Abstract
Bietti's crystalline corneoretinal dystrophy (BCD) is a recessive degenerative eye disease caused by germline mutations in the CYP4V2 gene. More than 80% of mutant alleles consist of three mutations, that is, two splice-site alterations and one missense mutation, c.992C>A, which translates to p.H331P. In the present study, we analyzed the expression of CYP4 family members in human tissues and conducted functional studies with the wild-type and p.H331P enzymes, to elucidate the link between CYP4V2 activity and BCD. Expression analysis of 17 CYP1 to CYP4 genes showed CYP4V2 to be a major cytochrome P450 in ARPE-19 cells (a human cell line spontaneously generated from normal human retinal pigmented epithelium) and the only detectable CYP4 transcript. Immunohistochemical analyses demonstrated that CYP4V2 protein was present in epithelial cells of the retina and cornea and the enzyme was localized to endoplasmic reticulum. Recombinant reconstituted CYP4V2 protein metabolized eicosapentaenoic acid and docosahexaenoic acid (an important constituent of the retina) to their respective ω-hydroxylated products at rates similar to those observed with purified CYP4F2, which is an established hepatic polyunsaturated fatty acid (PUFA) hydroxylase. The disease-associated p.H331P variant was undetectable in Western blot analyses of HepG2 cells stably transduced with lentiviral expression vectors. Finally, overexpression of functional CYP4V2 in HepG2 cells altered lipid homeostasis. We demonstrated that CYP4V2 protein is expressed at high levels in ocular target tissues of BCD, that the enzyme is metabolically active toward PUFAs, and that the functional deficit among patients with BCD who carry the H331P variant is most likely a consequence of the instability of the mutant protein.
Introduction
Bietti's crystalline corneoretinal dystrophy (BCD) is an autosomal recessive degenerative retinopathy that is characterized clinically by a progressive decline in central vision, night blindness, and constriction of the visual field. The average age of manifestation among affected family members is ∼30 years (Hu, 1983). BCD is defined morphologically on the basis of yellow-white crystalline lipid deposits in the retina and the cornea (Bietti, 1937), which ultimately lead to degeneration of the retina and sclerosis of the choroidal vessels. BCD is rare in white populations but is relatively common in Asian populations. After the genetic defect was initially linked to chromosome 4q35 (Jiao et al., 2000), Li et al. (2004) identified biallelic mutations in the “orphan” P450 enzyme CYP4V2 in 23 of 25 index patients from families with BCD. Numerous research groups (Gekka et al., 2005; Lin et al., 2005; Shan et al., 2005; Wada et al., 2005; Yokoi et al., 2010) confirmed the original genetic findings (Kelly et al., 2011). A recent study demonstrated that more than 95% of all patients with BCD for whom analyses were performed had germline mutations in the CYP4V2 gene (Xiao et al., 2011). Although more than 34 distinct mutations have been identified, variations in exons 6 to 9 account for >80% of all mutations, with at least three founder mutations (i.e., c.802-8_810del17insGC, c.992A>C, and c.1091-2A>G, accounting for 62.7, 7.4, and 6.4%, respectively of all mutated alleles) (Xiao et al., 2011).
Although complex lipid deposits are also found in the circulating lymphocytes and skin fibroblasts of patients with BCD (Wilson et al., 1989; Kaiser-Kupfer et al., 1994) and expression of CYP4V2 mRNA has been detected in most human tissues (Li et al., 2004), the clinical disease phenotype seems to be restricted to the eye. The composition of the crystalline lipids has not been elucidated, but early biochemical tracer studies indicated a cellular defect in the anabolism of ω-3-polyunsaturated fatty acids (PUFAs) (Lee et al., 2001). Analysis of total fatty acids in the plasma of patients with BCD, compared with control subjects, suggested a defect in the synthesis of oleic acid (Lai et al., 2010).
CYP4V2 is generally referred to as an orphan P450 because its substrate specificity is just beginning to be defined (Kelly et al., 2011). Typically, CYP4 enzymes are microsomal fatty acid ω-hydroxylases that function together with mitochondrial and peroxisomal α/β-oxidation enzymes to degrade cellular lipids. Although CYP4V2 is the most distantly related of all human CYP4 enzymes, with a sequence identity of only ∼35% (Rettie and Kelly, 2008), we found that the recombinantly expressed enzyme possessed typical CYP4 ω-hydroxylase activity toward medium-chain saturated fatty acids (Nakano et al., 2009). Therefore, it is tempting to speculate that genetic defects in the catalytic function of CYP4V2 prevent local ocular degradation of lipids, which subsequently accumulate in BCD.
For the aforementioned scenario to be possible, we postulated that CYP4V2 should be expressed locally in BCD target tissues and should be capable of metabolizing key ocular lipids. No quantitative data on CYP4V2 expression are available (Li et al., 2004), and no studies on protein expression in ocular tissues have been reported. Therefore, we analyzed the expression of CYP4V2 mRNA and protein in BCD target tissues and evaluated the functional activity of wild-type CYP4V2 and the most frequent coding-region BCD-associated mutation, p.H331P, with particular emphasis on the ability to process docosahexaenoic acid (DHA), which is found at high concentrations in the eye. We also stably expressed wild-type and mutant CYP4V2 enzymes in HepG2cells, to evaluate enzyme translation/stability and effects on endogenous lipid profiles.
Materials and Methods
Chemicals.
DHA (C22:6n-3), EPA (C20:5n-3), and arachidonic acid (C20:4n-6) were purchased from Cayman Chemical (Ann Arbor, MI). Fast Red was obtained from Thermo Fisher Scientific (Waltham, MA). Myristic acid (C14:0), palmitic acid (C16:0), palmitoleic acid (C16:1), stearic acid (C18:0), oleic acid (C18:1, cis-9), and 2,2-d2-stearic acid (C18:0-d2) were purchased from Sigma-Aldrich (St. Louis, MO). 22-Hydroxydocosanoic acid and 20-hydroxyeicosanoic acid were purchased from Larodan Fine Chemicals (Malmo, Sweden). Halt Protease Inhibitor Cocktail was obtained from Thermo Fisher Scientific. Polyethylenimine transfection reagent was purchased from Sigma-Aldrich, and an HIV1 helper plasmid, pCD/NL-BH, was kindly provided by Dr. Jakob Reiser (Louisiana State University, New Orleans, LA). G-418 (Geneticin) was purchased from Invitrogen (Carlsbad, CA). Laemmli buffer was obtained from Bio-Rad Laboratories (Hercules, CA) and Odyssey blocking buffer from LI-COR Biosciences (Lincoln, NE).
Enzyme, Antibodies, and Cell Sources.
CYP4 Supersomes were obtained from BD Biosciences (San Jose, CA), and ARPE-19 cells were obtained from American Type Culture Collection (Manassas, VA). PCR Master Mix, MultiScribe reverse transcriptase, random hexamers, and TaqMan expression system assay probes were obtained from Applied Biosystems (Foster City, CA). Recombinant NADPH-P450 oxidoreductase and cytochrome b5 expressed in Escherichia coli were prepared as described previously (Cheesman et al., 2003). Anticalreticulin antibody, anti-mouse IgG antibody conjugated with Cy5, and mouse monoclonal anti-β-actin were obtained from Abcam Inc. (Cambridge, MA). Anti-mouse IgG antibody conjugated with IR680 dye and anti-rabbit antibody conjugated with IR800 dye were purchased from Rockland Immunochemicals (Gilbertsville, PA). The human tissue panel (FDA992) was purchased from US Biomax Inc. (Rockville, MD). Human retina sections were purchased from Fred Hutchinson Cancer Research Center (Seattle, WA). Human kidney and small intestine microsomes were a gift from Dr. Ken Thummel (University of Washington, Seattle, WA). Human liver samples were obtained from the liver bank in the School of Pharmacy, University of Washington. HepG2 cells and Sf9 cells were purchased from American Type Culture Collection.
RT-PCR Analyses.
Total RNA from ARPE-19 cells was isolated by using TRI Reagent (Applied Biosystems), as described in the manufacturer's protocol. The quality and quantity of total RNA were determined spectrophotometrically. cDNA was synthesized from 1 μg of total RNA by using MultiScribe reverse transcriptase with random hexamers, in a total volume of 10 μl. Real-time RT-PCR assay mixtures contained 0.1 μg of cDNA, PCR Master Mix, and TaqMan gene expression probes (Applied Biosystems) for the following genes: human GusB, CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP2J2, CYP3A4, CYP4A11, CYP4B1, CYP4F3, CYP4F12, and CYP4V2. Amplification reactions were performed with an Applied Biosystems 7900HT sequence detection system for 40 cycles of 95°C for 15 s and 60°C for 60 s, and threshold cycle values were determined by using SDS 2.3 software. Expression levels were determined in relation to those of the housekeeping gene GusB, with CYP4V2 as a calibrator.
CYP4V2 Expression, Purification, and Quality Control.
Cloning and expression of CYP4V2 in insect cells were described previously (Nakano et al., 2009). Wild-type CYP4V2 protein was purified with nickel-nitrilotriacetic acid and hydroxyapatite columns as described elsewhere (Cheesman et al., 2003), with minor modifications. The final product was dialyzed twice against 100 mM potassium phosphate, pH 7.4, 20% glycerol, 0.1 mM EDTA, and was stored at −80°C. Purity was assessed through SDS-PAGE (9%) with Coomassie blue staining. Reduced carbon monoxide-bound spectra and absolute spectra were recorded with a Cary 300 UV/visible spectrometer (Nakano et al., 2009). The molecular mass of purified CYP4V2 was determined through liquid chromatography-MS, as described previously (Zheng et al., 2003).
CYP4F2 Expression and Purification.
The wild-type CYP4F2 gene was a kind gift from Drs. David E. Stec (University of Mississippi Medical Center, Jackson, MS) and Mark J. Rieder (University of Washington, Seattle, WA). To obtain the full-length cDNA with the hexahistidine tag, CYP4F2 was amplified through PCR with the specific primers 5′-GCGCGAATTCATGTCCCAGCTGAGCCTGTCCTGGC-3′ and 5′-GCGCGTCGACTCAATGATGATGATGATGATGGCTCAGGGGCTCCACCC-3′. A recombinant baculovirus containing CYP4F2 with a hexahistidine tag was generated by using the Bac-to-Bac baculovirus expression system (Invitrogen), as described elsewhere (Nakano et al., 2009). CYP4F2 expression and purification were performed as described above for CYP4V2, with minor modifications.
Antibody Production and Assessment of Cross-reactivity.
Anti-CYP4V2 antibody production was performed by R&R Research (Stanwood, WA), with purified CYP4V2 as the antigen. Two rabbits were immunized with the purified CYP4V2 protein supplemented with complete adjuvant, with three boosts (total of 1 mg). Twelve weeks after the initial immunization, the animals were killed and sera were isolated. Polyclonal IgG was isolated from the crude sera of animals before and after immunization, after fractionation with a saturated ammonium sulfate solution. The cross-reactivity of the anti-CYP4V2 IgG toward other CYP4 enzymes (Supersomes and purified CYP4B1 expressed in E. coli) was determined from Western blots with 0.2 pmol of purified CYP4V2, 1 pmol of Supersomes (CYP4A11, CYP4F2, CYP4F3A, CYP4F3B, and CYP4F12), and 1 pmol of purified rabbit CYP4B1.
Immunocytochemical Staining.
A detailed procedure can be found at http://www.abcam.com. After fixation and blocking, ARPE-19 cells were incubated first with anti-CYP4V2 or preimmune rabbit IgG and then with an anticalreticulin antibody. The secondary antibody incubation was performed with a mixture of anti-rabbit IgG antibody conjugated with fluorescein (Thermo Fisher Scientific) and anti-mouse IgG antibody conjugated with Cy5. The processed samples were analyzed by using a Zeiss LSM 510 META laser scanning microscope (Carl Zeiss Inc., Thornwood, NY).
Immunohistochemical Staining.
Tissue sections were first deparaffinized, rehydrated, and treated with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0). Sections were then blocked with secondary antibody host serum and were treated with anti-CYP4V2 antibody or preimmune IgG, followed by a secondary anti-rabbit IgG antibody conjugated with alkaline phosphatase. The sections were observed by using Fast Red and counterstaining with hematoxylin.
DHA and EPA Metabolic Incubations.
Purified CYP4V2 or CYP4F2 (200 pmol) was mixed with purified NADPH-cytochrome P450 reductase and cytochrome b5 at a molar ratio of 1:2:1 or 1:3:4, respectively (Powell et al., 1998; Nakano et al., 2009). 1,2-Dilauroyl-sn-glycero-3-phosphocholine (8 μg) and the substrates (dissolved in ethanol) were added and incubated on ice for 5 min. The mixture was brought to a total volume of 500 μl with 100 mM potassium phosphate buffer, pH 7.4, and was preincubated for 2 min at 37°C in a water bath. Metabolic reactions were initiated with the addition of 1 mM NADPH and were allowed to proceed for 30 min. Reactions were quenched with 500 μl of ice-cold 10% hydrochloric acid. Samples were spiked with the internal standards (i.e., 2.5 μg of 22-hydroxydocosanoic acid and 5 μg of 20-hydroxyeicosanoic acid) and were extracted twice with chloroform containing 0.01% butylated hydroxytoluene. Pooled organic extracts were dried under a N2 stream, reconstituted with ethyl acetate, and methylated with diazomethane. The methylated samples were dried under N2 and reconstituted with a small volume of ethyl acetate, N,O-bis(trimethylsilyl)trifluoroacetate was added, and the samples were heated at 60°C for 45 min before GC-MS analysis. Commercially available CYP4F3B Supersomes were used as a bioreactor to generate chromatographic standards for initial identification of the ω/ω-1-hydroxylated metabolites of DHA and EPA (Fer et al., 2008).
GC-CI/MS and GC-FID Analyses of EPA and DHA Metabolism.
Derivatized extracts were analyzed with a Shimadzu QP2010 gas chromatograph-quadrupole mass spectrometer (Shimadzu, Kyoto, Japan) fitted with a 12-m, fused-silica, capillary column (SPB-1; Sigma-Aldrich) and operating in the CI mode. Derivatized analytes were injected at a temperature of 80°C. After 1 min, the oven temperature was raised at 70°C/min to 200°C, held for 1 min, and then raised at 15°C/min to 300°C. Under these conditions, the derivatized ω- and ω-1-hydroxylated metabolites of DHA eluted at 8.8 and 9.2 min and those of EPA eluted at 7.9 and 8.3 min, respectively. To quantitate the formation of ω-hydroxylated metabolites of DHA and EPA, derivatized analytes were reinjected into a GC system with FID, equipped as described above for the GC-MS analysis. Because of a lack of authentic chemical standards, we constructed standard curves for quantitation with 22-hydroxydocosanoic acid and 20-hydroxyeicosanoic acid; these compounds possess the same carbon-chain lengths as ω-hydroxy-DHA and ω-hydroxy-EPA, respectively, and therefore exhibit equivalent signal intensities in GC-FID analyses. The ω-1-hydroxy-EPA and -DHA metabolites were not quantitated in GC-FID analyses because of low signal intensities.
HepG2 Cells Stably Expressing CYP4V2 and p.H331P.
The CYP4V2WT and CYP4V2H331P genes were amplified from pFastBac plasmids (Nakano et al., 2009) by using the primers 5′-ACGCCTCGAGGCCACCATGGCGGGGCTCTGGCTGG-3′ (forward) and 5′- GATCAAGTTGAAGAGGAGAAATGCAGATGAACGCTAAGGATCCGCGC-3′ (reverse). The PCR-amplified products were cloned into the puc2CL6IN lentiviral vector by using XhoI and BamHI in the multiple cloning sites. Next, HEK293T cells were transfected, by using polyethyleneimine transfection reagent, with 6 μg each of HIV1 helper plasmid pCD/NL-BH, an expression construct for HIV1 gag/pol/rev (Mochizuki et al., 1998), the envelope vector (pczVSV-G) (Pietschmann et al., 1999), and the vector plasmids puc2CL6IN, puc2CL6-4V2wtIN, and puc2CL6-4V2H331PIN, respectively. Viral supernatants were harvested 48 h after transfection, filtered through a 0.45-μm filter, and used to transduce HepG2 cells. After 24 h, the transduced cells were selected with 1 mg/ml G-418 for 7 to 10 days. The established cell lines were passaged at least four times before analysis.
Western Blot Analyses.
CYP4V2 samples (10 μg of microsomal protein from Sf9 cells or 50 μg of total membranes from HepG2 cells) were mixed with Laemmli buffer containing 56 mg/ml dithiothreitol and were boiled. Samples were separated on 9% SDS-PAGE gels and transferred to nitrocellulose membranes, and the membranes were blocked with Odyssey blocking buffer containing 3% goat serum. Membranes were incubated with anti-CYP4V2 and/or mouse monoclonal anti-β-actin, washed with phosphate-buffered saline containing 0.3% Tween 20, and incubated with anti-rabbit IgG antibody conjugated with IR800 dye and/or anti-mouse IgG antibody conjugated with IR680 dye. After washing with phosphate-buffered saline containing 0.3% Tween 20, the CYP4V2 and β-actin bands were observed and quantitated by using the Odyssey system (LI-COR Biosciences).
Lipid Extraction and Lipid Profiling through GC-Electron Impact Ionization/MS Analyses.
Harvested HepG2 cells were sonicated briefly. Cell lysates (500 μg) containing 1 μg of 2,2-d2-stearic acid (internal standard) were brought to a total volume of 500 μl with 100 mM potassium phosphate buffer, pH 7.4. Total lipids were extracted with methanol and methyl-t-butyl ether as described previously (Matyash et al., 2008). The extracted fraction was reconstituted with a small amount of ethyl acetate and was transesterified with methanolic sulfuric acid (10%, v/v; Sigma-Aldrich). Transesterified lipids were analyzed with a Shimadzu QP2010 gas chromatograph-quadrupole mass spectrometer fitted with a 60-m, fused-silica, capillary column (DB-1; Agilent Technologies, Santa Clara, CA). The analytes were injected at a temperature of 100°C. After 2 min, the oven temperature was raised at 30°C/min to 130°C and at 10°C/min to 180°C, held for 2 min, raised at 4°C/min to 210°C and at 10°C/min to 235°C, held for 3 min, raised at 4°C/min to 255°C and at 10°C/min to 310°C, and then held for 4 min. Under these conditions, the derivatized saturated and unsaturated fatty acids were eluted as follow: d2-C18:0, 21.30 min; C14:0, 13.86 min; C16:0, 17.77 min; C16:1, 17.36 min; C18:0, 21.30 min; C18:1, cis-9, 20.91 min; C18:1, cis-11, 21.01 min; C20:4n-6, 23.66 min; C20:5n-3, 23.57 min; C22:6n-3, 27.65 min (Supplemental Table 1). Quantitation was achieved through selected-ion monitoring of the characteristic [M−31]+ and [M−43]+ fragment ions found in electron ionization spectra of fatty acid methyl esters. Standard curves (r2 > 0.98) for each fatty acid were generated with the authentic chemical standards. Differences between two means within a group were tested with Student's t test (two-tail, two-sample, unequal-variance) by using Prism (GraphPad Software Inc., La Jolla, CA).
Results
Gene Expression of Cytochrome P450s in ARPE-19 Cells.
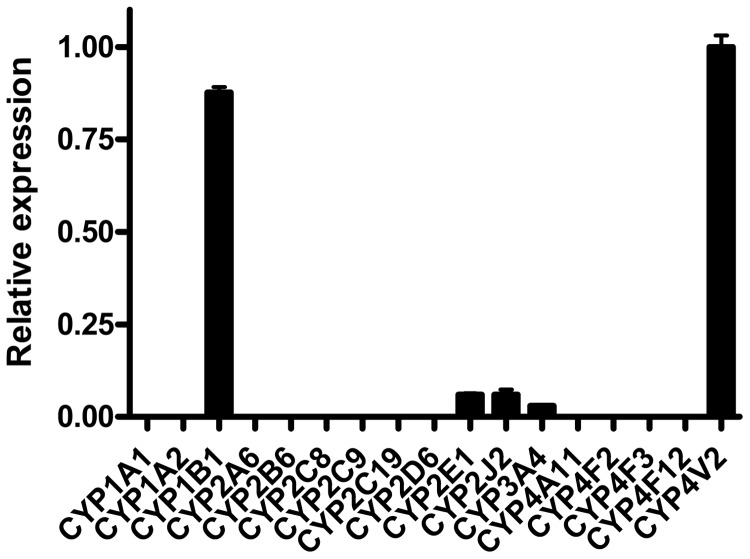
The ARPE-19 cell line is a human retinal pigmented epithelial cell line that arose spontaneously from tissue from a healthy individual (Dunn et al., 1996). Although ARPE-19 cells are a well established model system for studying the function of retinal pigmented epithelium, their expression of cytochrome P450 enzymes had not been determined previously. To characterize these cells more thoroughly for in vitro studies, we extracted total RNA from ARPE-19 cells and quantitated gene expression of CYP4V2 in relation to the known ocular P450 CYP1B1, the well established fatty acid hydroxylases CYP4A and CYP4F, and several other key human P450s. RT-PCR analysis of total RNA from ARPE-19 cells revealed threshold cycle values for CYP4V2 of 29.3 ± 0.08, compared with values of 26.6 ± 0.24 for the housekeeping gene GusB, which demonstrates robust CYP4V2 expression in this cell line (Fig. 1). In addition, we detected high levels of expression of CYP1B1, which was transcribed at a similar level (88% of CYP4V2 mRNA expression) in these cells. CYP2E1, CYP2J2, and CYP3A4 were transcribed at only low levels (5% of CYP4V2 mRNA expression), and transcripts for CYP4A11, CYP4B1, CYP4F2, CYP4F3, and CYP4F12 were not detectable (Fig. 1).
Fig. 1.
Real-time RT-PCR analysis of P450 expression in ARPE-19 cells. CYP4V2 was the dominantly expressed P450 of 17 genes examined. The relative expression was calculated by using the threshold cycle difference method, with CYP4V2 as a calibrator. Error bars, S.D. of triplicate experiments.
CYP4V2 Protein Expression in ARPE-19 Cells.
To determine whether CYP4V2 protein, as well as its mRNA, was abundant in ARPE-19 cells, we generated a highly specific polyclonal antibody against purified human CYP4V2 (see below). As proof of the specificity of the antibody, the cross-reactivity of the anti-CYP4V2 antibody with other human CYP4 enzymes was determined through Western blotting. We used rabbit CYP4B1 prepared in our laboratory (87% sequence identity with respect to human CYP4B1) because the human enzyme is not commercially available. As expected from the low level of sequence identity of CYP4V2 with respect to other CYP4 members (Rettie and Kelly, 2008), the rabbit anti-CYP4V2 antibody did not recognize 1 pmol each of CYP4A11, CYP4F2, CYP4F3A/B, CYP4F12, and rabbit CYP4B1, under conditions in which 0.2 pmol of CYP4V2 provided a strong signal (Fig. 2a).
Fig. 2.
Cross-reactivity of anti-CYP4V2 IgG with other CYP4 enzymes and CYP4V2 protein expression in ARPE-19 cell lysate. The anti-CYP4V2 antibody specifically reacted with CYP4V2. a, lanes 1, 5, and 9, 0.2 pmol of CYP4V2; lane 2, 1 pmol of rabbit CYP4B1; lane 3, 1 pmol of CYP4A11; lane 4, 1 pmol of CYP4F2; lane 6, 1 pmol of CYP4F3A; lane 7, 1 pmol of CYP4F3B; lane 8, 1 pmol of CYP4F12. b, left lanes, 330, 160, and 82 fmol of purified CYP4V2; right lanes, 10 and 30 μg of ARPE-19 cell lysate. CYP4V2 was expressed at a concentration of ∼6 pmol/mg of cell lysate. Band intensities were analyzed with an Odyssey system.
Next, we performed quantitative Western blot analyses of ARPE-19 cell lysates. As shown in Fig. 2b, CYP4V2 was readily detectable at levels that averaged 6 pmol/mg of protein. Immunocytochemical analyses revealed that CYP4V2 colocalized with calreticulin (Supplemental Fig. 1), a well characterized marker of endoplasmic reticulum. Therefore, CYP4V2 was expressed in ARPE-19 cells at significant levels and was correctly localized to microsomal membranes.
Immunohistochemical Analyses of Human Ocular Tissues.
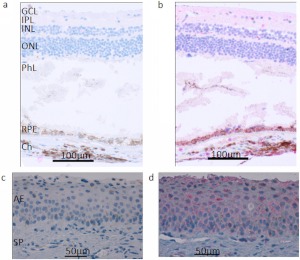
Because patients with BCD exhibit crystal deposits mainly in the retina and the cornea, we used the anti-CYP4V2 antibody to analyze the expression of CYP4V2 protein in these human ocular tissues from healthy individuals. As shown in Fig. 3, RPE cells stained strongly positive and corneal epithelium moderately positive for CYP4V2 expression, which demonstrates expression of CYP4V2 protein in the disease-targeted ocular tissues of BCD and thus confirms the mRNA expression data.
Fig. 3.
CYP4V2 expression in normal human retina and cornea. Immunohistochemical staining with the anti-CYP4V2 IgG showed strong positive staining of RPE cells in retinal tissue and weak staining of ganglion cells and internal/external nuclear layers in the retina and corneal epithelial cells. a, retina treated with preimmune IgG; b, retina treated with anti-CYP4V2 IgG; c, cornea treated with preimmune IgG; d, cornea treated with anti-CYP4V2 IgG. GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; ONL, outer nuclear layer; PhL, photoreceptor layer; Ch, choroid; AE, anterior epithelium; SP, substantia propria.
Tissue Distribution of CYP4V2 Protein.
Li et al. (2004) demonstrated the almost ubiquitous mRNA expression of CYP4V2 in human organs. Taking advantage of the newly generated anti-CYP4V2 antibody, our immunohistochemical analyses demonstrated that human liver, reproductive organs, and endocrine organs all stained strongly positive for the CYP4V2 protein. We found strong expression in tissues and cells with major roles in fatty acid and steroid metabolism, such as hepatocytes in liver, islet cells in pancreas, endothelial cells in hypophysis, prostate gland, adrenal gland, testis, breast, and parathyroid gland, and glandular epithelial cells in uterus (Supplemental Table 2). To analyze CYP4V2 expression specifically in drug-metabolizing tissues, microsomes were prepared from four individual kidneys, four small intestines, and nine livers and were examined in Western blot analyses. As shown in Western blot analyses (Supplemental Fig. 3), CYP4V2 protein was readily detectable in human liver, whereas only a negligible amount was found in human kidney, with no expression in small intestine.
Characterization of Purified CYP4V2.
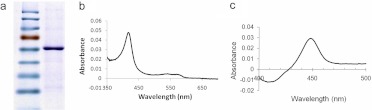
To characterize further the biochemical activity of CYP4V2, histidine-tagged CYP4V2 was overexpressed in insect cells and isolated through nickel affinity and hydroxyapatite chromatography. SDS-PAGE analysis revealed a single major band of apparent molecular mass of ∼60 kDa (Fig. 4a), which was confirmed as 62,163 Da through liquid chromatography-MS analysis (data not shown). These experimentally determined values closely matched the calculated molecular mass of CYP4V2 with its heme prosthetic group covalently bound (62,160 Da) (Cheesman et al., 2003). CYP4V2 was isolated mainly in its low-spin form, as indicated by the prominent Soret band in the absolute oxidized spectrum at 419 nm and the minor α and β bands at 571 and 535 nm, respectively (Fig. 4b). CYP4V2 displayed a typical reduced carbon monoxide-bound P450 spectrum, with a Soret maximum near 450 nm (Fig. 4c). No cytochrome P420 was observed in the final enzyme preparation, which had a specific content of 9.2 nmol/mg of protein, a finding that indicated that >50% was holoenzyme.
Fig. 4.
Characterization of purified His6-CYP4V2 through SDS-PAGE and UV spectrometric analyses. a, purified CYP4V2 migrated with an apparent molecular mass of ∼60 kDa. b, the absolute UV spectrum of CYP4V2 exhibited a Soret band at 419 nm and α and β bands at 571 and 535 nm. c, the reduced carbon monoxide-bound difference spectrum of purified CYP4V2 demonstrated the absence of cytochrome P420.
CYP4V2-Dependent ω-3-PUFA Metabolism.
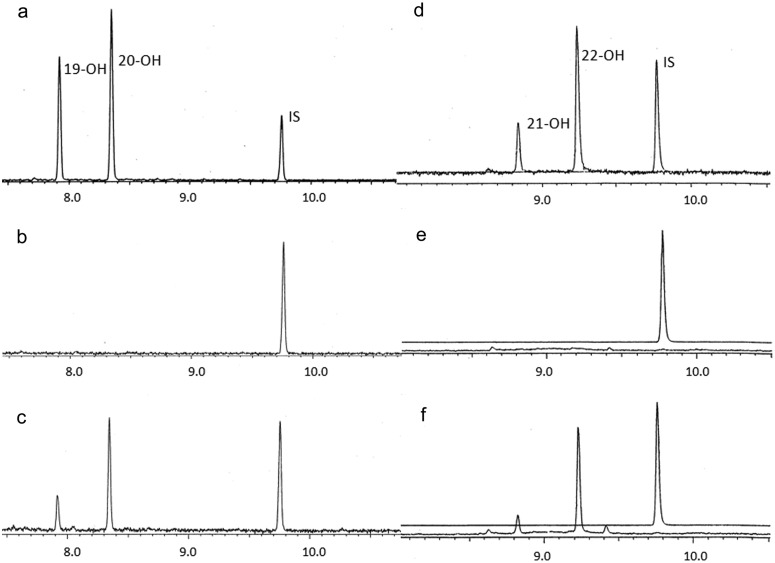
The main physiological function of normal RPE cells is to import PUFAs from the bloodstream and to recycle them to maintain lipid homeostasis in photoreceptors (Bazan, 2006). Therefore, we investigated the ability of purified CYP4V2 protein to metabolize the common ω-3-PUFAs EPA and DHA, relative to commercially available CYP4F3B Supersomes and purified CYP4F2. First, we used CYP4F3B Supersomes as a bioreactor (Harmon et al., 2006; Fer et al., 2008) to generate metabolite standards for ω-1-hydroxy-EPA, ω-1-hydroxy-DHA, ω-hydroxy-EPA, and ω-hydroxy-DHA (Fig. 5, a and d). Additional evidence for the assignment of peaks as 19-hydroxy-EPA, 20-hydroxy-EPA, 21-hydroxy-DHA, and 22-hydroxy-DHA was obtained from GC-CI/MS analysis of the N,O-bis(trimethylsilyl) trifluoroacetate-derivatized alcohols. These spectra yielded prominent fragment ions at m/z 389 ([M−15]+) and m/z 315 ([M−89]+) for the first and second peaks and at m/z 415 ([M−15]+) and m/z 341 ([M−89]+) for the third and fourth peaks (data not shown). Purified reconstituted CYP4V2 formed each of these metabolites in a NADPH-dependent manner (compare Fig. 5, b and c, with Fig. 5, e and f) and with a higher selectivity for ω/ω-1-hydroxylation, compared with CYP4F3B. Second, we quantitated the rates of formation of 22-hydroxy-DHA and 20-hydroxy-EPA by purified reconstituted CYP4V2 (and CYP4F2; data not shown) and found them to be 0.30 ± 0.03 nmol · min−1 · nmol−1 (CYP4F2, 0.23 ± 0.04 nmol · min−1 · nmol−1) and 0.19 ± 0.02 nmol · min−1 · nmol−1 (CYP4F2, 0.23 ± 0.01 nmol · min−1 · nmol−1), respectively. Finally, we attempted to quantitate the efficiency (Vmax/Km) of PUFA ω-hydroxylation by CYP4V2. Solubility problems and substrate inhibition at concentrations above 80 μM DHA/EPA complicated the analysis but, under comparable incubation conditions for the respective purified reconstituted enzymes, CYP4V2 metabolized DHA slightly more rapidly and metabolized EPA at an equivalent rate, compared with CYP4F2.
Fig. 5.
EPA and DHA metabolism catalyzed by purified CYP4V2 reconstituted with P450 reductase and cytochrome b5. a, EPA incubated with CYP4F3B Supersomes plus NADPH. b, EPA incubated with reconstituted CYP4V2 minus NADPH. c, EPA incubated with reconstituted CYP4V2 plus NADPH. d, DHA incubated with CYP4F3B Supersomes plus NADPH. e, DHA incubated with reconstituted CYP4V2 minus NADPH. f, DHA incubated with reconstituted CYP4V2 plus NADPH. Peaks were identified as 19-hydroxy-EPA, 20-hydroxy-EPA, 21-hydroxy-DHA, and 22-hydroxy-DHA. IS, internal standard.
Characterization of CYP4V2 p.H331P Variant Expression and Functional Activity.
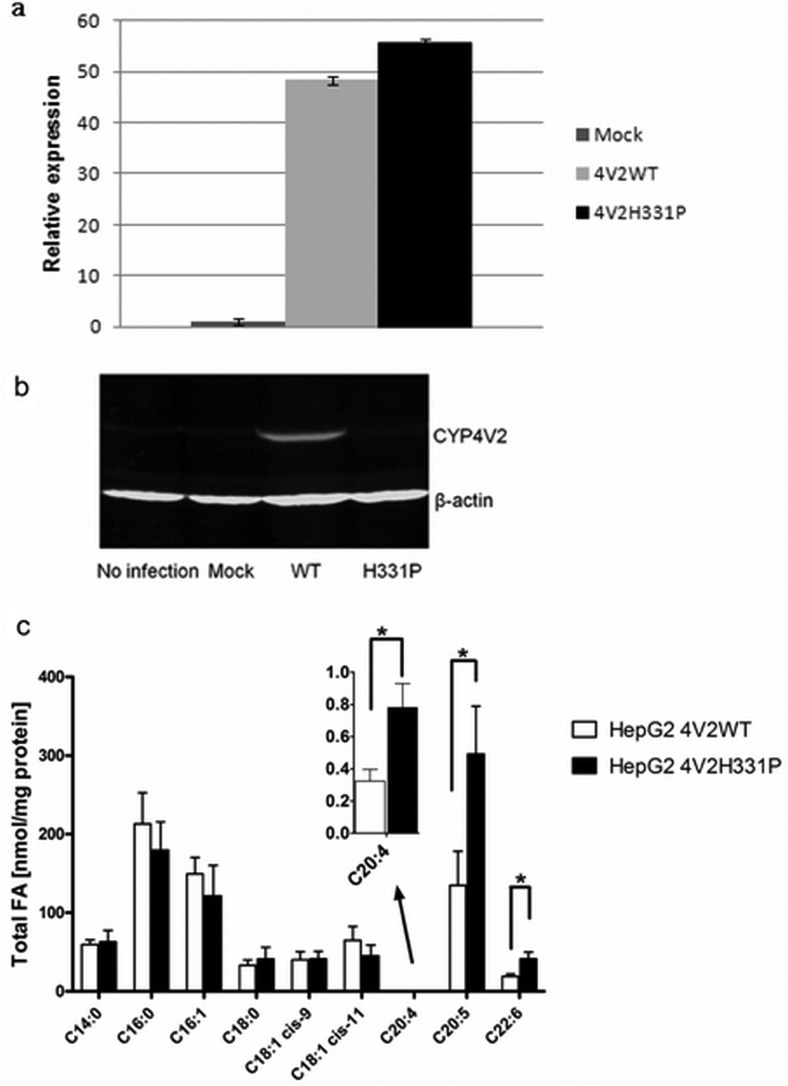
Finally, to evaluate the expression and functional consequences of the p.H331P mutation, relative to wild-type CYP4V2, we expressed both enzymes in HepG2 cells by using lentiviral expression vectors. mRNA analysis revealed comparable levels of expression for the wild-type (CYP4V2WT) and mutant (CYP4V2H331P) transcripts (Fig. 6a). However, steady-state protein expression levels were much higher for wild-type CYP4V2, compared with the mutant (Fig. 6b). Finally, lipid analysis revealed significantly lower levels of EPA and DHA (and arachidonic acid) in HepG2 cells expressing wild-type CYP4V2, compared with cells expressing the mutant enzyme (Fig. 6c). These lipid profiling data are consistent with reduced metabolism of PUFAs by the p.H331P variant.
Fig. 6.
Expression and functional consequences of the p.H331P variant of CYP4V2 stably transduced in HepG2 cells. a, real-time RT-PCR analysis of CYP4V2 wild-type and p.H331P mRNA in HepG2 cells. b, Western blot analysis of CYP4V2 wild-type and p.H133P protein in HepG2 cells. β-Actin was detected as a loading control. c, DHA, EPA, and arachidonic acid accumulation in p.H331P-transfected cells, compared with cells expressing the wild-type enzyme. Data are the mean and S.D. of triplicate experiments. WT, wild-type; FA, fatty acids. *, p < 0.02.
Discussion
Genetic defects in the cytochrome P450-dependent metabolism of endogenous substrates are recognized to underlie numerous diseases states. For example, steroid metabolism defects attributable to mutations in CYP7B1, CYP11A1, and CYP17A1 cause spastic paraplegia type 5, lipoid adrenal hyperplasia, and mineralocorticoid excess syndromes, respectively (Nebert and Russell, 2002; Russell et al., 2009). In terms of ocular disease, mutations in CYP1B1 are a well established cause of primary congenital glaucoma (Sarfarazi et al., 1995; Stoilov et al., 1997), although other loci, notably LTBP2, seem to be involved in the development of the disease (Ali et al., 2009). In contrast, mutations in CYP4V2 are singularly associated with BCD, a situation that provides experimental opportunities to decipher the endogenous substrates for the enzyme and perhaps also to shed light on the pathophysiological features of BCD.
If CYP4V2 plays a significant metabolic role in BCD, then it seems important that the enzyme be localized to target tissues for BCD. CYP2A6 and CYP2C8 mRNAs were identified previously in tissue homogenates of human retina/choroid through RT-PCR assays (Zhang et al., 2008) and expression of CYP4V2 mRNA in the retina and in RPE cells was reported (Li et al., 2004), but no quantitative data are available. In the present study, we found CYP4V2 and CYP1B1 to be by far the most highly expressed P450 mRNAs in ARPE-19 cells. In fact, of the 15 other P450s that were analyzed, only mRNAs for CYP2E1, CYP2J2, and CYP3A4 were quantifiable, and levels reached <10% of CYP4V2 levels. CYP4A and CYP4F are expressed at significant levels in human liver and especially kidney, where they are thought to have important roles in 20-hydroxyeicosatetraenoic acid formation (Lin et al., 1994; Zou et al., 1994; Carroll et al., 1997). In marked contrast, the mRNA data from ARPE-19 cells suggested that those more-commonly studied CYP4 enzymes may be functionally silent in these naturally transformed human RPE cells. To facilitate analysis of CYP4V2 protein expression, we expressed and purified the recombinant enzyme, to generate a highly selective polyclonal antibody for Western blotting and immunohistochemical analysis of target tissues in BCD. CYP4V2 protein was detected readily in human retinal epithelium and the endoplasmic reticulum of ARPE-19 cells and to a lesser extent in corneal epithelium. Therefore, CYP4V2 is clearly present in ocular cells that are affected in BCD and may be the dominant functional CYP4 enzyme at these sites. These studies establish ARPE-19 cells as an important tool for evaluating BCD-related biochemical changes in a disease-relevant cell line after knockdown of the endogenous enzyme.
We demonstrated previously that baculovirus-mediated expression of CYP4V2 in insect cells yielded a metabolically competent ω-hydroxylase for several saturated fatty acids that was sensitive to inhibition by N-hydroxy-N′-(4-butyl-2-methylphenyl)formamidine (HET0016) at nanomolar concentrations (Nakano et al., 2009). Therefore, despite its low level of sequence identity with other human CYP4 enzymes, CYP4V2 exhibits their prototypic functional characteristics. In the present study, we purified CYP4V2 to near-homogeneity and found that the recombinant enzyme was isolated from this expression system in the low-spin (ligand-free) state. To probe further the substrate specificity of the enzyme, we turned our attention to fatty acids that are present at high concentrations in the retina.
The physiological features of the retina have been studied extensively; photoreceptor cells (i.e., rods and cones) are especially well characterized. The rod outer segment contains rhodopsin-enriched disk membranes, and the lipids of these disk membranes are predominantly phosphatidylcholine and phosphatidylethanolamine. The majority of these lipids are composed of palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), and DHA (C22:6n-3), whereas EPA (C20:5n-3) is a minor retinal lipid (Anderson, 1970). In fact, DHA in the rod outer segments of rats represents ∼50% of the total fatty acids esterified to phosphatidylethanolamine and phosphatidylserine (Fliesler and Anderson, 1983). Rod outer segments together with RPE cells possess an efficient lipid-recycling system to regenerate disk membranes and to maintain high PUFA contents (Bazan, 2006). It is noteworthy that 10% of the rod outer segment is renewed daily in primates (Young, 1967, 1971), and the rates of synthesis and disposal of disk membranes in mice are approximately the same under normal conditions (Young, 1971; LaVail, 1973). Because DHA homeostasis is so tightly regulated in RPE cells, we compared the substrate specificity of CYP4V2 toward the ω-3-PUFAs EPA and DHA.
ω-3-PUFAs would be expected to undergo rate-limiting CYP4-dependant ω-oxidation followed by β-oxidation in peroxisomes and mitochondria (Hardwick, 2008). CYP4F family enzymes, principally CYP4F2 and CYP4F3B, were reported to be the main PUFA hydroxylases in human liver (Fer et al., 2008). The current study extends the repertoire of PUFA hydroxylases to CYP4V2, which we found to catalyze preferentially the ω- and ω-1-hydroxylation of EPA and DHA. Purified CYP4V2 and CYP4F2 metabolized these two PUFAs at similar rates but, because CYP4F enzymes were not expressed at significant levels in ARPE-19 cells (or corneal CRL11515 cells; data not shown), CYP4V2 is likely to dominate CYP4-dependent PUFA catalysis in BCD target tissues in the eye. Moreover, CYP4V2 is expressed at readily detectable levels in human liver and, for some subjects, the enzyme may contribute to the hepatic metabolism of ω-3-PUFAs. Additional studies are needed to test this hypothesis.
BCD is an autosomal recessive disease, and many discrete mutations of CYP4V2 have been identified, the most frequent of which is alternative splicing at the splice-acceptor site for exon 7 (Kaiser-Kupfer et al., 1994; Lee et al., 2001, 2005; Li et al., 2004; Lin et al., 2005; Shan et al., 2005). As a result, 17 base pairs at the 3′-end of intron 6 and the 5′-end of exon 7 are missing. Consequently, exon 7 is deleted and the resulting truncated enzyme presumably is nonfunctional. We focused here on the p.H331P mutation, the second most frequent mutation, to demonstrate unequivocally that a BCD-associated mutation abrogates enzyme activity, because of the relative ease of recombinant expression and subsequent analysis of this coding-region mutant. We initially expected some loss of function because of the critical location of His331 within the most conserved region of the I-helix, which constitutes part of the active site (Supplemental Fig. 4), because this might be expected to affect ligand binding or apoprotein formation. It is noteworthy that upon expression in HepG2 cells, the p.H331P mutant protein did not accumulate despite reasonable levels of mRNA expression. Therefore, the mechanism for loss of function in mammalian cells through expression of the p.H331P variant is post-transcriptional and likely involves either increased degradation secondary to reduced protein stability or impaired translation.
Finally, comparative lipid profiling studies with HepG2 cells stably transfected with wild-type CYP4V2 or the p.H331P variant revealed significant decreases in the total levels of several PUFAs in the former cell line (Fig. 6c). These studies complement the finding that DHA and EPA are substrates for reconstituted CYP4V2. It should be noted that CYP4V2 is not selective for DHA metabolism, compared with EPA metabolism, as assessed in either the catalytic studies with purified protein or the comparative lipidomic analyses. Another caveat is that the latter experiments were conducted with HepG2 cells and not retinal or corneal cell lines; therefore, the relevance of altered PUFA metabolism to the disease mechanism of BCD remains to be established.
In summary, we examined CYP4V2 expression and organ distribution with an emphasis on ocular tissues. We also evaluated the substrate specificity of the enzyme for PUFAs and characterized the functional deficit of the p.H331P mutant found in BCD. We demonstrated that 1) CYP4V2 is a major form of P450 expressed in retinal cells, 2) the enzyme possesses ω-hydroxylase activity toward ω-3-PUFAs, 3) the most common nonsynonymous single-nucleotide polymorphism found in BCD encodes a nonfunctional protein, and 4) overexpression of CYP4V2 in HepG2 cells alters lipid homeostasis. Because CYP4V2 seems to be the only CYP4 present at significant levels in retinal cells, it may be a prominent contributor to local metabolism of PUFAs in retinal cells.
Supplementary Material
Acknowledgments
We thank Dr. Jamil Haque for recombinant expression of CYP4F2 in insect cells and Chelsea Stewart for expression and purification of P450 coenzymes. We also thank Eric D. Kantor and Dr. Gail D. Anderson for assistance with GC-FID analyses and Kelly Hudkins for scoring of anti-CYP4V2-stained histological tissue sections.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM49054]; and the Drug Metabolism Transport and Pharmacogenetics Research Fund in the School of Pharmacy, University of Washington.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- BCD
- Bietti's crystalline corneoretinal dystrophy
- P450
- cytochrome P450
- DHA
- docosahexaenoic acid
- EPA
- eicosapentaenoic acid
- PUFA
- polyunsaturated fatty acid
- RPE
- retinal pigmented epithelium
- PCR
- polymerase chain reaction
- GC
- gas chromatography
- PAGE
- polyacrylamide gel electrophoresis
- RT
- reverse transcription
- CI
- chemical ionization
- MS
- mass spectrometry
- FID
- flame ionization detection
- HET0016
- N-hydroxy-N′-(4-butyl-2-methylphenyl)formamidine.
Authorship Contributions
Participated in research design: Nakano, Kelly, Hanenberg, and Rettie.
Conducted experiments: Nakano and Wiek.
Contributed new reagents or analytic tools: Nakano, Wiek, and Hanenberg.
Performed data analysis: Nakano, Kelly, and Rettie.
Wrote or contributed to the writing of the manuscript: Nakano, Kelly, Hanenberg, and Rettie.
References
- Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, et al. (2009) Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet 84:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RE. (1970) Lipids of ocular tissues. IV. A comparison of the phospholipids from the retina of six mammalian species. Exp Eye Res 10:339–344 [DOI] [PubMed] [Google Scholar]
- Bazan NG. (2006) Cell survival matters: docosahexaenoic acid signaling, neuroprotection and photoreceptors. Trends Neurosci 29:263–271 [DOI] [PubMed] [Google Scholar]
- Bietti G. (1937) Ueber familiaeres vorkommen von “retinitis punctata albescens” (verbunden mit“dystrophia marginalis cristallinea cormeae”), glitzern des glaskoerpers und anderen degenerativen augenveraenderungen [in German]. Klin Mbl Augenheilk 99:737–757 [Google Scholar]
- Carroll MA, Balazy M, Huang DD, Rybalova S, Falck JR, McGiff JC. (1997) Cytochrome P450-derived renal HETEs: storage and release. Kidney Int 51:1696–1702 [DOI] [PubMed] [Google Scholar]
- Cheesman MJ, Baer BR, Zheng YM, Gillam EM, Rettie AE. (2003) Rabbit CYP4B1 engineered for high-level expression in Escherichia coli: ligand stabilization and processing of the N-terminus and heme prosthetic group. Arch Biochem Biophys 416:17–24 [DOI] [PubMed] [Google Scholar]
- Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. (1996) ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res 62:155–169 [DOI] [PubMed] [Google Scholar]
- Fer M, Corcos L, Dréano Y, Plée-Gautier E, Salaün JP, Berthou F, Amet Y. (2008) Cytochromes P450 from family 4 are the main omega hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. J Lipid Res 49:2379–2389 [DOI] [PubMed] [Google Scholar]
- Fliesler SJ, Anderson RE. (1983) Chemistry and metabolism of lipids in the vertebrate retina. Prog Lipid Res 22:79–131 [DOI] [PubMed] [Google Scholar]
- Gekka T, Hayashi T, Takeuchi T, Goto-Omoto S, Kitahara K. (2005) CYP4V2 mutations in two Japanese patients with Bietti's crystalline dystrophy. Ophthalmic Res 37:262–269 [DOI] [PubMed] [Google Scholar]
- Hardwick JP. (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol 75:2263–2275 [DOI] [PubMed] [Google Scholar]
- Harmon SD, Fang X, Kaduce TL, Hu S, Raj Gopal V, Falck JR, Spector AA. (2006) Oxygenation of omega-3 fatty acids by human cytochrome P450 4F3B: effect on 20-hydroxyeicosatetraenoic acid production. Prostaglandins Leukot Essent Fatty Acids 75:169–177 [DOI] [PubMed] [Google Scholar]
- Hu D. (1983) Ophthalmic genetics in China. Ophthalmic Paediatr Genet 2:39–45 [Google Scholar]
- Jiao X, Munier FL, Iwata F, Hayakawa M, Kanai A, Lee J, Schorderet DF, Chen MS, Kaiser-Kupfer M, Hejtmancik JF. (2000) Genetic linkage of Bietti crystallin corneoretinal dystrophy to chromosome 4q35. Am J Hum Genet 67:1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser-Kupfer MI, Chan CC, Markello TC, Crawford MA, Caruso RC, Csaky KG, Guo J, Gahl WA. (1994) Clinical biochemical and pathologic correlations in Bietti's crystalline dystrophy. Am J Ophthalmol 118:569–582 [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Nakano M, Rohatgi P, Yarov-Yarovoy V, Rettie AE. (2011) Finding homes for orphan cytochrome P450s: CYP4V2 and CYP4F22 in disease states. Mol Interv 11:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai TY, Chu KO, Chan KP, Ng TK, Yam GH, Lam DS, Pang CP. (2010) Alterations in serum fatty acid concentrations and desaturase activities in Bietti crystalline dystrophy unaffected by CYP4V2 genotypes. Invest Ophthalmol Vis Sci 51:1092–1097 [DOI] [PubMed] [Google Scholar]
- LaVail MM. (1973) Kinetics of rod outer segment renewal in the developing mouse retina. J Cell Biol 58:650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jiao X, Hejtmancik JF, Kaiser-Kupfer M, Gahl WA, Markello TC, Guo J, Chader GJ. (2001) The metabolism of fatty acids in human Bietti crystalline dystrophy. Invest Ophthalmol Vis Sci 42:1707–1714 [PubMed] [Google Scholar]
- Lee KY, Koh AH, Aung T, Yong VH, Yeung K, Ang CL, Vithana EN. (2005) Characterization of Bietti crystalline dystrophy patients with CYP4V2 mutations. Invest Ophthalmol Vis Sci 46:3812–3816 [DOI] [PubMed] [Google Scholar]
- Li A, Jiao X, Munier FL, Schorderet DF, Yao W, Iwata F, Hayakawa M, Kanai A, Shy Chen M, Alan Lewis R, et al. (2004) Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am J Hum Genet 74:817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Abraham NG, Schwartzman ML. (1994) Cytochrome P450 arachidonic acid omega-hydroxylation in the proximal tubule of the rat kidney. Ann NY Acad Sci 744:11–24 [DOI] [PubMed] [Google Scholar]
- Lin J, Nishiguchi KM, Nakamura M, Dryja TP, Berson EL, Miyake Y. (2005) Recessive mutations in the CYP4V2 gene in East Asian and Middle Eastern patients with Bietti crystalline corneoretinal dystrophy. J Med Genet 42:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49:1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J. (1998) High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol 72:8873–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Rettie AE. (2009) Expression and characterization of CYP4V2 as a fatty acid omega-hydroxylase. Drug Metab Dispos 37:2119–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Russell DW. (2002) Clinical importance of the cytochromes P450. Lancet 360:1155–1162 [DOI] [PubMed] [Google Scholar]
- Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. (1999) Foamy virus capsids require the cognate envelope protein for particle export. J Virol 73:2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell PK, Wolf I, Jin R, Lasker JM. (1998) Metabolism of arachidonic acid to 20-hydroxy-5,8,11,14-eicosatetraenoic acid by P450 enzymes in human liver: involvement of CYP4F2 and CYP4A11. J Pharmacol Exp Ther 285:1327–1336 [PubMed] [Google Scholar]
- Rettie AE, Kelly EJ. (2008) The CYP4 family, in Cytochrome P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics (Ioannides C. ed) pp 385–407, Royal Society of Chemistry, London [Google Scholar]
- Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. (2009) Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem 78:1017–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfarazi M, Akarsu AN, Hossain A, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS. (1995) Assignment of a locus (GLC3A) for primary congenital glaucoma (buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics 30:171–177 [DOI] [PubMed] [Google Scholar]
- Shan M, Dong B, Zhao X, Wang J, Li G, Yang Y, Li Y. (2005) Novel mutations in the CYP4V2 gene associated with Bietti crystalline corneoretinal dystrophy. Mol Vis 11:738–743 [PubMed] [Google Scholar]
- Stoilov I, Akarsu AN, Sarfarazi M. (1997) Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6:641–647 [DOI] [PubMed] [Google Scholar]
- Wada Y, Itabashi T, Sato H, Kawamura M, Tada A, Tamai M. (2005) Screening for mutations in CYP4V2 gene in Japanese patients with Bietti's crystalline corneoretinal dystrophy. Am J Ophthalmol 139:894–899 [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Weleber RG, Klein ML, Welch RB, Green WR. (1989) Bietti's crystalline dystrophy. A clinicopathologic correlative study. Arch Ophthalmol 107:213–221 [DOI] [PubMed] [Google Scholar]
- Xiao X, Mai G, Li S, Guo X, Zhang Q. (2011) Identification of CYP4V2 mutation in 21 families and overview of mutation spectrum in Bietti crystalline corneoretinal dystrophy. Biochem Biophys Res Commun 409:181–186 [DOI] [PubMed] [Google Scholar]
- Yokoi Y, Nakazawa M, Mizukoshi S, Sato K, Usui T, Takeuchi K. (2010) Crystal deposits on the lens capsules in Bietti crystalline corneoretinal dystrophy associated with a mutation in the CYP4V2 gene. Acta Ophthalmol 88:607–609 [DOI] [PubMed] [Google Scholar]
- Young RW. (1967) The renewal of photoreceptor cell outer segments. J Cell Biol 33:61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. (1971) Shedding of discs from rod outer segments in the rhesus monkey. J Ultrastruct Res 34:190–203 [DOI] [PubMed] [Google Scholar]
- Zhang T, Xiang CD, Gale D, Carreiro S, Wu EY, Zhang EY. (2008) Drug transporter and cytochrome P450 mRNA expression in human ocular barriers: implications for ocular drug disposition. Drug Metab Dispos 36:1300–1307 [DOI] [PubMed] [Google Scholar]
- Zheng YM, Baer BR, Kneller MB, Henne KR, Kunze KL, Rettie AE. (2003) Covalent heme binding to CYP4B1 via Glu310 and a carbocation porphyrin intermediate. Biochemistry 42:4601–4606 [DOI] [PubMed] [Google Scholar]
- Zou AP, Imig JD, Ortiz de Montellano PR, Sui Z, Falck JR, Roman RJ. (1994) Effect of P-450 omega-hydroxylase metabolites of arachidonic acid on tubuloglomerular feedback. Am J Physiol 266:F934–F941 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.