Background: Stt3p is the catalytic subunit of oligosaccharyltransferase (OT) that catalyzes protein N-glycosylation.
Results: We report the first high resolution NMR structure of the acceptor-binding domain of yeast OT.
Conclusion: This work provides a structural basis for the function of the C-terminal domain of the Stt3p subunit.
Significance: Structure determination of this critical domain is an important step toward understanding the mechanisms of the eukaryotic N-glycosylation process.
Keywords: Glycosyltransferases, NMR, Protein Conformation, Protein Structure, Protein-Protein Interactions, N-Glycosylation, Stt3p, Oligosaccharyltransferase
Abstract
N-Glycosylation is an essential and highly conserved protein modification. In eukaryotes, it is catalyzed by a multisubunit membrane-associated enzyme, oligosaccharyltransferase (OT). We report the high resolution structure of the C-terminal domain of eukaryotic Stt3p. Unlike its soluble β-sheet-rich prokaryotic counterparts, our model reveals that the C-terminal domain of yeast Stt3p is highly helical and has an overall oblate spheroid-shaped structure containing a membrane-embedded region. Anchoring of this protein segment to the endoplasmic reticulum membrane is likely to bring the membrane-embedded donor substrate closer, thus facilitating glycosylation efficiency. Structural comparison of the region near the WWDYG signature motif revealed that the acceptor substrate-binding site of yeast OT strikingly resembles its prokaryotic counterparts, suggesting a conserved mechanism of N-glycosylation from prokaryotes to eukaryotes. Furthermore, comparison of the NMR and cryo-EM structures of yeast OT revealed that the molecular architecture of this acceptor substrate-recognizing domain has interesting spatial specificity for interactions with other essential OT subunits.
Introduction
Oligosaccharyltransferase (OT)2 catalyzes N-glycosylation, the most ubiquitous cotranslational protein modification in all eukaryotes. In the central reaction, a preassembled oligosaccharide moiety is transferred from a dolichol carrier (dolichol pyrophosphate-linked oligosaccharide) to an asparagine residue in the nascent polypeptide chain defined by the consensus sequon NX(T/S) (X ≠ P) (1, 2). The transferred N-glycans play essential roles in modulating protein function in many critical processes, including development, inflammation, cancer, and the immune response (3, 4). For many higher eukaryotes, protein N-glycosylation is a highly coordinated and complex process. In the most extensively studied eukaryotic system, Saccharomyces cerevisiae, OT contains nine non-identical integral membrane protein subunits: Ost1p–Ost6p, Stt3p, Wbp1p, and Swp1p (2). Among these, five subunits (Stt3p, Wbp1p, Swp1p, Ost1p, and Ost2p) are essential for the viability of the cell, whereas Ost3p and Ost6p are homologous interchangeable subunits (5). In contrast, a single protein chain defines the OT activity in prokaryotes, emphasizing the complexity of the glycosylation process in eukaryotes.
Although the detailed enzymatic reaction mechanism and the roles of the other subunits are not yet fully understood, a multitude of experimental evidence has suggested that Stt3p contains the catalytic site (6–8). As shown in supplemental Fig. S1, eukaryotic Stt3p is highly conserved. In fact, it is the most conserved subunit in the OT complex (6). The most convincing evidence favoring this conclusion has come from the composition of the OT in some lower eukaryotes, such as trypanosomatids (9), as well as a few prokaryotes in which N-glycosylation occurs. In these organisms, OT is a single integral membrane protein homologous to the Stt3p subunit, such as PglB proteins in the bacteria Campylobacter jejuni (10) and Campylobacter lari (11) or the AglB protein in the archaeon Pyrococcus furiosus (12). Further studies showed that when transferred into Escherichia coli or stt3-deficient yeast cells, these single-polypeptide OTs can either enable or complement N-glycosylation in these host cells (13, 14). Moreover, it has also been demonstrated that the introduction of Leishmania major Stt3p into yeast can replace the entire OT complex (15).
Despite the biological significance of N-glycosylation, very limited success has been achieved with regard to the structural investigation of eukaryotic OT. The scarcity of structural knowledge significantly hampers our understanding of the enzymatic mechanism of the OT complex. To date, there are only three structural reports at atomic resolution in terms of eukaryotic OT subunits: NMR structures of the minimembrane protein Ost4p from yeast (16) and humans (17) and the crystal structure of the N-terminal luminal domain of Ost3p/Ost6p (18). The low resolution structure (at 12 Å) of the yeast OT complex was also determined by EM techniques (19). In comparison with eukaryotic OT, the structures of prokaryotic OT are relatively well understood. Two crystal structures of the soluble C-terminal domain of the prokaryotic homologs of Stt3p have been determined in succession: the AglB protein of the archaeon P. furiosus (20) and the PglB protein of the bacterium C. jejuni (12). Recently, the crystal structure of a full-length bacterial Stt3p homolog, the PglB protein of C. lari, was determined in complex with an acceptor peptide (21). However, sequence alignment reveals that the bacterial and archaeal Stt3p homologs show very limited sequence similarities to eukaryotic Stt3p.
We have previously reported the preparation and NMR assignments of the 274-residue C-terminal domain of Stt3p (residues 466–718 plus 21 His-tagged residues) in detergent micelles (22, 23). Our results revealed that the C-terminal domain of yeast Stt3p is highly helical. Here, we present the NMR structure of this 31.5-kDa helical membrane protein fragment in detergent micelles. Our results represent the feasibility of de novo structure determination of a medium-to-high molecular weight helical membrane protein fragment using solution NMR methodologies. Structural comparison of the acceptor substrate-binding site of yeast OT with that of its prokaryotic homologs suggests an evolutionarily conserved N-glycosylation mechanism. Furthermore, fitting the NMR structure of the C-terminal domain of Stt3p to the EM model provides insight into the interaction between Stt3p and other OT subunits.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Samples prepared for side chain and NOE assignments included 13C,15N-double-labeled, partially deuterated (50%) 13C,2H,15N-triple-labeled, and perdeuterated 13C,2H,15N-triple-labeled protein. The labeled C-terminal domain of Stt3p samples were prepared as described previously (22, 23).
NMR Experiments and Resonance Assignments
All spectra were acquired at 55 °C using either a Bruker 600-MHz AVANCE spectrometer equipped with a triple-resonance cryoprobe in the Department of Chemistry and Biochemistry at Auburn University or a Varian Inova 900-MHz NMR spectrometer equipped with a cold probe in the Complex Carbohydrate Research Center at the University of Georgia. NMR data collected were subsequently processed using the NMRPipe program and analyzed using NMRView software (42).
NMR data collected for protein side chain assignments using either 13C,15N-double-labeled or highly deuterated 13C,2H,15N-triple-labeled protein samples were as follows: HBHA(CO)NH, HCCH-TOCSY and HNHA, HCC(CO)NH, and (H)CC(CO)NH, HSQC-TOCSY. For NOESY experiments, 15N-edited three-dimensional NOESY-HSQC, 13C-edited three-dimensional NOESY-HSQC, and 13C,15N-edited four-dimensional HSQC-NOESY-HSQC data were recorded. The mixing times for 15N-edited three-dimensional NOESY-HSQC, 13C-edited three-dimensional NOESY-HSQC, and 13C,15N-edited four-dimensional HSQC-NOESY-HSQC were set as 150, 110, and 150 ms, respectively.
ILV Methyl-protonated Sample Preparation and Assignment
The protocol of expression of the methyl-protonated (Ile (δ1 only), Leu (13CH3,12CD3), and Val (13CH3,12CD3)) U-13C,2H,15N-labeled sample of the C-terminal domain of Stt3p was same as for the expression of the U-13C,2H,15N-labeled sample, except that 1 h prior to induction, 2-keto-3,3-d2-1,2,3,4-[13C]butyrate sodium salt (Sigma-Aldrich) and 2-keto-3-methyl-d3-3-d1-1,2,3,4-[13C]butyrate sodium salt (Sigma-Aldrich) were added to the medium. Pulse programs for NMR experiments for site-specific methyl assignment were written in our laboratory, based mainly on the pulse schemes reported by Tugarinov and Kay (43). Both three-dimensional 13C-edited NOESY-HSQC and four-dimensional 13C,13C-edited HSQC-NOESY-HSQC spectra were recorded for the ILV methyl-protonated sample.
Topology Determination
16-Doxylstearic acid (16-DSA) was used as the hydrophobic paramagnetic spin probe to determine the transmembrane domain of the C-terminal domain of Stt3p in SDS micelles. Titrations were performed by stepwise addition of 16-DSA over a concentration range of 0–2 mm to a constant amount of U-15N-labeled protein sample (0.1 mm). 1H,15N HSQC experiments were carried out using the same parameters, except P1 (the 90° hard pulse) and shimming values. The peak intensities were measured at each titration point to assess the amount of paramagnetic-induced line broadening.
HSQC Titrations
HSQC titrations were carried out as described previously (22).
Structure Calculation and Refinement
Structure calculation was done with CYANA 3.0. NOESY spectra were peak-picked and integrated interactively. Dihedral angle constraints were determined by TALOS+ (31). Structure calculation of the C-terminal domain of Stt3p was done in two steps. In the first step, the structure of each secondary structural element was determined to high resolution. The structures of these elements were then fixed by medium-range NOE restraints and relatively tight backbone dihedral angle restraints. In the second step, these segments were folded together with long-range NOE restraints. 100 structures were calculated, and 10 structures of the lowest total energy were used to represent their ensemble conformation. In the final calculated structures, the percentages of residues that reside in the most favored, additionally allowed, and generously allowed regions of the Ramachandran diagram are 60.5, 28.6, and 8.1%, respectively.
RESULTS AND DISCUSSION
Structure Determination of the C-terminal Domain of Stt3p
De novo structure determination of membrane proteins remains a challenge. In x-ray crystallography, the presence of detergents often hinders sample crystallization (24); whereas in NMR spectroscopy, the slow tumbling of membrane proteins embedded in detergent micelles often dramatically broadens the resonance line widths due to their rapid transverse relaxation rates. The difficulty in structure determination for α-helical membrane proteins by NMR is even more striking in comparison with their β-barrel counterparts. This is mainly because unlike the β-barrel membrane proteins, where the long-range interstrand NOEs are abundant and readily provide adequate restraints to define the global folds, there are typically very few long-range NH-NH NOEs for α-helical membrane proteins (25). Additionally, the NMR spectral dispersion for the amide region is very narrow for α-helical membrane proteins, which results in severe overlapping of resonances, making unambiguous assignments much harder. Consequently, to date, successful cases of α-helical membrane protein structure determination using NMR methods have been largely limited to proteins of low molecular weights (25–30).
To achieve unambiguous assignments, we have used a combination of NOEs from 13C,15N-double-labeled, partially deuterated (50%) triple-labeled, and uniformly 13C, 2H,15N-triple-labeled samples, as well as an ILV methyl-protonated, otherwise uniformly 13C,2H,15N-triple-labeled sample, together with backbone dihedral angles from chemical shift analysis (TALOS+) (31). It is noteworthy that four-dimensional NOESY data proved to be very helpful in unambiguous identification of many medium-range NOEs, a characteristic for α-helices, together with many long-range NOEs. The detailed statistics of restraints used for the structure determination are provided in Table 1 under “Experimental Procedures.”
TABLE 1.
NMR and refinement statistics for the NMR ensemble of the C-terminal domain of Stt3p
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 1445 |
| Intraresidue | 0 |
| Interresidue | 1445 |
| Sequential (|i − j| = 1) | 665 |
| Medium-range (|i − j| < 4) | 597 |
| Long-range (|i − j| > 5) | 193 |
| Total dihedral angle restraints | 272 |
| φ | 136 |
| ψ | 136 |
| Structure statistics | |
| Violations (mean ± S.D.) | |
| Distance constraints (Å) | 0.27 ± 0.15 |
| Dihedral angle constraints | 4.24° |
| Maximum dihedral angle violation | 4.73° |
| Maximum distance constraint violation (Å) | 0.52 |
| Average pairwise r.m.s.d. (Å)a | |
| Heavy | 3.07 |
| Backbone | 2.3 |
a Pairwise r.m.s.d. (root mean square deviation) was calculated among 10 refined structures.
Overall Structure and Topology of the C-terminal Domain of Stt3p
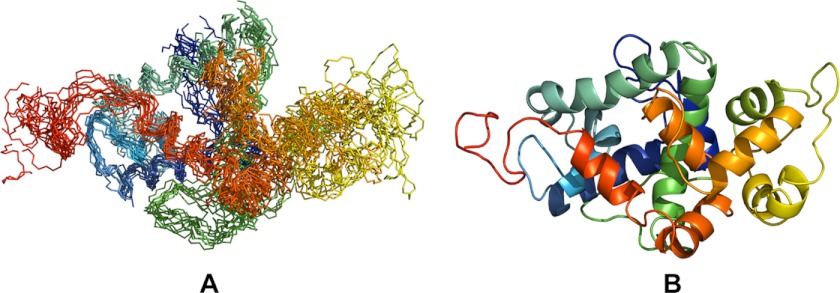
As presented in Fig. 1A, the C-terminal domain of Stt3p reveals a globally compact oblate spheroid-shaped structure, with a major axis of ∼680 Å and minor axis of ∼400 Å. This domain of Stt3p is primarily helical, containing 11 helices (α1–α11) and a disordered C terminus (Fig. 1B). These 11 helices are connected by short- or medium-length loops. This high helicity is consistent with our previous experimental data from far-UV CD and chemical shift index analysis (22, 23). Although both the TALOS+ program and chemical shift index predict the formation of a β-strand for Arg-592–Trp-598, we failed to unambiguously assign supportive NOEs to confirm this prediction, presumably due to peak overlapping. In fact, the absence of a β-strand in the C-terminal domain of eukaryotic Stt3p has been predicted by some structure predictions programs (20).
FIGURE 1.
Solution structure of the C-terminal domain of Stt3p. A, superposition of 10 conformers representing the final NMR structure. B, ribbon structure of the lowest energy conformer.
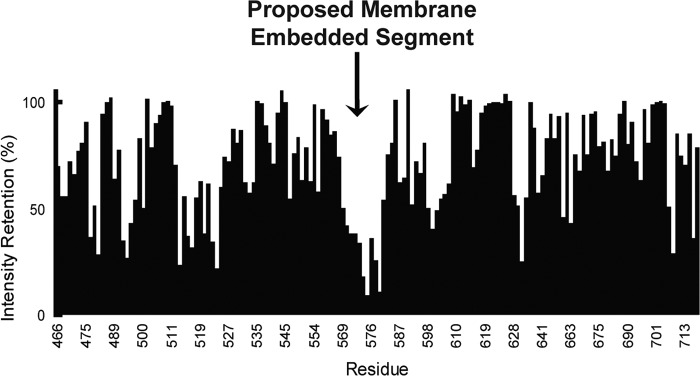
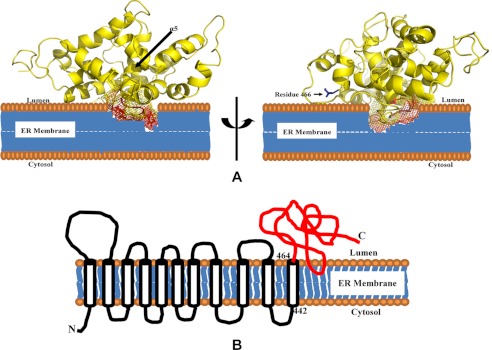
The topology of full-length yeast Stt3p has been proposed to be Ncyt-Clum with 11 transmembrane helices and a soluble C-terminal domain (32). However, some prediction programs predict that there is a transmembrane domain near the middle of the C-terminal domain (22, 33), whereas other programs predict that there is only a hydrophobic region at this position (32). Therefore, the detailed topology of the C-terminal domain of Stt3p remains controversial. In this study, we used a non-polar paramagnetic probe, 16-DSA, to investigate the topology of the C-terminal domain of Stt3p with respect to the membrane. This non-polar probe accelerates the relaxation of neighboring nuclear spins via dipole-dipole interactions of the residues located in the transmembrane domain, thus reducing the intensities of HSQC peak (34). The HSQC spectra were acquired in both the absence and presence of 2 mm 16-DSA (supplemental Fig. S2). Further analysis showed that the addition of 16-DSA resulted in a significant reduction in the peak intensities for residues 488–504, 511–526, 539–551, and 566–582 (Fig. 2). From the structure calculated in this study, we observed that, except for segment 566–582, which partially protrudes outward, all other segments could form a hydrophobic pocket. This explains the reason for the resonance intensity reduction in those segments because the hydrophobic probe 16-DSA can fit into a hydrophobic pocket. Taken together with the results obtained from transmembrane prediction programs (22), it is likely that the C-terminal segment encompassing residues 566–582 (α5) and the adjoining loop residues are membrane-embedded. Because it has been established that the C-terminal domain of Stt3p is located in the luminal side of the endoplasmic reticulum (ER) (32), we concluded that this membrane-embedded segment penetrates only the lipid bilayer but does not span it completely and therefore is not a transmembrane domain. We propose that the protein domain under study is monotopic. This is similar to the case of bacterial peptidoglycan glycosyltransferase, which was shown to be a monotopic membrane protein (35). As shown in Fig. 3, this topology model is consistent with the structure presented in this study, in which the highly hydrophobic C terminus of α5 and the adjoining loop protrude into the lipid bilayer. Accordingly, combined with existing results, we propose that full-length Stt3p has an Ncyt-Clum topology with 11 transmembrane domains in its N-terminal domain and a luminal monotopic C-terminal domain (Fig. 3B).
FIGURE 2.
Effects of the spin-labeled detergent 16-DSA on 1H,15N HSQC peak intensities for the U-15N-labeled C-terminal domain of Stt3p in SDS micelles at pH 6. 5 and 55 °C. The resonance intensity change of each residue upon the addition of 2 mm 16-DSA to the C-terminal domain of Stt3p in SDS micelles is plotted against the amino acid sequence. The chemical shift changes were calculated as combined 1H and 15N chemical shifts (Δδ(HN) = (Δδ(1H)2 + (0.2 × Δδ(15N))2)0.5).
FIGURE 3.
Proposed topology of the C-terminal domain of Stt3p. A, the C terminus of α5 and its adjoining loop (mesh) are proposed as the ER membrane-embedded region. Residue 466, which is known to be adjacent to the ER membrane, is depicted as blue sticks. B, topology of yeast Stt3p. The last transmembrane domain has been shown to be from residues 442 to 464 (32), suggesting that residue 466 is adjacent to the ER membrane.
We previously reported that different concentrations of detergent led to changes in the chemical shift positions of some residues in the 1H,15N HSQC spectra of the C-terminal domain of Stt3p (22). To confirm the topology of the C-terminal domain of Stt3p, we further analyzed the above data. Resonances in the HSQC spectra collected at different detergent concentrations were assigned (23). The effect of increasing detergent concentrations (50–200 mm) on the chemical shift positions of each residue in the protein is shown in supplemental Fig. S3A. Residues that underwent significant chemical shift perturbations (≥0.05 ppm) when mapped into the protein structure (supplemental Fig. S3B) revealed that the most of the perturbed residues are located at the proposed interface region between the protein and the ER membrane (shown in blue), except for a few that are located in a peripheral helix (shown in red). These results support our topology model of the C-terminal domain of Stt3p. Additionally, the last transmembrane segment of yeast Stt3p is predicted to encompass residues 442–464 (32), which suggests that the N-terminal residue of the protein under study (residue 466) should be located close to the ER membrane. Our model is in agreement with the above studies, showing close proximity between residue 466 and the lipid bilayer (Fig. 3A).
Comparison with the Crystal Structures of Prokaryotic Stt3p Homologs
Two crystal structures of the C-terminal soluble domain of prokaryotic Stt3p homologs have been reported previously: the AglB protein of P. furiosus (12) and the PglB protein of C. jejuni (20). Recently, the crystal structure of full-length PglB of C. lari was also determined (21). The C-terminal domain of these structures comprises mainly an α-helical “central core” domain and some β-sheet-rich domains either encircling or inserting into the central core domain. The central core domain contains the well conserved WWDYG motif and was therefore previously proposed as a catalytic domain (12, 20). However, recent crystal structural studies on full-length Stt3p of C. lari demonstrated that the WWDYG motif is indeed involved in binding of the signature sequon NX(T/S) (X ≠ P) on the acceptor peptide, but not directly in the catalysis process (21).
Comparison of the solution structure of the C-terminal domain of yeast Stt3p with the structures of its prokaryotic homologs reveals two major differences. First, the C-terminal domains of prokaryotic Stt3p homologs are water-soluble, lacking membrane-embedded segments, whereas our data suggest that the C-terminal domain of yeast Stt3p contains an ER membrane-embedded region, the C terminus of α5, and residues located in the adjoining loop. We postulate that the anchoring of the C-terminal domain of Stt3p, which contains the acceptor-binding site, to the ER membrane makes it closer to its donor substrate, dolichol-linked oligosaccharide, which is also embedded in ER membrane. This might potentially increase the effective local concentration of the donor substrate and hence facilitate the N-glycosylation process. Another striking difference is that the counterparts of the β-sheet-rich domains in AglB and PglB proteins are missing in the C-terminal domain of yeast Stt3p. It is thus reasonable to postulate that the C-terminal domain of Stt3p as a whole corresponds to the central core domain in the prokaryotic homologs, although it is larger and contains more helical elements. The function of these β-sheet-rich domains might be fulfilled by the other subunit(s) in the case of yeast OT.
Acceptor Substrate Binding Studies on the C-terminal Domain of Stt3p
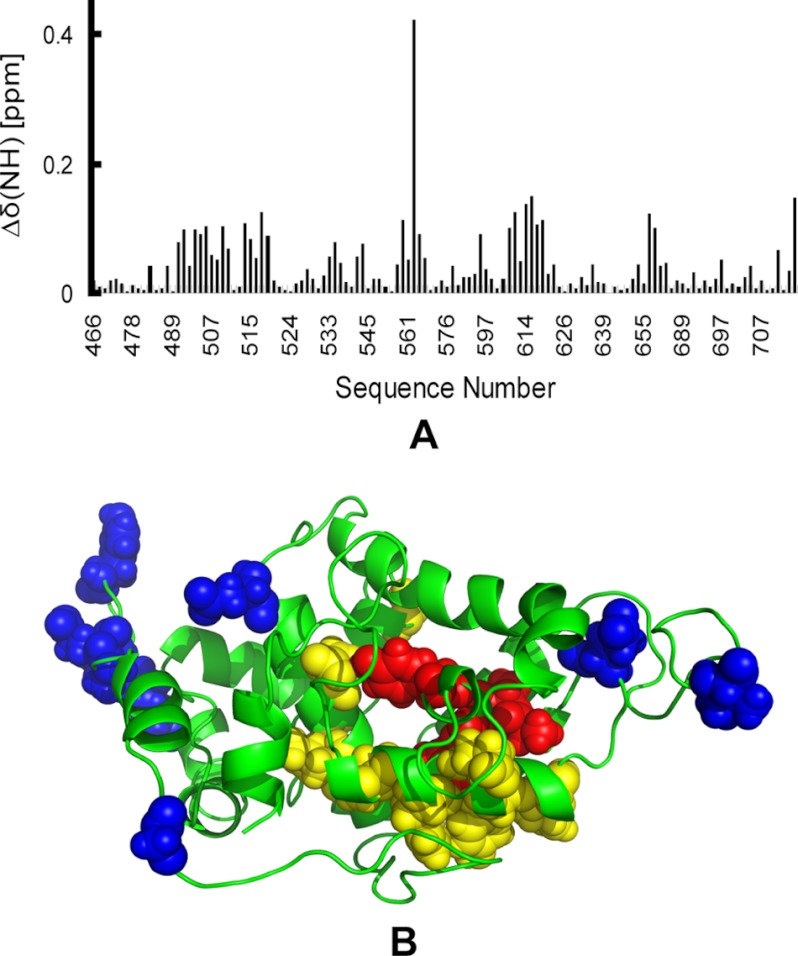
As mentioned above, the WWDYG motif, which is conserved across the three domains of life, has been shown to be the acceptor substrate-binding site of OT (21). To investigate the interaction between the C-terminal domain of Stt3p and the acceptor substrate, NMR titration studies were carried out using the Asn-Asp-Thr-NH2 acceptor peptide, which contains the consensus N-linked glycosylation sequon. Binding results were evaluated by monitoring the changes in chemical shift positions in the 1H,15N HSQC spectra of the C-terminal domain of Stt3p as described previously (22). As shown in Fig. 4A and supplemental Fig. S4, the resonance positions of some residues were perturbed upon the addition of the acceptor substrate peptide, confirming the interaction between the protein and peptide. Mapping of these residues within the structure of the C-terminal domain of Stt3p revealed that they can be divided into two groups either located in the peripheral loop regions (shown as blue spheres) or clustered around the WWDYG motif area (with the WWDYG motif and two previous Ala residues shown as red spheres and the adjacent residues shown as yellow spheres), as shown in Fig. 4B. Excluding the residues in the loop regions, other perturbed residues were found to be part of a pocket that is formed around the WWDYG motif. On the basis of the NMR mapping studies of the C-terminal domain of Stt3p with the acceptor peptide, we suggest that this pocket is the acceptor substrate-binding site located in the luminal C-terminal domain of Stt3p. As reported previously, the apparent K0.5 for the interaction between the substrate peptide and the C-terminal domain of Stt3p is ∼10 mm (22). The weak interactions could be due to the fact that we are not working with the whole OT complex. Moreover, in vitro experimental conditions can never be the same as within a cell. However, if the in vivo interaction is actually weak, it may provide the enzyme with a fast association and disassociation with the substrate, thus facilitating an efficient screening and glycosylation process of a nascent polypeptide chain.
FIGURE 4.
Interactions of the C-terminal domain of Stt3p with the acceptor peptide Asn-Asp-Thr-NH2. A, chemical shift plot of combined 1H and 15N chemical shift changes (Δδ(HN) = (Δδ(1H)2 + (0.2 × Δδ(15N))2)0.5) in the amide moieties of the C-terminal domain of Stt3p in SDS micelles induced by the addition of a 50-fold molar excess of ligand peptide. B, upon addition of peptide. The residues that underwent significant NMR resonance chemical shift perturbation are shown as spheres. The unspecific interactions in the peripheral regions are shown as blue spheres. The proposed acceptor substrate-binding pocket is formed by the WWDYG motif (red spheres) and its adjacent residues (yellow spheres).
Comparison of the Acceptor-binding Site of Stt3p and Its Prokaryotic Homologs
The limited sequence similarity between eukaryotic Stt3p and its prokaryotic homologs does not allow a meaningful sequence alignment. Because the architecture surrounding the WWDYG motif is highly relevant to understand the mechanisms of N-glycosylation, structural comparison of these regions of Stt3p and its prokaryotic homologs should provide information with respect to how well the mechanism of N-glycosylation is conserved throughout the three domains of life.
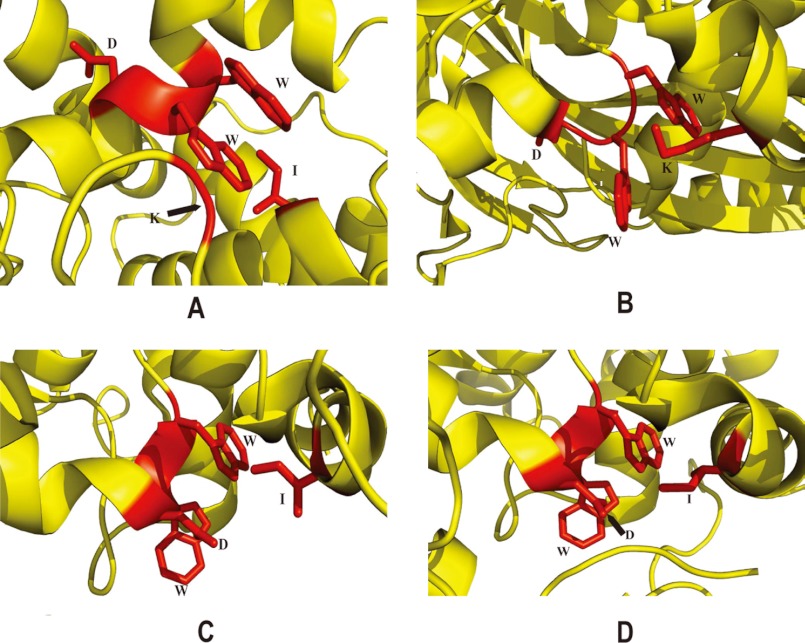
As shown in Fig. 5 (A–D), despite very low overall amino acid sequence similarities, the structural architecture of the region surrounding WWDYG motif of Stt3p is strikingly similar to that of its prokaryotic homologs: AglB protein from the archaeon P. furiosus and PglB proteins from the bacteria C. jejuni and C. lari. This observation suggests that OT shares a common mechanism of acceptor substrate binding in all three domains of life. In the binding sites of both eukaryotic and prokaryotic OTs (Fig. 5), the local helical structural feature of the WWDYG motif is strictly conserved, although the N-terminal part of the WWDYG motif in AglB adopts a rare left-handed helical conformation compared with the typical right-handed helix in the other three structures. Structural studies of PglB from the bacterium C. lari in complex with an acceptor peptide have revealed that the side chains of the two tryptophans in this motif directly interact with the β-hydroxyl group of Thr at position +2 of the acceptor sequon via hydrogen bonds (21). Additionally, an Ile residue, which is located adjacent to the WWDYG motif, was proposed to stabilize the interaction between the enzyme and the glycosylatable acceptor sequon through van der Waals contact (21). In our model, we also observed an Ile residue (Ile-572) in close proximity to the WWD residues (Fig. 5A). This observation appears to be reminiscent of PglB of C. lari, suggesting that Ile-572 might play a similar role in the binding of an acceptor substrate. In addition, sequence alignment showed that Ile-572 is absolutely conserved from yeast to human (supplemental Fig. S1).
FIGURE 5.
Comparison of the acceptor substrate-binding sites of the C-terminal domain of yeast Stt3p (A) and its prokaryotic homologs: AglB protein from the archaeon P. furiosus (B) and PglB proteins from the bacteria C. jejuni (C) and C. lari (D). B–D were prepared using Protein Data Bank codes 2ZAI, 3AAG, and 3RCE, respectively.
Close comparison of the acceptor-binding sites of these different models revealed that the aspartate residue side chain orientation within the WWDYG motif is different. In both yeast Stt3p and the archaeon P. furiosus AglB, the side chain of this aspartate residue orients away from the two Trp residues (Fig. 5, A and B). In contrast, in PglB from the bacteria C. jejuni and C. lari, the side chains of the aspartate residue in the WWDYG motif point toward the two Trp residues (Fig. 5, C and D). This observation implies that Asp-518 of Stt3p and AglB might play a different role from the bacterial homolog PglB, in which Asp-518, together with Trp-516 and Trp-517, directly interacts with the acceptor sequon via hydrogen bonds (21). Indeed, if Asp-518 in yeast OT also forms a hydrogen bond with the acceptor substrate, the OT activity would be expected to be largely unaffected upon its mutation to a glutamate residue because both aspartate and glutamate have comparable ability to form hydrogen bonds with the acceptor substrate due to their similar side chains. However, it has been shown that the D518E mutation in yeast Stt3p results in a lethal phenotype, rendering the enzyme completely nonfunctional (6). We proposed a structural role for Asp-518 in our earlier studies (22). Further structural and functional studies are needed to clarify the exact role of Asp-518 in the glycosylation process of higher organisms.
Based on phylogenetic tree analysis and structural studies of AglB protein from the archaeon P. furiosus, it was postulated that the acceptor substrate-binding site (which was tentatively proposed as the catalytic site of OT in this study) of eukaryotic OT was formed by the WWDYG motif and the so-called DK motif (20). Our model reveals that the DK motif of Stt3p (Asp-583–Lys-586) is indeed located in close proximity to the WWDYG motif (Fig. 5A), in agreement with the above proposal. However, the spatial orientation of the side chains of the residues in the DK motif cannot be precisely positioned in our model due to the missing or weak NMR resonances for these residues. For the same reason, we are uncertain about the local secondary structure of the DK motif. However, chemical shift index analysis and TALOS+ prediction based on the neighboring residues showed that it is very likely that Asp-582–Glu-587 form a small helix.
Structural Insights into the Interaction between the C-terminal Domain of Stt3p and Other OT Subunits
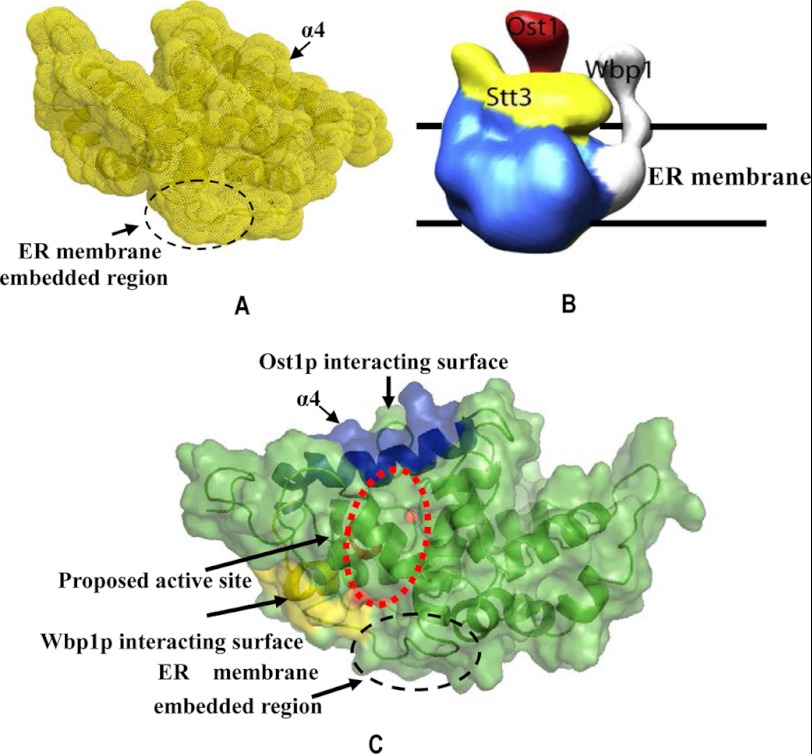
Arguably one of the most striking features of eukaryotic OT probably lies in its structural complexity. Yeast OT has two isoforms. Each isoform is composed of eight membrane protein subunits containing either Ost3p or Ost6p (36–38). In the absence of a high resolution structure of the whole OT complex, any information on the subunit-subunit interactions might help to understand the functional role of each subunit, as well as the mechanism of enzyme catalysis. The EM structure of the whole OT complex has been previously reported (19). Comparison of the NMR structure of the C-terminal domain of Stt3p with the corresponding region in the EM structure demonstrated a very good spatial fit despite the fact that protein expression and reconstitution methods were different (Fig. 6, A and B). Because the OT samples used for EM studies were enzymatically active, we conclude that the solution structure presented here represents the native fold of the C-terminal domain of Stt3p.
FIGURE 6.
Comparison of the NMR structure of the C-terminal domain of Stt3p with the corresponding region in the EM model of the OT complex (19). A, NMR model of the C-terminal domain of Stt3p. B, EM model of the OT complex. The corresponding region of the C-terminal domain of Stt3p is depicted in yellow in the EM model. Both the NMR and EM structures of the corresponding region present a very good spatial fit to each other. C, proposed interactions between the C-terminal domain of Stt3p and the other OT subunits. To clearly show the interacting surface, the protein orientation in C is different from that shown in A. The proposed surfaces that interact with Ost1p and Wbp1p are colored in blue and yellow, respectively, whereas the proposed active site is indicated by the dashed red circle.
Besides the catalytic Stt3p subunit, the EM model also demonstrates the spatial orientations of Ost1p, the proposed acceptor substrate-recognizing subunit (39), and Wbp1p, the proposed donor substrate-recognizing subunit (40). Based on the above information, our structure reveals that α4, which is aligned approximately parallel to the interface region of Ost1p, is responsible for the primary contact between the C-terminal domain of Stt3p and the luminal domain of Ost1p (Fig. 6C). More specifically, Asp-534, Asn-536, Thr-537, Asn-540, Thr-541, Ala-544, and Lys-548, which are oriented toward Ost1p, are very likely responsible for interactions between these two subunits. Sequence alignment of the C-terminal domain of Stt3p from yeast to human showed that these residues are all highly conserved (supplemental Fig. S1), suggesting their biological importance. With the exception of Ala-544, other polar residues can have electrostatic interactions or hydrogen bonding with the residues of Ost1p at the interface, thus providing hydrophilic environments to the groove formed between Stt3p and Ost1p. Further investigation will be needed to confirm the roles of those amino acid residues and to address the physiological significance of the hydrophilic properties of this proposed protein-conducting groove. Similarly, structural fitting of the C-terminal domain of Stt3p to the EM model allows us to identify the C terminus of α2 and the loop between α2 and α3 (loop L23) as the putative site for interactions with Wbp1p. Sequence alignment demonstrated that the C terminus of the α2 helix is highly conserved, whereas loop L23 is only partially conserved (supplemental Fig. S1). The presence of a disordered loop in the interface region between Stt3p and Wbp1p is in agreement with the dynamic feature of Wbp1p (19). This might provide a structural prerequisite for efficient N-glycosylation because it shows that the substrate of eukaryotic N-glycosylation reaction is in a flexible form (41).
In summary, we have presented the high resolution structure of the C-terminal domain of Stt3p, the proposed acceptor-binding domain of OT. Furthermore, considering the high sequence homology of eukaryotic Stt3p, determination of the first high resolution structure of the acceptor-binding domain of yeast OT is an important step toward understanding the mechanisms of the eukaryotic N-glycosylation process.
Supplementary Material
Acknowledgments
The 900-MHz NMR data were collected at the NMR Facility of the University of Georgia, funded by National Institutes of Health Grant GM66340. We thank Dr. Uma V. Katre for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK082397 (to S. M.). This work was also supported by United States Department of Agriculture PECASE Award 2003-35302-12930 and Grant 2011-65503-20030 and by National Science Foundation Grant IBN-0628064 (to S. M.).

This article contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (code 2LGZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- OT
- oligosaccharyltransferase
- HSQC
- heteronuclear single quantum correlation
- 16-DSA
- 16-doxylstearic acid
- ER
- endoplasmic reticulum
- TOCSY
- total correlation spectroscopy.
REFERENCES
- 1. Welply J. K., Shenbagamurthi P., Lennarz W. J., Naider F. (1983) Substrate recognition by oligosaccharyltransferase. Studies on glycosylation of modified Asn-X-Thr/Ser tripeptides. J. Biol. Chem. 258, 11856–11863 [PubMed] [Google Scholar]
- 2. Knauer R., Lehle L. (1999) The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta 1426, 259–273 [DOI] [PubMed] [Google Scholar]
- 3. Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 4. Sharon N., Lis H. (1995) Lectins–proteins with a sweet tooth: functions in cell recognition. Essays Biochem. 30, 59–75 [PubMed] [Google Scholar]
- 5. Silberstein S., Gilmore R. (1996) Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 10, 849–858 [PubMed] [Google Scholar]
- 6. Yan Q., Lennarz W. J. (2002) Studies on the function of oligosaccharyltransferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692–47700 [DOI] [PubMed] [Google Scholar]
- 7. Kelleher D. J., Karaoglu D., Mandon E. C., Gilmore R. (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell 12, 101–111 [DOI] [PubMed] [Google Scholar]
- 8. Nilsson I., Kelleher D. J., Miao Y., Shao Y., Kreibich G., Gilmore R., von Heijne G., Johnson A. E. (2003) Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 161, 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castro O., Movsichoff F., Parodi A. J. (2006) Preferential transfer of the complete glycan is determined by the oligosaccharyltransferase complex and not by the catalytic subunit. Proc. Natl. Acad. Sci. U.S.A. 103, 14756–14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glover K. J., Weerapana E., Numao S., Imperiali B. (2005) Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem. Biol. 12, 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwarz F., Lizak C., Fan Y. Y., Fleurkens S., Kowarik M., Aebi M. (2011) Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21, 45–54 [DOI] [PubMed] [Google Scholar]
- 12. Igura M., Maita N., Kamishikiryo J., Yamada M., Obita T., Maenaka K., Kohda D. (2008) Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., Morris H. R., Dell A., Valvano M. A., Aebi M. (2005) Engineering N-linked protein glycosylation with diverse O-antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hese K., Otto C., Routier F. H., Lehle L. (2009) The yeast oligosaccharyltransferase complex can be replaced by STT3 from Leishmania major. Glycobiology 19, 160–171 [DOI] [PubMed] [Google Scholar]
- 15. Nasab F. P., Schulz B. L., Gamarro F., Parodi A. J., Aebi M. (2008) All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zubkov S., Lennarz W. J., Mohanty S. (2004) Structural basis for the function of a minimembrane protein subunit of yeast oligosaccharyltransferase. Proc. Natl. Acad. Sci. U.S.A. 101, 3821–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gayen S., Kang C. (2011) Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex. Biochem. Biophys. Res. Commun. 409, 572–576 [DOI] [PubMed] [Google Scholar]
- 18. Schulz B. L., Stirnimann C. U., Grimshaw J. P., Brozzo M. S., Fritsch F., Mohorko E., Capitani G., Glockshuber R., Grütter M. G., Aebi M. (2009) Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. U.S.A. 106, 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H., Chavan M., Schindelin H., Lennarz W. J. (2008) Structure of the oligosaccharyltransferase complex at 12 Å resolution. Structure 16, 432–440 [DOI] [PubMed] [Google Scholar]
- 20. Maita N., Nyirenda J., Igura M., Kamishikiryo J., Kohda D. (2010) Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J. Biol. Chem. 285, 4941–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 22. Huang C., Mohanty S., Banerjee M. (2010) A novel method of production and biophysical characterization of the catalytic domain of yeast oligosaccharyltransferase. Biochemistry 49, 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C., Mohanty S. (2010) Challenging the limit: NMR assignment of a 31-kDa helical membrane protein. J. Am. Chem. Soc. 132, 3662–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostermeier C., Michel H. (1997) Crystallization of membrane proteins. Curr. Opin. Struct. Biol. 7, 697–701 [DOI] [PubMed] [Google Scholar]
- 25. Roosild T. P., Greenwald J., Vega M., Castronovo S., Riek R., Choe S. (2005) NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 26. MacKenzie K. R., Prestegard J. H., Engelman D. M. (1997) A transmembrane helix dimer: structure and implications. Science 276, 131–133 [DOI] [PubMed] [Google Scholar]
- 27. Rastogi V. K., Girvin M. E. (1999) Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature 402, 263–268 [DOI] [PubMed] [Google Scholar]
- 28. Van Horn W. D., Kim H. J., Ellis C. D., Hadziselimovic A., Sulistijo E. S., Karra M. D., Tian C., Sönnichsen F. D., Sanders C. R. (2009) Solution nuclear magnetic resonance structure of membrane integral diacylglycerol kinase. Science 324, 1726–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y., Cierpicki T., Jimenez R. H., Lukasik S. M., Ellena J. F., Cafiso D. S., Kadokura H., Beckwith J., Bushweller J. H. (2008) NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell 31, 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gautier A., Mott H. R., Bostock M. J., Kirkpatrick J. P., Nietlispach D. (2010) Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat. Struct. Mol. Biol. 17, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H., von Heijne G., Nilsson I. (2005) Membrane topology of the STT3 subunit of the oligosaccharyltransferase complex. J. Biol. Chem. 280, 20261–20267 [DOI] [PubMed] [Google Scholar]
- 33. Jaffee M. B., Imperiali B. (2011) Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyltransferases. Biochemistry 50, 7557–7567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hilty C., Wider G., Fernández C., Wüthrich K. (2004) Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem 5, 467–473 [DOI] [PubMed] [Google Scholar]
- 35. Lovering A. L., de Castro L. H., Lim D., Strynadka N. C. (2007) Structural insight into the transglycosylation step of bacterial cell wall biosynthesis. Science 315, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 36. Spirig U., Bodmer D., Wacker M., Burda P., Aebi M. (2005) The 3.4-kDa Ost4 protein is required for the assembly of two distinct oligosaccharyltransferase complexes in yeast. Glycobiology 15, 1396–1406 [DOI] [PubMed] [Google Scholar]
- 37. Yan A., Lennarz W. J. (2005) Two oligosaccharyltransferase complexes exist in yeast and associate with two different translocons. Glycobiology 15, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 38. Schwarz M., Knauer R., Lehle L. (2005) Yeast oligosaccharyltransferase consists of two functionally distinct subcomplexes, specified by either the Ost3p or Ost6p subunit. FEBS Lett. 579, 6564–6568 [DOI] [PubMed] [Google Scholar]
- 39. Yan Q., Prestwich G. D., Lennarz W. J. (1999) The Ost1p subunit of yeast oligosaccharyltransferase recognizes the peptide glycosylation site sequence, -Asn-X-(Ser/Thr)-. J. Biol. Chem. 274, 5021–5025 [DOI] [PubMed] [Google Scholar]
- 40. Lennarz W. J. (2007) Studies on oligosaccharyltransferase in yeast. Acta Biochim. Pol. 54, 673–677 [PubMed] [Google Scholar]
- 41. Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., Aebi M. (2006) N-Linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 [DOI] [PubMed] [Google Scholar]
- 42. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 43. Tugarinov V., Kay L. E. (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 125, 13868–13878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.