Abstract
Previously we demonstrated that aldehyde dehydrogenase (ALDH) 1a1 is the major ALDH expressed in mouse liver and is an effective catalyst in metabolism of lipid aldehydes. Quantitative real-time polymerase chain reaction analysis revealed a ≈2.5- to 3-fold induction of the hepatic ALDH1A1 mRNA in mice administered either acrolein (5 mg/kg acrolein p.o.) or butylated hydroxylanisole (BHA) (0.45% in the diet) and of cytosolic NAD+-dependent ALDH activity. We observed ≈2-fold increases in ALDH1A1 mRNA levels in both Nrf2(+/+) and Nrf2(−/−) mice treated with BHA compared with controls, suggesting that BHA-induced expression is independent of nuclear factor E2-related factor 2 (Nrf2). The levels of activator protein-1 (AP-1) mRNA and protein, as well as the amount of phosphorylated c-Jun were significantly increased in mouse liver or Hepa1c1c7 cells treated with either BHA or acrolein. With use of luciferase reporters containing the 5′-flanking sequence of Aldh1a1 (−1963/+27), overexpression of c-Jun resulted in an ≈4-fold induction in luciferase activity, suggesting that c-Jun transactivates the Aldh1a1 promoter as a homodimer and not as a c-Jun/c-Fos heterodimer. Promoter deletion and mutagenesis analyses demonstrated that the AP-1 site at position −758 and possibly −1069 relative to the transcription start site was responsible for c-Jun-mediated transactivation. Electrophoretic mobility shift assay analysis with antibodies against c-Jun and c-Fos showed that c-Jun binds to the proximal AP-1 site at position −758 but not at −1069. Recruitment of c-Jun to this proximal AP-1 site by BHA was confirmed by chromatin immunoprecipitation analysis, indicating that recruitment of c-Jun to the mouse Aldh1a1 gene promoter results in increased transcription. This mode of regulation of an ALDH has not been described before.
Introduction
The ALDH gene superfamily encodes enzymes that catalyze the NAD(P)+-dependent oxidation of aldehydes generated from a wide variety of endogenous and exogenous processes to their corresponding carboxylic acids. At present, 19 functional ALDH genes have been identified in the human genome (Marchitti et al., 2008). Among the ALDH isoforms, cytosolic mouse ALDH1A1 plays a critical role in oxidative metabolism of acrolein and 4-hydroxy-2-nonenal and protects hepatocytes from damage caused by these aldehydes (Makia et al., 2011). The Aldh1a1 gene is highly expressed in the liver, lung, lens, gonads, and retina (Hsu et al., 1999, 2000) and increased expression in the liver and lens has been postulated as the mechanism to protect these organs against oxidant and electrophile-induced cellular damage. Murine ALDH1A1 is important in retinoic acid (RA) biosynthesis by efficiently catalyzing the oxidation of all-trans- and 9-cis-retinal (Molotkov and Duester, 2003). It regulates normal growth, differentiation, and development of adult epithelia by synthesizing RA, a ligand for the nuclear RA receptor and retinoid X receptor. The Aldh1a1 gene is expressed at high levels in mouse hematopoietic stem cells and is critical for hematopoietic stem cell differentiation and function (Gasparetto et al., 2012) Because the substrate specificity and kinetics of murine and human ALDH1A1 are similar based on our studies and those of others (Xiao et al., 2009; Makia et al., 2011), we characterized the molecular regulation of murine Aldh1a1. Such information may assist interpretation of future research using transgenic models to study aldehyde toxicity in atherosclerosis and cardiovascular medicine.
Although the human and mouse Aldh1a1 genes have been cloned and the promoter region characterized, little is known about the molecular mechanisms that regulate Aldh1a1 expression in mouse liver (Hsu et al., 1999; Elizondo et al., 2009). Previous studies identified a putative RA response element located at position −91/−75 adjacent to the CCAAT box of the hALDH1A1 gene, which is conserved in mouse Aldh1a1. This RA response element is responsible for RA-mediated down-regulation of hALDH1A1 and mouse Aldh1a1 gene expression through interaction of the RA receptor and CCAAT/enhancer-binding protein β in human (HepG2) and mouse (Hepa1) hepatoma cells, respectively (Elizondo et al., 2000, 2009). The down-regulation of ALDH1A1 by elevated hepatic RA is a negative feedback pathway to control RA biosynthesis because ALDH1A1 is a major enzyme involved in the biosynthesis of RA. Alnouti and Klaassen (2008) also reported induction of the Aldh1a1 gene in mouse liver by constitutive androstane receptor activators, presumably operating through a constitutive androstane receptor/pregnane X receptor binding site in the proximal promoter of the Aldh1a1 gene (Alnouti and Klaassen, 2008). Aldh1a1 expression is enhanced by electrophiles and activators of Nrf2 (Thimmulappa et al., 2002; Lee et al., 2003; Hu et al., 2006; Leonard et al., 2006; Reddy et al., 2007). It is not yet known whether the Aldh1a1 gene is a direct target of Nrf2, because most activators of Nrf2 also induce AP-1 proteins. Nrf2 is hepatoprotective against oxidative and electrophilic stress by up-regulation of electrophile-detoxifying genes (Nguyen et al., 2003; Reisman et al., 2009a,b). Nrf2 activation is caused by electrophiles modifying sulfhydryl groups in Keap-1, which releases Nrf2, leading to nuclear translocation. In the nucleus, Nrf2 heterodimerizes with the small Maf proteins and binds to specific response elements termed antioxidant or electrophilic response elements (AREs) to coordinate expression of cytoprotective genes (Rushmore et al., 1991; Dinkova-Kostova et al., 2010). AP-1 proteins can also serve as binding partners for Nrf2 but predominantly modulate gene expression as either homodimers of Jun family proteins (c-Jun, JunD, and JunB) or heterodimers with members of the Fos family (c-Fos, FosB, Fra1, or Fra2). There is compelling evidence to indicate that AP-1 proteins are hepatoprotective, especially against oxidative and electrophilic stress (Weitzman et al., 2000; Lamb et al., 2003; Tsuji, 2005; Hasselblatt et al., 2007). To date, there has been little evidence for a role of AP-1 or Nrf2 in the regulation of ALDHs and detoxification of lipid-derived peroxidation products.
Our preliminary experiments with mouse liver after acrolein exposure or BHA feeding by microarray revealed a 2- to 3-fold up-regulation of Aldh1a1 expression along with that of other electrophile detoxification genes (N. L. Makia and R. A. Prough, data not shown). These results support a model in which high acrolein levels stimulate Aldh1a1 gene expression, thereby enhancing acrolein metabolism and detoxification. The purpose of this study was to investigate whether well known electrophiles modulate the expression of Aldh1a1 in mouse liver and hepatocyte-derived cells. Because the major function of ALDH1A1 is to detoxify highly toxic lipid aldehydes, the induction of Aldh1a1 gene expression in mouse liver might be a mechanism to alleviate toxicity associated with lipid peroxidation and environmental aldehydes.
Materials and Methods
Chemicals and Reagents.
BHA, tert-butylhydroquinone, propen-2-al (acrolein), 1,9-pyrazoloanthrone (SP600125; JNK inhibitor), 2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one (PD98059; MEK inhibitor), and okadaic acid (OA) were purchased from Sigma-Aldrich (St. Louis, MO). AIN-76A diet and AIN-76A diet containing BHA were obtained from Purina Test Diet (Richmond, IN). Anti-glyceraldehyde-3-phosphate dehydrogenase antibody (clone 6C5) was purchased from Millipore Bioscience Research Reagents (Temecula, CA). Rabbit polyclonal antibodies against c-Jun (N), Jun D (329), Jun B (N-17), JNK (FL), phospho-JNK (G-7), c-Jun (D), c-Fos (4), and Nrf2 (C-20) and horseradish peroxidase-conjugated goat anti-rabbit, and goat anti-mouse antibodies were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Phospho-c-Jun (Ser63) II antibody was obtained from Cell Signaling Technology (Danvers, MA).
Animal Genotyping and Treatment.
Wild-type C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Wild-type C57BL/6 (WT) and Nrf2 knockout [Nrf2(−/−)] male mice on a C57BL/6 background (22–27 g) were obtained from Professor Roberto Bolli, Division of Cardiovascular Medicine/Medicine, University of Louisville (Louisville, KY). Mice were maintained on AIN76A (Purina) diet or pair-fed an AIN-76A diet containing 0.45% BHA (Purina Test Laboratories) for 7 days. Acrolein (5 mg/kg b.wt.) was administered to mice fed a normal chow diet by gavage for 7 consecutive days. Mice were also treated with sulforaphane [1-isothiocyanato-4-methylsulfinylbutane (SFN)] (10 mg/kg) by gavage for 7 days. Control mice were either fed the AIN76A diet for BHA and SFN experiments or administered water by gavage for 7 days for acrolein experiments. The mice were euthanized 24 h after the last treatment; the livers were harvested and stored at −80°C until use. All procedures were performed in accordance with protocols approved by University of Louisville Institutional Animal Care and Use Committee.
RNA Isolation and qRT-PCR.
RNA was isolated from mouse liver or Hepa-1c1c7 cells using TRI reagent (Molecular Research Center, Inc., Cincinnati, OH) as described previously (Makia et al., 2011). In brief, ≈0.1 g of frozen liver tissue was homogenized in 1 ml of TRI reagent (Invitrogen, Carlsbad, CA). RNA was extracted using chloroform and purified using silica membrane spin columns (RNeasy reagent; QIAGEN, Valencia, CA). The mRNA levels of ALDH1A1, ALDH1A7, ALDH2, and ALDH1B1 and the Nrf2-regulated genes (HO-1, NQO-1, and GSTM1) were assessed by qRT-PCR using an absolute quantitation standard curve method. Total RNA isolated from mouse liver or Hepa-1c1c7 cells was reverse-transcribed to cDNA using the Advantage RT-for-PCR kit (Clontech Laboratories, Inc., Mountain View, CA) with random hexamer primers. RNase H was then used to degrade any residual RNA in the cDNA mix. qRT-PCR was performed with the ABI 7900HT Sequence Detector System (Applied Biosystems, Foster City, CA) using gene-specific FAM-labeled LUX primers (Invitrogen). The primer specificity was determined by BLAST analysis against the whole mouse genome to verify single annealing sites. A plot of the Ct versus quantity of RNA indicated a linear relationship between the amount of template cDNA added to the reaction and the Ct values determined. All qRT-PCR experiments were performed in triplicate using cDNA samples from independent RNA sets, and the gene expression levels were normalized with 18S rRNA as an endogenous control. The data are expressed as means ± S.D. and were analyzed by Student's t test. Values of p < 0.05 were considered to be significant.
Cytosolic ALDH Activity.
The liver cytosolic fractions were isolated following a protocol adapted for preparation of liver microsomes (Remmer et al., 1966; Makia et al., 2011). Glycerol was added to the 108,000g supernatant to a final concentration of 20%, and the supernatant stored at −80°C. The hepatic ALDH activity was spectrophotometrically measured in the cytosolic fractions isolated from mouse liver by monitoring the reduction of NAD+ or NADP+ at 340 nm as described previously by Lindahl and Petersen (1991). Enzyme activity was assayed at 25°C in 50 mM sodium phosphate buffer (pH 7.4) containing 1 mM EDTA with either 1 mM NAD+ or NADP+ as cofactor (Makia et al., 2011).
Western Blotting.
Western blotting was performed as described previously (Makia et al., 2011). Hepa-1c1c7 cells were treated with acrolein (20 μM) at various time intervals (0, 1, 2, and 4 h). Cellular extracts isolated from mouse liver treated with BHA or Hepa-1c1c7 cells were separated by SDS-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. Membranes were probed with antibodies against glyceraldehyde-3-phosphate dehydrogenase (1:10,000) at room temperature for 2 h or c-Jun (1:1000), phospho-c-Jun (1:1000), JNK (1:1000), and phospho-JNK (1:1000) at 4°C overnight. The membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:10,000) or goat anti-mouse secondary (1:10,000) antibody, and the transferred proteins were visualized with the Amersham ECL Plus Western blotting detection system.
Transcription Factor Binding Site Analysis.
The putative transcription factor binding sites of the murine Aldh1a1 5′-flanking region (−8000/+27) were analyzed by Genomatix MatInspector software with emphasis on the binding sites for Nrf2 (A/GTGACNNNGC) and AP-1 (ATGAC/GTCA). The annotated sequence of the 5′-flanking region of the Aldh1a1 is shown in Supplemental Fig. 1.
Plasmids and Cloning of Aldh1a1 Promoter Luciferase Constructs.
Mouse genomic DNA was purchased from Promega (Madison, WI) and used to generate a 2002-bp fragment of the mouse Aldh1a1 5′-flanking sequence. This region spans from position −1963 to +27 relative to the transcription start site. The upstream primer (5′-GGTACCAAATGGGCAGGCATGGTAAC-3′) introduced a Kpn1 site, whereas the downstream primer (5′-AGATCTTGGTTTGGCTCCTGGAACAC-3′) introduced a HindIII site. The PCR amplification product was cloned into a pCR2.1 vector (Invitrogen). The sequence identity of the product was confirmed by sequence analysis in the University of Louisville Center for Genetics and Molecular Medicine Nucleic Acid core facility. Kpn1 and HindIII enzymes were used to subclone the 2002-bp Aldh1a1 promoter region into the pGL3-Basic vector (Promega) to generate the luciferase construct, p2.0Aldh1a1Luc (−1963/+27Aldh1a1Luc). Three deletion constructs, 1534-bp (−1496/+27), 1043-bp (−1005/+27), and 518-bp (−480/+27) fragments of the mouse Aldh1a1 promoter, were generated by PCR using the −1963/+23 construct as template. The downstream primer for synthesis of the three deletion constructs was identical to that of the 2002-bp fragment. The following upstream primers, which introduced a Nhe1 site, were used to generate the PCR products for the deletion constructs: 1534 bp, 5′-GCTAGCCATGGATCTGGCTGGATCTG-3; 1043 bp, 5′-GCTAGCGGAGGTGGCAATTTCACTAC-3′; and 518 bp, 5′-GCTAGCGGTTTGCTGGTAGCCATGTT-3′.
Nhe1 and HindIII were used to subclone the 1534-, 1043-, and 518-bp Aldh1a1 promoter fragments into pGL3-Basic to generate p1.5Aldh1a1Luc (−1496/+27Aldh1a1-Luc), p1.0Aldh1a1Luc (−1005/+27Aldh1a1Luc), and p0.5Aldh1a1Luc (−480/+27Aldh1a1Luc) constructs, respectively. A far upstream region of the Aldh1a1 promoter (position −4673) that contains a putative ARE/TRE site was also generated using the upstream primer 5′-GGTACCACTCAAATGGCTGAGCCAATG-3′ and a downstream primer identical to that of the 2002-bp fragment. This 4673-bp PCR fragment was cloned into pGL3 Basic using the Kpn1 and HindIII enzymes to generate the mouse Aldh1a1 promoter luciferase constructs, p4.6Aldh1a1Luc.
The expression plasmids for c-Fos (pRSV-cfos) and c-Jun (pRSV-cjun) were provided by C. B. Pickett and T. Nguyen (Schering-Plough Research Institute, Kenilworth, NJ). The expression plasmids for JunD (pcDNA3.1-JunD) and JunB (pcDNA3.1-JunB) were kind gifts from S. K. Agarwal (Genetics and Endocrinology Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD). The Nrf2 expression plasmid (pCI-Nrf2neo) was provided by K. S. Ramos (University of Louisville, Louisville, KY), and the pcDNA3.1-TAM67 expression plasmid was provided by D. J. Samuelson (University of Louisville). The plasmid pGSTYa-ARELuc (0.164Ya-ARELuc) containing 164 bases of the rat GST-Ya minimal promoter with a consensus ARE cloned upstream of the luciferase reporter gene was previously used as a positive control for Nrf2-dependent regulation of gene expression (Falkner et al., 1998). A luciferase reporter construct containing the human collagenase TRE (pColTRELuc) was generated using a double-stranded oligonucleotide including ≈30 bp of the 5′-flanking region of the human collagenase gene centered on the defined AP-1 site. This plasmid is a functional positive control for AP-1 transcription activity; it is regulated similarly to the CAT construct reported by Nyguyen et al. (1994).
Transfection of HepG2 Cells and Luciferase Reporter Assays.
The human hepatoma cell line, HepG2 (American Type Culture Collection, Manassas, VA) was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan UT) and 1% antibiotic-antimycotic solution (Invitrogen). Cells were plated at a density of approximately 150,000 cells/ml per well in 12-well plates for 24 h. Cells were transfected using 1 μg/2.5 μl of Lipofectamine LTX and 1 μl of Plus (Invitrogen)with 250 ng/well of p4.6Aldh1a1Luc (−4673/+ 27Aldh1a1Luc), p2.0Aldh1a1Luc (−1963/+27Aldh1a1Luc), p1.5Aldh1a1Luc (−1496/+27Aldh1a1-Luc), p1.0Aldh1a1Luc (−1005/+27Aldh1a1Luc), p0.5Aldh1a1Luc (−480/+27Aldh1a1Luc), pGSTYa-ARELuc, or pColTRELuc luciferase constructs. The cells were cotransfected with varying amounts of pRSV-cFos (0–80 ng), pRSV-cJun (0–80 ng), pcDNA3.1-JunD (0–80 ng), pcDNA3.1-JunB (0–80 ng), pcDNA3.1-Fra1 (0–80 ng), pcDNA3.1-Fra2 (0–80 ng), or pCI-Nrf2neo (0–200 ng). All plasmids were diluted in Opti-MEM I reduced serum media (Invitrogen). The plasmid lipofectamine complexes were transferred into each well containing cells and DMEM containing 10% FBS. The total mass of DNA being transfected was kept constant by cotransfection with blank plasmids such as pRSV (transfection with pRSV-cJun or pRSV-cFos) or pcDNA3.1 (transfection with pcDNA3.1-JunD, or pcDNA3.1-JunB vector) to demonstrate the absence of squelching. After incubation at 37°C for 24 h, the DMEM containing 10% FBS was replaced with fresh DMEM and then cells were treated with either 0.1% DMSO or various compounds at concentrations indicated in the figure legends. After overnight incubation (∼17–24 h), the growth medium was aspirated, and the cells were washed one time with PBS. The crude cell lysates were prepared by adding 0.1 ml of luciferase cell lysis buffer (Promega). The β-galactosidase and luciferase activities were measured as described previously (Falkner et al., 1998). The data were expressed as luciferase activity relative to β-galactosidase activity to normalize for transfection efficiency. Cell lysates were measured for expression of c-Jun, c-Fos, JunB, and JunD by Western blot analysis. All transient transfection experiments were performed in triplicate, and experiments were repeated at least twice for confirmation.
Site-Directed Mutagenesis.
Mutagenesis of the two putative AP-1 sites located at position −1069 and −758 of Aldh1a1 proximal promoter was performed using GeneTailor Site-Directed Mutagenesis systems (Invitrogen) following the Invitrogen protocol. In brief, the p1.5Aldh1a1Luc (−1496/+27Aldh1a1Luc) luciferase construct as the template DNA was methylated using DNA methylase at 37°C for 1 h. The template strand was methylated to mark it for degradation by the host McrBC endonuclease. The methylated plasmid was then amplified by PCR with two overlapping primers, one of which contains the target mutation using Platinum Taq Polymerase High Fidelity (Invitrogen). The PCR conditions were as follows: 94°C for 2 min; 20 cycles of 94°C for 30 s; 55°C for 30 s; 68°C for 7 min (≈6.5 kb plasmid); and 68°C for 10 min. The primers for mutagenesis of AP-1 sites located at positions −1069 and −758 are as follows: −758: sense, 5′-TCGACACTGCTTAGAGTAATAATAAACAAGTGCACGC-3′; antisense, 5′ ATTACTCTAAGCAGTGTCGAAGGAAAGAAT-3′; −1069: sense, 5′-TATTTACAAATTGAGAAGCTAAATAAAGGCAAAAAGA-3′; antisense, 5′-AGCTTCTCAATTTGTAAATACAGAGAGGAA-3′.
The PCR product, linear and double-stranded, was analyzed by a 1% agarose gel stained with ethidium bromide. The mutagenesis mixture was then transformed into One-Shot MAX Efficiency DH5α-T1 Escherichia coli. The host E. coli circularized the linear mutated DNA and McrBC endonuclease in the host cell and then digested the methylated template DNA, leaving only the unmethylated and mutated product. The sequences of the recovered PCR products were analyzed by sequence analysis at the University of Louisville, Center for Genetics and Molecular Medicine Nucleic Acid core facility.
Preparation of Nuclear Extracts.
Nuclear extracts were prepared from HepG2 cells transfected with either pRSV (vector control; 2 μg/well) or pRSV-c-Jun (2 μg/well) for 48 h using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, Waltham, MA) according to the standard protocol. In brief, HepG2 cells (106) in 35-mm dishes were washed twice with ice-cold PBS and transferred to microcentrifuge tubes. Cells were then centrifuged at 1500g for 5 min. The supernatant was discarded, and the cell pellet was allowed to swell in ice-cold cytoplasmic extraction reagent I for 10 min with vigorous mixing of the tube to resuspend the cell pellet. Ice-cold cytoplasmic extraction reagent II was added, and the tube was again vigorously mixed for 1 min. The sample was then centrifuged at 16,000g for 5 min, and the cytoplasmic fraction was transferred to a clean tube. The insoluble pellet was resuspended in ice-cold nuclear extraction reagent and incubated on ice for 40 min with 15-s vortexing every 10 min. The tube was then centrifuged at 16,000g for 10 min; the nuclear extract (supernatant) was stored in aliquots at −80°C until used for electrophoretic mobility shift assay (EMSA). Protein concentration was determined by bicinchoninic acid protein assay (Thermo Fisher Scientific) using bovine serum albumin as standard.
EMSA.
A nonradioactive LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) was used to examine whether c-Jun/AP-1 binds to the putative AP-1 binding sites on the Aldh1a1 proximal promoter. The oligonucleotides used as probes or competitors in gel shift assays were end-labeled at their 5′ ends with biotin, and the sequences are as follows: AP-1 site B: sense, 5′-TGCTTAGAGTAATGATTCACAAGTGCACG-3′; antisense, 5′-CGTGCACTTGTGAATCATTACTCTAAGCA-3′; AP-1 mut B: sense, 5′-TGCTTAGAGTAATAATAAACAAGTGCACG-3′; antisense, 5′-CGTGCACTTGTTTATTATTACTCTAAGCA-3′; and AP-1 site C: sense, 5′-AATTGAGAAGCTCAGTCAAGGCAAAAAGA-3′; antisense, 5′-TCTTTTTGCCTTGACTGAGCTTCTCAATT-3′.
The complementary oligonucleotides were annealed using a thermocycler (GeneAmp PCR system 2400; Applied Biosystems). EMSA reactions containing 20 fmol of biotin end-labeled double-stranded probes and 5 μg of nuclear proteins were incubated for 20 min at room temperature in a 1× binding buffer with 5 mM MgCl2, 2.5% glycerol, 0.05% NP-40, and 50 ng/μl poly(deoxyinosinic-deoxycytidylic) acid. The specificity of binding was determined by addition of a 25- to 100-fold excess of unlabeled double-stranded probes in the binding reactions. To further confirm specificity of binding, a 100-fold excess of unlabeled double-stranded probe with mutations at the AP-1 binding site was included in the binding reactions. For EMSA analyses, the binding reactions were incubated with rabbit polyclonal antibodies against c-Jun, c-Fos, or Nrf2 for 30 min at 4°C. Loading buffer was added to the reactions, and the protein-DNA complexes were resolved by electrophoresis on 6% precast DNA retardation gel (Invitrogen) in 0.5× TBE buffer (90 mM Tris-HCl, 90 mM boric acid, and 2.5 mM EDTA) at 100 V for 1 h. After electrophoresis, the binding reactions were then electrophoretically transferred onto a nylon membrane (Hybond-N+, GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) using 0.5× TBE for 1 h at 100V. At the end of transfer, the transferred DNA-protein complexes were then cross-linked onto membranes using a UV cross-linker that was set at 120 mJ/cm2 for an exposure time of 2 min. The biotin-labeled DNA was detected using the chemiluminescent nucleic acid detection module (Pierce) following their standard procedure for detection of immobilized nucleic acids.
Chromatin Immunoprecipitation Assay.
C57BL/6 mice were fed an AIN76 diet (control) or a diet containing 0.45% BHA for 7 days. Animals were euthanized, and intact nuclei from the livers of control or BHA-treated mice were purified by a sucrose density gradient without disrupting the internal macromolecular interactions. The chromatin immunoprecipitation (ChIP) assay was performed using the MAGnify chromatin-immunoprecipitation system (Invitrogen) according to the manufacturer's protocol with minor modifications. The pure nuclei were suspended in PBS and fixed in 1% formaldehyde at room temperature for 20 min to cross-link the DNA binding proteins to cognate cis-acting elements. The cross-linking reaction was stopped by incubation with 125 mM glycine for 10 min at room temperature. The nuclei were harvested after 30 min, washed with PBS, and solubilized in buffer containing 50 mM Tris-Cl, pH 8.0, 1% SDS, 5 mM EDTA, 5 mM EGTA, and 0.5 mM phenylmethylsulfonyl fluoride with complete protease inhibitor mix (Roche Molecular Biochemicals, Indianapolis, IN). The homogenate was sonicated six to seven times on ice at 40% setting (Branson sonicator) to shear the chromosomal DNA into fragments of ≈500 bp in size. The insoluble material was removed by centrifugation at 20,000g at 4°C for 5 min, and the soluble chromatin supernatant (chromatin extract) was stored at −80°C until use. Before use, the chromatin extract was diluted 10-fold with dilution buffer containing 20 mM Tris-Cl, pH 8.0, 1% Triton X-100, 1.2 mM EDTA, and 150 mM NaCl. The cross-linked protein-DNA fragments were immunoprecipitated by overnight incubation of diluted chromatin with antibodies against IgG (control), c-Jun (2 μg), or c-Fos (2 μg) conjugated to Dynabeads Protein A/G. The immune complexes were washed sequentially with immunoprecipitation buffer 1 and 2, and the protein-DNA cross-links were reversed by heat treatment in reverse cross-linking buffer containing proteinase K. The non-cross-linked DNA pulled down by the different antibodies was purified using DNA purification magnetic beads. The purified DNA was analyzed by real-time PCR (ABI 7900HT Sequence Detector System; Applied Biosystems) with primers spanning the putative AP-1 sites B (position −758) and C (position −1069) in the proximal promoter of the Aldh1a1 gene. The sequences of the ChIP primers are as follows: AP-1 site B: forward, 5′-GTTCCTTCCATATCTTGTGCTGGG-3′; reverse, 5′-GAGGTGCGTGCACTTGTGAATCAT-3′; and AP-1 site C: forward, 5′-TCCTTCAAGGTCTGTGACCAAAGC-3′; reverse, 5′-AACAGGGACCTGAGGAGTGTGTTT-3′.
Statistical Analysis.
Data were analyzed with Student's t tests or one-way analysis of variance (ANOVA) followed by the Tukey test using SigmaStat software (SigmaStat 3.5; SPSS, Chicago, IL). Comparisons were made between appropriate groups, and differences were considered to be statistically significant at a p level of 0.05. Results are expressed as means ± SD.
Results
Regulation of Aldh1a1 Gene Expression by Acrolein and BHA in Mouse Liver.
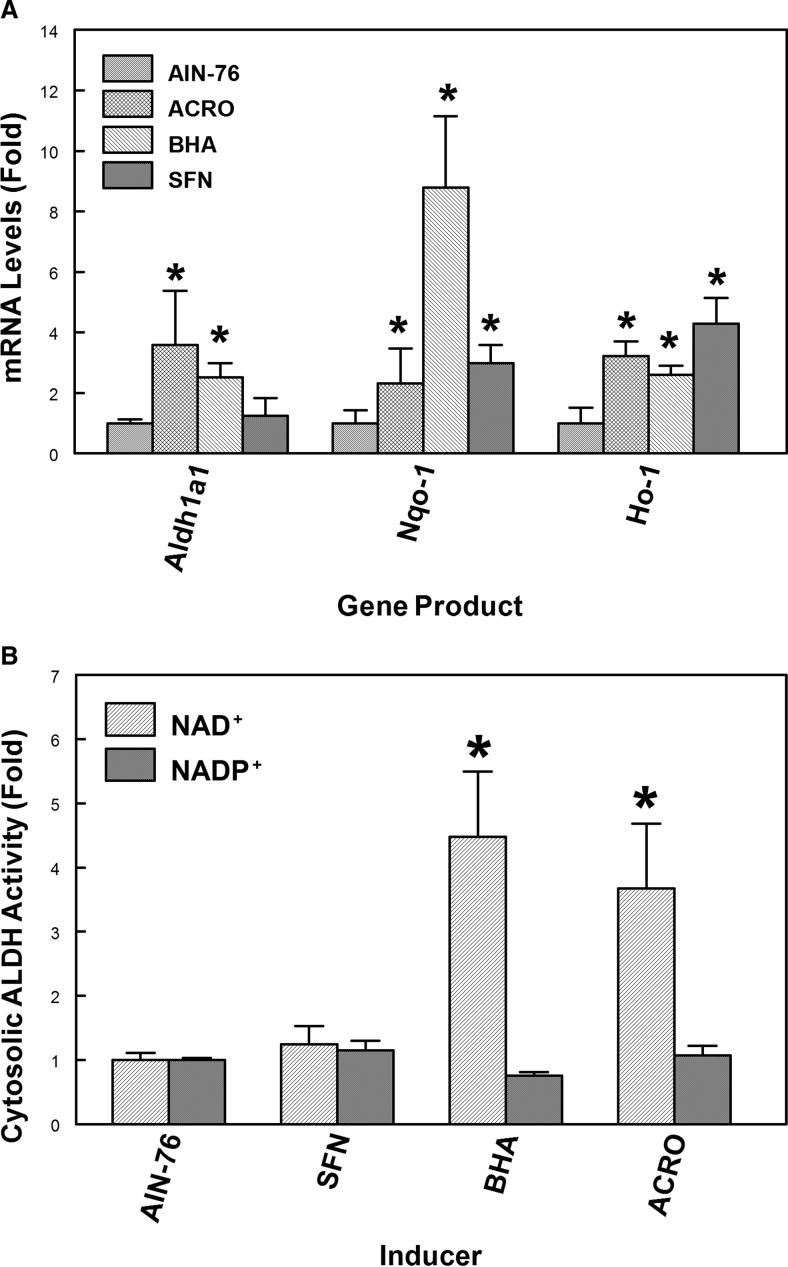
Previous studies demonstrated that the expression of the Aldh1a1 gene is modulated by a wide variety of endogenous and exogenous stimuli (Leonard et al., 2006; Alnouti and Klaassen, 2008). Initial microarray studies were performed in collaboration with C. A. Bradfield, University of Wisconsin (Madison, WI) using the EDGE cDNA microarray (Hayes et al., 2005). The results indicated that acrolein feeding increases Aldh1a1 (≈2-fold) gene expression in mouse liver. Hepatic Aldh1a1 mRNA levels were also induced (≈3-fold) in mice by treatment of mice with BHA that is rapidly metabolized to the electrophile/oxidant, t-butylhydroquinone. To confirm these microarray data, we examined whether acrolein or BHA induces the expression of the Aldh1a1 gene in mouse liver. C57BL/6 mice fed a laboratory chow diet were treated with water (control) or acrolein (5 mg/kg b.wt. by gavage daily for 7 days). The daily human consumption of unsaturated aldehydes is estimated to be ≈5 mg/kg b.wt. No hepatocellular damage was observed in mice exposed to this dose of acrolein (Conklin et al., 2010), indicating that 5 mg/kg b.wt. is a sublethal concentration of acrolein. Significant damage to the liver was observed when mice were administered an acrolein dose of 10 mg/kg. Mice fed an AIN76A diet were also administered SFN (10 mg/kg b.wt.) daily by gavage or fed an AIN-76A diet containing 0.45% BHA for 7 days. Aldh1a1, Nqo-1, and Ho-1 gene expression in mouse liver was analyzed by qRT-PCR (Fig. 1A). ALDH1A1 mRNA levels were significantly increased in mouse liver by acrolein (≈4-fold) or BHA (≈2.5-fold) treatment compared with those in controls. As expected, acrolein treatment induced the expression of antioxidant and electrophile detoxification genes, Nqo-1 (≈2.5-fold) and Ho-1 (≈3.5-fold) gene. Nqo-1 and Ho-1 expression was increased by BHA treatment as indicated by ≈9- and ≈2.5-fold increases in mRNA levels, respectively (Fig. 1A). The isothiocyanate, SFN, is believed to be a classic activator of only Nrf2 (Nguyen et al., 2009; Ahn et al., 2010), whereas BHA and acrolein modulate gene expression by activation of either Nrf2 or AP-1 (Choi and Moore, 1993; Lee and Murray, 2010). As anticipated, HO-1 (≈5-fold) and NQO-1 (≈3-fold) mRNA levels were significantly induced by SFN, but no significant induction of Aldh1a1 gene expression was observed with SFN (Fig. 1A). These results suggest that the mechanism of induction of prototypical Nrf2-regulated genes, Ho-1 and Nqo-1, by electrophiles differs from that of Aldh1a1.
Fig. 1.
The regulation of antioxidant and electrophile detoxification gene expression in mouse liver treated with either acrolein, BHA, or SFN. A, qRT-PCR analysis of expression of Aldh1a1 and other electrophile detoxification genes in mouse liver after treatment with either acrolein, BHA, or SFN. For the acrolein experiment, C57BL/6J mice fed a laboratory chow diet were treated with water (control) or 5 mg/kg acrolein by gavage daily for 7 days (n = 5). For BHA and SFN experiments, C57BL/6J mice fed an AIN-76 diet were fed a diet containing 0.45% BHA for 7 days or administered 10 mg/kg SFN daily for 7 days by gavage (n = 4). Total RNA was extracted from the liver and reverse-transcribed to cDNA, and qRT-PCR analysis of Aldh1a1, NQO-1, and HO-1 gene expression was performed as described under Materials and Methods. *, p < 0.05, significantly different compared with controls (Student's t test). B, increased liver cytosolic NAD+-dependent ALDH activity after treatment with either BHA or acrolein. Treatment by BHA, acrolein, or SFN was as indicated above. Liver cytosolic ALDH activity was measured as described under Materials and Methods using 1 mM propionaldehyde as substrate and either NAD+ or NADP+ as cofactor. Specific activities (nanomoles per minute per milligram of protein) for cytosolic fractions were 2.22 ± 0.24 (NAD+) and 15.8 ± 0.51 (NADP+) for controls; 9.97 ± 2.28 (NAD+) and 11.95 ± 0.88 (NADP+) for BHA-treated animals. *, p < 0.05, significantly different from controls (Student's t test).
Consistent with the changes in ALDH1A1 mRNA levels noted above, hepatic cytosolic NAD+-dependent ALDH activity was increased in mice treated with either acrolein (≈4-fold) or BHA (≈4.5-fold) using 1 mM propionaldehyde and NAD+ as substrate and cofactor, respectively (Fig. 1B). The basal NADP+-dependent activity was unaffected by treatment with either acrolein, BHA, or SFN. From our previous studies, we demonstrated that propionaldehyde is a good substrate for ALDH1A1 and that ALDH1A1 only uses NAD+ as cofactor in oxidative metabolism of this aldehyde (Makia et al., 2011). Even though the Vmax for ALDH3A1 relative to ALDH1A1 is large, the mRNA levels of endogenous Aldh3a1 gene in normal mouse liver are negligible, suggesting that the major isoform that contributes to NAD+-dependent ALDH activity induced by acrolein or BHA in mouse liver is most likely ALDH1A1. These results validate our microarray data, which demonstrate the induction of hepatic Aldh1a1 gene expression and other electrophile detoxification genes by BHA or acrolein. Because neither NAD+- nor NADP+-dependent cytosolic ALDH activity was increased in mouse liver by SFN, even at a concentration of 10 mg/kg, these data suggest that Nrf2 is not likely to be involved in electrophile-induced Aldh1a1 expression.
Induction of Aldh1a1 Gene Expression by BHA in Nrf2(+/+) and Nrf2(−/−) C57BL/6 Mouse Liver.
To more directly assess the role of Nrf2 in electrophile-induced transcription of Aldh1a1, Nrf2(+/+) and Nrf2(−/−) mice on a C57BL/6 background were fed 0.45% BHA in an AIN76 diet for 7 days, and mRNA levels of Aldh1a1 and known Nrf2 target genes, such as Gstm1, Nqo-1 and Ho-1 were determined by qRT-PCR. The exposure of mice to BHA resulted in a ≈2-fold increase in mRNA levels of hepatic Aldh1a1 expression in both Nrf2(+/+) and Nrf2(−/−) mice compared with those in untreated controls of the same genotype (Table 1) (p < 0.05 significance by two-tailed t test). However, the basal expression of Aldh1a1 was significantly reduced in Nrf2(−/−) mice compared with that in Nrf2(+/+) mice. The up-regulation of Nqo-1 (8.8-fold), Gstm1 (10-fold), and Ho-1 (2.6-fold) expression by BHA in Nrf2(+/+) mice was significantly reduced in Nrf2(−/−) mice, consistent with BHA-induced expression of these genes being mediated by Nrf2. However, the fold induction of Aldh1a1 gene expression by BHA in Nrf2(+/+) mice was not statistically different from that observed in Nrf2(−/−) mice, suggesting that BHA-induced expression of Aldh1a1 is probably independent of Nrf2 and due to other transcription factors.
TABLE 1.
Effects of Nrf2 genotype on changes in mRNA levels of antioxidant and electrophile detoxification genes in response to dietary BHA
qRT-PCR analysis of gene expression in the liver of Nrf2(+/+) and Nrf2(−/−) mice on C57BL/6 background in response to dietary administration of the phenolic antioxidant, BHA. Values are average fold change per genotype in the liver of Nrf2(+/+) and Nrf2(−/−) mice. The Nrf2(+/+) mice were fed either an AIN-76A diet (control, n = 4) or a diet supplemented with 0.45% BHA (treated, n = 4) for 7 days. The Nrf2(−/−) mice were fed chow diet only (control, n = 3) or diet supplemented with 0.45% BHA (treated, n = 3). RNA extracted from the liver was reversed-transcribed to cDNA. The mRNA levels of Aldh1a1, Aldh1a7, Aldh2, Aldh1b1, Ho-1, Nqo1, and Gstm1 gene expression were analyzed by qRT-PCR. For each sample, a single mRNA was analyzed in triplicate, and the average is shown.
| Gene | Constitutive |
Inducible |
Fold Induction |
|||
|---|---|---|---|---|---|---|
| Nrf2(+/+) | Nrf2(−/−) | Nrf2(+/+) | Nrf2(−/−) | Nrf2(+/+) | Nrf2(−/−) | |
| Aldh1a1 | 1.0 | 0.22 | 2.51 | 0.43 | 2.5* | 2.0** |
| Aldh1a7 | 1.0 | 0.38 | 2.07 | 0.66 | 2.1* | 2.2** |
| Aldh2 | 1.0 | 0.23 | 1.28 | 0.31 | 1.3 | 1.4 |
| Aldh1b1 | 1.0 | 0.53 | 1.06 | 1.29 | 1.1 | 2.4** |
| Gstm1 | 1.0 | 0.1 | 10.8 | 0.27 | 10.8* | 2.6*** |
| Nqo1 | 1.0 | 0.11 | 8.8 | 0.48 | 8.8* | 4.6*** |
| Ho-1 | 1.0 | 0.77 | 2.84 | 1.09 | 2.6* | 1.4*** |
P < 0.05, significantly increased compared with untreated Nrf2(+/+) mice (two-tailed t test).
P < 0.05, significantly increased compared with untreated Nrf2(−/−) mice.
P < 0.05 decreased fold-induction compared with Nrf2(+/+) mice.
Regulation of AP-1 Gene Expression by Acrolein or BHA in Mouse Liver.
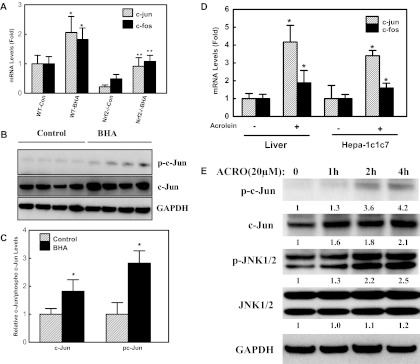
The AP-1 proteins are activated by increased gene expression, increased phosphorylation, and DNA binding capacity. To examine whether electrophile-induced expression of the Aldh1a1 gene is dependent on AP-1, we assessed the mRNA levels of c-jun and c-fos in the livers of BHA-treated mice. We observed significant induction of c-jun (≈2.0 fold) and c-fos (≈1.8 fold) expression in Nrf2(+/+) mice after BHA treatment (Fig. 2A). In Nrf2(−/−) mice, basal expression of c-fos and c-jun was markedly lower than that in Nrf2(+/+) mice, suggesting that basal c-jun and c-fos gene expression is also modulated by Nrf2. The expression of AP-1 genes in Nrf2(+/+) and Nrf2(−/−) mice parallels that of the Aldh1a1 gene (Fig. 2A; Table 1) and, thus, the low basal expression of Aldh1a1 in Nrf2(−/−) mice could be due to an indirect effect of Nrf2 on expression of AP-1 genes in these mice. We observed ≈4.0- and 2.0-fold induction in the mRNA levels of c-jun and c-fos genes, respectively, in mouse liver after acrolein treatment per os (Fig. 2D). The acrolein-induced expression of c-jun and c-fos gene in Hepa-1c1c7 cells was consistent with that observed in mouse liver (Fig. 2D).
Fig. 2.
Effect of electrophiles on AP-1 gene expression in mouse liver. A, BHA induces c-jun and c-fos gene expression in mouse liver. Nrf2(+/+) (n = 4) or Nrf2(−/−) (n = 3) mice on a C57BL/6 background were fed an AIN-76A diet (control) or a diet supplemented with 0.45% BHA (treated) for 7 days. qRT-PCR analysis of mRNA levels of c-jun and c-fos in mouse liver. *, p < 0.05, significantly different compared with Nrf2(+/+) control mice; **, p < 0.05, significantly different compared with BHA-treated Nrf2(+/+) mice (one-way ANOVA followed by the Tukey test.) B, C57BL/6J mice were fed either an AIN-76A diet (control, n = 4) or a diet supplemented with 0.45% BHA (fed, n = 4) for 7 days. Western blot procedures are described under Materials and Methods. The membranes were incubated with antibodies against c-Jun (1:1000) and phospho (p)-c-Jun (1:1000; Ser63) overnight at 4°C. Western blot was repeated three times, and the figure is a representative blot. C, densitometry analysis of c-Jun protein and phospho-c (pc)-Jun (Ser63) levels in mouse liver in response to BHA. *, p < 0.05, significantly different compared with control (Student's t test). D, acrolein induces c-jun and c-fos gene expression in mouse liver and Hepa-1c1c7 cells. C57BL/6J mice were treated with water (control, n = 5) or 5 mg/kg acrolein (treated, n = 5) by gavage daily for 7 days. Hepa1c1c7 cells were exposed to 20 μM acrolein for 6 h. The mRNA levels of c-jun and c-fos were assessed by qRT-PCR. *, p < 0.05, significantly different from DMSO control (Student's t test). E, time-dependent activation of c-Jun by acrolein in Hepa-1c1c7 cells. Cells were treated with 20 μM acrolein at various time intervals (0–4 h). After separation by electrophoresis, the proteins were transferred to membranes and the membranes were probed with antibodies against phosph-c-Jun (Ser63), c-Jun, JNK1/2, and phospho-JNK1/2. The Western blot experiment was repeated three times with similar results and the figure shown is a representative blot. The numbers between blots represent the densitometry analysis expressed relative to zero time expressed as fold change by the number below each blot.
To further test our hypothesis that Aldh1a1 gene expression is regulated by AP-1 proteins, we assessed the protein levels and phosphorylation status of c-Jun in cellular extracts from Nrf2(+/+) mice exposed to BHA. Western blot analysis with antibodies against c-Jun and phospho-c-Jun was performed in extracts from the livers of Nrf2(+/+) mice exposed to a control diet or a diet containing BHA (Fig. 2, B and C). We observed significant increases in both protein levels and phosphorylation status of c-Jun in BHA-treated mice livers compare with those in controls. The effect of acrolein on c-Jun activation was assessed in Hepa-1c1c7 cells. Hepa-1c1c7 cells were treated with acrolein (20 μM) at various time intervals and the activities of JNK and c-Jun were analyzed by Western blot with antibodies against phospho-c-Jun and phospho-JNK1/2 (Fig. 2E). We noticed increased phosphorylation of c-Jun 2 h after treatment of cells with 20 μM acrolein. The kinetics of c-Jun phosphorylation were similar to that of its upstream kinase, JNK1/2. Thus, the results of increased levels of c-Jun and increased phosphorylation of c-Jun and JNK1/2 implicate the JNK signaling pathway and its downstream substrate c-Jun in electrophile-induced expression of the Aldh1a1 gene.
Aldh1a1 Transcriptional Activity Is Not Induced by Nrf2.
To confirm that the mechanism of Aldh1a1 transcription by acrolein or BHA is not mediated by Nrf2, luciferase reporter gene assays using constructs containing portions of the Aldh1a1 5′-flanking sequence (positions −1963 to +27 relative to the transcription start site) were used. These sites were conserved in the rat and human ALDH1A1. An Oct1 binding site and a CCAAT box were identified at position −68 and −87 relative to the transcriptional start site, respectively. This promoter region of the Aldh1a1 gene contains putative AP-1 (positions −1516, −1069, −758, and −60) and Nrf2 (positions −665, −1068, and −1753) binding sites, whose sequence closely resembles the canonical TRE (TGACTCA) and ARE (TGACNNNGCA), respectively (Supplemental Fig. 1). HepG2 cells were transfected with the Aldh1a1 luciferase construct (p2.0Aldh1a1Luc) containing the Aldh1a1 promoter cloned upstream of the luciferase gene with increasing concentrations of Nrf2 expression plasmids (0–200 ng). As a control for electrophile-induced transcription of ARE genes dependent on Nrf2, cells were transfected with the pGSTYa-ARE luciferase construct that contains the consensus ARE sequence and the minimal promoter of the rat glutathione transferase A2 gene cloned upstream of a luciferase gene (Falkner et al., 1998). As anticipated, we noticed a concentration-dependent increase in GSTA2 ARE-driven transcriptional activity (normalized to β-galactosidase activity) by Nrf2 compared with the vector control (Supplemental Fig. 2). In contrast, overexpression of Nrf2 failed to stimulate transcriptional activity of the Aldh1a1 promoter construct. We also assessed the effect of Nrf2 on an Aldh1a1 luciferase construct containing a 4.6-kb fragment of the Aldh1a1 promoter. This construct also did not demonstrate any significant difference in promoter activity by Nrf2 compared with the vector control. These transient transfection experiments are consistent with our observations in Nrf2(−/−) and Nrf2(+/+) mice treated with SFN, suggesting that electrophile-induced expression of Aldh1a1 is independent of Nrf2, but dependent on JNK/c-Jun activation.
Increased Aldh1a1 Promoter Luciferase Activity by AP-1.
We examined the effect of AP-1 proteins on Aldh1a1 transcriptional activity by promoter luciferase reporter assay in HepG2 cells. Cells were cotransfected with either p2.0Aldh1a1Luc or the AP-1-positive control (pColTRE) luciferase constructs and with c-Jun, c-Fos, or both c-Jun and c-Fos expression plasmids. The pColTRE construct contains the TRE sequence from the human collagenase gene cloned upstream of a luciferase gene and is a positive control for AP-1-dependent gene transcription (Angel et al., 1987). Overexpression of c-Jun resulted in an ≈4-fold induction in Aldh1a1 transcriptional activity (Fig. 3A). The overexpression of c-Jun in HepG2 cells was verified by Western blot analysis (Supplemental Fig. 2). The regulation of Aldh1a1 gene promoter activity by c-Jun and c-Fos was different from that of the prototypic AP-1 responsive gene, the human collagenase gene. As shown in Fig. 3A, both c-Jun and c-Fos are required for induction of collagenase TRE luciferase activity. This synergism between c-Jun and c-Fos for induction of collagenase TRE activity was absent in the Aldh1a1 gene, suggesting that c-Jun induced Aldh1a1 transcription as a homodimer but not as a c-Jun/c-Fos heterodimer.
Fig. 3.
Differential regulation of mouse Aldh1a1 and human collagenase transcriptional activity by AP-1. A, the Aldh1a1 promoter activity was transactivated by c-Jun proteins. HepG2 cells were transiently cotransfected with p2.0Aldh1a1Luc (spanning −1963 to +27 of the Aldh1a1 promoter), pGL3 Basic or pColTRELuc (containing human collagenase TRE) luciferase constructs, and 40 ng of c-Jun, c-Fos, or both expression plasmids. The normalized luciferase activity was determined, and data are means ± S.D. from at least three independent experiments. *, p < 0.05, significantly different from vector-transfected cells; **, p < 0.05, significant difference compared with c-Jun-transfected cells (one-way ANOVA followed by the Tukey test.) B, induction of Aldh1a1 transcription is mediated by the c-Jun homodimer and not by the c-Jun/c-Fos heterodimer. HepG2 cells were transiently cotransfected with either pColTRE-Luc (TRE control) or p2.0Aldh1a1Luc luciferase construct with a constant concentration of c-Jun (40 ng) and increasing concentrations of c-Fos (0–40 ng) expression plasmid. Luciferase activity was normalized to β-galactosidase activity. The results are expressed as the fold induction from at least three independent experiments compared with that for control vector transfected cells. *, p < 0.05, significantly different compared with c-Jun-transfected cells (one-way ANOVA followed by the Tukey test). C, inhibition of c-Jun-mediated transactivation of Aldh1a1-luciferase activity by c-Jun dominant-negative (TAM67) protein. HepG2 cells were transiently cotransfected with the p2.0Aldh1a1Luc reporter with c-Jun, TAM67, or both expression plasmid and normalized luciferase activity (mean ± S.D. from at least three independent experiments) was measured. *, p < 0.05, significantly different compared with vector-transfected cells; **, p < 0.05, significantly different compared with acrolein-treated cells; ***, p < 0.05, significantly different from c-Jun-transfected and acrolein-treated cells (one-way ANOVA followed by the Tukey test).
To test the effect of c-Fos on Aldh1a1 promoter activity, we transiently cotransfected HepG2 cells with p2.0Aldh1a1Luc or pColTRELuc luciferase constructs and constant amounts of c-Jun (40 ng) with increasing additions of c-Fos to the cells (0–40 ng) (Fig. 3B). Although increasing c-Fos resulted in a concentration-dependent increase in the transcriptional activity of collagenase TRE, increasing concentrations of c-Fos caused a concentration-dependent decrease in Aldh1a1 promoter activity, suggesting that c-Fos displays a dominant-negative effect on c-Jun-mediated transactivation of Aldh1a1 promoter activity (Fig. 3B). These results indicated that transcriptional activation of Aldh1a1 promoter was mediated by the c-Jun homodimer and that increased expression of c-Fos caused inhibition of c-Jun homodimer formation. We also assessed the role of c-Jun dominant-negative protein (TAM67) on acrolein-mediated Aldh1a1 transcription (Fig. 3C). HepG2 cells were cotransfected with p2.0Aldh1a1 and either c-Jun, TAM67, or both c-Jun/TAM67 expression plasmids. The c-Jun-mediated increases in Aldh1a1 promoter activity were enhanced by acrolein treatment. However, acrolein or c-Jun-induced transcription of Aldh1a1 was abrogated by overexpression of TAM67, supporting the importance of c-Jun in electrophile-induced transcription of Aldh1a1.
Jun AP-1 Family Proteins Activate Aldh1a1 Transcriptional Activity.
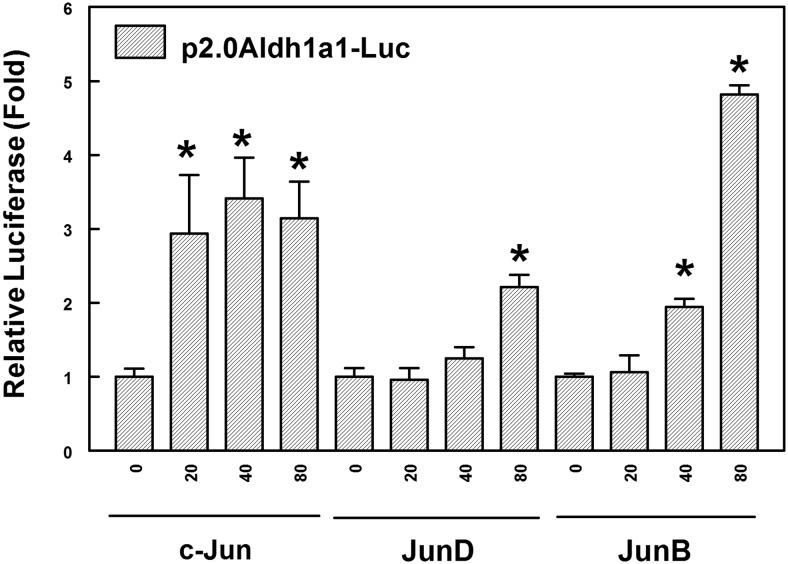
Previous studies indicated that the Jun family members (c-Jun, JunD, and JunB) induce the expression of genes as either homodimers or heterodimers with members of the Fos (c-Fos, FosB, Fra-1, and Fra-2) family. We tested the effect of increasing concentrations of c-Jun, JunD, or JunB on Aldh1a1 transcriptional activity in HepG2 cells. As shown in Fig. 4, c-Jun (≈3-fold), JunD (≈2.5-fold), or JunB (≈4.5-fold) at a concentration of 80 ng significantly transactivates Aldh1a1 promoter activity compared with that of controls. However, at lower concentrations (20 ng), only c-Jun showed significant induction of Aldh1a1 luciferase activity, suggesting increased potency of the c-Jun homodimer compared with that of homodimers of other Jun family members. This discrepancy may be due to the differences in the promoters of the backbone plasmids with CMV promoter for the other family members (JunD and JunB) and c-Jun promoter driven by RSV. However, Western blot analysis of HepG2 cell lysates from cells transfected with expression vectors for either c-Jun, c-Fos, JunB, or JunD demonstrated highly elevated levels of the respective transcription factors expression under the conditions of the luciferase reporter analysis (Supplemental Fig. 2).
Fig. 4.
The transactivation of Aldh1a1 promoter activity by Jun family proteins. HepG2 cells were transiently cotransfected with the p2.0Aldh1a1Luc construct with increasing concentrations (0, 20, 40, and 80 ng) of either c-Jun, JunD, or JunB expression plasmid. Luciferase activities from at least three independent experiments are expressed as fold induction ± S.D. compared with that for vector-transfected cells. *, p < 0.05, significantly different compared with vector-transfected cells (one-way ANOVA followed by the Tukey test).
Previous studies showed that Nrf2 induces the expression of cytoprotective genes by forming active heterodimers with AP-1 proteins, in particular c-Jun (Lee and Murray, 2010). We examined whether there is cross-talk between c-Jun and Nrf2 in Aldh1a1 transcriptional activation. Cells were transfected with p2.0Aldh1a1Luc with varying concentrations of c-Jun (0–40 ng) and Nrf2 (0–80 ng) (Supplemental Fig. 4). Luciferase activity normalized to β-galactosidase activity was assessed in these cells 48 h after transfection. Overexpression of Nrf2 had no effect on c-Jun-mediated transactivation of Aldh1a1 promoter activity (Supplemental Fig. 3), demonstrating that Nrf2 does not regulate Aldh1a1 expression either by itself or as heterodimer of c-Jun/Nrf2.
Role of Mitogen-Activated Protein Kinases in Electrophile-Mediated Transcription of Aldh1a1.
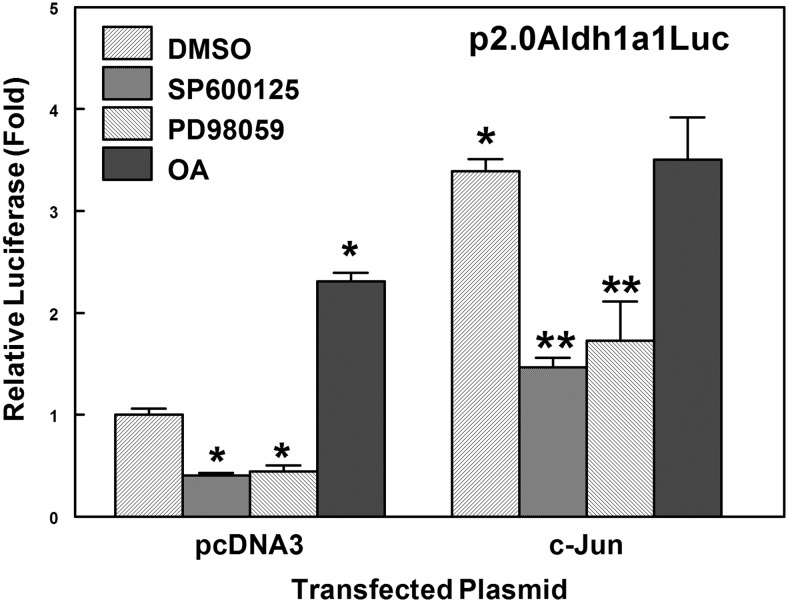
The activities of AP-1 proteins are regulated by mitogen-activated protein kinases (MAPKs), such as p38, extracellular signal-regulated kinase 1/2, and JNK. The activation of JNK by oxidative stress or electrophile is known to promote phosphorylation of its downstream target c-Jun, thereby increasing transcriptional activity of c-Jun (Eferl and Wagner, 2003). HepG2 cells were cotransfected with p2.0Aldh1a1 luciferase construct and c-Jun expression plasmid, followed by treatment with 0.1% DMSO, 50 μM SP600125 (JNK inhibitor), 50 μM PD98059 (MEK inhibitor), or 40 nM OA (phosphatase 2a inhibitor) for 17 h. Aldh1a1 promoter activity was diminished when cells were treated with JNK (SP600125) or MEK (PD98059) inhibitors (Fig. 5), suggesting that inhibition of the JNK signaling pathway also abrogated the c-Jun-mediated transactivation of Aldh1a1 promoter activity. Presumably, OA inhibits phosphatase 2a, affecting the phosphorylation state of c-Jun and increasing basal Aldh1a1 promoter activity, but displayed little or no effect on c-Jun-mediated transcriptional activation of the Aldh1a1 gene after c-Jun induction and phosphorylation has occurred. These results indicate that the JNK signaling pathway plays a critical role in electrophile-mediated transcriptional activation of the Aldh1a1 gene.
Fig. 5.
The role of MAPKs in transactivation of Aldh1a1 promoter activity. Aldh1a1 transcriptional activity is significantly reduced by inhibition of JNK (SP600125) and MEK (PD98059) kinase and increased by inhibition of protein phosphatase 2A [okadaic acid (OA)] activity in HepG2 cells. HepG2 cells were cotransfected with p2.0Aldh1a1Luc (−1963/+27Aldh1a1) construct and a c-Jun expression plasmid. Cells were treated with either 0.1% DMSO, 50 μM SP600125, 50 μM PD98059, or 40 nM OA for 17 h. Data are mean luciferase activities ± S.D. from at least three independent experiments and are presented as fold change over control vector cells. *, p < 0.05, significantly different compared with vector transfected cells; **, p < 0.05, significantly different from c-Jun-transfected cells (one-way ANOVA followed by the Tukey test).
Characterization of AP-1-Like Elements Responsible for c-Jun-Mediated Transactivation of Aldh1a1 Promoter Activity by Deletion and Mutagenesis Analysis.
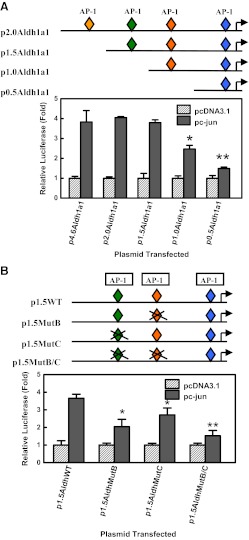
The analysis of the proximal promoter of Aldh1a1 by Genomatix MatInspector Professional software revealed four putative AP-1-like elements at positions −1516, −1069, −758, and −60 (Supplemental Fig. 1). The p2.0Aldh1a1 construct containing the −1963 to +27 fragment of the Aldh1a1 proximal promoter was used as a template for the generation of three deletion constructs: p1.5Aldh1a1 (−1496/+27Aldh1a1Luc), p1.0Aldh1a1 (−1005/+27Aldh1a1Luc), and p0.5Aldh1a1 (−480/+27Aldh1a1Luc) with progressive deletion of the AP-1-like elements. HepG2 cells were cotransfected with the Aldh1a1 promoter deletion mutant luciferase constructs and a plasmid encoding c-Jun protein. The deletion of the AP-1-like element at position −1516 had no significant effect on c-Jun-mediated transactivation of Aldh1a1 luciferase activity (Fig. 6A). The deletion of the putative AP-1 sites at positions −1069 and −758 significantly decreased transactivation of Aldh1a1 luciferase activity by c-Jun. Thus, deletion analyses indicate that the region containing these two AP-1-like elements at positions −758 (AP-1 site B) and −1069 (AP-1 site C) are important for c-Jun-dependent transactivation of Aldh1a1 transcriptional activity.
Fig. 6.
Characterization of the AP-1-like responsive element in the proximal promoter of the Aldh1a1 gene. A, deletion analysis to identify the AP-1-like responsive element in the proximal promoter of the Aldh1a1 gene. HepG2 cells were transiently cotransfected with 250 ng of p4.6Aldh1a1Luc (−4673/+27Aldh1a1), p2.0Aldh1a1Luc (−1963/+27Aldh1a1), p1.5Aldh1a1Luc (−1496/+27Aldh1a1), p1.0Aldh1a1Luc (−1005/+27Aldh1a1), or p0.5Aldh1a1Luc (−480/+27Aldh1a1) luciferase constructs and either pcDNA3.1 vector or c-Jun (40 ng) expression plasmid. Normalized luciferase activity was measured, and the data are expressed as mean activity ± S.D. from at least three independent experiments. *, p < 0.05, significantly different compared with p2.0Aldh1a1-transfected cells; **, p < 0.05, significantly different compared with p1.0Aldh1a1-transfected cells (one-way ANOVA followed by the Tukey test). B, mutagenesis of the putative AP-1-like elements located at positions −1069 (C site) and −758 (B site) on Aldh1a1 promoter activity. HepG2 cells were transiently cotransfected with 250 ng of p1.5Aldh1a1Luc (−1496/+27Aldh1a1), p1.5Aldh1a1MutB, p1.5Aldh1a1MutC, or p1.5Aldh1a1MutB/C luciferase constructs and either pcDNA3.1 or c-Jun (40 ng) expression plasmids. Normalized luciferase activity was expressed as mean activity± S.D. from at least three independent experiments, and data are presented as fold induction over that for vector-transfected cells. *, p < 0.05, significantly different compared with p1.5AldhWT transfected cells; **, p < 0.05, significantly different compared with p1.5Aldh1a1MutC-transfected cells (one-way ANOVA followed by the Tukey test).
To investigate the importance of the AP-1-like elements at positions −1069 and −758 in the activation of the Aldh1a1 luciferase construct by c-Jun, these putative AP-1 elements were mutated by site-directed mutagenesis. The mutation of the proximal putative AP-1 element at position −758 (p1.5Aldh1a1MutB) resulted in an ≈50% reduction in c-Jun-mediated activation of Aldh1a1 promoter activity compared with that for the p1.5Aldh1a1WT (WT) constructs (Fig. 6B). However, mutation of the distal AP-1 sequence at position −1069 (p1.5Aldh1a1MutC) only modestly (≈26%) decreased Aldh1a1 luciferase activity compared with the WT plasmid. Double mutation of the putative AP-1 elements at sites B and C in the promoter of the Aldh1a1 gene further attenuated c-Jun responsiveness, indicating that both AP-1-like elements at sites B and C contribute to c-Jun-dependent transactivation of the Aldh1a1 gene, and the AP-1 at site B apparently plays a more dominant role than that at site C.
EMSA Analysis to Assess Nuclear Protein Binding to AP-1-Like Elements in the Proximal Promoter of the Aldh1a1 Gene.
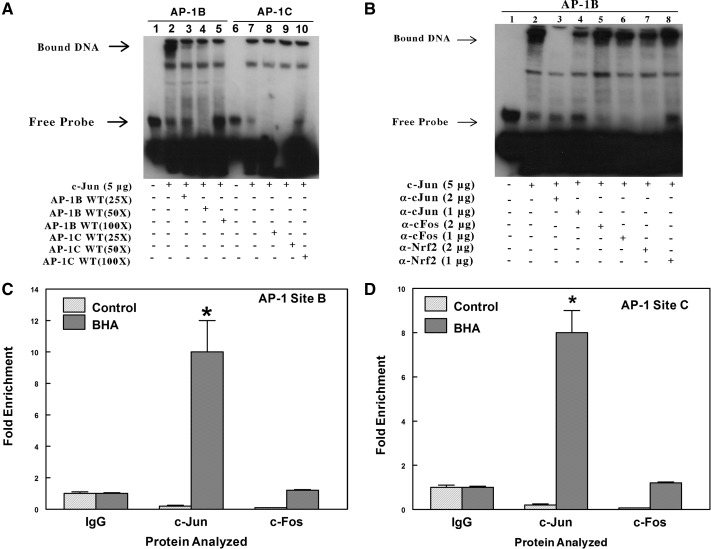
The nuclear extract isolated from HepG2 cells transfected with plasmid encoding c-Jun was incubated with biotin-labeled double-stranded probes containing AP-1-like elements B (position −758) and C (position −1069). We observed formation of a DNA-protein complex when labeled probe spanning AP-1-like element B was incubated with nuclear extract but not with site C (Fig. 7A). The specificity of nuclear proteins binding to the AP-1-like element B was examined by competition with unlabeled WT probe or unlabeled mutant probe containing mutations at the AP-1-like element at site B. The nuclear proteins isolated from HepG2 cells overexpressing c-Jun were allowed to form complexes with unlabeled WT (25- to 100-fold) or mutant probe (100-fold) before incubation with biotin-labeled probes spanning the AP-1-like element at site B. The results demonstrate that a 100-fold excess concentration of unlabeled WT probe completely competed with the biotin-labeled probe containing site B for AP-1-binding proteins (Fig. 7A). Thus, excess unlabeled specific WT probe effectively inhibited the formation of protein complexes with the biotin-labeled probe containing AP-1 element at site B. However, preincubation with unlabeled mutant probe resulted in little or no loss of the DNA-protein complex with the biotin-labeled AP-1B site for protein binding (Supplementary Fig. 5). These results indicate that the nuclear proteins from HepG2 do not form DNA complexes under the conditions used in this assay, with the AP-1-like element at site C, but bind instead to site B. Transfection data support the conclusion that the B site is a stronger affinity response element for AP-1 than the C site. The C site may require other yet unknown protein factors to bind before AP-1 binding and act synergistically to facilitate AP-1-dependent gene transcription.
Fig. 7.
EMSA analysis indicates the formation of a nuclear protein complex with the AP-1-like element at site B (position −758) but not with the AP-1-like element at site C (position −1069). A, biotin-labeled double-stranded probes were incubated for 20 min with 5 μg of nuclear extracts from HepG2 transfected with c-Jun expression plasmids. In the competition experiments, a 25- to 100-fold excess of unlabeled double-stranded probes was included in the binding reactions. The protein-DNA complexes were resolved by electrophoresis and detected using chemiluminescence. The result shown in this figure was identical to that of three other independent experiments. B, analysis using precipitating antibodies confirms that a protein in the nuclear extract that binds to the AP-1 site B is c-Jun and not c-Fos or Nrf2. EMSAs were performed by incubating nuclear extracts from HepG2 cells overexpressing c-Jun with a biotin-labeled probe containing the AP-1B site of the mouse Aldh1a1 promoter. In SuperShift experiments with antibodies, the binding reactions were incubated with either 1 or 2 μg of rabbit polyclonal c-Jun or c-Fos or Nrf2 antibodies for 30 min at 4°C. The figure is a representative of three other independent experiments. C, ChIP analysis to examine in vivo binding of c-Jun and c-Fos to the AP-1 at site B. Chromatin was prepared from the liver of control or BHA-treated C57BL/6J mice. The complexes containing cross-linked DNA binding proteins to their cognate cis-element were pulled down with antibodies against IgG, c-Jun, or c-Fos conjugated to Dynabeads protein A/G. The purified DNA was analyzed by real-time PCR with primers spanning AP-1 sites B or C in the proximal promoter of the Aldh1a1 gene. *, p < 0.05, significantly different compared with nonimmune immunoglobulin (Student's t tests). D, ChIP analysis to examine in vivo binding of c-Jun and c-Fos to the AP-1 at site C. *, p < 0.05, significantly different compared with nonimmune immunoglobulin (Student's t test).
To identify the composition of the nuclear proteins that form complexes with the AP-1-like element at site B (position −758), assays were performed when binding reactions were incubated with antibodies against c-Jun, c-Fos, or Nrf2 at 4°C for 30 min (Fig. 7B). Incubation of nuclear protein-DNA complexes with antibodies against c-Jun, but not against c-Fos or Nrf2, blocked the formation of the nuclear protein-DNA complex. Similar experiments have been performed with the TRE from the human collagenase gene, which forms a DNA-protein complex at positions similar to the AP-1 B site. Antibodies against c-Jun caused a significant loss of the DNA-protein complex formed with that TRE, as well (I. Amunom and R. A. Prough, unpublished results). These results indicate that c-Jun is a component of the nuclear protein complex on the AP-1 site B (position −758).
ChIP Experiment to Assess In Vivo Binding of Nuclear Proteins to Putative AP-1 Sites in the Proximal Promoter of Aldh1a1 Gene.
ChIP experiments were used to assess whether BHA promotes recruitment of c-Jun to these putative AP-1 binding sites at B (position −758) and C (position −1069) in the proximal promoter of Aldh1a1. Using c-Jun or c-Fos antibodies, the complexes containing cross-linked AP-1 proteins to the cognate AP-1 elements were pulled down from chromatin extracts prepared from the livers of control or BHA-treated mice. The DNA fragments were then analyzed by qRT-PCR with primers spanning the AP-1-like elements at B or C in the proximal promoter of the Aldh1a1 gene. As shown in Fig. 7C, liver extracts from BHA-treated mice showed ≈10-fold increases in recruitment of c-Jun to the AP-1-like element at site B compared with the IgG control. The recruitment of c-Fos to the AP-1-like element at site B in BHA-treated mice, although higher than in control mice, was comparable with that in the IgG control. These results confirm that BHA promotes enhanced binding of c-Jun to the AP-1 B site, and this correlates with increased transcriptional activity. The recruitment of c-Jun to the AP-1-like element at site C was comparable with that of site B (Fig. 7D). However, this result contradicts our EMSA experiment, in which we demonstrated the absence of nuclear protein complexes with the AP-1-like sequence at site C. This disparity might be due to the close proximity of the AP-1-like elements at sites B and C, which are only 300 bp apart. The sonication method used for DNA shearing in the ChIP experiment generated DNA fragments of ≈500 bp. Thus, inefficient shearing of the DNA might produce a single DNA fragment containing both AP-1-like elements.
Discussion
BHA, known to be rapidly metabolized to the electrophile t-butylhydroquinone, increases the expression of many cytoprotective genes by direct activation of the redox- and electrophile-sensitive transcription factor, Nrf2, or by indirect activation of the protein kinase signaling pathways, such as MAPK (Choi and Moore, 1993; Wu et al., 2006; Yuan et al., 2006; Lee and Murray, 2010). BHA-induced expression of Nrf2 target genes, Nqo-1, Ho-1, and Gstm1 in Nrf2(+/+) mice is significantly reduced in Nrf2(−/−) mice, implicating Nrf2 in BHA-induced expression of many of these genes (Table 1). Studies using keap1-knockdown (kd) mice with constitutively active hepatic Nrf2 (Reisman et al., 2009b) demonstrated that mRNA levels encoded by the Nqo-1 and Gstm1 genes were significantly increased in keap1-kd mice compared with those in WT mice [Nrf2(+/+)], suggesting that the activation of Nrf2 is important for induced expression of these genes. However, Reisman et al. (2009b) also noted that expression of several genes was not altered, implying that other signaling systems may be involved in increased gene expression due to BHA administration.
We show in this report that the fold induction of Aldh1a1 mRNA expression by BHA in Nrf2(+/+) mice was not significantly different from that in Nrf2(−/−) mice, suggesting that BHA-mediated transcription of the Aldh1a1 gene is independent of Nrf2. Aldh1a1 mRNA expression in the livers of keap1-kd mice was comparable with that of WT mice (Reisman et al., 2009b), supporting the hypothesis that inducible Aldh1a1 gene expression by electrophiles is regulated by an Nrf2-independent mechanism. Analysis of the mouse gene expressing ALDH1A1 by the Genomatix MatInspector software demonstrated in the 5′-flanking region between position −1963 to +27 relative to the transcriptional start site contains AP-1-like (−1516, −1069, −758, and −60) and Nrf2-like (−665, −1068, and −1753) binding sites. Although there was a concentration-dependent activation of GSTYa-ARE luciferase activity by Nrf2, increasing concentrations of Nrf2 failed to stimulate Aldh1a1 promoter luciferase activity (Supplemental Fig. 2) in the absence or presence of expressed c-Jun. In addition, the Aldh1a1-luciferase construct containing the 4.6-kilobase region of Aldh1a1 promoter was also unresponsive to Nrf2. The basal and BHA-induced expression of c-jun and c-fos genes were significantly reduced in Nrf2(−/−) mice compared with that in the Nrf2(+/+) control mice, suggesting that the Nrf2 may modulate the basal and BHA-induced expression of the AP-1 genes. Studies in Nrf2(−/−) mouse embryonic fibroblast documented the regulation of AP-1 gene expression by Nrf2 (Yang et al., 2005). Thus, the low basal expression of c-jun and c-fos possibly explains the reduced expression of Aldh1a1 gene observed in Nrf2(−/−) mouse liver but not the fold induction of gene expression caused by BHA administration. Using deletion and mutation analysis of the 5′-flanking region of Aldh1a1 defined two AP-1 sites at positions −1069 and −758 of the 5′-flanking region, that seem to be involved in Aldh1a1 regulation. Subsequent experiments using EMSA and ChIP analysis demonstrate that the AP-1 site at position −758 and to a lesser extent the site at position −1069 are responsive elements responsible for increased gene expression caused by increased levels of c-Jun. Use of inhibitors of protein kinase and protein phosphatase 2 demonstrated that phosphorylated c-Jun is most likely essential for increased Aldh1a1 gene expression. Other members of the c-Jun superfamily also activate the gene, but c-Fos overexpression suppresses Aldh1a1 reporter expression, demonstrating that c-Jun homodimers regulate this gene. Nrf2 had no effect on expression, in contrast to the results with CYP2J2 (Lee and Murray, 2010).
The role of AP-1 transcription factors in induction of genes involved in oxidative metabolism has been less well characterized to date. Aldehydes such as acrolein or 4-hydroxy-2-nonenal have been reported (Forman, 2010) to regulate several signaling pathways, including AP-1 and Nrf2. Lipid-derived aldehydes were also shown to induce the expression of electrophile-detoxifying genes by activation of MAPKs such as JNK (Wu et al., 2006). For example, oxidative stress-induced expression of human ferritin H gene was shown to be dependent on activation of JunD (Tsuji, 2005). Abrogation of c-Jun-dependent gene activation by c-Fos was previously reported by Marden et al. (2003), who demonstrated that whereas c-Jun homodimers strongly activated CYP2J2 expression, heterodimers formed between c-Fos and c-Jun were not active. The up-regulation of CYP2J2 gene transcription by BHA is dependent on the binding of both c-Jun and Nrf2 involving an atypical AP-1-like element in the proximal promoter of CYP2J2 (Lee and Murray, 2010). The c-Jun-mediated activation of the human atrial natriuretic peptide promoter is also inhibited by overexpression of c-Fos (Kovacic-Milivojevic and Gardner, 1992). Thus, the murine Aldh1a1, atrial natriuretic peptide and human CYP2J2 genes show a similar pattern of regulation by c-Jun homodimers. Because CYP2J2, Aldh1a1, CYP2c29 (I. Amunom and R. A. Prough, unpublished results), and CYP2C9 (N. L. Makia, R. A. Prough, and Goldstein, unpublished results) all metabolize aldehydes such as acrolein and 4-hydroxy-2-nonenal and are regulated by AP-1 transcription factors, it is interesting to speculate that these enzymes may form a battery of genes regulated via AP-1 signaling to protect cells from toxic aldehydes therefore representing a previously underappreciated mode of regulation of genes encoding aldehyde-oxidizing enzymes. This area of research will require continued analysis of these genes by molecular and informatics analysis.
In summary, we have provided evidence that Aldh1a1 gene expression is induced by environmental pollutants and exogenous electrophiles, such as acrolein and BHA and that increased expression of Aldh1a1 gene by electrophiles is dependent on c-Jun proteins but not on Nrf2 or c-Fos proteins. We also described an AP-1-like sequence at position −758 that plays a critical role in Aldh1a1 gene expression by electrophiles. The induction of Aldh1a1 gene expression in the mouse liver probably represents a mechanism of the liver to protect itself against electrophile and oxidant-induced damage generated from endogenous and exogenous compounds or during inflammatory disease conditions. Because the mouse and human enzyme display very similar substrate specificity and enzyme kinetics (Xiao et al., 2009; Makia et al., 2011) and the 5′-flanking regions are significantly conserved between the species, normal and transgenic mice may allow the testing of the protective effects of ALDH1A1 against lipid aldehyde toxicity associated with atherosclerosis and cardiotoxity of endogenous and exogenous aldehydes.
Supplementary Material
Acknowledgments
We thank the following individuals for providing transgenic animals or critical molecular biological expression vectors: Roberto Bolli, Cecil Pickett, Sunita Agrawal, Kenneth Ramos, and David Samuelson.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported in part by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL89380] (to D.J.C.); the National Institutes of Health National Institute for Environmental Health Sciences [Grant ES11860]; and the Preston Pope Joyes Research Endowment.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- ALDH
- aldehyde dehydrogenase
- RA
- retinoic acid
- Nrf2
- nuclear factor E2-related factor 2
- AP-1
- activator protein 1
- Keap-1/keap-1
- Ketch-like ECH-associated protein 1
- ARE
- antioxidant response element
- BHA
- BHA, tert-butylated hydroxyanisole
- JNK
- c-Jun NH2-terminal kinase
- MEK
- mitogen-activated protein kinase kinase
- OA
- okadaic acid
- SP600125
- 1,9-pyrazoloanthrone
- PD98059
- 2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one
- WT
- wild type
- SFN
- sulforaphane
- TRE
- TPA response element
- q
- quantitative
- RT
- real-time
- PCR
- polymerase chain reaction
- JNK
- c-Jun NH2-terminal kinase
- bp
- base pair(s)Y
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- DMSO
- dimethyl sulfoxide
- PBS
- phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay
- ChIP
- chromatin immunoprecipitation assay
- ANOVA
- analysis of variance
- MAPK
- mitogen-activated protein kinase
- kd
- knockdown.
Authorship Contributions
Participated in research design: Makia, Conklin, Surapureddi, and Prough.
Conducted experiments: Makia, Amunom, Falkner, and Surapureddi (ChIP).
Contributed new reagents or analytic tools: Conklin.
Performed data analysis: Makia, Surapureddi, Goldstein, and Prough.
Wrote or contributed to the writing of the manuscript: Makia, Surapureddi, Goldstein, and Prough.
References
- Ahn YH, Hwang Y, Liu H, Wang XJ, Zhang Y, Stephenson KK, Boronina TN, Cole RN, Dinkova-Kostova AT, Talalay P, et al. (2010) Electrophilic tuning of the chemoprotective natural product sulforaphane. Proc Natl Acad Sci USA 107:9590–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes MRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101:51–64 [DOI] [PubMed] [Google Scholar]
- Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P, Karin M. (1987) Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729–739 [DOI] [PubMed] [Google Scholar]
- Choi HS, Moore DD. (1993) Induction of c-fos and c-jun gene expression by phenolic antioxidants. Mol Endocrinol 7:1596–1602 [DOI] [PubMed] [Google Scholar]
- Conklin DJ, Barski OA, Lesgards JF, Juvan P, Rezen T, Rozman D, Prough RA, Vladykovskaya E, Liu S, Srivastava S, et al. (2010) Acrolein consumption induces systemic dyslipidemia and lipoprotein modification. Toxicol Appl Pharmacol 243:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Talalay P, Sharkey J, Zhang Y, Holtzclaw WD, Wang XJ, David E, Schiavoni KH, Finlayson S, Mierke DF, et al. (2010) An exceptionally potent inducer of cytoprotective enzymes: elucidation of the structural features that determine inducer potency and reactivity with Keap1. J Biol Chem 285:33747–33755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. (2003) AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868 [DOI] [PubMed] [Google Scholar]
- Elizondo G, Corchero J, Sterneck E, Gonzalez FJ. (2000) Feedback inhibition of the retinaldehyde dehydrogenase gene ALDH1 by retinoic acid through retinoic acid receptor α and CCAAT/enhancer-binding protein β. J Biol Chem 275:39747–39753 [DOI] [PubMed] [Google Scholar]
- Elizondo G, Medina-Díaz IM, Cruz R, Gonzalez FJ, Vega L. (2009) Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPβ interaction. Biochem Pharmacol 77:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner KC, Rushmore TH, Linder MW, Prough RA. (1998) Negative regulation of the rat glutathione S-transferase A2 gene by glucocorticoids involves a canonical glucocorticoid consensus sequence. Mol Pharmacol 53:1016–1026 [PubMed] [Google Scholar]
- Forman HJ. (2010) Reactive oxygen species and α,β-unsaturated aldehydes as second messengers in signal transduction. Ann NY Acad Sci 1203:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparetto M, Sekulovic S, Brocker C, Tang P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF, et al. (2012) Aldehyde dehydrogenases are regulators of hematopoietic stem cell numbers and B-cell development. Exp Hematol 40:318–329 [DOI] [PubMed] [Google Scholar]
- Hasselblatt P, Rath M, Komnenovic V, Zatloukal K, Wagner EF. (2007) Hepatocyte survival in acute hepatitis is due to c-Jun/AP-1-dependent expression of inducible nitric oxide synthase. Proc Natl Acad Sci USA 104:17105–17110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KR, Vollrath AL, Zastrow GM, McMillan BJ, Craven M, Jovanovich S, Rank DR, Penn S, Walisser JA, Reddy JK, et al. (2005) EDGE: a centralized resource for the comparison, analysis, and distribution of toxicogenomic information. Mol Pharmacol 67:1360–1368 [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chang WC, Hoffmann I, Duester G. (1999) Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J 339 (Pt 2):387–395 [PMC free article] [PubMed] [Google Scholar]
- Hsu LC, Chang WC, Yoshida A. (2000) Mouse type-2 retinaldehyde dehydrogenase (RALDH2): genomic organization, tissue-dependent expression, chromosome assignment and comparison to other types. Biochim Biophys Acta 1492:289–293 [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. (2006) Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci 79:1944–1955 [DOI] [PubMed] [Google Scholar]
- Kovacic-Milivojević B, Gardner DG. (1992) Divergent regulation of the human atrial natriuretic peptide gene by c-jun and c-fos. Mol Cell Biol 12:292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Ventura JJ, Hess P, Flavell RA, Davis RJ. (2003) JunD mediates survival signaling by the JNK signal transduction pathway. Mol Cell 11:1479–1489 [DOI] [PubMed] [Google Scholar]
- Lee AC, Murray M. (2010) Up-regulation of human CYP2J2 in HepG2 cells by butylated hydroxyanisole is mediated by c-jun and Nrf2. Mol Pharmacol 77:987–994 [DOI] [PubMed] [Google Scholar]
- Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. (2003) Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem 278:12029–12038 [DOI] [PubMed] [Google Scholar]
- Leonard MO, Kieran NE, Howell K, Burne MJ, Varadarajan R, Dhakshinamoorthy S, Porter AG, O'Farrelly C, Rabb H, Taylor CT. (2006) Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J 20:2624–2626 [DOI] [PubMed] [Google Scholar]
- Lindahl R, Petersen DR. (1991) Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharmacol 41:1583–1587 [DOI] [PubMed] [Google Scholar]
- Makia NL, Bojang P, Falkner KC, Conklin DJ, Prough RA. (2011) Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem Biol Interact 191:278–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4:697–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden NY, Fiala-Beer E, Xiang SH, Murray M. (2003) Role of activator protein-1 in the down-regulation of the human CYP2J2 gene in hypoxia. Biochem J 373:669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Duester G. (2003) Genetic evidence that retinaldehyde dehydrogenase raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem 278:36085–36090 [DOI] [PubMed] [Google Scholar]
- Nguyen T, Nioi P, Pickett CB. (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284:13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Rushmore TH, Pickett CB. (1994) Transcriptional regulation of a rat liver glutathione S-transferase Ya subunit gene. Analysis of the antioxidant response element and its activation by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. J Biol Chem 269:13656–13662 [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. (2003) Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol 43:233–260 [DOI] [PubMed] [Google Scholar]
- Reddy NM, Kleeberger SR, Yamamoto M, Kensler TW, Scollick C, Biswal S, Reddy SP. (2007) Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol Genomics 32:74–81 [DOI] [PubMed] [Google Scholar]
- Reisman SA, Csanaky IL, Aleksunes LM, Klaassen CD. (2009a) Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol Sci 109:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. (2009b) Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol Sci 108:35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmer H, Schenkman J, Estabrook RW, Sasame H, Gillette J, Narasimhulu S, Cooper DY, Rosenthal O. (1966) Drug interaction with hepatic microsomal cytochrome. Mol Pharmacol 2:187–190 [PubMed] [Google Scholar]
- Rushmore TH, Morton MR, Pickett CB. (1991) The antioxidant responsive element. activation by oxidative stress and identification of the DNA Consensus sequence required for functional activity. J Biol Chem 266:11632–11639 [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. (2002) Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res 62:5196–5203 [PubMed] [Google Scholar]
- Tsuji Y. (2005) JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene 24:7567–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman JB, Fiette L, Matsuo K, Yaniv M. (2000) JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell 6:1109–1119 [DOI] [PubMed] [Google Scholar]
- Wu CC, Hsieh CW, Lai PH, Lin JB, Liu YC, Wung BS. (2006) Upregulation of endothelial heme oxygenase-1 expression through the activation of the JNK pathway by sublethal concentrations of acrolein. Toxicol Appl Pharmacol 214:244–252 [DOI] [PubMed] [Google Scholar]
- Xiao T, Shoeb M, Siddiqui MS, Zhang M, Ramana KV, Srivastava SK, Vasiliou V, Ansari NH. (2009) Molecular cloning and oxidative modification of human lens ALDH1A1: implication in impaired detoxification of lipid aldehydes. J Toxicol Environ Health A 72:577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. (2005) Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-κB and AP-1. Mol Cell Biol 25:5933–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Xu C, Pan Z, Keum YS, Kim JH, Shen G, Yu S, Oo KT, Ma J, Kong AN. (2006) Butylated Hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog 45:841–850 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.