Background: Gluconeogenic enzymes are degraded in the vacuole via the Vid pathway during glucose re-feeding.
Results: Fructose-1,6-bisphosphatase is in the extracellular fraction during glucose starvation and decreases levels during glucose re-feeding.
Conclusion: VPS34 is required for the decline of extracellular fructose-1,6-bisphosphatase during glucose re-feeding.
Significance: Learning how gluconeogenic enzymes are secreted is critical for understanding the non-classical secretory pathway.
Keywords: Cell Biology; Endocytosis; Phosphatidylinositol 3-Kinase; Protein Degradation; Yeast Metabolism; VPS34; Fructose-1,6-bisphosphatase; Vacuole Import and Degradation
Abstract
When Saccharomyces cerevisiae are starved of glucose for a prolonged period of time, gluconeogenic enzymes such as fructose-1,6-bisphosphatase (FBPase), malate dehydrogenase, isocitrate lyase, and phosphoenolpyruvate carboxykinase are induced. However, when glucose is added to prolonged-starved cells, these enzymes are degraded in the vacuole via the vacuole import and degradation (Vid) pathway. The Vid pathway merges with the endocytic pathway to remove intracellular and extracellular proteins simultaneously. Ultrastructural and cell extraction studies indicate that substantial amounts of FBPase were in the extracellular fraction (periplasm) during glucose starvation. FBPase levels in the extracellular fraction decreased after glucose re-feeding in wild-type cells. The decline of FBPase in the extracellular fraction was dependent on the SLA1 and ARC18 genes involved in actin polymerization and endocytosis. Moreover, the reduction of extracellular FBPase was also dependent on the VPS34 gene. VPS34 encodes the PI3 kinase and is also required for the Vid pathway. Vps34p co-localized with actin patches in prolonged-starved cells. In the absence of this gene, FBPase and the Vid vesicle protein Vid24p associated with actin patches before and after the addition of glucose. Furthermore, high levels of FBPase remained in the extracellular fraction in the Δvps34 mutant during glucose re-feeding. When the Asn-736 residue of Vps34p was mutated and when the C-terminal 11 amino acids were deleted, mutant proteins failed to co-localize with actin patches, and FBPase in the extracellular fraction did not decrease as rapidly. We suggest that VPS34 plays a critical role in the decline of extracellular FBPase in response to glucose.
Introduction
The vacuole of Saccharomyces cerevisiae is important for numerous cellular processes such as osmoregulation, protein degradation, and pH maintenance (1–7). In addition, the function of the vacuole requires the targeting of specific vacuole resident proteins into this organelle (1, 3–7). For instance, aminopeptidase I is transported from the cytosol to the vacuole for maturation by the cytoplasm to the vacuole (Cvt) pathway (8–10). Similarly, another vacuole resident protein, carboxypeptidase Y, is transported to the vacuole by the vacuole protein sorting (Vps)2 pathway (3–7). Extracellular and plasma membrane proteins can be internalized and delivered to the vacuole by endocytosis (11, 12). Moreover, organelles such as peroxisomes can also be targeted to the vacuole for degradation by pexophagy (13, 14).
Under nitrogen starvation conditions in S. cerevisiae, a non-selective macroautophagic pathway targets proteins and organelles to the vacuole for degradation (15–19). Our laboratory studies a selective autophagic pathway that delivers specific cytosolic proteins to the vacuole in response to nutrient replenishment (13). Gluconeogenic enzymes, fructose-1,6-bisphosphatase (FBPase), malate dehydrogenase (MDH2), isocitrate lyase, and phosphoenolpyruvate carboxykinase are induced when cells are grown in glucose-depleted medium (20–23). These proteins are inactivated after replenishment of cells with fresh glucose. This is referred to as “catabolite inactivation” (20, 24). The inactivation and degradation of FBPase has been studied extensively (20, 25–34). FBPase is targeted to the proteasome (32–36) or to the vacuole for degradation (22, 23) depending on growth conditions. For example, upon replenishing cells with glucose after starvation for 1 day, FBPase is degraded in the proteasome (37). However, after glucose starvation for 3 days and subsequent replenishment with glucose, FBPase is degraded in the vacuole (37). We have characterized several VID (vacuole import and degradation) genes, which play a role in the vacuole dependent degradation of FBPase (21, 30, 38, 39). Interestingly, some of these VID genes also mediate the degradation of FBPase in the proteasomal pathway (37).
For the vacuolar pathway, FBPase associates with unique vesicles called Vid vesicles (40). We have determined that Vid22p, cyclophilin A, and the heat shock protein Ssa2p are required for the import of FBPase into the Vid vesicles (26, 29, 30). Moreover, the biogenesis of Vid vesicles requires the UBC1 gene (22). Vid24p and COPI coatomer proteins such as Sec28p have been identified as peripheral proteins on Vid vesicles (31, 38). Vid30p is also localized to Vid vesicles and interacts with Vid24p and Sec28p (41).
More recently we demonstrated that the endocytic pathway merges with the Vid pathway to target cargo proteins to the vacuole (27). In yeast, actin polymerization is required for early steps of endocytosis (11, 42, 43). The Vid vesicle proteins, Vid24p and Sec28p, co-localize with actin patches during glucose starvation and after glucose replenishment for up to 30 min (27). However, co-localization is diminished after glucose re-addition for 60 min (27). Moreover, cargo proteins such as FBPase and malate dehydrogenase (MDH2) are associated with actin patches after glucose re-addition for 30 min, and co-localization diminishes by the 60-min time point (27). In addition, we have determined that VID30 is required for the association of Vid vesicles and actin patches (39). In the absence of this gene, FBPase and Vid24p failed to co-localize with actin patches (39).
Although our present understanding of the Vid pathway indicates that this pathway integrates with the endocytic pathway, essential questions remain to be answered. How does the Vid pathway converge with the endocytic pathway? Is FBPase secreted during glucose starvation? Is it internalized during glucose re-feeding? To address some of these questions, we examined FBPase distribution at the ultra-structural level in wild-type cells. Substantial amounts of FBPase were in the periplasm in glucose-starved wild-type cells, suggesting that FBPase is secreted during glucose starvation.
Vps34p is involved in multiple protein and membrane trafficking events, which include sorting of vacuolar proteins, vacuole segregation, endocytosis, multivesicular body formation, starvation induced macroautophagy, and the Cvt (cytoplasm to the vacuole) pathway (44–47). This gene is also required for the Vid pathway. This gene encodes a class III phosphatidylinositol 3-kinase (PI3K) that functions to phosphorylate phosphatidylinositol at the 3′ hydroxyl position to produce phosphatidylinositol 3-phosphate (44). This function is conserved from yeast to human (44). For the Vps pathway, Vps34p is recruited from the cytosol to the Golgi/endosome via interaction with the membrane-associated protein kinase Vps15p, which also stimulates the phosphatidylinositol 3-kinase activity of Vps34p (48, 49). The C-terminal 11 amino acids of Vps34p (amino acids 864–875) are implicated in the protein association on the membrane (50). Furthermore, the deletion of the C-terminal 11 residues reduces PI3K activity (49, 50).
In prolonged-starved cells, Vps34p co-localized with actin patches. In the absence of this gene, FBPase and Vid24p were associated with actin patches before and after glucose replenishment. A high abundance of FBPase was in the extracellular fraction (periplasm) in prolonged-starved Δvps34 strain. However, after glucose replenishment, significant amounts of FBPase remained in the extracellular fraction in the Δvps34 strain. When glucose was added to prolonged-starved cells, FBPase in the extracellular fraction decreased rapidly. The decline of extracellular FBPase was dependent on the SLA1 and ARC18 genes involved in actin polymerization and endocytosis. The N736K point mutation of Vps34p and the deletion of the C-terminal 11 amino acids retarded the degradation of FBPase. Furthermore, both mutant proteins caused a delay in the reduction of FBPase in the extracellular fraction after glucose addition. In summary, our results suggest that VPS34 plays a critical role in the decline of extracellular FBPase in response to glucose re-feeding.
EXPERIMENTAL PROCEDURES
Strains, Media, Plasmids, and Antibodies
Strains used in the study are listed in Table 1. The deletion strains derived from BY4742 were from Euroscarf. The Scw4p-GFP strain was purchased from Invitrogen. For most experiments, cells were grown in glucose-deficient medium (YPKG) (1% yeast extracts, 2% peptone, 1% potassium acetate, and 0.5% glucose) medium for 3 days at 30 °C. Cells were then transferred to glucose-rich medium (YPD; 1% yeast extracts, 2% peptone, and 2% glucose) medium for the indicated times. In some experiments cells were grown in YPD for 1 day or in YPKG for 1, 2, and 3 days. Cells were extracted, and proteins were separated into the extracellular (E) and intracellular (I) fractions. The distribution of proteins was determined by Western blotting. Anti-FBPase antibodies were produced in rabbits using purified FBPase. Mouse monoclonal anti-HA was purchased from Roche Applied Science, and mouse monoclonal anti-V5 was purchased from Invitrogen. Anti-GAPDH and anti-GFP were purchased from Protein Tech and Abcam, respectively. Pil1p antibodies were gifts from Dr. Dickson (University of Kentucky). Primers used to tag FBPase, Vps34p, and Vid24p are listed in Table 2. Plasmid containing GFP-Vid24p was produced as described (27).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| HLY635 | MATα ura3–52 LEU2 trp1Δ63 his3Δ200 GAL2 |
| Δvps34 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vps34::kanMX4 |
| Δvps15 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vps15::kanMX4 |
| Δvps30 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vps30::kanMX4 |
| HLY2753 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 VPS34-GFP-URA3 |
| Δsla1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sla1::kanMX4 |
| Δarc18 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 arc18::kanMX4 |
| HLY2862 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 sla1::kanMX4 VPS34-GFP-URA3 |
| HLY2879 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 arc18::kanMX4 VPS34-GFP-URA3 |
| HLY2793 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 vps15::kanMX4 VPS34-GFP-URA3 |
| HLY1418 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 FBP1-GFP-HIS3 |
| HLY2639 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 vps34::kanMX4 FBP1-GFP-HIS3 |
| HLY2998 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3 GFP-VID24-URA3 |
| HLY2878 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3 vps34::kanMX4 GFP-VID24-URA3 |
| Scw4p-GFP | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ SCW4-GFP-HIS3 |
| HLY2239 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ LST8-GFP-HIS3 |
| HLY2218 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ TOR1-GFP-HIS3 |
| HLY2588 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ VID30-GFP-HIS3 |
| HLY229 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ HA-VID24-URA3 |
| HLY2658 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ VPS34-V5-His-URA3 |
| HLY3031 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3 N736K-VPS34-GFP-URA3 |
| HLY3030 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3 ΔC11-VPS34-GFP-URA3 |
TABLE 2.
Primers used in this study
| Vps34p-V5-His | |
| P200 forward | TCCATCTCGAGTGAATCGGAAACTTCCGGGACAGAATCGCTACCA |
| P201 reverse | GTTAATTAAGGTCCG CCAGTATTGTGCCAGATTATGTAAATGATC |
| Vps34p-GFP | |
| P200 forward | TCCATCTCGAGTGAATCGGAAACTTCCGGGACAGAATCGCTACCA |
| P202 reverse | GACGTCTTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTAC |
| FBPase-GFP | |
| P121 forward | ATTTGGTTGGGTTCTTCAGGTGAAATTGACAAATTTTTAGACCATATTGGCAAGTCACAGCGGATCCCCGGGTTAATTAA |
| P121 reverse | CCATCCCATTCCATTCGCTACTTCCTTTCTCTTTTCCTAAGAATTTTCATTATTAGAAGGGAATTCGAGCTCGTTTAAAC |
| C11 deletion | |
| P221 forward | CTGCCTATCGTGATTGATCGGATCCCCGGG |
| P222 reverse | CCCGGGGATCGGATCAATCACGATAGGCAG |
| N736K mutation | |
| P223 forward | CGATACGCATTTAGACAAGTTACTAGTCACGCCAGAT |
| P224 reverse | ATCTGGCGTGACTAGTAACTTGTCTAAATGCCTATCG |
Fluorescence Microscopy
Proteins were tagged with GFP via primers using PCR-based reactions. For actin staining, yeast cells were grown under starvation conditions for 3 days in 2 ml of YPKG. At each time point, samples of the cells (300 μl) were taken and fixed with 22 μl of formaldehyde for 5 min at room temperature. Cells were harvested by centrifugation at 1500 × g for 2.5 min. After the removal of the supernatant, cells were washed in 400 μl of PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4 (pH 7.4)). After further centrifugation at 1500 × g for 2.5 min, the supernatants were removed, and −20 °C acetone (800 μl) was added dropwise while vortexing the sample. Cells were then incubated for 5 min at −20 °C. Cells were washed in 400 μl of PBS and resuspended in 80 μl of PBS. Cells were finally stained with 1 μl of rhodamine-conjugated phalloidin at 0.2 units/μl in methanol and incubated for 30 min in the dark at room temperature. GFP and actin were visualized by fluorescence microscopy at 26 °C with FLUAR 100× objective lens (1.30 NA) using FITC and rhodamine filters, respectively. The cells were imaged using a Zeiss Axiovert S100 inverted microscope with an Axiocam MRm CCD camera and Axiovision v. 4.5 Software. The percent co-localization of GFP fusion proteins with actin was determined from three images and was represented as the mean and S.D.
Whole Cell Immunoelectron Microscopy
Wild-type and the Δvps34 cells were grown in 2 ml of YPKG culture for 3 days and then transferred to medium containing high glucose for the indicated time points. To examine FBPase distribution before starvation, wild-type cells were grown in 2 ml of YPD for 1 day and harvested. The cells were fixed with 3% paraformaldehyde and 0.2% glutaraldehyde overnight. The following day cells were washed by centrifugation (4500 × g) sequentially using 1 ml of each of the following buffers: 0.5 m sorbitol in 0.08 m potassium phosphate buffer (pH 6.7), 0.25 m sorbitol in 0.08 m potassium phosphate buffer (pH 6.7), and 0.08 m potassium phosphate buffer (pH 6.7). After centrifugation of the last wash, cells were resuspended in 1 ml of sodium metaperiodate (1% w/v) and were incubated at room temperature for 20 min. Thereafter, cells were resuspended in 1 ml of 50 mm ammonium chloride and incubated for 15 min at room temperature. Cells were centrifuged at 4500 × g and subjected to serial dehydration by adding an ethanol series (1 ml each) to the cell pellet. Samples were incubated for 5 min in 50% cold ethanol, 70% cold ethanol, and 80% cold ethanol followed by 10 min of incubation in 85% cold ethanol, 90% cold ethanol, 95% cold ethanol, and 100% cold ethanol twice. Finally, 1 ml of fresh 100% ethanol was added to the cell pellet and incubated for 5 min at room temperature. After dehydration, cells were sequentially incubated in 1 ml each of the following concentrations of resin (LR White): 2:1 ethanol:resin at room temperature rotator for 2 h; 1:1 ethanol:resin at room temperature rotator overnight; 1:2 ethanol:resin at room temperature rotator for 2 h; 100% resin at room temperature rotator for 2 h three times. Cells were then transferred to gelatin capsules and were then filled with extra resin to create a proper seal. These capsules were dried and cut into 10-nm-thin sections. Sections were placed onto grids and incubated with purified FBPase antibodies followed by goat anti-rabbit antibodies conjugated with 10-nm gold particles.
Cell Extraction Assay
For most experiments, cells were grown in 10 ml of YPKG medium for 3 days and transferred to YPD media for the indicated time points. In some experiments, cells were grown in YPD for 1 day or in YPKG for 1, 2, or 3 days. Cells (10 optical density) were collected and pelleted. Cell extraction was performed as described (51). Briefly, cells (10 optical density) were resuspended with 100 μl of cell extraction buffer (0.1 m Tris-Base (pH 9.4) and 10 mm β-mercaptoethanol) and incubated in a 37 °C shaker at 200 rpm for 15 min. After incubation, cells were pelleted, and supernatants were transferred to microcentrifuge tubes. After extraction, the supernatant fraction was centrifuged at 16,000 × g for 30 s at room temperature and transferred to microcentrifuge tubes. Proteins from the supernatant were precipitated using 15% trichloroacetic acid, washed, and resuspended in SDS-PAGE buffer. Cell pellet fractions were also lysed and resuspended in SDS-PAGE buffer. Both pellet and supernatant fractions were examined by Western blotting with anti-FBPase, anti-V5, anti-GFP, anti-Pil1p, and anti-HA antibodies.
N736K and ΔC11 Mutations
Vps34p-GFP was amplified by PCR reaction using P200 and P202 (Table 2) and cloned into TOPO plasmid (B548). To produce the N736K and the ΔC11 mutations, site-directed mutagenesis was performed according to the manufacturer's suggestions (Stratagene). The ΔC11-Vps34p-GFP mutation was produced using the Vps34p-GFP template (B548) and the P221 and P222 (Table 2) primers. The N736K-Vps34p-GFP mutation was generated by PCR using the Vps34p-GFP (B548) template and the P223 and P224 (Table 2) forward and reverse primers, respectively. The resulting mutations were confirmed by DNA sequencing at the Core Facility of the Penn State University College of Medicine.
RESULTS
VPS34 Is Required for the Degradation of FBPase in Prolonged-starved Cells
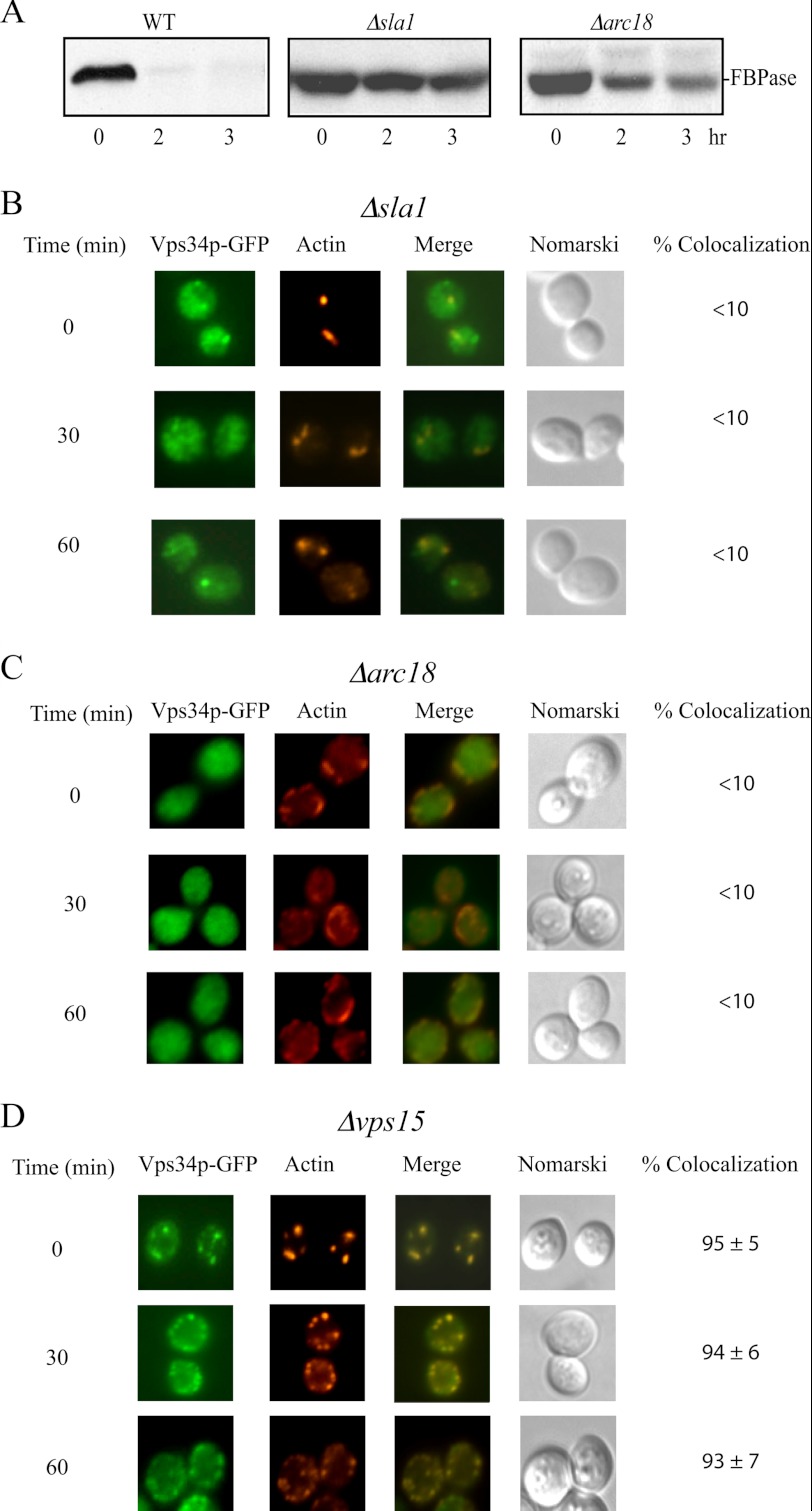
The yeast VPS34 gene is involved in multiple protein-targeting pathways to the vacuole. We determined whether or not the VPS34 gene also has a role in the Vid pathway. We have shown previously that FBPase is degraded in the proteasome when glucose is added to cells that are starved of glucose for 1 day (37). In contrast, when glucose is added to cells that are starved of glucose for 3 days, FBPase is degraded in the vacuole (37). Wild-type and the Δvps34 mutant cells were starved of glucose for 1 or 3 days, transferred to medium containing 2% glucose for 0, 2, and 3 h, and examined for FBPase degradation (Fig. 1). FBPase was degraded in 1 day-starved wild-type and Δvps34 cells (Fig. 1A), suggesting that VPS34 is not involved in the degradation of FBPase in the proteasome. FBPase was degraded in 3 day-starved wild-type cells (Fig. 1B). In contrast, FBPase degradation was inhibited in the 3-day-starved Δvps34 mutant (Fig. 1B). These results suggest that the VPS34 gene is required for the degradation of FBPase in the vacuole. For the Vps pathway, Vps15p forms a complex with Vps34p and activates the PI3K activity of Vps34p (48, 52). FBPase degradation was also impaired in the Δvps15 mutant that was starved of glucose for 3 days and replenished with glucose (Fig. 1C). Vps34p, Vps15p, and Vps30p are common subunits of two distinct phosphatidylinositol 3-kinase complexes; Complex I is involved in the autophagic pathway, whereas Complex II is required for the Vps pathway (45, 54, 55). We next determined whether or not VPS30 is also involved in the Vid pathway by examining FBPase degradation in the Δvps30 strain. FBPase was degraded in the Δvps30 strain that was glucose-starved for 3 days and re-fed with glucose, suggesting that Complex I and Complex II are not involved in the Vid pathway. In this study we focused on the role of VPS34 in the vacuole-dependent pathway. Therefore, most of our experiments were performed in cells that had been starved of glucose for 3 days and transferred to medium containing high glucose for the indicated time points.
FIGURE 1.
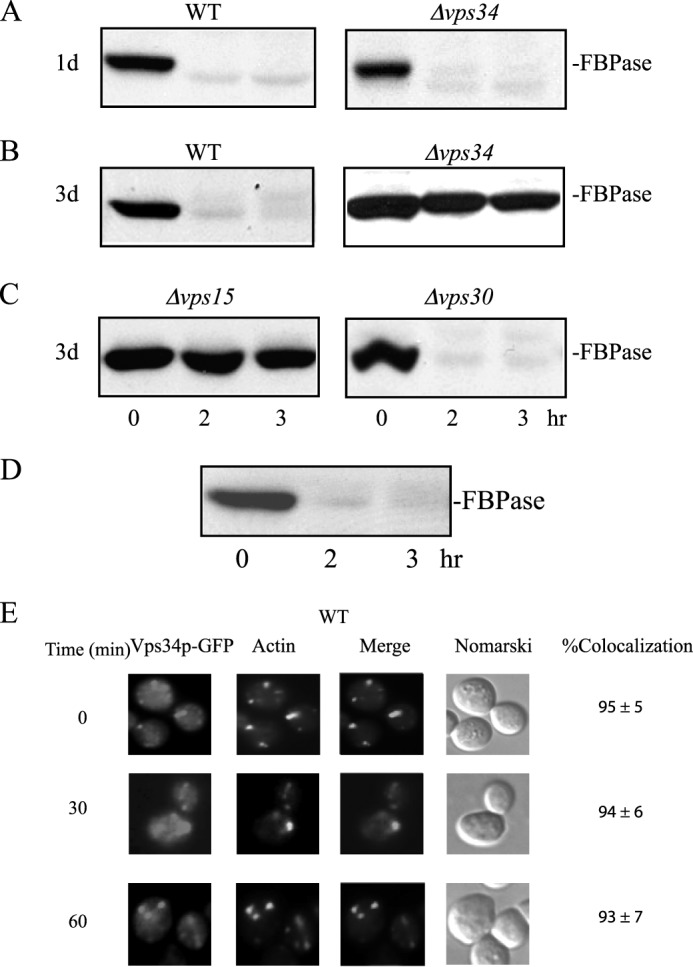
FBPase degradation in the vacuole requires the VPS34 gene. A and B, wild-type and cells lacking the VPS34 gene were starved of glucose for 1 day (A) and 3 days (B). Cells were transferred to medium containing high glucose for 0, 2, and 3 h and examined for FBPase degradation. C, cells lacking VPS15 and VPS30 were starved of glucose for 3 days, transferred to medium containing high glucose for 0, 2, and 3 h, and examined for FBPase degradation. D, wild-type cells expressing Vps34p-GFP were starved of glucose for 3 days, re-fed with glucose for the indicated time points, and examined for the degradation of FBPase. E, the distribution of Vps34p-GFP and actin patches was determined using fluorescence microscopy.
Vps34p Is Associated with Actin Patches
We have shown recently that the Vid pathway merges with the endocytic pathway (27, 31). Vid vesicle proteins such as Vid24p, Sec28p, and Vid30p associate with actin patches initially, but they dissociate later (27, 41). We determined whether or not Vps34p was associated with actin patches in glucose-starved wild-type cells using a protocol that gives consistent and reliable results regarding the distribution of GFP tagged proteins with actin patches (27, 31, 41). Wild-type cells expressing Vps34p-GFP were starved of glucose for 3 days and then shifted to glucose for the indicated time points. FBPase was degraded in cells expressing Vps34p-GFP in response to glucose (Fig. 1D), suggesting that the GFP tag does not interfere with FBPase degradation. A high percentage of Vps34p-GFP signals were in punctate structures that colocalized with actin patches that were stained with phalloidin during glucose starvation (Fig. 1E). After the addition of glucose, Vps34p-GFP still co-localized with actin patches up to the 60-min time point (Fig. 1E). This is different from FBPase, Vid24p, and Vid30p in that they showed less co-localization with actin patches at the 60-min time point (27, 41).
Vps34p Distribution Is Affected in the Δsla1 and Δarc18 Mutants
Given that Vps34p shows co-localization with actin patches before and after glucose addition, Vps34p distribution may be affected when the polymerization of actin is disrupted. In yeast grown in rich medium, actin is assembled in a stepwise manner (11, 42, 56, 57). Sla1p, End3p, and Pan1p are required early in the process of assembly, whereas the Arp2/3 complex is required at later steps (11, 42, 56, 57). Arc18p is the subunit of Arp2/3 complex involved in the nucleation and formation of short actin filaments (11, 42, 56, 57). FBPase degradation was retarded in cells lacking the SLA1 gene and the ARC18 gene (Fig. 2A), suggesting that these genes are required for the Vid pathway.
FIGURE 2.
Vps34p distribution is affected in cells lacking the SLA1 and ARC18 genes. A, wild-type and cells lacking the SLA1 and ARC18 genes were starved of glucose for 3 days and replenished with glucose. FBPase degradation was examined after the addition of glucose for 0, 2, and 3 h. B–D, Vps34p-GFP was expressed in the Δsla1 (B), the Δarc18 (C), and the Δvps15 (D) strains. These cells were starved of glucose for 3 days and shifted to glucose for the indicated times. The distribution of Vps34p-GFP and actin patches was determined by fluorescence microscopy.
We determined the distribution of Vps34p-GFP in the Δsla1, the Δarc18, and the Δvps15 strains that expressed Vps34p-GFP. For the Vps pathway, the protein kinase Vps15p recruits Vps34p to membranous structures, resulting in the activation of the PI3K activity (48, 58). These strains were starved of glucose for 3 days and transferred to media containing glucose for the indicated time points. In the Δsla1 strain, Vps34p-GFP appeared to be diffused, and some GFP puncta were also observed. However, the majority of the Vps34p-GFP did not co-localize with phalloidin in the Δsla1 mutant before and after the addition of glucose (Fig. 2B). In the Δarc18 mutant, the distribution of Vps34p-GFP was diffused before and after the addition of glucose (Fig. 2C). Therefore, Vps34p localization is affected in cells lacking the SLA1 and ARC18 gene. In prolonged-starved Δvps15 mutant, Vps34p-GFP showed co-localization with actin patches during glucose starvation and after the addition of glucose for up to 60 min (Fig. 2D). Thus, the absence of Vps15p does not appear to affect the distribution of Vps34p and actin patches.
FBPase and Vid24p Are Associated with Actin Patches in Cells Lacking the VPS34 Gene
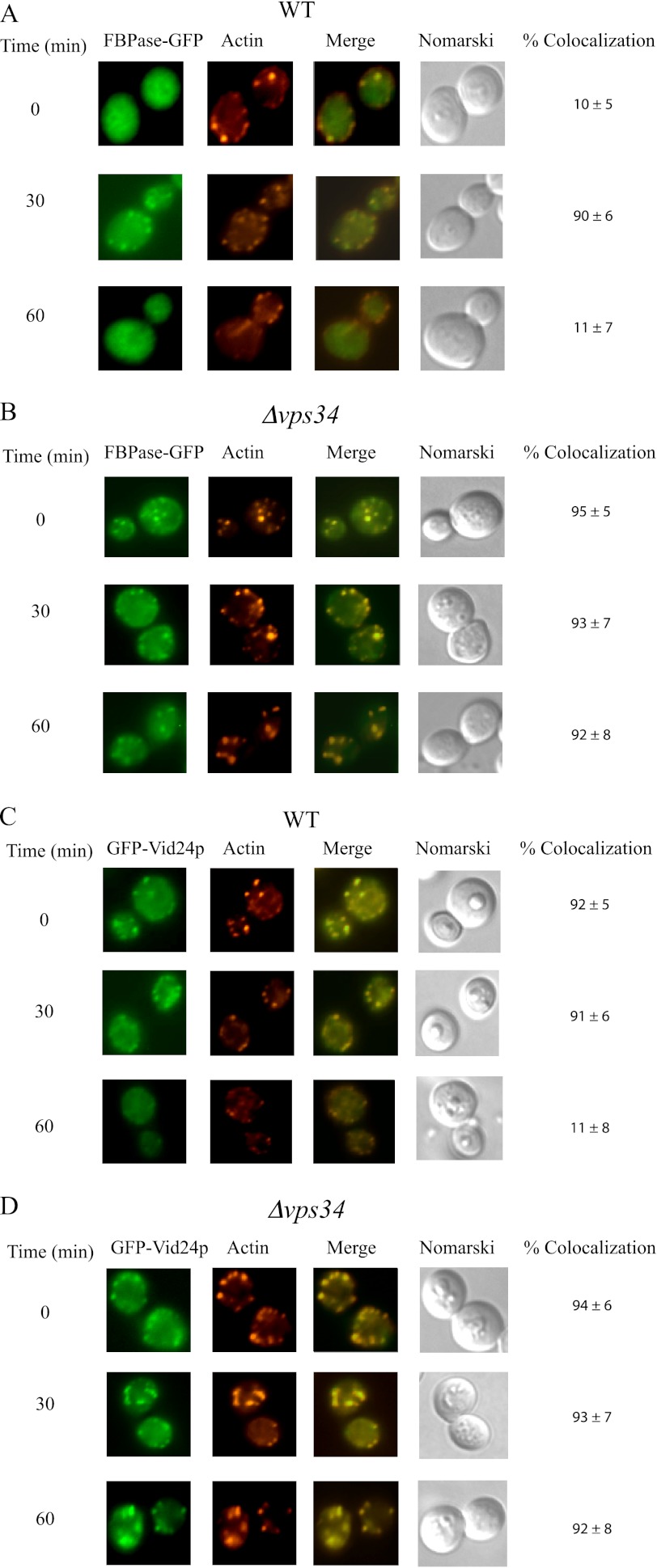
We next determined whether or not Vps34p has a role in the co-localization of FBPase and Vid24p with actin patches. Vid24p is a peripheral protein on Vid vesicles (38) and has been used to follow the trafficking of Vid vesicles in the Vid pathway (27). FBPase-GFP and GFP-Vid24p were expressed in wild-type and the Δvps34 mutant strain that were starved of glucose and then transferred to media containing fresh glucose. In wild-type cells, the association of FBPase with actin patches was low at t = 0 min (Fig. 3A). However, the co-localization of FBPase with actin patches increased at t = 30 and then decreased at t = 60 min (Fig. 3A). In the Δvps34 mutant, a high percentage of FBPase was associated with actin patches before and after the addition of glucose for up to 60 min (Fig. 3B). In wild-type cells, GFP-Vid24p was associated with actin patches during glucose starvation and after the addition of glucose for 30 min (Fig. 3C). However, this protein showed less co-localization with actin patches at the 60-min time point (Fig. 3C). In the Δvps34 strain, GFP-Vid24p co-localized with actin patches during glucose starvation and after the addition of glucose for up to 60 min (Fig. 3D). Hence, the association of FBPase and Vid24p with actin patches persists in the absence of the VPS34 gene.
FIGURE 3.
FBPase and Vid24p are associated with actin patches in the Δvps34 mutant before and after the addition of glucose. Wild-type (A) and the Δvps34 mutant (B) strains expressing FBPase-GFP were starved of glucose for 3 days and replenished with glucose for the indicated time points. The distribution of FBPase and actin was determined by fluorescence microscopy. Wild-type (C) and the Δvps34 (D) strains expressing GFP-Vid24p were glucose-starved, re-fed with glucose for the indicated time points, and examined for the distribution of GFP-Vid24p and actin patches by fluorescence microscopy.
FBPase Is in the Periplasm in Prolonged-starved Wild-type Cells
To gain a better understanding of how the Δvps34 mutant affected the FBPase degradation pathway, we used immunoelectron microscopy to examine the distribution of FBPase at the ultra-structural level. Wild-type and Δvps34 mutant cells were starved of glucose for 3 days and transferred to medium containing 2% glucose for the indicated time points. Cells were fixed and embedded. Thin sections of embedded cells were incubated with purified FBPase antibodies followed by goat anti-rabbit secondary antibodies conjugated with 10-nm gold particles. In wild-type cells grown in medium containing high glucose, FBPase levels were low (Fig. 4A). In 3-day-starved wild-type cells, significant amounts of FBPase were in the periplasm (Fig. 4B). When glucose was added to starved cells for 15 min, high levels of FBPase were in intracellular structures (Fig. 4C). These structures resembled the Vid/endosomes that we have characterized previously (27). In cells that were re-fed with glucose for 2 h, amounts of FBPase decreased (Fig. 4D). In 3-day-starved Δvps34 mutants, high levels of FBPase were in the periplasm (Fig. 5A). In the Δvps34 mutant that was re-fed with glucose for 2 h, significant amounts of FBPase remained in the periplasm (Fig. 5B).
FIGURE 4.
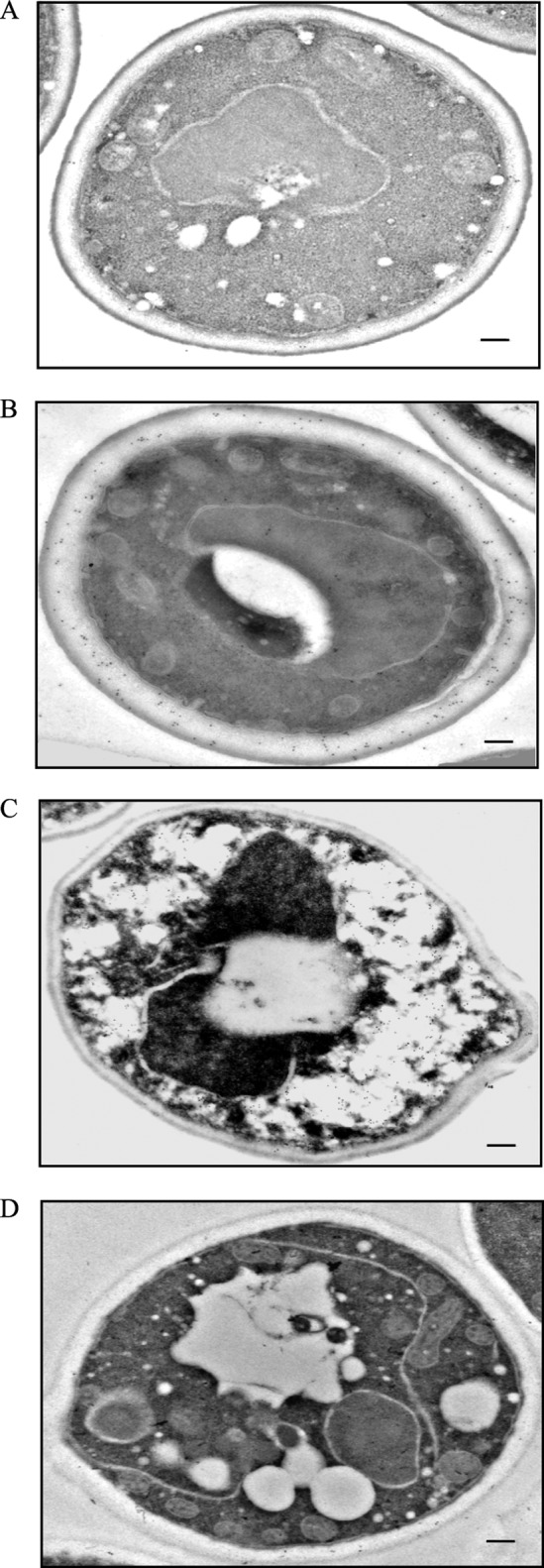
FBPase is in the periplasm in prolonged-starved wild-type cells. FBPase distribution was determined in wild-type cells that were grown in glucose-rich medium (A), in cells starved of glucose for 3 days (B), and in cells that were starved and then re-fed with glucose for 15 min (C) and 2 h (D). Cells were processed and embedded as described under “Experimental Procedures.” Thin sections were incubated with FBPase antibodies followed by goat anti-rabbit antibodies conjugated with 10-nm colloid gold particles and then visualized with transmission electron microscopy. Bars, 200 nm. The number of gold particles in the cytoplasmic and periplasmic space was 6 and 17 before starvation (A), 62 and 156 after 3 days of starvation (B), 260 and 16 after re-feeding for 15 min (C), and 8 and 16 after re-feeding for 120 min (D).
FIGURE 5.
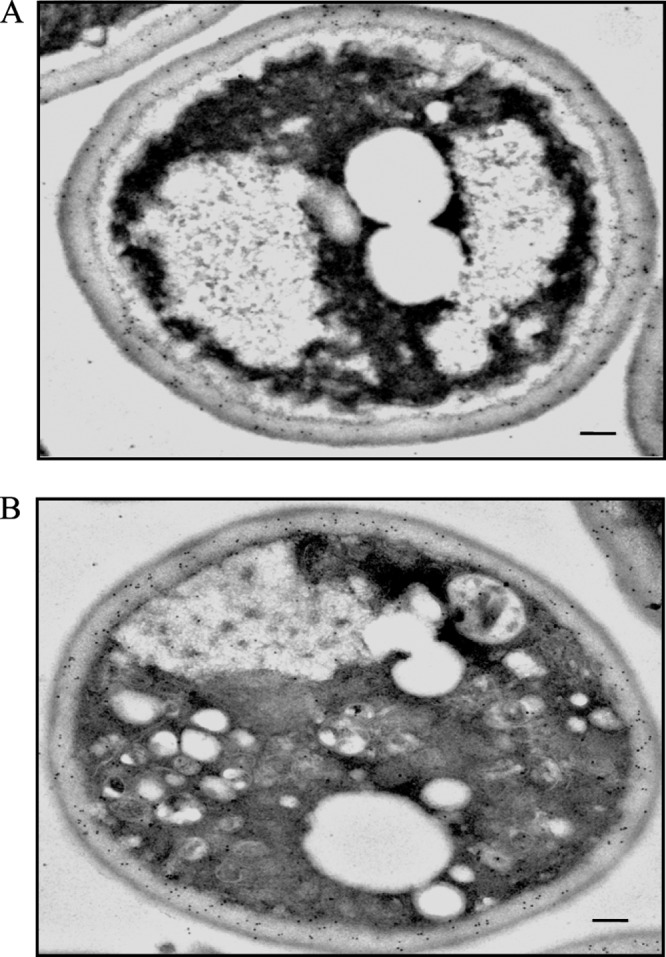
FBPase is in the periplasm in the Δvps34 mutant during glucose starvation and after glucose re-feeding. FBPase distribution was determined in the Δvps34 mutant that was starved of glucose for 3 days (A) and transferred to medium containing glucose for 2 h (B). FBPase distribution was examined by incubating thin sections with FBPase antibodies followed by goat anti-rabbit antibodies conjugated with 10-nm colloid gold particles. Bars, 200 nm. The number of gold particles in the cytoplasmic and periplasmic space in glucose-starved Δvps34 mutant was 51 and 268 (A). The number of gold particles in glucose re-fed Δvps34 mutant was 72 and 182 (B).
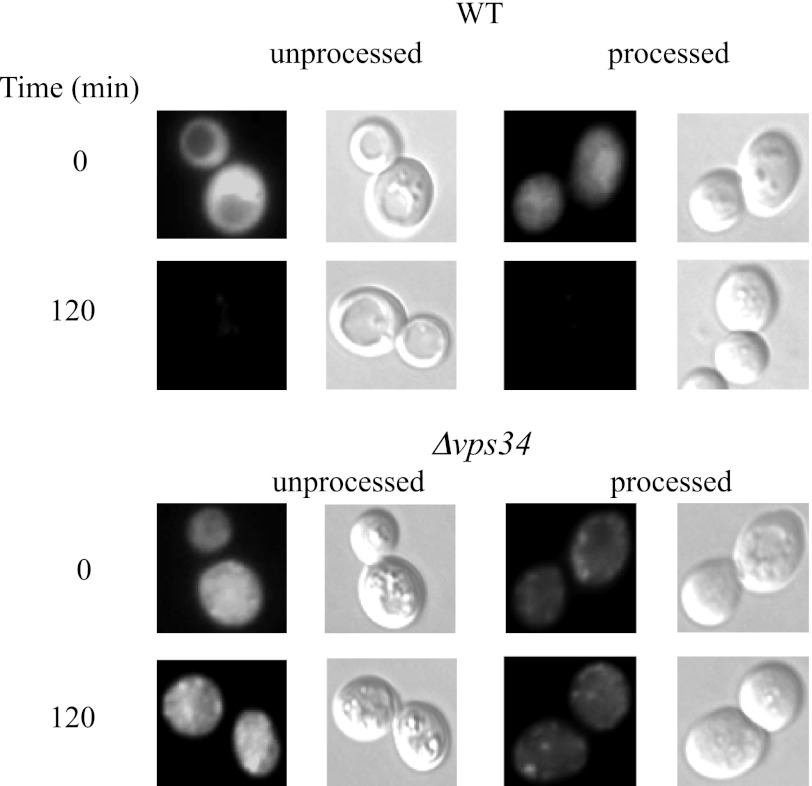
We determined the distribution of FBPase-GFP in unprocessed and processed wild-type and Δvps34 mutant cells that were glucose-starved and then re-fed with glucose (Fig. 6). We detected strong FBPase-GFP signals in unprocessed wild-type cells that were glucose-starved. In processed wild-type cells that were glucose-starved, FBPase-GFP signals were weaker. FBPase is degraded in response to glucose. As such, FBPase-GFP signals were low in both unprocessed and processed wild-type cells that were re-fed with glucose for 120 min. In the Δvps34 mutant, FBPase-GFP signals were stronger in unprocessed cells than in processed cells at t = 0 min and at t = 120 min. Lower fluorescence signals in processed wild-type and the Δvps34 mutant cells may result from the loss of intracellular pool of FBPase-GFP, the loss of extracellular pool of FBPase-GFP, or both.
FIGURE 6.
FBPase-GFP in unprocessed and processed wild-type and Δvps34mutant. FBPase-GFP was expressed in wild-type and Δvps34 cells that were starved of glucose for 3 days and transferred to medium containing high glucose for 120 min. FBPase-GFP in unprocessed and processed wild-type and Δvps34 mutants were visualized by fluorescence microscopy.
To further examine the presence of FBPase in the extracellular fraction, we utilized a protocol that detected the secretion of mammalian galectin-1 expressed in S. cerevisiae (51). This protocol uses the combination of high pH and reducing agents such as β-mercaptoethanol or DTT to release periplasmic proteins that are linked by ionic interactions and by disulfide bonds, respectively.
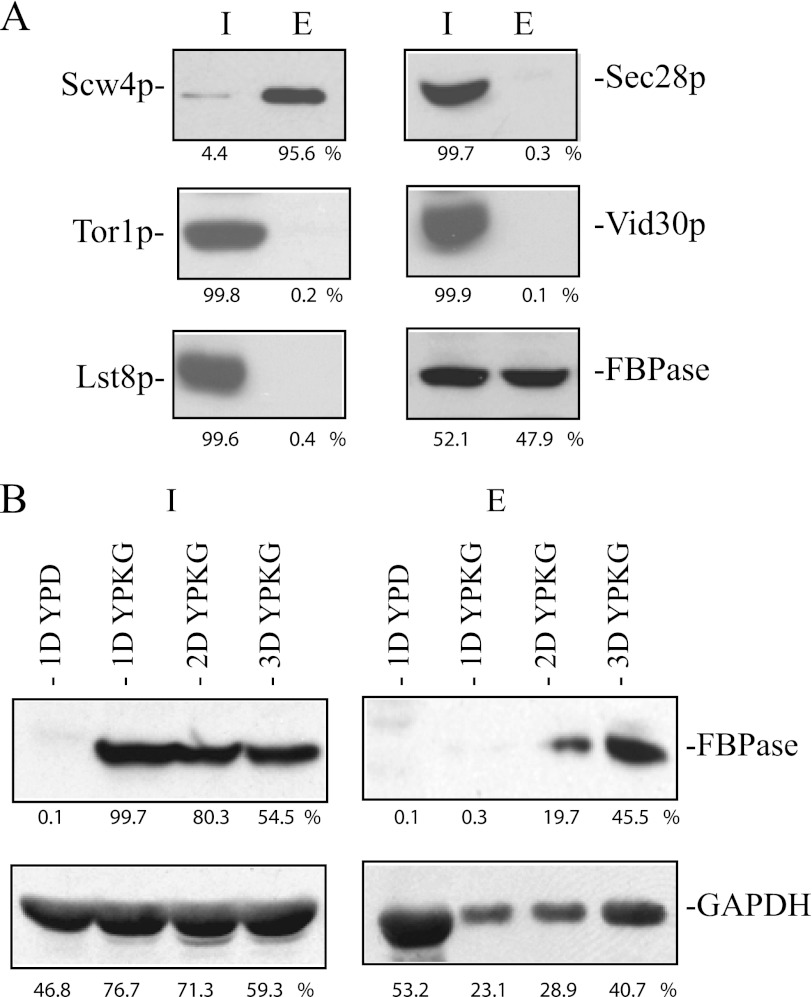
We first determined whether or not this protocol extracted known periplasmic proteins into the extracellular fraction. Scw4p is a soluble protein of the cell wall (59). Wild-type cells expressing Scw4p-GFP were starved of glucose for 3 days and extracted. After the extraction protocol, extracellular proteins were precipitated with TCA and solubilized in SDS sample buffer. This fraction is called the extracellular fraction (E) in this study. After extraction, cells were lysed and solubilized in SDS buffer. This fraction is called the cell-associated fraction/intracellular fraction (I) in this study. More than 90% of the Scw4p-GFP was in the extracellular fraction after the extraction (Fig. 7A), suggesting that this protocol extracts most of the Scw4p into the extracellular fraction.
FIGURE 7.
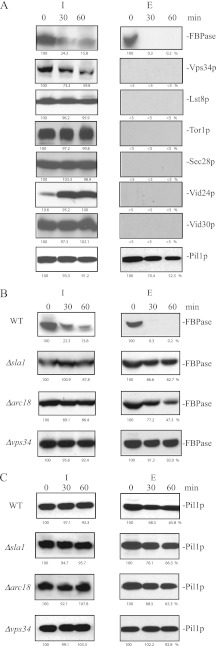
FBPase is in the extracellular fraction in prolonged-starved wild-type cells. A, wild-type cells expressing Scw4p-GFP, Tor1p-GFP, Lst8p-GFP, and Vid30p-GFP were starved of glucose for 3 days and subjected to the extraction protocol. Levels of Scw4p, Tor1p, Lst8p, Sec28p, Vid30p, and FBPase in the intracellular (I) and extracellular (E) fractions were examined. Proteins were quantified using NIH ImageJ program. Relative percentages of proteins in the intracellular and extracellular fractions were determined. B, wild-type cells were grown in YPD for 1 day or in YPKG for 1 (1D), 2 (2D), and 3 (3D) days. Cells were extracted, and proteins were separated into the intracellular (I) and extracellular (E) fractions. Levels of FBPase and GAPDH in the intracellular and intracellular fractions were determined and quantified using NIH ImageJ program. Relative percentages of FBPase and GAPDH in the intracellular and extracellular fractions under each growth condition were determined.
We next determined the distribution of proteins that are known to play important roles in the Vid pathway. The Target of Rapamycin complex 1 is involved in the Vid pathway (20) and comprises multiple subunits including Lst8p, Kog1p, Tco89p, and Tor1p (60–62). Cells expressing Lst8p-GFP and Tor1p-GFP were starved of glucose for 3 days, extracted, and examined for the distribution of these proteins (Fig. 7A). After extraction, the majority of the Lst8p-GFP and Tor1p-GFP were in the intracellular fraction, and minimal amounts were in the extracellular fraction (Fig. 7A). Sec28p and Vid30p are also involved in the Vid pathway (31, 41). Again, most of the Sec28p and Vid30p were in the intracellular fraction, and little was in the extracellular fraction (Fig. 7A). By contrast, FBPase was in both intracellular and extracellular fractions in 3-day-starved cells (Fig. 7A). Therefore, molecules involved in the Vid pathway are retained inside the cells, whereas cargo protein is present in both fractions.
We used this protocol to determine the conditions that led to the presence of FBPase in the extracellular fraction. Wild-type cells were grown in YPD or in YPKG for 1, 2, and 3 days. Cells were harvested and subjected to the extraction protocol. Proteins were separated into intracellular, and extracellular fractions and examined for the distribution of FBPase (Fig. 7B). FBPase was not expressed in cells grown in YPD and was not detected in the extracellular fraction. In cells grown in YPKG for 1 day, FBPase was expressed in the intracellular fraction but was not detected in the extracellular fraction. In 2-day-starved cells grown in YPKG, low amounts of FBPase were detected in the extracellular fraction. Higher amounts of FBPase were present in the extracellular fraction in 3-day-starved cells grown in YPKG. Thus, the presence of FBPase in the extracellular fraction is dependent on the duration of starvation.
It has been reported that the glycolytic enzyme GAPDH is on the cell surface of S. cerevisiae grown in YPD (63). Therefore, this protein should also be detected in the extracellular fraction in cells grown in YPD. Consistent with previous reports, levels of GAPDH in the extracellular fraction were high in cells grown in YPD (Fig. 7B). Levels of GAPDH in the extracellular fraction were low in cells grown in YPKG for 1 and 2 days and increased after growth in YPKG for 3 days (Fig. 7B). Thus, amounts of FBPase and GAPDH in the extracellular fraction vary depending on the growth conditions.
FBPase Levels in the Extracellular Fraction Decrease in Response to Glucose in Wild-type Cells
We determined whether or not levels of FBPase in the extracellular fraction changed when glucose-starved cells are replenished with glucose. Wild-type cells were starved of glucose for 3 days and transferred to medium containing 2% glucose for 0, 30, and 60 min. Cells were subjected to the extraction protocol, and the distribution of FBPase in the intracellular and extracellular fractions was determined (Fig. 8A and Table 3). In wild-type cells, levels of FBPase in the extracellular fraction were high at t = 0 min and decreased dramatically after the addition of glucose (Fig. 8A). Under the same conditions, most of the Vps34p, Lst8p, Tor1p, Sec28p, Vid24p, and Vid30p were in the intracellular fraction, and minimal levels were detected in the extracellular fraction (Fig. 8A). However, Pil1p, which is a component of eisosomes, was detectable in the intracellular as well as extracellular fractions in wild-type cells that were starved and then re-fed with glucose (Fig. 8A).
FIGURE 8.
Levels of extracellular FBPase decrease in response to glucose in wild-type cells, but levels of extracellular FBPase remained high in the Δvps34 mutant in response to glucose. A, wild-type cells expressing Vps34p-V5-His, Lst8p-GFP, Tor1p-GFP, Vid24p-HA, and Vid30p-GFP were starved of glucose for 3 days and re-fed with glucose for 0, 30, and 60 min. FBPase degradation was normal in cells expressing these tags. Cells were subjected to the extraction protocol, and levels of these proteins in the intracellular (I) and extracellular (E) fractions were determined by Western blotting. Proteins were quantified with NIH ImageJ program, and the percentages of proteins remaining in the intracellular fraction after the addition of glucose were determined using t = 0 min as 100%. The amount of Vid24p in the intracellular fraction at t = 60 min was used as 100%. B and C, wild-type cells and cells lacking SLA1, ARC18, and VPS34 were starved of glucose for 3 days and replenished with glucose for the indicated time points. Cells were extracted, and proteins were separated into the intracellular (I) and extracellular (E) fractions. The distribution of FBPase (B) and Pil1p (C) was determined by Western blotting. The percentages of FBPase (B) and Pil1p (C) remaining in both intracellular and extracellular fractions after the addition of glucose were determined using the amounts of FBPase and Pil1p at t = 0 min as 100%. Relative distribution of proteins in the intracellular (I) and extracellular (E) fractions in glucose-starved wild-type and mutant cells was determined and listed in Table 3.
TABLE 3.
Relative distribution of FBPase and Pil1p in the intracellular and extracellular fractions in glucose-starved wild type and mutant strains
| Figures | Proteins and strains | Intracellular | Extracellular |
|---|---|---|---|
| Fig. 8A | FBPase in WT | 58.5 | 41.5 |
| Vps34p in WT | >95 | <5 | |
| Lst8p in WT | >95 | <5 | |
| Tor1p in WT | >95 | <5 | |
| Sec28p in WT | >95 | <5 | |
| Vid24p (t60) in WT | >95 | <5 | |
| Vid30p in WT | >95 | <5 | |
| Pil1p in WT | 53.3 | 46.7 | |
| Fig. 8B | FBPase in WT | 56.1 | 43.9 |
| FBPase in the Δsla1 mutant | 55.2 | 44.8 | |
| FBPase in the Δarc18 mutant | 50.6 | 49.4 | |
| FBPase in the Δvps34 mutant | 53.5 | 46.7 | |
| Fig. 8C | Pil1p in WT | 49.7 | 50.3 |
| Pil1p in the Δsla1 mutant | 48.3 | 51.7 | |
| Pil1p in the Δarc18 mutant | 49.7 | 50.3 | |
| Pil1p in the Δvps34 mutant | 53.4 | 46.6 | |
| Fig. 9C | FBPase in WT | 56.7 | 43.3 |
| FBPase in the N736K mutant | 48.8 | 51.2 | |
| Fig. 9D | Pil1p in WT | 51.1 | 48.9 |
| Pil1p in the N736K mutant | 49.8 | 50.2 | |
| Fig. 10C | FBPase in WT | 53.8 | 46.2 |
| FBPase in the ΔC11 mutant | 51.6 | 48.4 | |
| Fig. 10D | Pil1p in WT | 52.5 | 47.5 |
| Pil1p in the ΔC11 mutant | 51.7 | 48.3 |
The Δsla1, Δarc18, and Δvps34 Mutants Delay the Decrease of FBPase in the Extracellular Fraction in Response to Glucose
We next determined whether or not levels of extracellular FBPase changed in response to glucose in the Δsla1 and the Δarc18 mutants that block endocytosis. The Δsla1 and the Δarc18 mutants were starved of glucose for 3 days, re-fed with glucose, and examined for FBPase distribution (Fig. 8B). In these strains, FBPase levels in the extracellular fraction did not decrease as rapidly as wild-type cells.
Our immunoelectron microscopy results showed that high amounts of FBPase remained in the periplasm after the addition of glucose to the Δvps34 mutant for 2 h. We determined whether or not high levels of FBPase in the extracellular fraction remained in the Δvps34 mutant that was replenished with glucose. The Δvps34 mutant was starved of glucose for 3 days and re-fed with glucose for up to 60 min. Cells were subjected to the extraction protocol, and the distribution of FBPase in the intracellular (I) and extracellular (E) fractions was examined (Fig. 8B). FBPase was detected in the extracellular fraction during glucose starvation in the Δvps34 mutant. After the addition of glucose to the Δvps34 mutant, levels of FBPase in the extracellular fraction did not decrease as rapidly as wild-type cells. Thus, the absence of the VPS34 gene appears to retard the decrease of FBPase in the extracellular fraction in response to glucose. Pil1p was detectable in both intracellular and extracellular fractions after glucose re-feeding in strains lacking the SLA1, ARC18 and VPS34 genes (Fig. 8C).
N736K and ΔC11 Mutants Delayed the Decline of FBPase in the Extracellular Fraction
The N736K mutation of Vps34p affects a number of vacuole targeting pathways (44–47). In addition, the last 11 amino acids of the C terminus (residues 864–875) are implicated in membrane binding (49, 50). We determined whether or not the N736K mutation and the deletion of the C-terminal 11 amino acids blocked the Vid pathway. We produced the N736K and the ΔC11 mutants using site-directed mutagenesis. Vps34p-GFP was used as the template for site-directed mutagenesis because FBPase degradation was not affected in wild-type cells expressing Vps34p-GFP (see Fig. 1D).
Cells expressing N736K-GFP were starved of glucose for 3 days, transferred to medium containing 2% glucose for 0, 2, and 3 h, and examined for FBPase degradation (Fig. 9A). FBPase degradation was retarded in cells expressing N736K-GFP. We next determined whether or not the N736K mutant co-localized with actin patches. Cells expressing N736K-GFP were starved of glucose for 3 days, transferred to medium containing 2% glucose, and examined for the distribution of GFP and actin (Fig. 9B). In prolonged-starved cells expressing N736K-GFP, most of the GFP signals appeared to be in punctate structures, and some GFP signals were diffused. However, there was a low percentage of co-localization of GFP signals with actin patches (Fig. 9B). We next determined whether or not the N736K mutant affected the decline of FBPase in the extracellular fraction in response to glucose. In cells expressing N736K-GFP that were starved of glucose for 3 days and re-fed with glucose, levels of FBPase in the extracellular fraction did not decrease as rapidly as the wild-type control (Fig. 9C). Pil1p was detectable in the extracellular fraction after the addition of glucose to wild-type and the N736K mutant (Fig. 9D).
FIGURE 9.
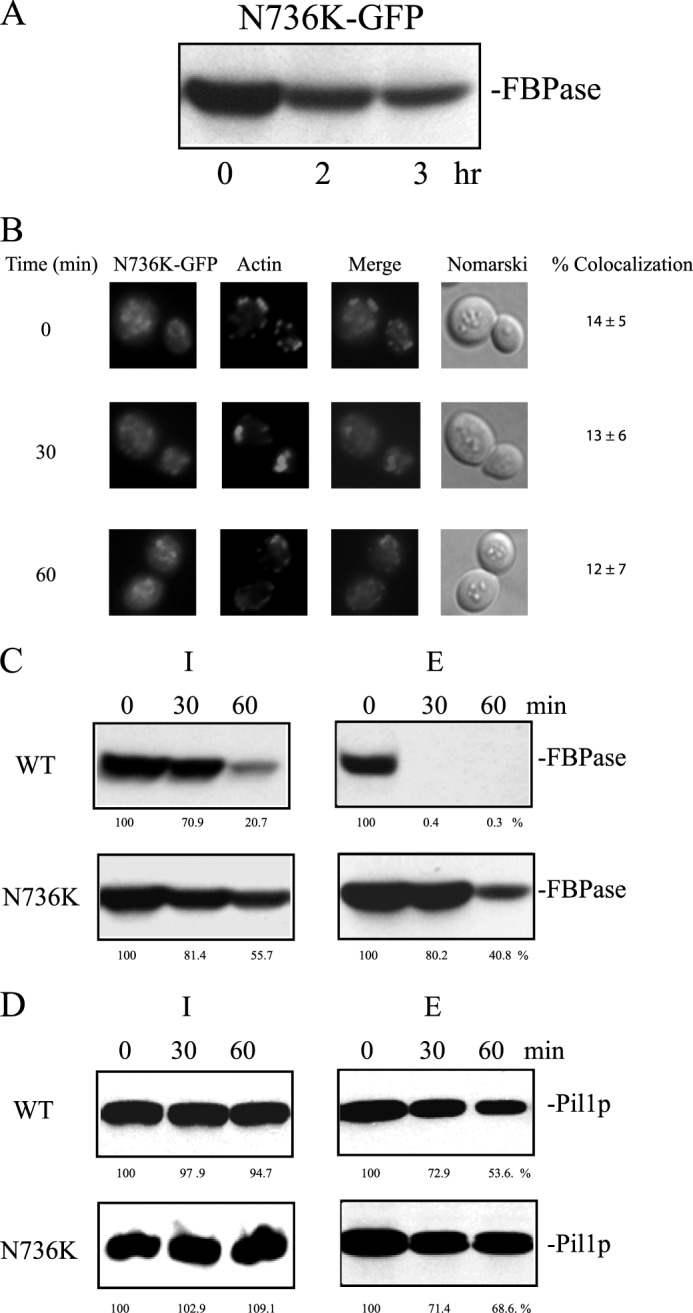
The N736K mutant impairs FBPase degradation, association with actin patches, and internalization of extracellular FBPase. A, FBPase degradation was examined in cells expressing N736K-GFP. B, cells expressing N736K-GFP were starved of glucose for 3 days and transferred to medium containing glucose. The distribution of N736-K-GFP and actin patches was examined by fluorescence microscopy. Levels of FBPase (C) and Pil1p (D) in the intracellular and extracellular fractions were determined in the N736K-GFP cells that were starved of glucose for 3 days and transferred to media containing high glucose for 0, 30, and 60 min. The percentages of FBPase and Pil1p remaining in intracellular and extracellular fractions after glucose re-feeding were determined using t = 0 min as 100%. Relative distribution of FBPase and Pil1p in the intracellular (I) and extracellular (E) fractions in glucose-starved wild-type and N736K cells was determined and listed in Table 3.
Finally, we determined whether or not the deletion of the last 11 amino acids affected the Vid pathway. FBPase degradation was inhibited in cells expressing the ΔC11 mutation (Fig. 10A). In prolonged-starved cells expressing ΔC11-GFP, the GFP signals were mostly diffused and did not show significant co-localization with actin before and after the addition of glucose (Fig. 10B). Furthermore, the ΔC11 mutant also delayed the decline of FBPase in the extracellular fraction after glucose re-feeding (Fig. 10C). Pil1p was detectable in the intracellular and extracellular fractions in wild-type and the ΔC11 mutant that were starved and re-fed with glucose (Fig. 10D). Hence, both N736K and ΔC11 mutants inhibited the degradation of FBPase, retarded the decrease of FBPase in the extracellular fraction in response to glucose, and failed to co-localize with actin patches.
FIGURE 10.
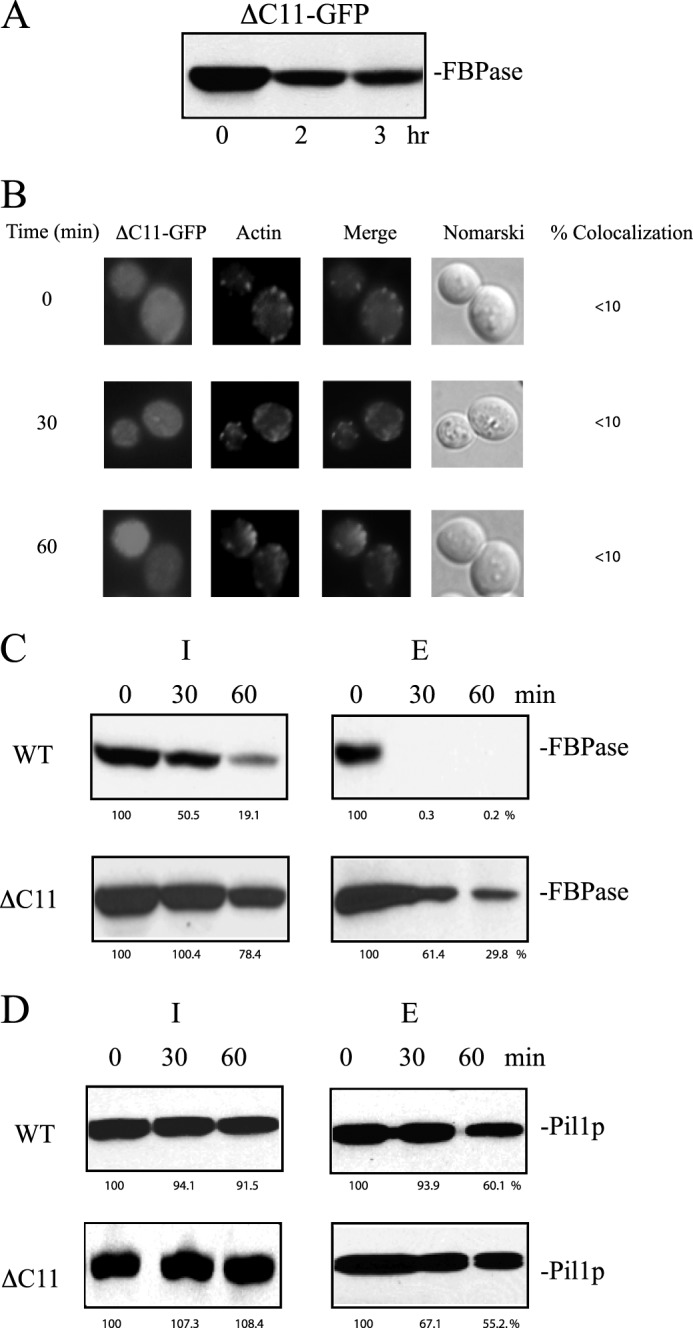
The ΔC11 mutant blocks FBPase degradation, localization with actin patches, and FBPase internalization. A, cells expressing ΔC11-GFP were starved of glucose for 3 days, transferred to media containing high glucose for the indicated time points, and examined for FBPase degradation. B, prolonged-starved cells expressing ΔC11-GFP were transferred to medium containing glucose and examined for the distribution of GFP and actin patches by fluorescence microscopy. C and D, FBPase and Pil1p in the intracellular (I) and extracellular (E) fractions were determined in cells expressing ΔC11-GFP that were starved of glucose and transferred to media containing high glucose for the indicated time points. The percentages of FBPase and Pil1p remaining after the addition of glucose were determined using t = 0 min as 100%. Relative distribution of FBPase and Pil1p in the intracellular and extracellular fractions in glucose-starved wild-type and ΔC11 cells was determined and listed in Table 3.
DISCUSSION
In this paper we demonstrate that Vps34p plays an important role in the Vid pathway. In the absence of this gene, FBPase and Vid24p associated with actin patches before and after addition of glucose. These characteristics are different from those seen in wild-type cells in which the association of FBPase, Vid24p, Sec28p, and Vid30p with actin all decreased at the 60-min time point. Given that actin polymerization is required for endocytosis, prolonged association of FBPase and Vid24p with actin patches after the addition of glucose in the Δvps34 mutant is consistent with the idea that endocytosis is affected in the absence of the VPS34 gene. Immunoelectron microscopy data and cell extraction data indicated that substantial amounts of FBPase were in the periplasm or extracellular fraction in glucose-starved wild-type and the Δvps34 mutant. These results suggest that FBPase is secreted in glucose-starved cells and that the VPS34 gene is not involved in the secretion of FBPase during glucose starvation.
The appearance of FBPase in the extracellular fraction is dependent on the duration of starvation. In cells starved of glucose for 1 day, FBPase was expressed but was not detected in the extracellular fraction. In 2-day-starved cells grown in YPKG, low levels of FBPase were in the extracellular fraction, and higher levels were detected in the extracellular fraction in 3-day-starved cells. GAPDH is a glycolytic enzyme and is associated with the cell surface of S. cerevisiae grown in rich medium (53). Interestingly, levels of GAPDH in the extracellular fraction also varied. For example, levels of GAPDH in the extracellular fraction were high in cells grown in YPD but were low in cells grown in YPKG for 1 and 2 days. Levels of GAPDH increased in cells grown in 3-day YPKG. Hence, levels of FBPase and GAPDH in the extracellular fraction vary depending on the physiological states of the cells.
Why is FBPase secreted during prolonged glucose starvation? We speculate that when cells are starved of glucose for a shorter period of time (1 day), FBPase is needed for gluconeogenesis inside the cells and, therefore, is not secreted. However, as cells are starved of glucose for a longer period of time (3 days), substrates for gluconeogenesis may be depleted, and the need for FBPase inside cells decreases. Therefore, more FBPase is secreted into the periplasm in 3-day-starved cells. Interestingly, when glucose was added to glucose-starved wild-type cells, levels of FBPase in the extracellular fraction decreased rapidly. Because FBPase is degraded in the vacuole, extracellular FBPase may be internalized in response to glucose. This idea is consistent with the findings that when glucose was added to the Δsla1 and Δarc18 mutants that block actin polymerization and endocytosis, the decline of FBPase in the extracellular fraction was delayed. Under the same conditions, the majority of Vps34p, Lst8p, Tor1p, Vid24p, Sec28p, and Vid30p were in the intracellular fraction. The decline of extracellular FBPase may also result from the release of this protein into the medium or the degradation of this protein in the extracellular space.
The decrease of FBPase in the extracellular fraction was delayed in the Δvps34 mutant. Furthermore, the N736K and the ΔC11 mutants also retarded the decline of FBPase in the extracellular fraction and impaired the association of Vps34p with actin patches. Vps34p distribution was affected in the Δsla1 and Δarc18 mutants that also delayed the reduction of FBPase in the extracellular space. Taken together, we suggest that Vps34p localization to actin patches is important for its function in the reduction of FBPase in the extracellular fraction. At present, it is not known how Vid vesicles associate with actin patches and how they dissociate. Vid vesicles may aggregate in the cytoplasm, and actin is assembled on clusters of Vid vesicles. They then move to sites of internalization on the plasma membrane. Alternatively, actin may mark the sites for Vid vesicles to aggregate near the plasma membrane. In the Δvps34 mutant, a high percentage of Vid24p remained associated with actin patches at t = 60 min, suggesting that the dissociation process is blocked in the absence of this gene. We suggest that the dissociation of Vid vesicles/actin is tightly linked to the decrease of extracellular FBPase. Dissociation may occur after cargo proteins are internalized. If this model is true, the Δvps34 mutant may inhibit the internalization process and hence Vid vesicles, and actin cannot dissociate. This model is consistent with the findings that the decline of extracellular FBPase is nearly complete after the addition of glucose for 30 min, whereas the dissociation of Vid vesicles and actin is observed at the 60-min time point. If the decline of extracellular FBPase results from the degradation of this protein in the extracellular space or the release of this protein into the medium, this would suggest that VPS34, SLA1, and ARC18 have roles in processes that are unrelated to endocytosis.
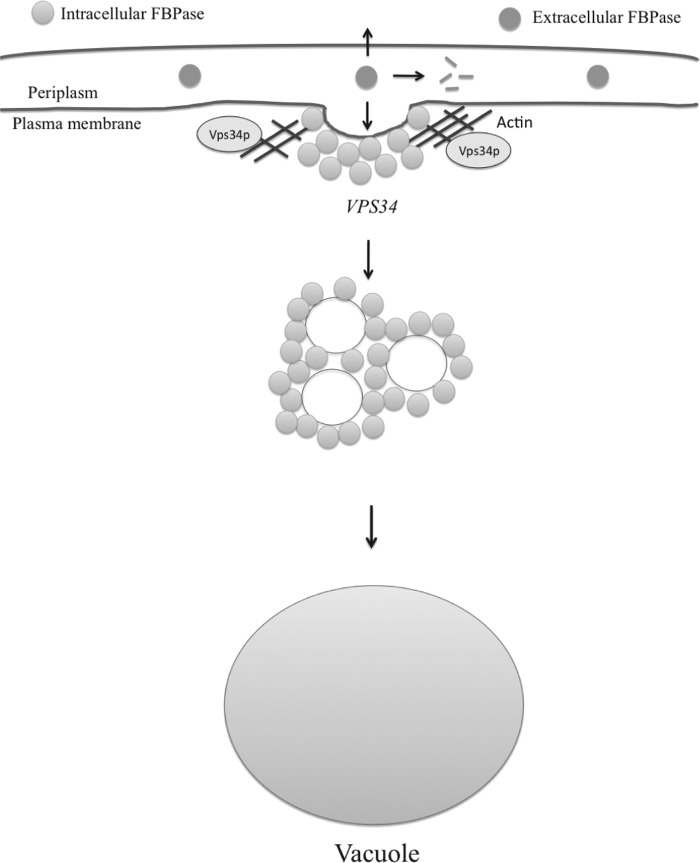
Based on our findings, we propose the following model (Fig. 11). FBPase is present in two pools in prolonged starved cells. High levels of FBPase were observed in the extracellular fraction (periplasm) during prolonged starvation. Because FBPase does not contain the N-terminal signal for the ER-Golgi pathway, this protein is likely to be secreted via a non-classical pathway. After the addition of glucose, levels of extracellular FBPase decrease. The decline in extracellular FBPase may result from internalization, degradation in the extracellular space, or release of this protein into the medium. Further experiments will be required to sort out these possibilities.
FIGURE 11.
The Vid pathway model. During prolonged glucose starvation, FBPase is in both the intracellular and extracellular fractions. After the addition of glucose, levels of extracellular FBPase decrease in a process dependent on the SLA1, ARC18, and VPS34 genes. Vps34p association with actin patches is linked to the decline of extracellular FBPase. When the Asn-736 residue was mutated or when the C-terminal 11 amino acids were deleted, Vps34p association with actin patches was impaired, and the reduction of extracellular FBPase was inhibited. The decrease of extracellular FBPase was also inhibited in the Δsla1 and Δarc18 mutants that affected Vps34p distribution. The decline of extracellular FBPase may result from internalization of FBPase into the cells, release into the medium, or degradation in the extracellular space.
Acknowledgments
Primers were synthesized at the Core Facility of the Penn State University College of Medicine. Pil1p antibodies were gifts from Dr. Dickson (University of Kentucky).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM59480. This work was also supported by a Tobacco Settlement Fund (to H.-L. C.).
- Vps
- vacuolar protein sorting
- FBPase
- fructose-1,6-bisphosphatase
- Vid
- vacuole import and degradation.
REFERENCES
- 1. Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. (1988) Organelle assembly in yeast. Characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107, 1369–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aplin A., Jasionowski T., Tuttle D. L., Lenk S. E., Dunn W. A., Jr. (1992) Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell Physiol. 152, 458–466 [DOI] [PubMed] [Google Scholar]
- 3. Banta L. M., Vida T. A., Herman P. K., Emr S. D. (1990) Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol. Cell. Biol. 10, 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bryant N. J., Piper R. C., Weisman L. S., Stevens T. H. (1998) Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol. 142, 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bryant N. J., Stevens T. H. (1998) Vacuole biogenesis in Saccharomyces cerevisiae. Protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62, 230–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conibear E., Stevens T. H. (1998) Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta 1404, 211–230 [DOI] [PubMed] [Google Scholar]
- 7. Klionsky D. J., Herman P. K., Emr S. D. (1990) The fungal vacuole. Composition, function, and biogenesis. Microbiol. Rev. 54, 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y., Klionsky D. J. (2011) The regulation of autophagy. Unanswered questions. J. Cell Sci. 124, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Z., Klionsky D. J. (2009) An overview of the molecular mechanism of autophagy. Curr. Top Microbiol. Immunol. 335, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klionsky D. J. (2007) Monitoring autophagy in yeast. The Pho8Δ60 assay. Methods Mol. Biol. 390, 363–371 [DOI] [PubMed] [Google Scholar]
- 11. Engqvist-Goldstein A. E., Drubin D. G. (2003) Actin assembly and endocytosis. From yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287–332 [DOI] [PubMed] [Google Scholar]
- 12. Riballo E., Herweijer M., Wolf D. H., Lagunas R. (1995) Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J. Bacteriol. 177, 5622–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiang H. L., Schekman R., Hamamoto S. (1996) Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J. Biol. Chem. 271, 9934–9941 [DOI] [PubMed] [Google Scholar]
- 14. Tuttle D. L., Lewin A. S., Dunn W. A., Jr. (1993) Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J. Cell Biol. 60, 283–290 [PubMed] [Google Scholar]
- 15. Cecconi F., Levine B. (2008) The role of autophagy in mammalian development. Cell makeover rather than cell death. Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms. Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 17. García-Arencibia M., Hochfeld W. E., Toh P. P., Rubinsztein D. C. (2010) Autophagy, a guardian against neurodegeneration. Semin. Cell Dev. Biol. 21, 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alvers A. L., Fishwick L. K., Wood M. S., Hu D., Chung H. S., Dunn W. A., Jr., Aris J. P. (2009) Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8, 353–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown C. R., Hung G. C., Dunton D., Chiang H. L. (2010) The TOR complex 1 is distributed in endosomes and in retrograde vesicles that form from the vacuole membrane and plays an important role in the vacuole import and degradation pathway. J. Biol. Chem. 285, 23359–23370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffman M., Chiang H. L. (1996) Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics 143, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shieh H. L., Chen Y., Brown C. R., Chiang H. L. (2001) Biochemical analysis of fructose-1,6-bisphosphatase import into vacuole import and degradation vesicles reveals a role for UBC1 in vesicle biogenesis. J. Biol. Chem. 276, 10398–10406 [DOI] [PubMed] [Google Scholar]
- 23. Shieh H. L., Chiang H. L. (1998) In vitro reconstitution of glucose-induced targeting of fructose-1,6-bisphosphatase into the vacuole in semi-intact yeast cells. J. Biol. Chem. 273, 3381–3387 [DOI] [PubMed] [Google Scholar]
- 24. Cui D. Y., Brown C. R., Chiang H. L. (2004) The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-bisphosphatase in the vacuole. J. Biol. Chem. 279, 9713–9724 [DOI] [PubMed] [Google Scholar]
- 25. Brown C. R., Chiang H. L. (2009) A selective autophagy pathway that degrades gluconeogenic enzymes during catabolite inactivation. Commun. Integr. Biol. 2, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown C. R., Cui D. Y., Hung G. G., Chiang H. L. (2001) Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J. Biol. Chem. 276, 48017–48026 [DOI] [PubMed] [Google Scholar]
- 27. Brown C. R., Dunton D., Chiang H. L. (2010) The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J. Biol. Chem. 285, 1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown C. R., Liu J., Hung G. C., Carter D., Cui D., Chiang H. L. (2003) The Vid vesicle to vacuole trafficking event requires components of the SNARE membrane fusion machinery. J. Biol. Chem. 278, 25688–25699 [DOI] [PubMed] [Google Scholar]
- 29. Brown C. R., McCann J. A., Chiang H. L. (2000) The heat shock protein Ssa2p is required for import of fructose-1,6-bisphosphatase into Vid vesicles. J. Cell Biol. 150, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown C. R., McCann J. A., Hung G. G., Elco C. P., Chiang H. L. (2002) Vid22p, a novel plasma membrane protein, is required for the fructose-1,6-bisphosphatase degradation pathway. J. Cell Sci. 115, 655–666 [DOI] [PubMed] [Google Scholar]
- 31. Brown C. R., Wolfe A. B., Cui D., Chiang H. L. (2008) The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J. Biol. Chem. 283, 26116–26127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schork S. M., Bee G., Thumm M., Wolf D. H. (1994) Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 349, 270–274 [DOI] [PubMed] [Google Scholar]
- 33. Schork S. M., Thumm M., Wolf D. H. (1995) Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 270, 26446–26450 [DOI] [PubMed] [Google Scholar]
- 34. Schüle T., Rose M., Entian K. D., Thumm M., Wolf D. H. (2000) Ubc8p functions in catabolite degradation of fructose-1, 6-bisphosphatase in yeast. EMBO J. 19, 2161–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Regelmann J., Schüle T., Josupeit F. S., Horak J., Rose M., Entian K. D., Thumm M., Wolf D. H. (2003) Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae. A genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horak J., Regelmann J., Wolf D. H. (2002) Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J. Biol. Chem. 277, 8248–8254 [DOI] [PubMed] [Google Scholar]
- 37. Hung G. C., Brown C. R., Wolfe A. B., Liu J., Chiang H. L. (2004) Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J. Biol. Chem. 279, 49138–49150 [DOI] [PubMed] [Google Scholar]
- 38. Chiang M. C., Chiang H. L. (1998) Vid24p, a novel protein localized to the fructose-1,6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J. Cell Biol. 140, 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alibhoy A. A., G. B. J., Dunton D. D., Chiang H. L. (2012) Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy 8, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang P. H., Chiang H. L. (1997) Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J. Cell Biol. 136, 803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alibhoy A. A., Giardina B. J., Dunton D. D., Chiang H. L. (2012) Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy 8, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaksonen M., Sun Y., Drubin D. G. (2003) A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487 [DOI] [PubMed] [Google Scholar]
- 43. Idrissi F. Z., Grötsch H., Fernández-Golbano I. M., Presciatto-Baschong C., Riezman H., Geli M. I. (2008) Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J. Cell Biol. 180, 1219–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herman P. K., Emr S. D. (1990) Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burda P., Padilla S. M., Sarkar S., Emr S. D. (2002) Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115, 3889–3900 [DOI] [PubMed] [Google Scholar]
- 47. Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., Emr S. D. (1993) Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91 [DOI] [PubMed] [Google Scholar]
- 48. Stack J. H., DeWald D. B., Takegawa K., Emr S. D. (1995) Vesicle-mediated protein transport. Regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Budovskaya Y. V., Hama H., DeWald D. B., Herman P. K. (2002) The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 277, 287–294 [DOI] [PubMed] [Google Scholar]
- 50. Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B. T., Shokat K. M., Williams R. L. (2010) Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 327, 1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cleves A. E., Cooper D. N., Barondes S. H., Kelly R. B. (1996) A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 133, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stack J. H., Herman P. K., Schu P. V., Emr S. D. (1993) A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delgado M. L., O'Connor J. E., Azorín I., Renau-Piqueras J., Gil M. L., Gozalbo D. (2001) The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2, and TDH3 genes are also cell wall proteins. Microbiology 147, 411–417 [DOI] [PubMed] [Google Scholar]
- 54. Obara K., Sekito T., Ohsumi Y. (2006) Assortment of phosphatidylinositol 3-kinase complexes. Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao Y., Klionsky D. J. (2007) Physiological functions of Atg6/Beclin 1. A unique autophagy-related protein. Cell Res. 17, 839–849 [DOI] [PubMed] [Google Scholar]
- 56. Winter D., Podtelejnikov A. V., Mann M., Li R. (1997) The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 7, 519–529 [DOI] [PubMed] [Google Scholar]
- 57. Winter D. C., Choe E. Y., Li R. (1999) Genetic dissection of the budding yeast Arp2/3 complex. A comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. U.S.A. 96, 7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stack J. H., Horazdovsky B., Emr S. D. (1995) Receptor-mediated protein sorting to the vacuole in yeast. Roles for a protein kinase, a lipid kinase, and GTP-binding proteins. Annu. Rev. Cell Dev. Biol. 11, 1–33 [DOI] [PubMed] [Google Scholar]
- 59. Cappellaro C., Mrsa V., Tanner W. (1998) New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180, 5030–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762 [DOI] [PubMed] [Google Scholar]
- 61. Wedaman K. P., Reinke A., Anderson S., Yates J., 3rd, McCaffery J. M., Powers T. (2003) Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 63. Gozalbo D., Gil-Navarro I., Azorín I., Renau-Piqueras J., Martínez J. P., Gil M. L. (1998) The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66, 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]