Background: The IL1RL1/ST2 gene encodes the receptor for IL-33, which is important for Th2 responses.
Results: GATA2 knockdown reduced the expression of human IL1RL1/ST2 in KU812 and LAD2 cells and in human primary peripheral basophils.
Conclusion: GATA2, but not GATA1, is a critical transcription factor for expression of human IL1RL1/ST2 in mast cell/basophil lineages.
Significance: GATA2 and GATA1 exhibit distinctive roles in the expression of human IL1RL1/ST2.
Keywords: Allergy, GATA, Interleukin, Mast Cell, Transcription Factors, IL1RL1/ST2, IL-33 Receptor
Abstract
The IL1RL1/ST2 gene encodes a receptor for IL-33. Signaling from IL1RL1/ST2 induced by IL-33 binding was recently identified as a modulator of the Th2 response. The target cells for IL-33 are restricted in some hematopoietic lineages, including mast cells, basophils, eosinophils, Th2 cells, natural killer cells, and dendritic cells. To clarify the molecular mechanisms of cell type-specific IL1RL1/ST2 expression in mast cells and basophils, transcriptional regulation of the human IL1RL1/ST2 promoter was investigated using the mast cell line LAD2 and the basophilic cell line KU812. Reporter assays suggested that two GATA motifs just upstream of the transcription start site in the ST2 promoter are critical for transcriptional activity. These two GATA motifs possess the capacity to bind GATA1 and GATA2 in EMSA. ChIP assay showed that GATA2, but not GATA1, bound to the ST2 promoter in LAD2 cells and that histone H3 at the ST2 promoter was acetylated in LAD2 cells, whereas binding of GATA1 and GATA2 to the ST2 promoter was detected in KU812 cells. Knockdown of GATA2 mRNA by siRNA reduced ST2 mRNA levels in KU812 and LAD2 cells and ST2 protein levels in LAD2 cells; in contrast, GATA1 siRNA transfection up-regulated ST2 mRNA levels in KU812 cells. The ST2 promoter was transactivated by GATA2 and repressed by GATA1 in coexpression analysis. When these siRNAs were introduced into human peripheral blood basophils, GATA2 siRNA reduced ST2 mRNA, whereas GATA1 siRNA up-regulated ST2 mRNA. These results indicate that GATA2 and GATA1 positively and negatively control human ST2 gene transcription, respectively.
Introduction
IL-33, which is expressed in various cell types, including fibroblasts, epithelial cells, and endothelial cells, is localized in the nucleus of cells at steady state and is released upon cell lysis following proinflammatory stimulation (1, 2). IL-33 causes Th2-type immune responses with the enhancement of IL-4, IL-5, and IL-13 production through binding to the IL-33 receptor designated IL1RL1/ST2. Several studies have demonstrated that not only Th2 cells but also mast cells and basophils express IL1RL1/ST2 and are key target cells of IL-33 (1, 3–8).
Human IL1RL1/ST2 possesses two promoters: the distal promoter functions in hematopoietic cells, including mast cells and basophils, and the proximal promoter is transactivated in fibroblasts (9, 10). The transcriptional regulation mechanism of IL1RL1/ST2 has been analyzed using the mouse IL1RL1/ST2 gene (9, 11). In these studies, GATA motifs were identified as cis-elements, which are involved in the function of the mouse ST2 distal promoter in mouse mast cells (9) and mouse T cells (11), and GATA1 and GATA3 were identified in nuclear proteins from mast cells and T cells, respectively, as GATA motif-binding proteins.
In this study, the regulation mechanism of the human ST2 promoter in mast cells and basophil lineages was analyzed. Reporter assays showed that two GATA motifs just upstream of the transcription start site are critical for the function of the human ST2 distal promoter, and one of these was not previously known. Although these GATA motifs exhibited binding activity against GATA1 and GATA2 in EMSA, the occupancy of the ST2 promoter by GATA1 and GATA2 in living cells was different between the human mast cell line LAD2 and the basophilic cell line KU812. GATA2 knockdown by siRNA reduced ST2 expression, whereas GATA1 siRNA up-regulated ST2 mRNA levels in KU812 cells but had no effect on ST2 mRNA levels in LAD2 cells, thus demonstrating the critical role of GATA2 in ST2 expression. Furthermore, this may explain the degree of ST2 expression in the mast cell/basophil lineages.
EXPERIMENTAL PROCEDURES
Cells
The LAD2 (human mast cell leukemia; a kind gift from Dr. Arnold Kirshenbaum) (12) and KU812 (human basophilic leukemia) cell lines were maintained as described previously (13, 14). Human basophils were purified from the peripheral blood of volunteers using the MACS® separation system with a basophil isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) as described (15). This study was approved by the ethics committee of the Juntendo University School of Medicine.
mRNA Quantification by Real-time PCR
Total RNA was extracted from LAD2 or KU812 cells or from peripheral blood basophils using an RNeasy micro kit (Qiagen) and was reverse-transcribed using a High Capacity cDNA reverse transcription kit (Applied Biosystems). The mRNA levels of IL1RL1/ST2, GATA1, and GATA2 were quantified using an ABI7500 system (Applied Biosystems) with TaqMan gene expression assays (Hs00545033_m1 for IL1RL1/ST2, Hs01085823_m1 for GATA1, and Hs00231119_m1 for GATA2; Applied Biosystems) and TaqMan Universal Master Mix (Applied Biosystems). mRNA levels were evaluated as a ratio to the housekeeping gene GAPDH (4326317E, Applied Biosystems) by calculation of cycle threshold values as described previously (16, 17).
Luciferase Assay
Various lengths of human IL1RL1/ST2 gene promoter regions were obtained from human genomic DNA purified from the peripheral blood of volunteers using a QIAamp DNA blood midi kit (Qiagen) based on a PCR method. Briefly, the −1092/+84 and −611/+84 regions were amplified using the primers listed in supplemental Table I and were inserted into the multicloning site of pGL4-Basic (Promega) after confirmation of nucleotide sequences of amplified DNA fragments. With regard to reporter plasmids carrying the −244/+84, −100/+84, or +9/+84 region, 5′-sites were removed by XhoI digestion and self-ligation after introduction of an XhoI recognition sequence at the appropriate site on the −1092/+84 region using a QuikChange II site-directed mutagenesis kit (Stratagene) with the synthesized oligonucleotides listed in supplemental Table I. Mutant plasmids lacking one or both of the GATA motifs were also generated by site-directed mutagenesis using the synthesized oligonucleotides listed in supplemental Table II as primers and the −100/+84 region as a template. Reporter plasmid (2 μg) and pRL-null (0.5 ng; Promega) were introduced into LAD2 and KU812 cells by electroporation using a Gene Pulser II system (Bio-Rad), and the relative luciferase activity in cells harvested 20–24 h after transfection was determined as described previously (16).
EMSA
EMSA was performed as described previously (18–21). GATA1 and GATA2 proteins were prepared with a TnT T7 quick-coupled transcription/translation system (Promega) using the expression plasmids pCR-GATA1 (22), pCR-GATA2 (20), and their empty vector pCR3.1 (Invitrogen) as templates. Nuclear proteins were extracted from cells using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Double-stranded DNA probes were prepared by annealing synthesized oligonucleotides and their complementary oligonucleotides, all of which were FITC-labeled at the 5′-end. Abs against GATA1 (N6, sc-265) and GATA2 (H-116, sc-9008) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Electrophoresis of the probe/protein mixture and detection of fluorescence were performed as described previously (16, 18–20).
ChIP Assay
ChIP assay was performed in the same way as described in our previous studies (16, 18, 19, 21). The following primers and TaqMan probes were used to quantify the amount of chromosomal DNA including the ST2 promoter (−74/+32): forward (−74/−50), 5′-GTGATCATCGGGTTCAGCTTATC-3′; reverse (+32/+7), 5′-GCTTTACTAAATACAACAGCCAGCCT-3′; and probe (−47/−30), 5′-FAM-TAACCTGGTTCCTGTCTC-MGB-3′. The same Abs used for EMSA were used to precipitate GATA1 and GATA2. Rabbit anti-acetyl-histone H3 IgG (06-599) was purchased from Upstate Biotechnology (Lake Placid, NY). Rat IgG (BD Biosciences) and rabbit IgG (02-6102, Invitrogen) were used as control Abs for anti-GATA1 Ab and for anti-GATA2 and anti-acetyl-histone H3 Abs, respectively.
Western Blotting
Western blotting was performed as reported previously (21). The same Abs used for EMSA and ChIP assay were used to detect GATA1 and GATA2. Monoclonal Abs against β-actin (AC-15 (A1978), Sigma-Aldrich) were also used.
Knockdown of mRNA Expression by siRNA
GATA1 siRNA (Stealth Select RNAiTM, HSS-142151), GATA2 siRNA (HSS-104004), and control siRNA (Stealth RNAiTM negative universal control, 46-2000) were purchased from Invitrogen. A 10 (or 1)-μl aliquot of 20 μm siRNA was introduced into 2 × 106 (or 2 × 105) cells with a NeonTM 100-μl kit (or a Neon 10-μl kit) using an electroporator, a NeonTM transfection system (Invitrogen) set at program 16. The transfection efficiency of this experimental condition was confirmed to be >98% (KU812 and LAD2 cells) or >80% (peripheral blood basophils) using a FACSCalibur flow cytometer (BD Biosciences) by monitoring with BLOCK-iTTM Alexa Fluor® Red Fluorescence Oligo (14750-100, Invitrogen). Peripheral blood basophils were maintained in the presence of 10 ng/ml recombinant human IL-3 (PeproTech, London, United Kingdom) and were harvested at 48 h after transfection.
Flow Cytometric Analysis
LAD2 cells (1 × 106) harvested at 48 h after siRNA transfection were incubated with 0.5 μg of phycoerythrin-conjugated anti-human ST2 Ab (2A5) or phycoerythrin-conjugated mouse IgG1 (2E12) for 30 min at 4 °C. After washing with PBS, cells were resuspended in PBS and analyzed using the FACSCalibur flow cytometer and FlowJo software (Tree Star Inc., Ashland, OR). Relative mean fluorescence intensity (MFI)3 was calculated as follows: MFI = ((ST2 MFI on target cells/isotype MFI on target cells − 1)/(ST2 MFI on control cells/isotype MFI on control cells)) × 100.
Coexpression Reporter Analysis
In the coexpression analysis, 3 μg of expression plasmid (pCR3.1, pCR-GATA1, or pCR-GATA2) was introduced into 1 × 106 cells with 5 μg of reporter plasmid and 25 ng of pRL-null using the Neon transfection system.
Statistical Analysis
Statistical analysis was performed using Student's two-tailed t test for paired or unpaired data, and p values <0.05 were considered to be significant.
RESULTS
Identification of cis-Enhancing Elements in the Human IL1RL1/ST2 Promoter
To examine promoter regulation in the human IL1RL1/ST2 gene, a series of reporter plasmids carrying various lengths of the 5′-flanking region of the ST2 promoter were generated. Luciferase activity driven by the −1092/+84 region was significantly higher than that of the promoterless construct in both cell lines (Fig. 1A). Other shorter promoter regions from −611/+84 to −100/+84, which had deletions at the 5′-site, showed comparable activity to the −1092/+84 region in KU812 cells. In the case of LAD2 cells, the −100/+84 region showed significant promoter activity, although deletions of −1092/−612 and −611/−245 markedly reduced this activity. When the 5′-site of the −100/+84 region was further deleted to +9/+84, promoter activity was reduced to the level driven by the promoterless basic plasmid in both cell lines. These results indicate that the cis-enhancing elements critical for ST2 promoter function are located between −100 and +8, which are commonly functional in KU812 and LAD2 cells. The graded reduction of promoter activity from −1092/+84 to −244/+84 in LAD2 cells suggests the presence of additional cis-enhancing elements that function in LAD2 cells, but not in KU812 cells.
FIGURE 1.
Determination of critical cis-elements of the human ST2 promoter by reporter assay using mast cell and basophilic cell lines. A, reporter assay with deletion constructs. Luc, luciferase. B, nucleotide sequences of the murine and human ST2 promoters. GATA family protein-binding consensus sequences, (A/T)GATA(A/G), are boxed. +1 is the transcription start site. C, reporter assay with mutant constructs. Relative luciferase activity is displayed as the ratio of luciferase activity to that seen in cells transfected with the promoterless construct pGL3-Basic (A and C). Data represent means ± S.E. of three independent experiments performed in triplicate samples (A and C).
A search for transcription factor-binding motifs in the −100/+8 region of the human ST2 promoter confirmed two GATA motifs, GATA-A (at −54/−49) and GATA-B (at −24/−19), in the human gene, and these are conserved in the mouse ST2 promoter (Fig. 1B). An additional GATA motif, GATA-C, that was reported to be essential for mouse ST2 promoter function is not present in the human ST2 gene. Thus, to evaluate the effects of the GATA motifs on promoter activity, mutant reporter plasmids lacking on or both of the two GATA motifs were generated by site-directed mutagenesis. Mutants A and B, lacking GATA-A and GATA-B, respectively, exhibited moderately decreased promoter activity compared with the wild-type promoter, and the transcriptional activity of Mutant AB, lacking both GATA-A and GATA-B, was comparable to that of the promoterless construct (Fig. 1C). These results indicate that both GATA motifs, GATA-A and GATA-B, are involved in human ST2 promoter function.
Ability of the Two GATA Motifs to Bind Directly to GATA1 and GATA2
Two motifs at −54/−49 and at −24/−19 containing GATA sequence were found to be essential for the transcriptional activity of the human ST2 promoter in the abovementioned reporter assays. To examine whether these sequences are recognizable by GATA family proteins, EMSA was performed with GATA1 and GATA2, which are major GATA family proteins in mast cells and basophils. When double-stranded DNA containing the GATA motif at −24/−19 (Fig. 2A) or at −54/−49 (Fig. 2B) was mixed with GATA1 protein, a specific band shift was seen on the gel (lane 1 in Fig. 2, A and B), and this was supershifted in the presence of anti-GATA1 Ab (lane 2). Similarly, a specific band shift was observed upon loading GATA2 protein and probe (lane 3), and this disappeared upon the addition of anti-GATA2 Ab (lane 4). In EMSA using nuclear extracts from KU812 and LAD2 cells (supplemental Fig. S2), a band with a slightly higher mobility compared with those of in vitro transcribed/translated GATA1 and GATA2, whose molecular weights were slightly higher than those of the native proteins because of the fusion of the FLAG tag at the N terminus, was detected. The intensity of this band and of an additional two bands showing slower mobility was decreased in the presence of anti-GATA1 Ab, thus suggesting that GATA1 is expressed predominantly in KU812 cells and forms complexes with other nuclear proteins. Similarly, in LAD2 cells, GATA2 is dominant and possibly forms complexes with other proteins. Taken together, these observations suggest that the two GATA motifs identified in reporter assays are directly bound to GATA1 and GATA2 in vitro.
FIGURE 2.
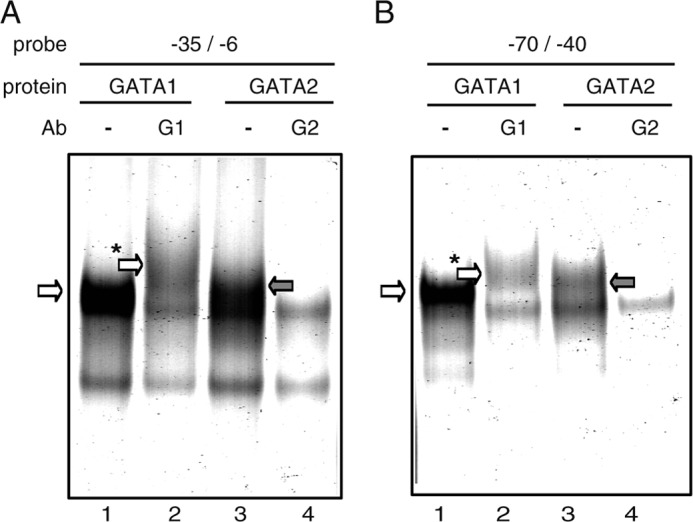
GATA1 and GATA2 bind to two GATA motifs essential for human ST2 promoter transactivation. A and B, EMSA profiles using probes −35/−6 and −70/−40, respectively. EMSA data are representative results obtained in one of three independent experiments. In vitro transcription/translation was performed using pCR-GATA1 (GATA1) or pCR-GATA2 (GATA2) as a template. −, without Ab; G1, with anti-GATA1 Ab; G2, with anti-GATA2 Ab. Specific bands corresponding to probe·GATA1 and probe·GATA2 complexes are marked with white and gray arrows, respectively. Supershifted bands corresponding to probe·GATA1·anti-GATA1 Ab complexes are marked with a white arrow with an asterisk.
Profiles of Binding of GATA1 and GATA2 to the Human ST2 Promoter Differ in KU812 and LAD2 Cells
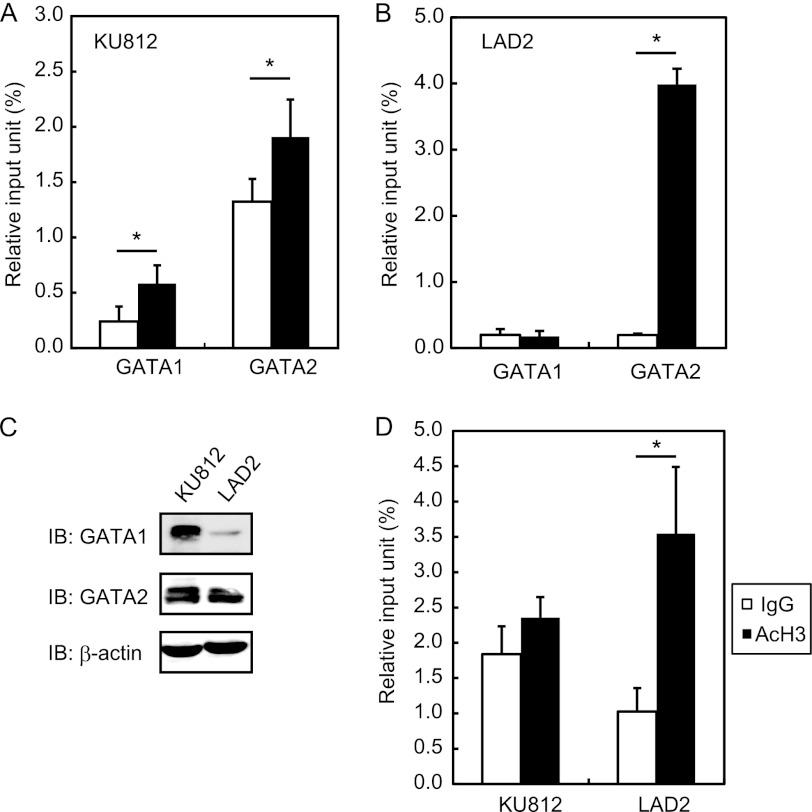
EMSA data showed that the two GATA motifs in the human ST2 promoter possess the ability to bind GATA1 and GATA2 in vitro. To evaluate the function of these GATA motifs in the binding of GATA1 and GATA2 in live ST2-positive cells, ChIP assay targeting the ST2 promoter region was performed. The amount of DNA immunoprecipitated by the anti-GATA2 Ab was markedly higher than that immunoprecipitated by the control Ab, whereas significant binding of GATA1 to the ST2 promoter was not detected in LAD2 cells (Fig. 3B). In contrast, both GATA1 and GATA2 bound significantly to the ST2 promoter in KU812 cells, although the degree of binding was less marked than that of GATA2 in LAD2 cells (Fig. 3A). These results demonstrate that GATA2, but not GATA1, is recruited to the ST2 promoter in LAD2 cells, whereas the ST2 promoter is occupied by GATA1 and GATA2 in KU812 cells.
FIGURE 3.
Quantitative analyses of GATA1 and GATA2 binding to the ST2 promoter and of the degree of histone H3 acetylation at the ST2 promoter by ChIP assays. ChIP assays were performed to examine GATA1 and GATA2 binding to the ST2 promoter in KU812 (A) and LAD2 (B) cells. C, Western blot analysis with anti-GATA1, anti-GATA2, or anti-β-actin Ab. IB, immunoblot. D, ChIP assay with anti-acetylated histone H3 (AcH3) Ab. Data represent means ± S.D. of triplicate samples. Black bars, specific Ab; white bars, control Ab. *, p < 0.05.
Expression Levels of GATA1 and GATA2 and Histone Acetylation at the ST2 Promoter in KU812 and LAD2 Cells
ChIP assay was used to determine the differences in the binding of GATA1 and GATA2 to the ST2 promoter in KU812 and LAD2 cells. We performed Western blotting to determine the protein expression levels of GATA1 and GATA2 in KU812 and LAD2 cells and found that the expression levels of GATA1 protein in KU812 cells were markedly higher than those in LAD2 cells, whereas a significant difference were not observed in GATA2 expression levels between these two cell lines (Fig. 3C). The high levels of GATA1 in KU812 cells may be an explanation for the occupancy of the ST2 promoter by GATA1 in KU812 cells, but not in LAD2 cells.
In addition, histone modification at the ST2 promoter was determined by ChIP assay using anti-acetylated H3 Ab. Histone H3 at the ST2 promoter was significantly acetylated in LAD2 cells in contrast to KU812 cells, in which acetylation was not detected (Fig. 3D). This suggests that the higher acetylation of histone H3 causes the binding of higher amounts of GATA2 to the ST2 promoter in LAD2 cells compared with KU812 cells and that the chromatin status at the ST2 promoter tends to be transcribed in LAD2 cells.
Effects of GATA1 and GATA2 siRNAs on ST2 Expression
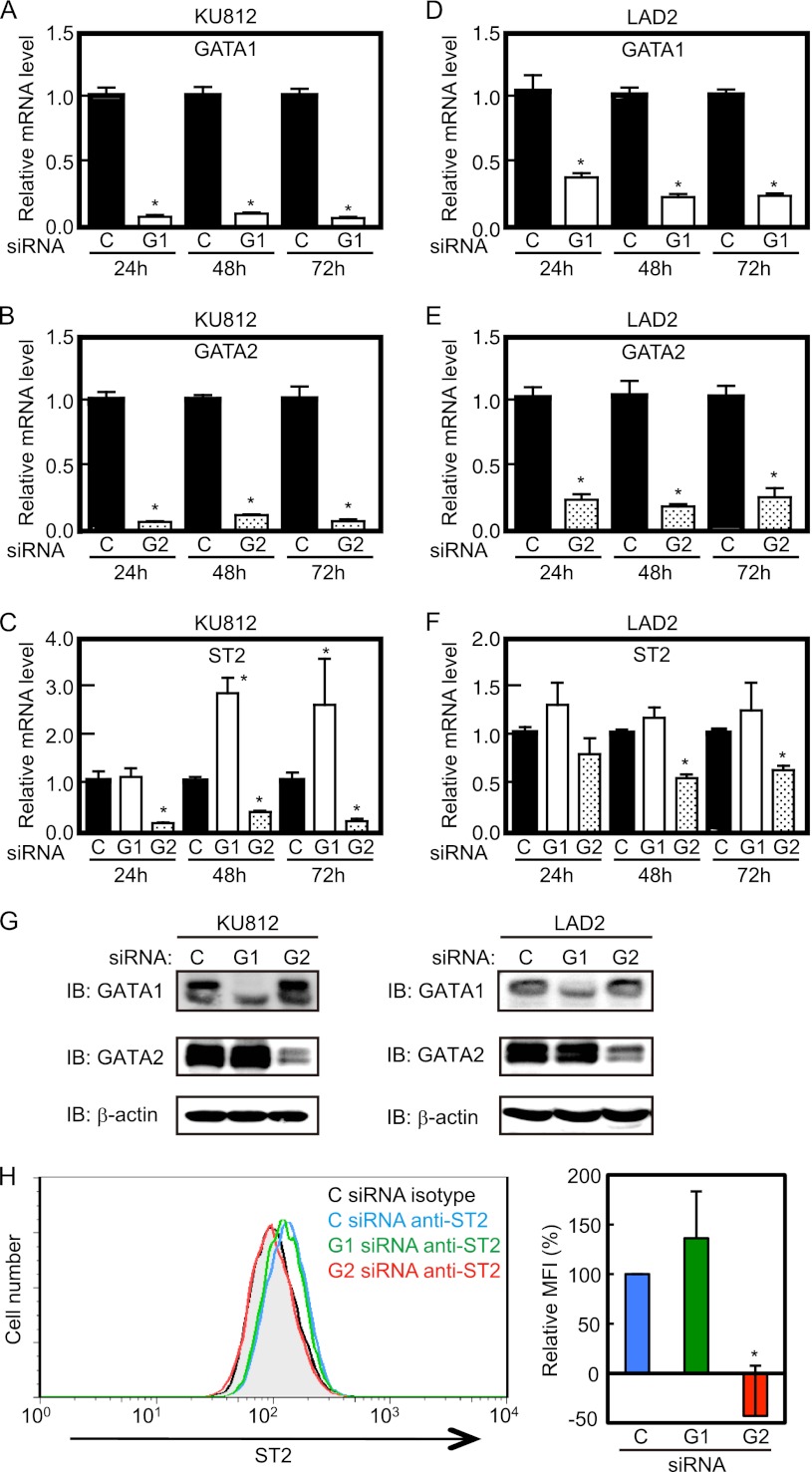
The GATA motifs, as critical cis-elements in the ST2 promoter, bound GATA1 and GATA2 in vitro, and the degree of GATA1 and GATA2 binding to the ST2 promoter differed between the two cell lines. Therefore, the involvement of GATA1 and GATA2 in ST2 expression was investigated by knockdown of GATA1 or GATA2 expression using siRNA. The mRNA levels of GATA1 and GATA2 decreased to ∼10% of control levels upon introduction of GATA1 and GATA2 siRNAs, respectively, in KU812 cells, which continued from 24 to 72 h after transfection (Fig. 4, A and B). In the case of LAD2 cells, GATA1 and GATA2 siRNAs reduced target mRNA levels by ∼75% (Fig. 4, D and E). The effects of GATA1 and GATA2 siRNAs on the expression levels of target proteins were confirmed by Western blot analysis (Fig. 4G). Under these experimental conditions, ST2 mRNA levels were significantly reduced by GATA2 siRNA from 24 to 72 h after transfection in KU812 cells and from 48 to 72 h in LAD2 cells (Fig. 4, C and F), thus suggesting that GATA2 is involved in transactivation of the ST2 gene. In contrast, interestingly, GATA1 siRNA introduction caused up-regulation of ST2 mRNA levels in KU812 cells, whereas GATA1 siRNA had no effect on ST2 mRNA levels in LAD2 cells. ST2 protein levels were verified by monitoring ST2 expression on the LAD2 cell surface using flow cytometry to evaluate the effects of knockdown of GATA1 and GATA2 on ST2 expression because ST2 was detected on the LAD2 cell surface, but not on KU812 cells. As shown in Fig. 4H, the amount of ST2 protein on the LAD2 cell surface was significantly reduced by GATA2 knockdown, whereas surface ST2 expression levels on GATA1 siRNA-introduced cells were comparable with those on control cells. These results indicate that GATA2 is a positive regulator of ST2 gene expression and that GATA1 acts as a negative regulator for ST2 transcription in KU812 cells, but not in LAD2 cells.
FIGURE 4.
Effects of GATA1 and GATA2 siRNAs on ST2 expression. Shown are the mRNA expression levels of GATA1 (A and D), GATA2 (B and E), and ST2 (C and F) in cells transfected with control siRNA (black bars; C), GATA1 siRNA (white bars; G1), and GATA2 siRNA (stippled bars; G2). Cells were harvested at 24, 48, or 72 h after siRNA introduction. Relative mRNA levels are displayed as a ratio to those seen in control siRNA-introduced cells. Results are expressed as means ± S.D. of triplicate samples. *, p < 0.05. G, protein expression levels of GATA1 and GATA2 in siRNA-introduced cells. IB, immunoblot. H, cell surface expression levels of ST2 in LAD2 cells. Representative results are shown in the left panel, and means ± S.E. of three independent experiments are shown in the right panel. The MFI of ST2 is shown as a ratio to that in control siRNA-introduced LAD2 cells (100%). The MFI of isotype controls was not affected by each siRNA introduction (data not shown). Cells were harvested at 48 h after siRNA introduction (G and H).
Effects of Exogenous GATA1 and GATA2 on ST2 Promoter Activity
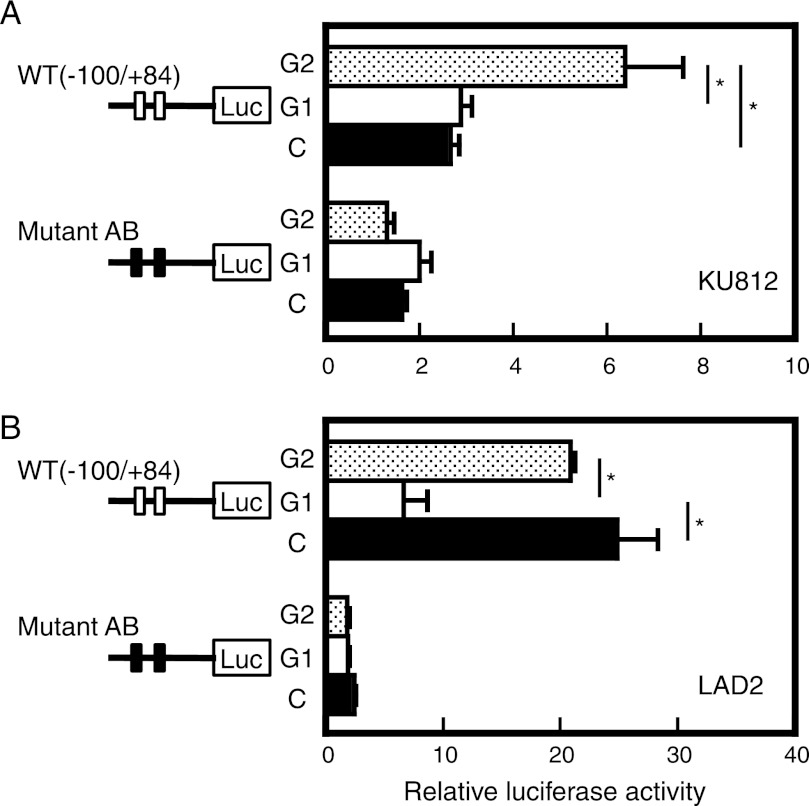
The results from the knockdown experiments suggested the roles of GATA2 and GATA1 as a transactivator and a repressor, respectively. To evaluate the direct effects of GATA1 and GATA2 on promoter activity, coexpression reporter assay was performed. When GATA2 expression plasmid was introduced into KU812 cells, the transcriptional activity of the ST2 promoter (−100/+84) was up-regulated, whereas introduction of GATA1 expression plasmid did not affect promoter activity (Fig. 5A). In the case of LAD2 cells, coexpression of GATA1 repressed ST2 promoter activity (Fig. 5B). The ST2 promoter lacking the two GATA motifs (Mutant AB) was not affected by exogenous GATA1 and GATA2 in both cell lines. These results suggest that GATA1 and GATA2 regulate the ST2 promoter in positive and negative manners, respectively, via both GATA motifs.
FIGURE 5.
Direct effects of GATA1 and GATA2 on ST2 promoter activity. Relative luciferase activity is displayed as the ratio of luciferase activity to that seen in cells transfected with pGL3-Basic. Data represent means ± S.D. of two independent experiments performed in triplicate. *, p < 0.05. The reporter plasmid was cotransfected with pCR3.1 (black bars; C), pCR-GATA1 (white bars; G1), and pCR-GATA2 (stippled bars; G2). Luc, luciferase.
Involvement of GATA1 and GATA2 in ST2 Expression in Human Peripheral Blood Basophils
Finally, to confirm the involvement of GATA1 and GATA2 in ST2 expression in primary cells, GATA1 or GATA2 mRNA in purified human peripheral blood basophils was knocked down by siRNA. In this experiment, human IL-3 was added to the culture medium to maintain purified basophils for 48 h after transfection. Using the Neon system, we confirmed that GATA1 and GATA2 siRNAs reduced target mRNA levels by ∼80% (Fig. 6A). ST2 mRNA levels were significantly up-regulated and down-regulated by GATA1 and GATA2 siRNAs, respectively, under these conditions (Fig. 6B). These results suggest that expression of ST2 mRNA in human primary basophils is positively and negatively regulated by GATA2 and GATA1, respectively, as in the case of KU812 cells.
FIGURE 6.
Effects of GATA1 and GATA2 knockdown on ST2 expression in primary basophils. Shown are the mRNA levels of GATA1 and GATA2 (A) and ST2 (B) in cells transfected with control siRNA (black bars; C), GATA1 siRNA (white bars; G1), and GATA2 siRNA (stippled bars; G2). Cells were harvested at 48 h after siRNA introduction. Results are expressed as means ± S.D. of triplicate samples. *, p < 0.05.
DISCUSSION
In this study, reporter assays demonstrated that two GATA motifs at −54/−49 and −24/−19 are critical for activation of the human ST2 distal promoter in KU812 and LAD2 cells. Although GATA1 and GATA2, which are major GATA family proteins in mast cells and basophils, directly bound to these GATA motifs in vitro in EMSA, the ST2 chromosomal DNA region around these GATA motifs was occupied only by GATA2 in LAD2 cells, whereas GATA1 and GATA2 bond the same region in KU812 cells. Finally, GATA2 siRNA introduction reduced ST2 mRNA levels in both cell lines and in purified peripheral blood human basophils; in contrast, GATA1 siRNA up-regulated ST2 mRNA levels in KU812 cells and primary basophils but had no effect in LAD2 cells. On the basis of these results, we concluded that GATA2 is a critical transcription factor for human ST2 gene expression in mast cell and basophil lineages.
Studies on the transcriptional regulation mechanism of the mouse ST2 distal promoter have been reported (9, 11). Gächter et al. (9) identified three GATA motifs in the ST2 promoter as cis-elements functional in bone marrow-derived mouse mast cells: mGATA at −76/−81 (GATA-B in Fig. 1B), pGATA at −53/−58 (GATA-C in Fig. 1B), and dGATA at −210 (9). Of these GATA motifs, mGATA (GATA-B) was the most important for transcriptional activity, and GATA1 was found to be involved in ST2 transcription. Hayakawa et al. (11) identified two GATA motifs, GATA-b at −84/−79 (GATA-B in Fig. 1B) and GATA-a at −53/−58 (GATA-A in Fig. 1B). In coexpression analysis, the absence of GATA-b significantly reduced GATA3-mediated transactivation of the ST2 promoter, whereas the effects of nucleotide replacement at GATA-a on promoter activity were slight. The results of our study of human ST2 gene transcription are consistent with those of previous mouse studies with regard to the importance of GATA-B (Fig. 1B). However, GATA-C, which was the subject of studies into the mouse ST2 gene, does not exist in the human genome. Furthermore, GATA-A, which is conserved between humans and mice, was identified as a novel cis-element in addition to GATA-B. Interestingly, the effects of a single mutation of GATA-A or GATA-B were moderate, whereas double mutation of GATA-A and GATA-B reduced promoter activity to background levels. A similar effect of a double mutation was observed in the longer promoter region, −611/+84 (data not shown). These results suggest that one of the two GATA motifs is functional, even when the other is absent, and that these two GATA motifs are essential for ST2 promoter activity in mast cells and basophils.
ST2 mRNA levels in LAD2 cells were >10-fold higher than those in KU812 cells (supplemental Fig. S1). This difference in mRNA levels may be due to the high occupancy of the ST2 promoter by GATA2 in LAD2 cells compared with KU812 cells, in which GATA1 and GATA2 bound to the ST2 promoter (Fig. 3). siRNA experiments showed that GATA2 knockdown decreased the amount of ST2 mRNA in both cell lines and that GATA1 knockdown increased ST2 mRNA levels in KU812 cells but did not affect ST2 mRNA levels in LAD2 cells (Fig. 4). On the basis of these results, we hypothesize that the degree of GATA2 binding to the ST2 promoter correlates with the ST2 transcription level. The chromatin status at the ST2 promoter may also explain the higher ST2 mRNA levels in LAD2 cells. Briefly, acetylated histone H3 was detected at the ST2 promoter in LAD2 cells, but not in KU812 cells (Fig. 3D). In contrast to GATA2 as a transactivator, GATA1 does not appear to affect ST2 transcription or to function as a suppressive transcription factor on the promoter. We speculate that the ST2 gene is strongly transactivated by GATA2 with high occupancy or the promoter in LAD2 cells, whereas transactivation of the ST2 promoter by GATA2 is partially suppressed by GATA1, which competes with GATA2 for binding to the ST2 promoter. It is known that ST2 mRNA levels in cultured human mast cells are higher than those in peripheral blood basophils (5). The difference in ST2 mRNA levels in the primary mast cells and basophils may also be explained by the dynamics of GATA1 and GATA2 on the ST2 promoter. Actually, GATA1 siRNA up-regulated ST2 mRNA levels in peripheral blood basophils, whereas GATA2 siRNA reduced ST2 mRNA levels (Fig. 6B). Considering that GATA1 and GATA2 are expressed in mast cells and basophils, an unknown mechanism that regulates recruitment of GATA1 and GATA2 to the ST2 promoter is required to cause the different binding profiles. Further detailed analysis will be necessary to clarify the regulation mechanisms of the ST2 gene.
Western blot analysis showed that GATA1 expression levels in KU812 cells were higher than those in LAD2 cells (Fig. 3C). This is one of the possible explanations for the significant binding of GATA1 to the ST2 promoter in KU812 cells and the high occupancy of the ST2 promoter by GATA2 in LAD2 cells. The different ratios of GATA1 and GATA2 may also have led to the differences in observed in KU812 and LAD2 cells in coexpression analysis. Briefly, the transactivation activity of GATA2 on the ST2 promoter was detected in KU812 cells, and suppressive effects of GATA1 on the promoter were observed in LAD2 cells (Fig. 5). Differences in the amount of endogenous GATA1 or GATA2 proteins may have direct effects of exogenous proteins on the promoter.
It has previously been reported that single nucleotide polymorphisms in the ST2 distal promoter are associated with atopic dermatitis (23), suggesting that the degree of function of the ST2 distal promoter can affect the risk and/or pathology of allergic diseases. The regulation of the ST2 distal promoter may therefore be a target for controlling allergic reactions to prevent or treat allergic diseases. IL-3 is known to up-regulate ST2 mRNA levels and subsequently induce cell surface expression of ST2 protein to detectable levels in human primary basophils (5, 7). A recent study demonstrated that basophils from eosinophilic esophagitis patients express higher amounts of ST2 on the cell surface, and TSLP (thymic stromal lymphopoietin) is considered to be a candidate that affects ST2 expression levels in basophils in an IL-3-independent manner (24). Further experiments to evaluate the effects of stimulation of signaling on the ST2 promoter, possibly using a system in which cells respond to IL-3 or TSLP, may be beneficial. In this study, we found that GATA1 and GATA2 are involved in ST2 transcription levels in primary basophils maintained in the presence of IL-3 (Fig. 6B). The dynamics and function of GATA2 and GATA1, which were identified to be a critical transactivator and suppressor, respectively, for the ST2 promoter in steady-state basophils, may therefore be affected by stimulation signaling.
Supplementary Material
Acknowledgments
We are grateful to the staff of the Atopy Research Center and the Departments of Pediatrics and Immunology for helpful discussions. We thank Drs. Nobuhiro Nakano, Nao Kitamura, Maya Kamijo, Kentaro Ishiyama, and Ryusaku Matsuda for helpful suggestions and Michiyo Matsumoto for secretarial assistance.
This work was supported by the Funding Program for Next Generation World-Leading Researchers from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to C. N.).

This article contains supplemental Figs. S1 and S1 and Tables I and II.
- MFI
- mean fluorescence intensity.
REFERENCES
- 1. Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T. K., Zurawski G., Moshrefi M., Qin J., Li X., Gorman D. M., Bazan J. F., Kastelein R. A. (2005) IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T-helper type 2-associated cytokines. Immunity 23, 479–490 [DOI] [PubMed] [Google Scholar]
- 2. Liew F. Y., Pitman N. I., McInnes I. B. (2010) Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat. Rev. Immunol. 10, 103–110 [DOI] [PubMed] [Google Scholar]
- 3. Allakhverdi Z., Smith D. E., Comeau M. R., Delespesse G. (2007) Cutting edge: The ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J. Immunol. 179, 2051–2054 [DOI] [PubMed] [Google Scholar]
- 4. Xu D., Jiang H. R., Kewin P., Li Y., Mu R., Fraser A. R., Pitman N., Kurowska-Stolarska M., McKenzie A. N., McInnes I. B., Liew F. Y. (2008) IL-33 exacerbates antigen-induced arthritis by activating mast cells. Proc. Natl. Acad. Sci. U.S.A. 105, 10913–10918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzukawa M., Iikura M., Koketsu R., Nagase H., Tamura C., Komiya A., Nakae S., Matsushima K., Ohta K., Yamamoto K., Yamaguchi M. (2008) An IL-1 cytokine member, IL-33, induces human basophil activation via its ST2 receptor. J. Immunol. 181, 5981–5989 [DOI] [PubMed] [Google Scholar]
- 6. Pushparaj P. N., Tay H. K., H'Ng S. C., Pitman N., Xu D., McKenzie A., Liew F. Y., Melendez A. J. (2009) The cytokine interleukin-33 mediates anaphylactic shock. Proc. Natl. Acad. Sci. U.S.A. 106, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Pecaric-Petkovic T., Didichenko S. A., Kaempfer S., Spiegl N., Dahinden C. A. (2009) Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood 113, 1526–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider E., Petit-Bertron A. F., Bricard R., Levasseur M., Ramadan A., Girard J. P., Herbelin A., Dy M. (2009) IL-33 activates unprimed murine basophils directly in vitro and induces their in vivo expansion indirectly by promoting hematopoietic growth factor production. J. Immunol. 183, 3591–3597 [DOI] [PubMed] [Google Scholar]
- 9. Gächter T., Moritz D. R., Gheyselinck J., Klemenz R. (1998) GATA-dependent expression of the interleukin-1 receptor-related T1 gene in mast cells. Mol. Cell. Biol. 18, 5320–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwahana H., Yanagisawa K., Ito-Kosaka A., Kuroiwa K., Tago K., Komatsu N., Katashima R., Itakura M., Tominaga S. (1999) Different promoter usage and multiple transcription initiation sites of the interleukin-1 receptor-related human ST2 gene in UT-7 and TM12 cells. Eur. J. Biochem. 264, 397–406 [DOI] [PubMed] [Google Scholar]
- 11. Hayakawa M., Yanagisawa K., Aoki S., Hayakawa H., Takezako N., Tominaga S. (2005) T-helper type 2 cell-specific expression of the ST2 gene is regulated by transcription factor GATA3. Biochim. Biophys. Acta 1728, 53–64 [DOI] [PubMed] [Google Scholar]
- 12. Kirshenbaum A. S., Akin C., Wu Y., Rottem M., Goff J. P., Beaven M. A., Rao V. K., Metcalfe D. D. (2003) Characterization of novel stem cell factor-responsive human mast cell lines LAD1 and LAD2 established from a patient with mast cell sarcoma/leukemia: activation following aggregation of FcϵRI or FcγRI. Leuk. Res. 27, 677–682 [DOI] [PubMed] [Google Scholar]
- 13. Niyonsaba F., Ushio H., Hara M., Yokoi H., Tominaga M., Takamori K., Kajiwara N., Saito H., Nagaoka I., Ogawa H., Okumura K. (2010) Antimicrobial peptides human β-defensins and cathelicidin LL-37 induce the secretion of a pruritogenic cytokine IL-31 by human mast cells. J. Immunol. 184, 3526–3534 [DOI] [PubMed] [Google Scholar]
- 14. Nishiyama C., Hasegawa M., Nishiyama M., Takahashi K., Akizawa Y., Yokota T., Okumura K., Ogawa H., Ra C. (2002) Regulation of human FcϵRI α-chain gene expression by multiple transcription factors. J. Immunol. 168, 4546–4552 [DOI] [PubMed] [Google Scholar]
- 15. Nishiyama C., Akizawa Y., Nishiyama M., Tokura T., Kawada H., Mitsuishi K., Hasegawa M., Ito T., Nakano N., Okamoto A., Takagi A., Yagita H., Okumura K., Ogawa H. (2004) Polymorphisms in the FcϵRIβ promoter region affecting transcription activity: a possible promoter-dependent mechanism for association between FcϵRIβ and atopy. J. Immunol. 173, 6458–6464 [DOI] [PubMed] [Google Scholar]
- 16. Maeda K., Nishiyama C., Tokura T., Nakano H., Kanada S., Nishiyama M., Okumura K., Ogawa H. (2006) FOG-1 represses GATA1-dependent FcϵRI β-chain transcription: transcriptional mechanism of mast cell-specific gene expression in mice. Blood 108, 262–269 [DOI] [PubMed] [Google Scholar]
- 17. Nakano N., Nishiyama C., Kanada S., Niwa Y., Shimokawa N., Ushio H., Nishiyama M., Okumura K., Ogawa H. (2007) Involvement of mast cells in IL-12/23 p40 production is essential for survival from polymicrobial infections. Blood 109, 4846–4855 [DOI] [PubMed] [Google Scholar]
- 18. Kanada S., Nakano N., Potaczek D. P., Maeda K., Shimokawa N., Niwa Y., Fukai T., Sanak M., Szczeklik A., Yagita H., Okumura K., Ogawa H., Nishiyama C. (2008) Two different transcription factors discriminate the −315C>T polymorphism of the FcϵRIα gene: binding of Sp1 to −315C and of a high mobility group-related molecule to −315T. J. Immunol. 180, 8204–8210 [DOI] [PubMed] [Google Scholar]
- 19. Kanada S., Nishiyama C., Nakano N., Suzuki R., Maeda K., Hara M., Kitamura N., Ogawa H., Okumura K. (2011) Critical role of transcription factor PU.1 in the expression of CD80 and CD86 on dendritic cells. Blood 117, 2211–2222 [DOI] [PubMed] [Google Scholar]
- 20. Maeda K., Nishiyama C., Ogawa H., Okumura K. (2010) GATA2 and Sp1 positively regulate the c-kit promoter in mast cells. J. Immunol. 185, 4252–4260 [DOI] [PubMed] [Google Scholar]
- 21. Kitamura N., Yokoyama H., Yashiro T., Nakano N., Nishiyama M., Kanada S., Fukai T., Hara M., Ikeda S., Ogawa H., Okumura K., Nishiyama C. (2012) Role of PU.1 in MHC class II expression through transcriptional regulation of class II transactivator pI in dendritic cells. J. Allergy Clin. Immunol. 129, 814–824.e6 [DOI] [PubMed] [Google Scholar]
- 22. Nishiyama C., Yokota T., Okumura K., Ra C. (1999) The transcription factors Elf-1 and GATA1 bind to cell-specific enhancer elements of human high affinity IgE receptor α-chain gene. J. Immunol. 163, 623–630 [PubMed] [Google Scholar]
- 23. Shimizu M., Matsuda A., Yanagisawa K., Hirota T., Akahoshi M., Inomata N., Ebe K., Tanaka K., Sugiura H., Nakashima K., Tamari M., Takahashi N., Obara K., Enomoto T., Okayama Y., Gao P. S., Huang S. K., Tominaga S., Ikezawa Z., Shirakawa T. (2005) Functional SNPs in the distal promoter of the ST2 gene are associated with atopic dermatitis. Hum. Mol. Genet. 14, 2919–2927 [DOI] [PubMed] [Google Scholar]
- 24. Siracusa M. C., Saenz S. A., Hill D. A., Kim B. S., Headley M. B., Doering T. A., Wherry E. J., Jessup H. K., Siegel L. A., Kambayashi T., Dudek E. C., Kubo M., Cianferoni A., Spergel J. M., Ziegler S. F., Comeau M. R., Artis D. (2011) TSLP promotes interleukin-3-independent basophil hematopoiesis and type 2 inflammation. Nature 477, 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.