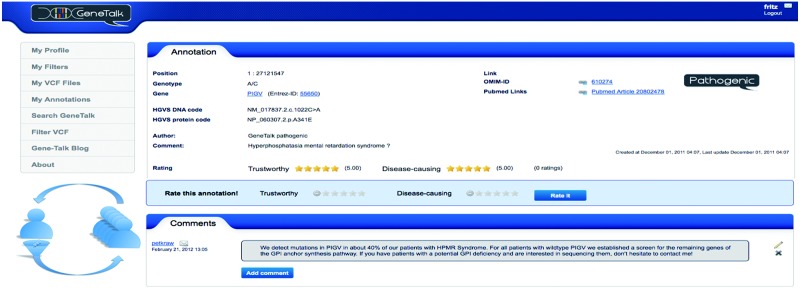
Fig. 1.
GeneTalk, a communication platform for sequence variants. A user filters sequence variants down to a small set of potentially disease relevant mutations. She then searches for detailed information annotated by the GeneTalk community for these variants. In GeneTalk, users may annotate and comment genetic variants. Annotations and comments may link to the relevant literature or discuss experimental and clinical findings. Based on this locus-specific information, GeneTalk users may rate the trustworthiness of an annotation and the potential of a mutation to be disease causing. This screenshot is taken from fritz’ account who is looking at the annotation of a mutation in the gene PIGV. The GeneTalk community finds this annotation trustworthy and rates the described mutation as highly likely to cause a syndrome called hyperphosphatasia with mental retardation. The user petkraw left a comment for this variant. He seems to have some expertise in this disease and might be an interesting person to contact for fritz