GA signaling, mutations in certain members of this superfamily result in dwarfing of plants, others result in overgrowth and on rare occasions different alleles of the same gene cause either dwarfing or overgrowth (Lee et al., 2008). Additionally, application of external GA is able to rescue the effects of loss of function mutations in another member of the GRAS family, SCR, in relation to asymmetric cell division during root development to restore normal development of endodermis, middle cortex and cortex cells. GRAS proteins have been localized in the nucleus and are implicated in transcription-related functions that are not clearly understood in terms of mechanisms (Cui et al., 2011). In large part this lack of understanding of the GRAS family functions stems from a previously published sequence analysis study that erroneously reported a relationship between them and metazoan STAT proteins (Richards et al., 2000). Other erroneous relationships have also been reported for the GRAS family and include the purported presence of a leucine zipper or a SH2 domain (Di Laurenzio et al., 1996; Peng et al., 1999). These erroneous relationships have contributed to unsupported assumptions that they must be conventional transcription factors. Indeed, there have been several experimental studies that have built on this and attempted to show DNA binding for certain GRAS proteins, although this cannot be confirmed for other related proteins (Hirsch et al., 2009). Thus, a unified mechanism for the action of the GRAS proteins could considerably help in understanding their apparently opposite roles with respect to GA signaling. Through sequence profile searches, structural comparisons and phylogenetic analysis, we establish that the GRAS family belongs to the Rossmann fold methyltransferase superfamily that first emerged in bacteria. We further show that ancestral bacterial GRAS proteins are likely to function as active small-molecule methylases. In contrast, the majority of plant GRAS domains show disruptions of their AdoMet (SAM)-binding residues while preserving their substrate-binding residues, suggesting that they might function as small-molecule-binding domains.
2 RESULTS AND DISCUSSION
2.1 GRAS proteins belong to the Rossmann fold methyltransferase superfamily
To understand better the affinities of the GRAS family, we first investigated the veracity of previous claims regarding their sequence relationships. Using a PSI-BLAST search of the non-redundant protein database (NR) (Altschul et al., 1997) initiated with the Arabidopsis thaliana Scarecrow protein (SCR, gi: 1 497 987), we obtained a comprehensive collection of GRAS family sequences. An examination of the multiple alignment of these GRAS proteins revealed a globular region of 385–400 amino acids (referred to hereinafter as the GRAS domain) with no other clear globular regions in the remainder of the protein. Some versions had two tandem copies of the GRAS domain (e.g. gi: 8778540 from A. thaliana), while others had poorly structured N-terminal extensions with a few α-helical segments that have been termed the DELLA domain (Murase et al., 2008). Secondary structure prediction using an alignment of the globular GRAS domain revealed a regular succession of α-helices and β-strands reminiscent of the pattern observed in three-layered α/β sandwiches, such as the domains possessing the Rossmann fold. This indicated that the GRAS family is unlikely to possess a STAT-type DNA-binding domain or a SH2 domain as previously claimed. The STAT-type DNA-binding domains adopt a cytochrome f-like β-sandwich fold, whereas the SH2 domain adopts a β-barrel structure (Andreeva et al., 2008), both of which are incompatible with the predicted secondary structure of the GRAS domain. It also ruled out the presence of the previously proposed leucine zipper in the GRAS family. We further performed profile–profile comparisons using a HMM derived from the multiple alignment of the GRAS domain with the HHpred program against a panel of HMMs derived using PFAM models and PDB structures as search seeds (Soding et al., 2005). These did not recover any statistically significant hits to STAT-like DNA-binding domains, SH2 domains or leucine zippers, strongly excluding the possibility of any relationship between GRAS and these domains.
Our sequence profile searches recovered homologous sequences from angiosperms, gymnosperms, lycopodiophytes (club-mosses, i.e. Selaginella) and bryophytes (classical mosses, i.e. Physcomitrella), but not any of the basal members of the green-plant lineage, such as chlorophyte algae. Interestingly, outside of the land plant clade, the GRAS domain was also observed in several bacteria, namely, representatives of deltaproteobacteria, firmicutes and cytophagaceae (e.g. gi:383452287 from deltaproteobacterium Corallococcus coralloides recovered in iteration 2 with e = 10−15). Detection of the bacterial versions of the GRAS domain offered a new handle to understand its affinities, especially given that the versions from plants are very closely related, being part of a relatively recent lineage-specific expansion. Accordingly, we searched the NR database with the GRAS protein from the deltaproteobacterium Stigmatella aurantiaca (gi: 115 373 923) and restricted the search to bacteria in order to avoid obscuring of the search by the over 2500 hits from land plants. In this search, in addition to the bacterial GRAS proteins, we also recovered significant hits to several Rossmann fold methyltransferase domains (e.g. the small-molecule methylase ubiG from Rhodobacter sphaeroides with e = 10−5) and the pairwise alignment revealed a match to the GXGXG signature seen in the Rossmann fold of the methylases (Schubert et al., 2003). This relationship between the GRAS domains and the methyltransferases was further confirmed using profile–profile comparisons using the HHpred program initiated with a HMM based on the alignment of the GRAS domain, which recovered profiles based on the structures of several small molecule methylases as the best hits (e.g. histamine N-methyltransferase, PDB: 2aot, probability 99.7%; YecO, PDB: 1im8, probability 98%; rebeccamycin biosynthesis methyltransferase, PDB: 3bus, probability 97%). This strongly suggested that the GRAS domain is indeed a version of the Rossmann fold methylase domain.
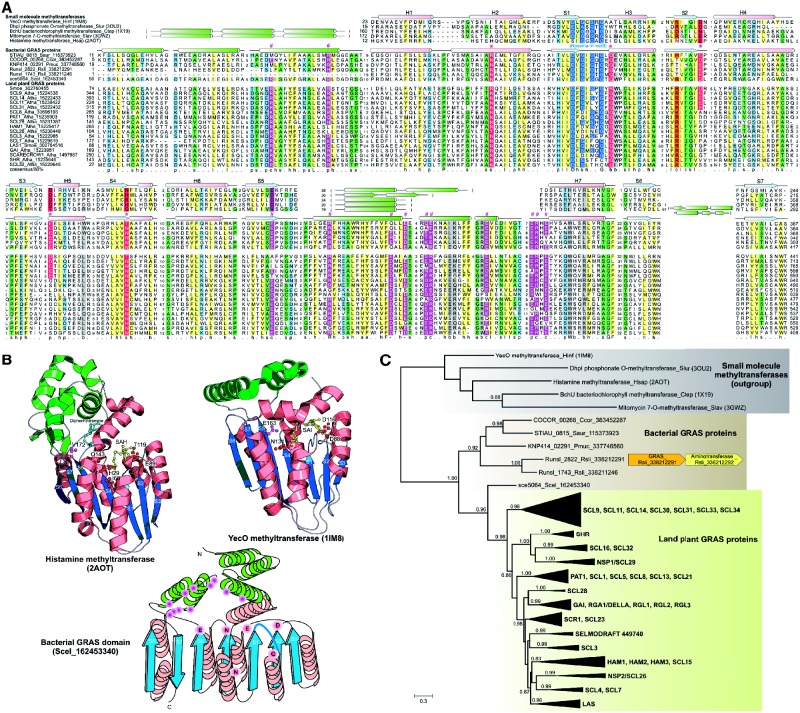
We generated a multiple alignment that included bacterial and plant GRAS domains along with several experimentally and structurally characterized methyltransferase domains and refined this alignment based on their three-dimensional structures (Fig. 1A). This revealed that GRAS domain contains a core typical of the Rossmann fold methylases with seven strands forming a central sheet, with the last two strands (6 and 7) forming a β-hairpin. The GRAS domain shares with several small-molecule and protein arginine methylases an α-helical extension, N-terminal to strand-1 of the Rossmann fold, which forms a cap over the active site (Fig. 1B), and plays an important role in contacting the substrate and shielding the active site from the bulk solvent (Horton et al., 2005; Lim et al., 2001). The GRAS domain also shows a well-developed α-helical insert between the strand-5 and the downstream helix of the core Rossmann fold. This insert occupies a position similar to the bi-helical insert observed in the histamine N-methyltransferase, YecO (PDB: 1im8) and phosphoethanolamine methyltransferase (PDB: 3ujc), and along with the N-terminal helical extension, is likely to play a role in binding the substrate (Fig. 1B). Examination of the multiple alignment showed that bacterial GRAS domains contain a conserved glutamate at the end of strand-2 (Fig. 1B) that in other methyltransferases is known to form hydrogen bonds with the 2′- and 3′-hydroxyls of the ribose moiety of the AdoMet (SAM) substrate (Horton et al., 2005; Schubert et al., 2003). Similarly, they display a conserved glutamate just at the beginning of helix-5 which corresponds to a polar residue in several methyltransferases that interacts with the adenine ring of SAM (Horton et al., 2005; Lim et al., 2001; Schubert et al., 2003). The end of strand-4 in bacterial GRAS domains is characterized by a polar residue, typically asparagine, which in other methylases plays a catalytic role in the methyl-transfer reaction (Horton et al., 2005; Schubert et al., 2003). These features, in conjunction with the intact SAM-binding loop at the C-terminus of strand-1, indicate that the bacterial GRAS proteins are likely to function as active SAM-dependent methylases. The GRAS proteins show a conserved glutamate at the end of strand-5, which by analogy to other methyltransferases is likely to correspond to the position that contacts the substrate. Likewise, the GRAS proteins show conserved charged residues both in the α-helical insert downstream of strand 5 and the N-terminal α-helical extension, which, based on the precedence of other methylase domains, are likely to be critical for substrate interaction. Importantly, the residues specific for binding SAM have been partially or entirely substituted in the majority of plant GRAS domains barring certain representatives of the clade including SCL14 and its cognates from basal land plants such as Selaginella (Fig. 1A). However, the predicted substrate-binding residues appear to be intact in majority of the plant proteins (Fig. 1A). This observation suggests that, unlike the bacterial versions, majority of plant versions are likely to lack methyltransferase activity but still bind a similar substrate as the bacterial versions.
Fig. 1.
(A) Multiple sequence alignment of GRAS domains with known small-molecule methyltransferases. The conserved loop of the Rossmann fold, SAM-binding and substrate-binding residues are labeled and highlighted in blue, red and purple background, respectively. α-Helical extensions and inserts within the core Rossmann fold are colored green. (B) Representative structures of two small-molecule methyltransferases (PDB ids: 2aot and 1im8) and the predicted topology of GRAS domains. (C) Bacterial origin of GRAS domains. The tree was reconstructed using an approximately maximum-likelihood method implemented in the FastTree 2.1 program (Price et al. 2009) under default parameters. Only bootstrap values >80% are shown (details in Supplementary Material). Species abbreviations: Atha, Arabidopsis thaliana; Ccor, Corallococcus coralloides; Ctep, Chlorobaculum tepidum; Hinf, Haemophilus influenza; Hsap, Homo sapiens; Pmuc, Paenibacillus mucilaginosus; Rsli, Runella slithyformis; Saur, Stigmatella aurantiaca; Scel, Sorangium cellulosum; Slav, Streptomyces lavendulae; Slur, Streptomyces luridus; Smoe, Selaginella moellendorffii. Chemical name abbreviations: SAH, S-adenosyl-l-homocysteine; SAI, S-adenosyl-l-homoselenocysteine
2.2 Phylogenetic analysis with inclusion of bacterial GRAS proteins suggests a single transfer to the lineage leading to land plants
We utilized the identification of bacterial GRAS proteins to better characterize the evolutionary history of the GRAS proteins, which were hitherto seen as suddenly appearing in the land plant lineage. The above analysis indicated that the bacterial versions are catalytically active methylases nested within a larger radiation of active methyltransferases in bacteria. A phylogenetic tree including bacterial, bryophyte, lycopodiophyte and angiosperm GRAS domains (Fig. 1C) revealed that the plant versions are nested within the bacterial radiation, and of the currently available bacterial sequences appear closest to versions from deltaproteobacteria. As, excepting versions from the basal-most plant-specific SCL14-like clade, majority of plant versions are inactive, the bacterial versions are likely to represent the ancestral condition and to be precursors of the plant versions. The plant versions can be divided into 13 distinct well-supported clades (bootstrap values >80%) that contained at least one representative from bryophytes, lycopodiophytes and angiosperms. These lines of evidence suggest that there was a single transfer of a GRAS domain from a bacterial source to the common ancestor of land plants (Fig.1C). These 13 distinct clades include those prototyped by the A. thaliana versions such as Scarecrow, Shortroot, SCL3, GAI/RGA/DELLA and Hairy Meristem, among others, suggesting that major functional diversification of the GRAS family happened very early in land plant evolution. These indicate that separate GRAS-dependent GA signaling mechanisms, which are cognates of shoot elongation, root growth and root cortex differentiation in angiosperms, were already established at the base of the extant land plant lineages. Beyond this, much of the evolution of the GRAS family is characterized by extensive lineage-specific expansions within each of the 13 primary clades. In particular, the SCL14 clade has undergone extensive independent lineage-specific expansions in each of the plant clades considered in our analysis.
2.3 Functional inference for the GRAS family based on the methyltransferase domain
The profile–profile comparisons indicated that the GRAS domain is closest to various N- and O-methylases of small molecules, such as those modifying histamine, rebeccamycin sugar moiety, phosphoethanolamine, ubiquinone and trans-aconitate, as opposed to known nucleic acid and protein methylases (Aravind et al., 2011; Horton et al., 2005; Iyer et al., 2011). This suggests that bacterial and a subset of plant GRAS proteins could also be small-molecule methylases. Importantly, they share with certain small-molecule methylases the presence of the ‘cap-like’ structure formed by the N-terminal extension and the α-helical inserts between strand-5 and strand-6. This is consistent with the bacterial versions from Runella slithyformis being encoded in predicted operons with genes coding for enzymes that operate on small molecules (a small-molecule phosphotriesterase and aminotransferase superfamily protein) (Fig. 1C). The substitution in key SAM-interacting residues of the methyltransferase domain in majority of the plant versions suggests that there was a functional shift after the transfer of the ancestral GRAS gene from bacteria to plants. Nevertheless, conservation of the predicted target substrate-binding site (as opposed to the SAM-binding site) suggests that they might interact with similar ligands.
As many plant members of the GRAS family have been reported to function as nuclear proteins affecting transcriptional responses, it is plausible that they bind particular covalent modifications on chromatin proteins. However, this appears less likely due to number of reasons. First, GRAS proteins do not display any of the multidomain architectures typical of chromatin proteins. Second, the majority of characterized members show a strong association with GA signaling, and the genetic and molecular evidence shows that they function both as positive and negative regulators in this signaling system. This dedicated coupling with GA signaling is again not consistent with a general role of binding a chromatin protein modification. Similarly, different functionally characterized plant GRAS proteins appear to be associated with both repressive and active chromatin modifications; this is not entirely consistent with the prediction of similar ligand-binding properties across the family. Given that the strong common denominator for the GRAS proteins is the link with GA signaling, it is possible that they actually bind GAs themselves or GA derivatives. GRAS proteins have not been implicated as GA receptors and a known GA receptor is the α/β hydrolase domain protein GID1. However, the previous erroneous reports on the affinities of the GRAS proteins have resulted in them being labeled as transcription factors despite the lack of any evidence from sequence analysis to support such a contention. This appears to have biased their analysis with most studies attempting to interpret all aspects of their function in terms of them being transcription factors. Hence, we suspect that the alternative hypothesis of them being binders of GAs or GA-derivatives has not been seriously pursued. Given that there are many GA molecules in plants, including multiple bioactive forms, and methylated, oxidized and hydroxylated ones which are supposed to be inactive or have negative effects on GA signaling, it is possible that all the GA-binding factors have not be exhausted. Thus, at least a subset of the GRAS proteins might modify GAs or their derivatives by methylation or represent alternate binders of these molecules. In particular, it would be of interest to investigate if some of them might bind the methylated inactive ester derivatives of GAs.
3 CONCLUSION
In this work, we falsify the previously published relationships that were proposed for the GRAS domain and highlight the presence of bacterial versions. We show that the GRAS domain contains a version of the Rossmann fold methyltransferase, which is likely to bind or modify GA or derivative molecules. The evolutionary history of the GRAS family parallels that of the GID1-like α/β hydrolases, which also appear to have been horizontally transferred from bacteria to plants close to the origin of land plants. In a more general sense, these findings are in line with our recent studies that suggest that several components of eukaryotic regulatory systems have their ultimate origins in bacterial small-molecule metabolism systems which have been repeatedly acquired by eukaryotes at different points in their evolutionary history (Aravind et al., 2012). Importantly, our finding provide a new way of looking at this group of plant regulatory proteins that have been often described as being at the heart of the ‘Green revolution’.
Funding: Intramural funds of the National Library of Medicine, National Institutes of Health, USA.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36(Database issue):D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, et al. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- Aravind L, et al. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front. Cell. Inf. Microbio. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, et al. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Cui H, et al. Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol. 2011;157:1221–1231. doi: 10.1104/pp.111.183178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Heo JO, et al. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA. 2011;108:2166–71. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, et al. Structural basis for inhibition of histamine N-methyltransferase by diverse drugs. J. Mol. Biol. 2005;353:334–344. doi: 10.1016/j.jmb.2005.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, et al. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- Kalo P, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Koornneef M, et al. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 1985;65:33–39. [Google Scholar]
- Lee MH, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- Lim K, et al. Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine. Proteins. 2001;45:397–407. doi: 10.1002/prot.10004. [DOI] [PubMed] [Google Scholar]
- Murase K, et al. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Peng J, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Richards DE, et al. Plant GRAS and metazoan STATs: one family? Bioessays. 2000;22:573–577. doi: 10.1002/(SICI)1521-1878(200006)22:6<573::AID-BIES10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Schubert HL, et al. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, et al. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Soding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.