Background: Phosphoisoprenoid stimulation of Vγ9Vδ2 T cells can be modulated by anti-BTNA3 antibodies.
Results: Agonist and antagonist antibodies associate differently with BTN3A structurally and biophysically.
Conclusion: Differential binding of antibodies to BTN3A modulates its oligomerization on the cell surface.
Significance: Defining how γδ T cells recognize antigen is critical for understanding their functions in the immune response.
Keywords: Antibodies, Immunology, Receptor Structure-Function, Stress Response, T Cell
Abstract
Human Vγ9Vδ2 T cells are well known for their rapid and potent response to infection and tumorigenesis when in the presence of endogenous or exogenous phosphoisoprenoids. However, the molecular mechanisms behind the activation of this γδ T cell population remains unclear. Evidence pointing to a role for the CD277/butyrophilin-3 (BTN3A) molecules in this response led us to investigate the structures of these molecules and their modifications upon binding to an agonist antibody (20.1) that mimics phosphoisoprenoid-mediated Vγ9Vδ2 activation and an antagonist antibody (103.2) that inhibits this reactivity. We find that the three BTN3A isoforms: BTN3A1, BTN3A2, and BTN3A3, have high structural homology to the B7 superfamily of proteins and exist as V-shaped homodimers in solution, associating through the membrane proximal C-type Ig domain. The 20.1 and 103.2 antibodies bind to separate epitopes on the BTN3A Ig-V domain with high affinity but likely with different valencies based on their binding orientation. These structures directly complement functional studies of this system that demonstrate that BTN3A1 is necessary for Vγ9Vδ2 activation and begin to unravel the extracellular events that occur during stimulation through the Vγ9Vδ2 T cell receptor.
Introduction
γδ T cells are an innate-like lineage of T cells that demonstrate a preactivated or memory phenotype and are capable of secreting a diverse array of cytokines corresponding to either an immunostimulatory or an immunoregulatory phenotype (1, 2). In addition, some γδ T cells can directly kill target cells through induction of apoptosis and/or cytolysis. Most γδ T cells in mice and humans reside in the epithelial tissues; however, there is a smaller percentage that circulates in the blood (1). In humans, γδ T cells comprise between 0.5 and 20% of blood-circulating T cells, the majority of which express a rearranged T cell receptor (TCR)3 composed of Vγ9 and Vδ2 domains; thus, this population is referred to as Vγ9Vδ2. This γδ T cell population responds to small phosphate-based antigens (phosphoisoprenoids (PiPs)) that derive from microbes (such as hydroxymethyl but-2-enyl pyrophosphate) or are endogenous intermediates of the mevalonate pathway (such as isoprenylpyrophosphate) that accumulate in virally infected and transformed cells (3, 4). Thus, this population can survey for both microbial infection and cellular transformation, making PiPs and their derivatives attractive targets for immunotherapy (5).
The direct role of the Vγ9Vδ2 T cell receptor in PiP-mediated recognition has been demonstrated through TCR transfer (6); mutagenesis has further defined that PiP specific stimulation is mediated by an assemblage of the complementary determining region (CDR) loops of the Vγ9Vδ2 TCR (7). Although a crystal structure of a Vγ9Vδ2 TCR revealed a basic patch formed by the CDR loops suitable for PiP binding (8), there is no evidence of direct association between the two. Instead, several lines of evidence point to a cell surface protein or protein complex that directly mediates this interaction. In particular, cell-cell contact is required for Vγ9Vδ2 stimulation (9), trypsin treatment of target cells impairs Vγ9Vδ2 T cell activation (10), and macaque but not mouse target cells (10–12) are recognized in the presence of PiP. Together these data implicate the presence of a primate-specific, cell surface molecule or molecules that are recognized in the presence of high concentrations of PiPs.
Recent characterization of an antibody that can mimic the stimulatory effect of PiPs on Vγ9Vδ2 T cells (13) has focused attention on a class of cell surface proteins belonging to the B7 superfamily. B7 and related molecules contain two immunoglobulin-like domains: an N-terminal Ig-V-like and a C-terminal Ig-C-like domain. These proteins have important functions in immune regulation, acting as co-stimulatory molecules (B7-1 and B7-2) for T cell activation and potent inhibitory molecules (PD-L1) to down-regulate T cell responses (14). The B7-related butyrophilin and butyrophilin-like proteins have recently been more fully characterized, revealing a range of immune and nonimmune related functions (15–19). Application of an antibody specific to a subclass of BTN proteins, CD277/butyrophilin-3 (BTN3A) (15), induced strong stimulation of Vγ9Vδ2 T cells (13). This agonist antibody, 20.1, binds to the extracellular domain of BTN3A molecules and induces Vγ9Vδ2 activation with similar kinetics and potency as with addition of PiPs. A second antibody, 103.2, has been characterized that inhibits this effect.
Three BTN3A genes have been characterized in humans, BTN3A1, BTN3A2, and BTN3A3, which are members of a large family of butyrophilin genes located in the telomeric end of the major histocompatibility complex class I region (20–22) and encode cell surface-expressed proteins that have high similarity in their extracellular domains yet differ in the domain structure of their intracellular domains. BTN3A1 and BTN3A3 both contain an intracellular B30.2 domain, whereas BTN3A2 does not. Although crystal structures exist of B7 family members such as B7-1 (CD80) (23), B7-2 (CD86) (24), and PD-L1 (25, 26), none currently exist of the butyrophilin proteins. Therefore, to understand the molecular basis for the role of BTN3A in Vγ9Vδ2 reactivity, we have determined the three-dimensional structures of the extracellular domains of the three BTN3A isoforms and studied their binding by single-chain versions (scFvs) of the 20.1 and 103.2 antibodies. Along these lines, we co-crystallized BTN3A1 with 20.1 scFv and 103.2 scFv and determined their complex structures to 3.15 and 3.50 Å, respectively. Our biophysical studies of antibody interaction with BTN3A and their corresponding structures provide insight into the structures and conformations of these proteins that serve to modulate Vγ9Vδ2 T cell activation.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization of BTN3A Proteins
The extracellular domains of BTN3A1, 2, and 3 were subcloned into the baculovirus transfer vector pAcGP67A (BD Biosciences) with a C-terminal His6 tag and expressed in High Five insect cells with the baculovirus expression system (BestBac linearized baculovirus DNA; Expression Systems). The proteins were purified with nickel-nitrilotriacetic acid resin and eluted with buffer containing 200 mm imidazole. The proteins were further purified by gel filtration via a Superdex 200 column (GE Healthcare) equilibrated with HBS (HEPES-buffered saline: 10 mm HEPES, pH 7.2, 150 mm NaCl, 0.02% azide). Carbohydrate residues added by N-linked glycosylation were removed by endoglycosidase F followed by purification over anion exchange column (GE Healthcare). Peak fractions were pooled and concentrated to 10 mg/ml. Crystallization conditions for the three proteins were as follows: 1) BTN3A1: 0.1 m citric acid, pH 4, 1.6 m ammonium sulfate; 2) BTN3A2: 1 m lithium sulfate, 0.1 m sodium citrate, pH 5.6, 0.5 m ammonium sulfate; 3) BTN3A3: 0.1 m citric acid, pH 4, 0.8 m ammonium sulfate, 0.15 m sodium fluoride.
Cloning and Expression of the Antibodies
Total RNA for the agonist and antagonist antibodies were provided by Dr. Daniel Olive (INSERM). A RT-PCR was performed on RNA according to the manufacturer's protocol (SuperScript One-Step RT-PCR with Platinum Taq; Invitrogen), using degenerate primers described by Wang et al. (38) Individual heavy and light chains of both the antibodies were cloned into the TopoTA vector (Invitrogen). Single-chain constructs of the antibodies containing a His6 tag at the C terminus were cloned into the pak400 expression vector. The antibodies were expressed by periplasmic secretion in Escherichia coli (39) and purified over a nickel-nitrilotriacetic acid column; the His tag was removed by overnight treatment with carboxypeptidase A (Sigma). The proteins were further purified by gel filtration via a Superdex 200 column (GE Healthcare) equilibrated with HBS.
Co-crystallization of BTN3A1-Antibody Complexes
The BTN3A1 protein and scFv antibodies were expressed and purified as described above except that the BTN3A1 protein was treated overnight with carboxypeptidase A after nickel-nitrilotriacetic acid purification. The two proteins were mixed in a 1:1 ratio, and the complex was purified by over a Superdex 200 gel filtration column equilibrated in HBS buffer. For crystallization, the protein complex was concentrated to 10 mg/ml. The BTN3A1-20.1 complex crystallized in the following conditions: 0.15 m KF, 14% PEG 3350. BTN3A1-103.2 complex crystals were optimized in 0.1 m HEPES, pH 7.5, 1 m NaCl, 15% PEG 8000.
Data Collection and Processing
X-ray data sets were measured on a MAR300 CCD at Beamline 23 ID-D for the BTN3A1 protein and BTN3A1-103.2 complex, Beamline 21 ID-G for the BTN3A2 and 3 proteins, and Beamline 23 ID-B for the BTN3A1-20.1 complex at the Advanced Photon Source at Argonne National Laboratory. The crystals were cryoprotected with 20% glycerol before cooling to 100 K. The data sets were collected with 1° oscillations at a 1.033 × 10−10 m wavelength for the BTN3A1 protein. The BTN3A2 and BTN3A3 structures were collected as BTN3A1 but at a wavelength of 0.99857 × 10−10 m. HKL2000 was used to index, integrate, and scale the data (30).
Structure Determination and Refinement
A turkey telokin domain (31) and a constant domain of PD-L1 protein (26) were used as search models in molecular replacement with Phaser (32) and located the single BTN3A1 molecule per asymmetric unit. Rigid body refinement was performed with Phenix (33), followed by manual building with Coot (34) and individual site and B-factor refinement with Phenix. The stereochemistry of the model was improved via a backbone optimization procedure incorporating a torsional statistical potential contingent on amino acid type, its secondary structure, as well as the identity of its two neighbors (35), followed by real space refinement with a two-step algorithm developed by Haddadian et al. (36). The refined BTN3A1 protein was used as search model for the other two BTN3A proteins, as well as the complex structures, and refinement was performed as described above. The variable domains from a IgG2a mouse antibody (Protein Data Bank code 1IGT) were used as a search model for the 20.1 single chain. The 103.2 single chain search model was built using the heavy chain variable domain from a mouse monoclonal antibody (Protein Data Bank code 3DIF) and the light chain variable domain of different structure (Protein Data Bank code 1XGP). The 103.2-BTN3A1 complex data set suffered from severe anisotropy; therefore, the structure factors were processed using the UCLA MBI-Diffraction Anisotropy Server (37), which significantly improved the density map. All of the structural figures were generated with the program PyMOL (DeLano Scientific). Ramachandran statistics (% preferred region, allowed region, and outliers) for the final models were: BTN3A1: 97.6, 2.4, and 0; BTN3A2: 99, 1, and 0; BTN3A3: 98.1, 1.9, and 0; BTN3A1-20.1 complex: 94.1, 5.9, and 0; and BTN3A1-103.2 complex: 93.7, 6.2, and 0.1.
Multiangle Light Scattering (MALS)
For molecular weight analysis, size exclusion chromatography of the BTN3A proteins alone and in complex with the agonist and the antagonist was performed on a Superdex 200 column (GE Healthcare) equilibrated with HBS. The system was connected to a 18 angle DAWN-Heleos multiangle laser light scattering detector equipped with a K5 flow cell and a 658-nm laser diode and to an Optilab rEX refractometer operating with a 685-nm laser diode at 25 °C (Wyatt Technology, Santa Barbara, CA). The data were collected at scattering angles θ ranging from 22.5 to 147°. Astra software version 5.3.4 was used to analyze the data and determine the molecular weight of the proteins.
Surface Plasmon Resonance
All of the surface plasmon resonance was measured at 25 °C on a BiacoreTM 3000 with a nitrilotriacetic acid chip (BiacoreTM). HBS-P buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm imidazole, and 0.005% (v/v) surfactant Tween 20) was used for all measurements. Approximately 179, 125, and 160 response units of histidine-tagged BTN3A1, 2, and 3 proteins, respectively, were immobilized. Sensograms were fit locally with BiacoreTM 3000 analysis software (BIAevaluation, version 4.1) using 1:1 Langmuir binding with drifting base line.
FRET Measurements
A C-terminal cysteine was engineered into the BTN3A1 and 103.2 single chain constructs. Proteins were expressed and purified as described previously. Of note, ∼90% of the BTN3A1 C-terminal cysteine appeared as a disulfide-linked homodimer when run on nonreducing SDS-PAGE gel. To reduce the disulfide bond, the samples were concentrated to 30 μm in HBS, 0.2 mm DTT was added, and the samples were incubated for 30 min at room temperature. 300 μm dyes, either Atto532 maleimide (68499; Sigma-Aldrich), Cy5 maleimide (PA25031; GE Healthcare), or a 1:1 combination of both dyes, were added to each sample and then incubated for 2 h at room temperature. The samples were then diluted from 100 μl to 400 μl in HBS and dialyzed in a 20-kDa molecular mass cutoff cassette (66005; Thermo Scientific) against 1 liter of HBS for 16 h to remove free dye. The samples were diluted to 400 nm in PBS for spectral measurements. Fluorescence measurements were made on a Fluorolog-3 instrument with the excitation wavelength set to 520 nm. Emission spectra were collected from 665 to 750 nm. Excitation and emission slits were set to 2 and 10 nm, respectively. Background measurements were conducted with singly labeled components. Equal molar amounts of the acceptor-labeled BTN3A1 and donor-labeled 103.2 were mixed for the intermolecular FRET measurements. The 20.1 single chain was added to a 10-fold molar excess for each experiment. FRET efficiency was calculated from measurements of the enhancement of acceptor fluorescence as described previously (28). In short, FRET efficiency was calculated as the ratio of acceptor signal after excitation of the donor at 520 nm to direct excitation of acceptor at 650 nm after background subtraction from 665 to 750 nm. The estimated Förster radius (R0) was calculated as R0 = (8.79 × 10−5 × κ2 × n−4 × QD × J(λ))(1/6) where the orientation factor κ2 is assumed to be two-thirds, the index of refraction n assumed to be 1.4, the quantum yield of the donor (QD) reported by the manufacturer was 0.9, and the spectral overlap J(λ) was determined in units of nm4/(Mcm).
Antibody Reagents
Purified mouse anti-CD277 mAbs and Fab (clones 20.1 and 103.2) were generated, produced, and purified as described (15). The following monoclonal antibodies were from BD Biosciences: phycoerythrin-cyanin 5.1-labeled anti-CD107a (H4A3), phycoerythrin-labeled anti-IFN-γ (B27). Monensin A was purchased from Sigma-Aldrich. Live/dead cell marker and Alexa 647-labeled goat anti-mouse IgG were from Invitrogen. Recombinant human IL-2 (rhIL-2) was from Chiron Therapeutics.
Cells
Human Vγ9Vδ2 (polyclonal line GUI) and CD8pos αβ (clone 13, HCV-1/HLA-A2) T cells were maintained in complete RPMI 1640 medium with 10% FCS, 2 mm l-glutamine, 10 μg/ml streptomycin, 100 IU/ml penicillin, supplemented with 300 IU/ml recombinant human IL-2.
Functional Assays
Human Vγ9Vδ2 T cells were activated with anti-CD277 (20.1) mAb, scFv, or Fab at the indicated concentrations. Cytolytic activity of γδ T cells was analyzed by measuring CD107a surface mobilization by flow cytometry. In this assay, T cells were activated at 37 °C in complete RPMI 1640 medium containing 10 μm monensin and a fluorochrome-labeled CD107a specific antibody. After 4 h, T cells were harvested and stained with fluorochrome-labeled TCR-specific antibodies and a viability marker (live/dead cell marker, 0.3 μm). Intracellular staining of IFN-γ was performed within the same samples. T cells stained for cell surface markers and viability were next fixed at room temperature for 20 min with 2% paraformaldehyde, permeabilized for 20 min with BD PhosFlow Perm/Wash buffer (BD Biosciences), and incubated for 30 min with fluorochrome-conjugated specific antibodies. The data were collected on FACSCantoII (BD Biosciences). For blocking experiments, human Vγ9Vδ2 T cells were activated by grading doses of bromohydrin pyrophosphate/phosphostim, kindly provided by Innate Pharma, in the presence of 103.2 anti-CD277 mAb or scFv, at 10 μg/ml.
RESULTS
Crystal Structures of the Three BTN3A Extracellular Domains
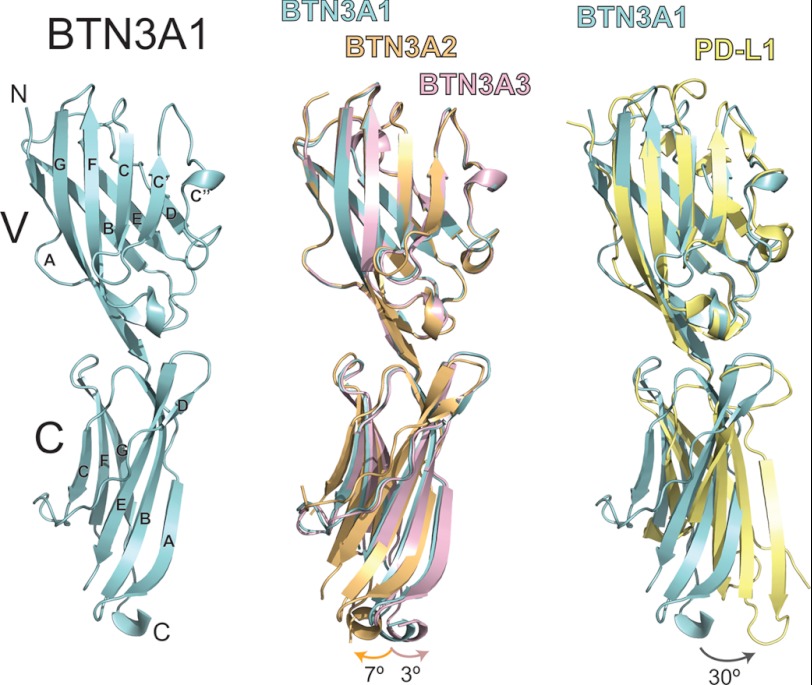
The extracellular domains of the three BTN3A isoforms were expressed recombinantly in insect cells for use in crystallization trials. Each isoform readily crystallized in related conditions producing large diamond-shaped plates that diffracted well. Data sets were collected for each isoform, and their structures were determined to 1.9, 2.4, and 2.4 Å resolution for BTN3A1, BTN3A2, and BTN3A3, respectively. The crystallographic and refinement statistics are summarized in Table 1. One molecule was present in the asymmetric unit for each of the BTN3A isoforms. The general domain structure of BTN3A is shown in Fig. 1 (left panel). BTN3A molecules have two Ig-like domains: an N-terminal Ig-V like domain and a C-terminal Ig-C type domain, similar to other B7 family members. The closest structural homologue, as assessed through the DALI server was programmed cell death 1 ligand 1 (PD-L1) (21% amino acid identity and 3.4 Å2 root mean square deviation (RMSD)). However, the Ig-V like domain of BTN3A had higher structural homology (1.0Å RMSD) and amino acid identity (45%) than the single Ig domain containing protein, myelin-oligodendrocyte glycoprotein (27).
TABLE 1.
Data collection and refinement statistics (molecular replacement)
| BTN3A1 | BTN3A2 | BTN3A3 | 20.1-BTN3A1 | 103.2-BTN3A1 | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21221 | P21221 | P21221 | P4 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) | 30.06, 77.99, 98.69 | 33.00, 77.58, 100.97 | 39.84, 78.43, 98.41 | 165.13, 165.13, 53.81 | 87.74, 118.42, 122.84 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 19.9–1.9 | 42.313–2.379 | 30.963–2.382 | 45.8–3.15 | 42.6–3.5 |
| Rmerge | 0.071 (0.382) | 0.073 (0.292) | 0.069 (0.306) | 0.102 (0.705) | 0.087 (0.558) |
| I/σ | 27.3 (3.2) | 25.5 (2.5) | 22.5 (2.9) | 19.9 (2.4) | 16.4 (2.3) |
| Completeness (%) | 95.2 (89.9) | 93.2 (50.0) | 94.7 (66.9) | 99.9 (99.1) | 99.2 (99.6) |
| Redundancy | 5.0 (4.2) | 5.4 (2.9) | 5.5 (3.1) | 4.9 (4.1) | 6.5 (5.4) |
| Refinement | |||||
| Resolution (Å) | 1.9 | 2.4 | 2.4 | 3.15 | 3.5 |
| No. of reflections | 23011 | 10267 | 12256 | 25881 | 16002 |
| Rwork/Rfree | 0.22/0.25 | 0.23/0.27 | 0.22/0.24 | 0.26/0.30 | 0.37/0.41 |
| No. of atoms | |||||
| Protein | 1582 | 1586 | 1591 | 6629 | 5513 |
| Ligand/ion | 0 | 0 | 0 | 28 | 0 |
| Water | 124 | 60 | 79 | 0 | 0 |
| B-factors | |||||
| Protein | 30.878 | 44.313 | 34.406 | 86.4 | 146.0 |
| Ligand/ion | 0 | 0 | 0 | 120.6 | 0 |
| Water | 36.435 | 40.806 | 36.9 | 0 | 0 |
| RMSDs | |||||
| Bond lengths (Å) | 0.011 | 0.012 | 0.011 | 0.008 | 0.015 |
| Bond angles (°) | 1.231 | 1.321 | 1.163 | 1.330 | 1.409 |
FIGURE 1.
Crystal structures of the BTN3A isoforms. The structure of BTN3A1 is shown colored in cyan in ribbon format (left panel). BTN3A molecules are composed of a membrane-distal Ig-V domain and a membrane-proximal Ig-C domain. The canonical Ig β-strand designations (A–G) are shown for both. The middle panel shows superposition of the BTN3A1 (cyan), BTN3A2 (orange), and BTN3A3 (pink) structures determined in this study. The three structures are superimposed via their highly conserved V domain, demonstrating subtle shifts in the orientation of the C domain. The angle of rotation is shown at the bottom of the structures, with color-coded arrows corresponding to the particular isoform. The right panel shows structural comparison of BTN3A1 with PD-L1, the closest structural homologue to the BTN3A molecules. PD-L1 is shown in yellow. The two molecules were superimposed via their V domains, revealing a C domain shift of ∼30° between the two molecules.
Comparing within the BTN3A structures, BTN3A1, BTN3A2, and BTN3A3 were highly similar, differing from each other by backbone Cα RMSD of no more than 0.89 Å2. These RMSD differences were primarily due to slight shifts in the orientation of the C domain in relation to the V domain between the three isoforms. Aligning the three isoforms via their V domains and using BTN3A1 as a reference, the C-terminal domain of BTN3A2 is shifted ∼7° clockwise, whereas the C-terminal domain of BTN3A3 is shifted ∼3° counterclockwise (Fig. 1, middle panel). There are no amino acid differences at the V-C domain interface between the three isoforms that would explain these shifts; therefore it is likely that rotational flexibility of the V-C domain interface combined with subtle differences in crystal packing between the three isoforms explains these alternative orientations. Alignment of BTN3A1 with PD-L1 (Protein Data Bank code 3BIK) via their V domains revealed a more exaggerated shift between the V-C domains of these two proteins. The C domain of PD-L1 is shifted ∼30° counterclockwise (Fig. 1, right panel), explaining the higher RMSD between these two proteins. Indeed, the RMSD differences by domain between BTN3A and PD-L1 were 1.6 and 2.0 Å2 for the Ig-V and Ig-C type domains, respectively.
Dimerization of the BTN3A Molecules
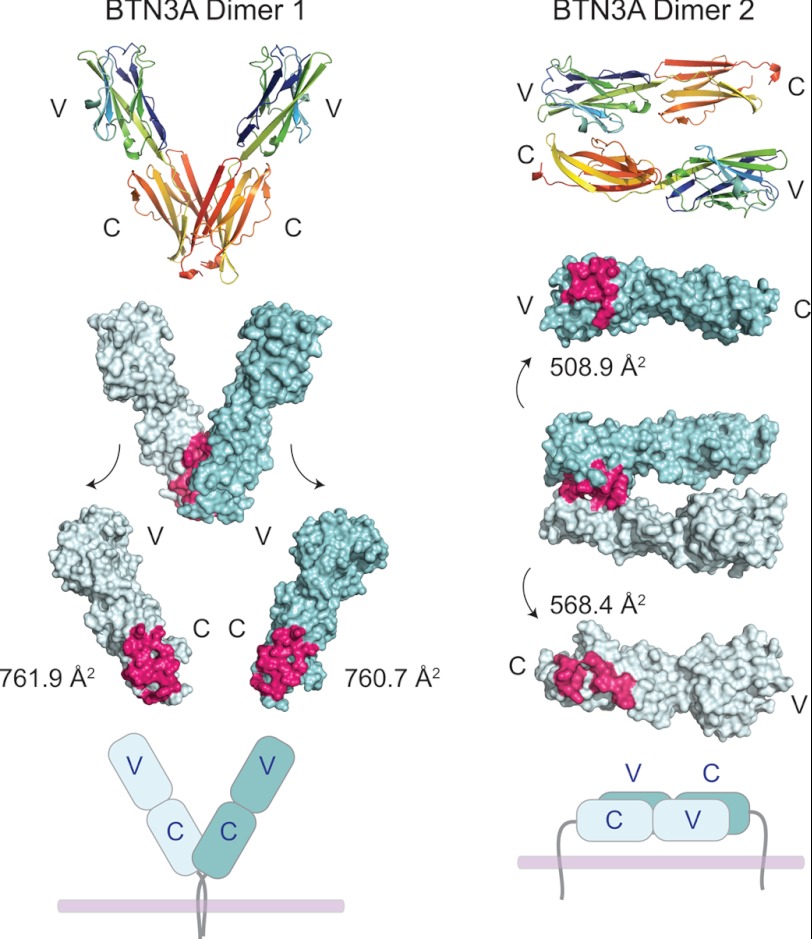
In our crystal structures of BTN3A1, BTN3A2, and BTN3A3, two separate dimer forms were noted in the crystal lattice (Fig. 2). Dimer 1 consists of two BTN3A molecules in a V-shape orientation, associated by a symmetrical interface at the membrane proximal region of the C domain (Fig. 2, left panel). This interface would orient the BTN3A molecules in an upright position away from the membrane when modeled onto the cell surface with the transmembrane regions located close to each other. Dimer 2, in contrast, associates two BTN3A in a nonsymmetrical interface in a head to tail orientation, joining a V domain from one BTN3A with a C domain from a second BTN3A (Fig. 2, right panel). This association would require the BTN3A dimer to be oriented parallel to the cell membrane (Fig. 2, bottom of right panel), and the transmembrane domains would be separated by ∼80 Å, the approximate length of one BTN3A molecule. Both interfaces occupy considerable surface area, with a combined buried surface area (BSA) of ∼1523 Å2 for Dimer 1 and ∼1077 Å2 for Dimer 2. Both interfaces are mediated by hydrogen bonding and van der Waals contacts (supplemental Table S1). The hydrogen-bonding network of the Dimer 1 interface is predominantly main chain/main chain contacts that connect the Ig β-sheets together by two symmetric, parallel, β-strand pairings of strands A and G (supplemental Fig. S1, top panel). Only three residues contribute specific side chain interactions to this interface: Asp-120, Asp-124, and Thr-203, and these contacts comprise only seven of the sixteen H-bonds between the two monomers. The Dimer 2 interface incorporates more side chain H-bonding interactions (five of the seven H-bonds are side chain-mediated), although the overall numbers of contacts is less than that of Dimer 1. The C′ and C″ strands from the V domain dominate the interface of Dimer 2, pairing with residues from the A and D strands from the C domain of the opposing monomer. Neither Dimer 1 or Dimer 2 have been observed in the crystal structures of other B7 family members; although they are homodimeric molecules, each appears to have a unique dimerization interface (23–25).
FIGURE 2.
Dimerization of the BTN3A molecules in the crystal lattice. Two potentially physiologically relevant dimerization states were noted in the crystal lattices of each BTN3A isoform. BTN3A1 is shown as the representative. Top panel, BTN3A1 is in rainbow colors from the N terminus to the C terminus to clarify domain orientation of the two dimerization states. The Dimer 1 contact surface is symmetric via the A and G β-strands of the C domains of BTN3A, whereas the Dimer 2 contact surface is asymmetric, via a head-to-tail orientation via the C, C′, and C″ β-strands of the V domain with the A and D β-strands of the C domain. Middle panel, both dimers are shown as surface representations, with the dimerization interface color-coded in hot pink. The BSA in Å2 is indicated next to both dimer forms. Bottom panel, a cartoon representation of how these dimers would exist on the cell surface based on the location of the C-terminal transmembrane domains.
Examination of the residues that constitute these two dimers across primate species, and between the three BTN3A isoforms, reveals striking conservation at these positions across evolutionary time (supplemental Fig. S2). The interface of Dimer 1 shows less conservation, however the majority of contacts at this interface are main chain interactions. The three residues contributing specific side chain interactions, Asp-120, Asp-124, and Thr-204 are either conserved across species and isoforms or have only semi-conservative or conservative changes at this position (D120N and D124E). The Dimer 2 interface is completely conserved in the V domain (supplemental Fig. S2) and has moderate conservation in the C domain, although many of these differences are also observed in the other BTN3A isoforms. Because Dimer 2 was observed in the crystal structures of all three BTN3A isoforms, these amino acid polymorphisms are clearly tolerated in the Dimer 2 interface.
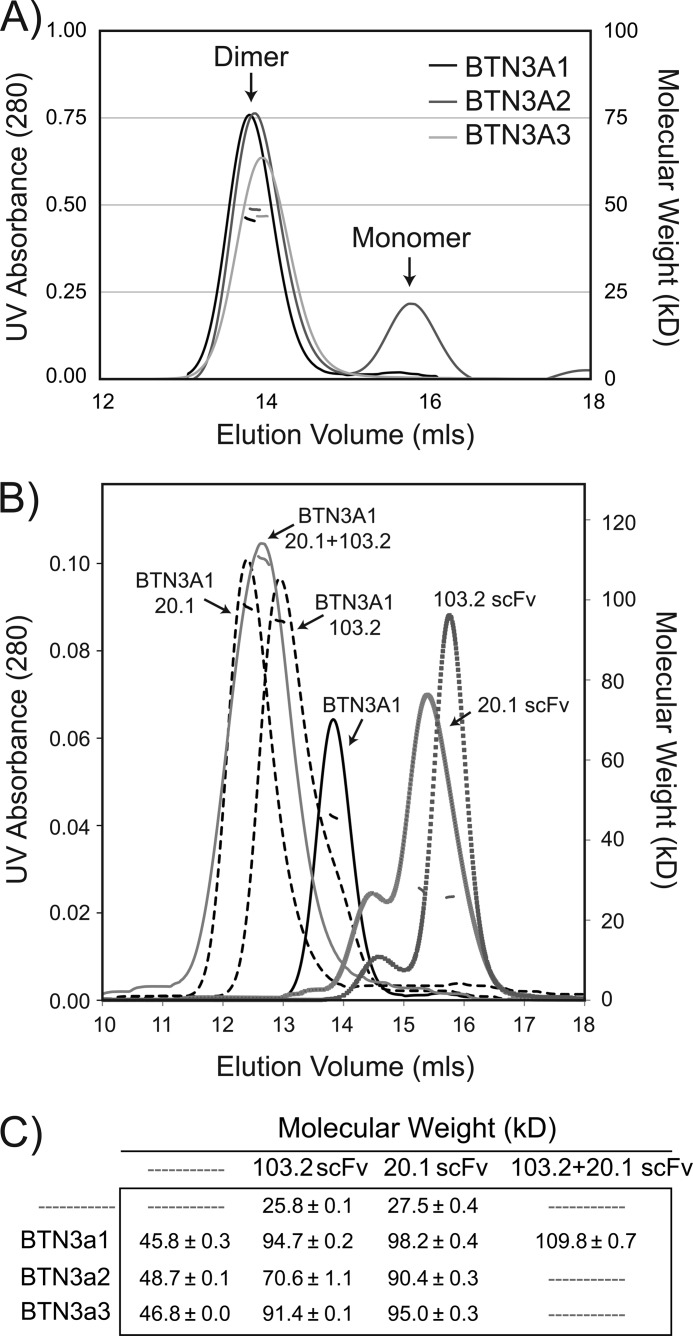
To determine whether BTN3A molecules exist as a dimer in solution, we subjected our recombinant BTN3A molecules to MALS to determine the molecular weight of the species in solution. Fig. 3 (A and C) shows the UV absorbance profile of the three isoforms and the calculated molecular weight of these peaks, respectively. Each isoform produced a symmetric peak at elution volume ∼14 ml; the molecular weight of this peak was determined to be ∼46–48 kDa depending on the BTN3A isoform. The theoretical molecular weight of a BTN3A extracellular domain monomer is ∼24 kDa, consistent with SDS-PAGE mobility of these proteins under denaturing and nonreducing conditions; therefore our MALS data support BTN3A molecules as noncovalently linked homodimers in solution because all three monomeric BTN3A isoforms lack free cysteines. The BTN3A2 isoform produced two peaks, the second of which was in the minority. This peak likely represents a population of monomer protein, a species not observed in the other isoforms.
FIGURE 3.
MALS establishes that BTN3A molecules exist as dimers in solution. A, MALS data of each BTN3A isoform reveals a clear dimer peak in solution with an estimated molecular masses of ∼46, ∼49, and ∼47 kDa for isoforms A1, A2, and A3, consistent with a BTN3A dimer. The left axis is UV absorbance from S200 size exclusion chromatography, and the right axis is the calculated molecular mass (kDa) from the light scattering. B, MALS data of BTN3A1 alone (solid black line) and in complex with the 20.1 scFv (∼98 kDa), the 103.2 scFv (∼95 kDa) (dashed lines), and both 20.1 and 103.2 (∼110 kDa) (solid gray line). Shown also are the profiles for the 20.1 and 103.2 scFvs alone, indicating estimated molecular masses of ∼26 and ∼28 kDa, respectively. C, molecular masses of BTN3A isoforms, individually and in complex, calculated from the MALS data.
Recognition of BTN3A by the 20.1 and 103.2 Antibodies
Two antibodies, 20.1 and 103.2, were shown to have opposing effects on the activation of Vγ9Vδ2 T cells by target cells (13). 20.1 produced an agonist effect, mimicking the addition of PiPs on target cells and stimulating Vγ9Vδ2 expressing effector T cells. In contrast, 103.2 was able to antagonist the effect of 20.1 and PiPs, inhibiting the activation of Vγ9Vδ2. To study the association of the 20.1 and 103.2 antibodies with BTN3A and to determine the structural basis of their binding, we generated single-chain versions of the heavy and light chain variable regions from each antibody. These scFv constructs were used to measure stoichiometry, determine binding kinetics and affinities, and determine structure.
To first determine the stoichiometry of binding of the scFvs to BTN3A, we performed MALS to measure the molecular weight of the scFvs alone and in complex with BTN3A. Shown in Fig. 3B are the MALS profiles of BTN3A1, 20.1 scFv, 103.2 scFv, and their complexes. This analysis was also performed for the BTN3A2 and BTN3A3 isoforms with similar results, confirming that the 20.1 and 103.2 antibodies bound to all three BTN3A isoforms. Only the MALS profiles for BTN3A1 are shown here. The molecular weights of all species are summarized in Fig. 3C. The scFvs had a molecular weight consistent with monomers: ∼28 and 26 kDa for the 20.1 scFv and 103.2 scFv, respectively. When associated with the BTN3A molecules, the calculated molecular weights of these species were consistent with two scFvs per BTN3A dimer (∼98 kDa and ∼95 kDa for 20.1-BTN3A1 and 103.2-BTN3A1, respectively), suggesting that each BTN3A monomer has one antibody epitope. In addition, we also tested whether there is competition in binding between the antibodies, suggestive of overlapping epitopes. The higher molecular weight of the BTN3A1-20.1scFv-103.2scFv complex (∼110 kDa) was consistent with both scFvs able to bind one BTN3A concurrently, our first line of evidence that these two antibodies recognize distinct epitopes on the BTN3A molecule. Although the measured molecular weight was less than that expected for the full BTN3A-20.1-103.2 complex (∼150 kDa), we interpret this measured value to represent an average of saturated and unsaturated complexes; however it still provides evidence that both antibodies can associate with BTN3A concurrently.
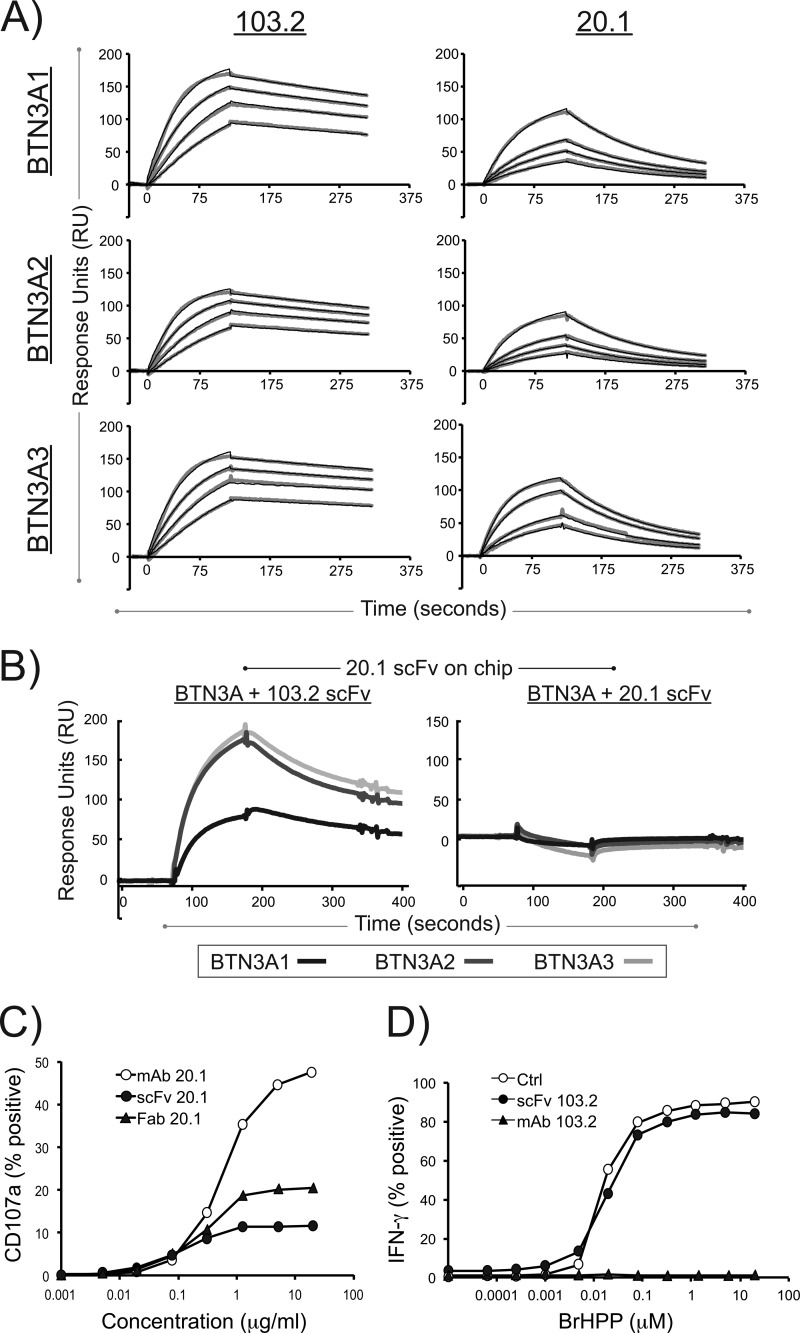
To quantitate the strength of binding of the 20.1 and 103.2 scFvs to the BTN3A isoforms and determine whether each BTN3A isoform is recognized with the same affinity, we performed surface plasmon resonance on each BTN3A isoform with each scFv. Shown in Fig. 4A are the concentration-dependent sensograms used to determine the kinetics of binding reported in Table 2. The binding kinetics and affinities are similar across the BTN3A isoforms, suggesting that these antibodies recognize epitopes conserved across the BTN3A isoforms. In addition, the association rate (Ka) is similar between 103.2 and 20.1 (ranging from 1.2 to 2.4 × 105 1/Ms); however, 20.1 dissociates approximately five times faster than 103.2, leading to an overall lower affinity of the agonist antibody scFv (∼50 nm for 20.1 compared with ∼10 nm for 103.2). These measured affinities are most likely an underestimation of the affinity of the full-length antibodies because of the avidity effect contributed by the bivalent nature of the antibodies. To further confirm that the antibodies bind to separate epitopes on the BTN3A molecule, we performed competition surface plasmon resonance, where the 20.1 scFv antibody was immobilized on the chip, and either the BTN3A-103.2 complex (Fig. 4B, left panel) or the BTN3A-20.1 complex (Fig. 4B, right panel) was flowed over this chip to assess binding. Clear binding was observed in the BTN3A-103.2 complex and not with the BTN3A-20.1 complex, establishing that 103.2 does not inhibit 20.1 binding. Therefore, the antagonist effect of 103.2 is not simply due to blocking of the 20.1 epitope.
FIGURE 4.
Surface plasmon resonance of the 20. 1 and 103.2 antibodies with BTN3A1, BTN3A2, and BTN3A3 reveal high affinities and noncompetitive binding. A, representative sensograms are shown for the surface plasmon resonance analysis of 20.1 scFv and 103.2 scFv binding to the three BTN3A isoforms. Each BTN3A isoform was immobilized on an individual flow cell on a sensor chip, and the scFv were flowed as analyte at concentrations ranging from 10 to 80 nm. The data curves are shown in gray, and the modeled fit is shown as black lines. B, competition assay between the 20.1 and 103.2 scFv for BTN3A isoforms. The 20.1 scFv was immobilized on the sensor chip surface and complexes of either BTN3A-103.2scFv (left panel) or BTN3A-20.1 scFv (right panel) were flowed as analyte. Clear binding is observed with the BTN3A-103.2 scFv complex, whereas binding is blocked with the BTN3A-20.1 scFv, indicating that the two scFvs bind different epitopes on the BTN3A molecules. C, CD107a expression on the human Vγ9Vδ2 T cell line GUI following incubation with either 20.1 full-length antibody (open circles), 20.1 Fab fragments (filled triangles), or 20.1 scFv (filled circles) used at the indicated concentrations. The data are presented as the percentages of CD107a+ γδ T cells and are representative of more than three experiments. D, IFN-γ expression in human Vγ9Vδ2 T cell line GUI following activation by increasing doses of bromohydrin pyrophosphate/phosphostim (BrHPP) in the presence of the 103.2 full-length antibody (filled triangles), 103.2 scFv (filled circles), or no antibody (open circles). The data are presented as the percentages of IFN-γ+ γδ T cells and are representative of more than three experiments.
TABLE 2.
Surface plasmon resonance analysis of the interaction between the BTN3A isoforms and the 20.1 and 103.2 scFvs
| Complex | Ka | Kd | KD |
|---|---|---|---|
| 1/Ms | 1/s | nm | |
| BTN3A1, 20.1 scFv | 1.21 × 105 | 7.98 × 10−3 | 66 |
| BTN3A2, 20.1 scFv | 2.41 × 105 | 7.82 × 10−3 | 33 |
| BTN3A3, 20.1 scFv | 1.70 × 105 | 8.28 × 10−3 | 48 |
| BTN3A1, 103.2 scFv | 1.08 × 105 | 1.63 × 10−3 | 15 |
| BTN3A2, 103.2 scFv | 2.38 × 105 | 1.67 × 10−3 | 7 |
| BTN3A3, 103.2 scFv | 1.31 × 105 | 1.06 × 10−3 | 8 |
We also evaluated the biological effects of the scFv antibodies on Vγ9Vδ2 T cell activation. Fig. 4C shows the effect of the 20.1 scFv relative to that of full-length 20.1 antibody and 20.1 Fab. The 20.1 scFv can clearly activate Vγ9Vδ2 as measured by CD107a expression, but less well than that of the full-length or Fab versions of the antibody, suggesting that the bivalent nature of the full-length antibody may function to enhance BTN3A-mediated activation of Vγ9Vδ2 T cells, either through increased avidity or through multimerization of BTN3A molecules on the cell surface. We also tested whether the scFv version of the 103.2 antibody could antagonize Vγ9Vδ2 activation (Fig. 4D). Although full-length 103.2 antibody was highly effective in blocking PiP activation of Vγ9Vδ2 cells, the scFv had no effect, suggesting an even more important role for the bivalent structure of this antibody for its antagonist function.
Complex Structures of BTN3A1 with the 20.1 and 103.2 Antibodies
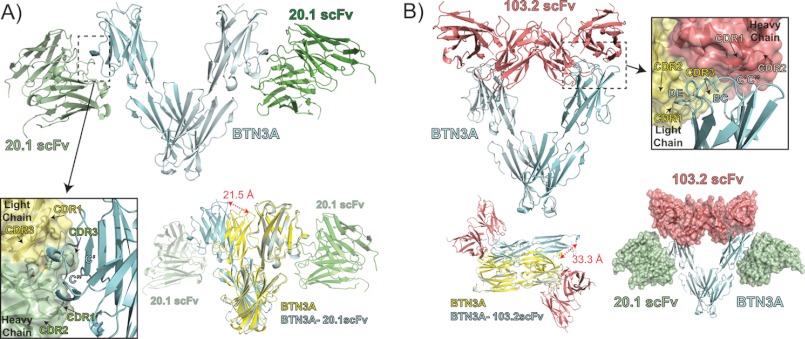
To fully understand the effect of antibody binding to the BTN3A molecules and to glean insight into their role in Vγ9Vδ2 stimulation, we determined the structures of the complexes between each scFv and the extracellular domain of the BTN3A1 isoform. The 20.1-BTN3A1 complex crystals diffracted to moderate resolution, and the structure of this complex was refined to 3.15 Å resolution with good statistics (Table 1). The 103.2-BTN3A1 crystals, however, were difficult to reproduce, diffracted to low resolution, and suffered from severe anisotropy; hence we report our preliminary model of this complex to 3.5Å resolution (Table 1). Although this resolution does not allow for discrimination of most side chains, it reveals the overall docking position of the 103.2 scFv on the BTN3A1 molecule. Together, these two complex structures shed light on the different association modes of the two scFv antibodies and allow us to generate a model of how the full-length antibody recognizes BTN3A molecules on the cell surface and their effect on BTN3A dimerization and multimerization.
In the 20.1 scFv-BTN3A1 complex structure, there are two 20.1scFv-BTN3A complexes in the asymmetric unit. These are related via a 2-fold symmetry axis based on a dimerization interface of the two BTN3A1 molecules, similar in fashion to Dimer 1 noted in the uncomplexed BTN3A structures (Fig. 5A). The complexed dimer, however, is shifted along the dimerization axis resulting in a ∼20 Å shift as measured from the most membrane-distal tip of the V domains. The relatively nonspecific main chain H-bonding of the Dimer 1 interface (supplemental Fig. S1, top panel) allows for this rotation of the V domains. The 20.1 scFvs bind to the outer region of the V domain, predominantly interfacing with the C′ and C″ strands of the V domain with a footprint almost exactly overlapping with that of the V domain interface of Dimer 2, explaining why this dimerization interface is not noted in this crystal lattice. Of the 37 V domain contacts, 33 are identical to that seen in the Dimer 2 interface (supplemental Table S2 and supplemental Fig. S2, shown as green stars); thus, binding of the 20.1 antibody to the V domain of BTN3A would directly compete with the Dimer 2 state. The heavy chain CDR loops of 20.1 scFv establish the majority of contacts and contribute ∼85% of the BSA of the interface. The numbers of contacts are spread equitably across the CDR1, CDR2, and CDR3 heavy chain loops, with H-bonds, salt bridges, and van der Waals interactions well represented. The light chain contributes only ∼15% of the BSA with seven of the eight contacts mediated by the CDR3 loop. The light chain CDR1 loop contributes one van der Waals contact to the interaction.
FIGURE 5.
Complex structures of BTN3A1 with the 20.1 scFv and 103.2 scFv. A, ribbon diagram showing the complex between the BTN3A1 isoform (cyan) and the 20.1 scFv (green). The 20.1 scFv binds BTN3A1 on the side of the V domain, contacting the C′ and C″ β-strands (see inset), with the binding interface dominated by heavy chain CDR1, CDR2, and CDR3 loop contacts (shown in green; light chain is shown in yellow for contrast). The BTN3A1 dimer present in the complex structure (shown in cyan with 20.1scFv in green) is similar in its general orientation to that of Dimer 1 in the uncomplexed BTN3A (shown in yellow); however, it is shifted at the dimerization interface, resulting in an ∼20 Å displacement as measured between V domains, as indicated by the red arrow. B, complex between the 103.2 scFv and BTN3A1, shown as ribbons in red and cyan, respectively. The 103.2 scFv binds at the top of the V domain, contacting the DE, BC, and C′C″ loops (see inset). The dimerization interface of the BTN3A molecules is similar to that of the unliganded BTN3A molecules, where Dimer 1 is strictly conserved and Dimer 2 is shifted and rotated ∼33 Å as measured from the C termini of the two dimer forms (lower left panel). The 20.1 (green) and 103.2 (red) scFvs bind to separate epitopes on BTN3A as shown from their superposition on the BTN3A1 Dimer 1 (lower right panel).
Our low resolution structure of the 103.2 scFv-BTN3A1 complex reveals that the 103.2 antibody binds to the top of the BTN3A V domain via the BC, C′C″, and DE loops (Fig. 5B), which are the equivalent of the CDR1, CDR2, and HV4 loops of an antibody or TCR. This is a distinct epitope from that of the 20.1 scFv, confirming our MALS and surface plasmon resonance data that demonstrated both antibodies could bind in tandem. The 103.2 scFv interface with BTN3A1 is robust, with an estimated 880Å2 BSA. The epitope is biased toward the heavy chain, with ∼60% of the BSA caused by heavy chain CDR loops. Although our resolution precludes evaluating side chain involvement, it appears the heavy chain CDR1 and CDR2 loops are predominantly contacting BTN3A, although clear electron density for side chains of the heavy chain CDR3 loop were missing. The light chain, however, contacts BTN3A through all three CDR loops, comprising an estimated 330 Å2 of the complex BSA.
Dimer 1 Is the Primary Conformation of BTN3A in Solution
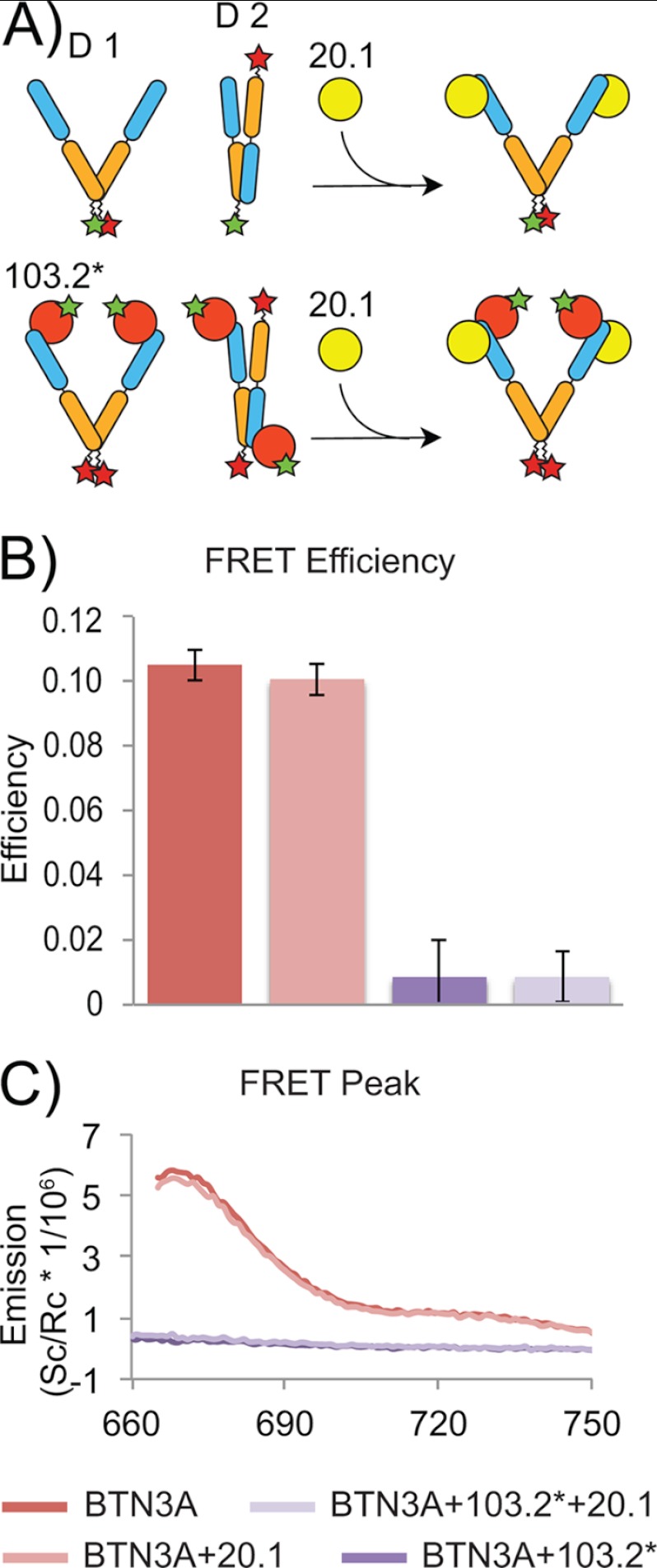
The two BTN3A dimer interfaces identified in the crystal lattice have robust interfaces suggestive of physiologically relevant dimer forms. Furthermore our MALS data verified that at least one dimer form was present in solution. Because the nature of the physiological dimer interface is important for generating a model by which to understand how agonist versus antagonist antibody binding modulates BTN3A activity in the context of Vγ9Vδ2 activation, we devised a FRET strategy to test whether one or both dimers were present in solution. To accomplish this, we engineered single cysteine mutations into the C terminus of BTN3A1 for covalent coupling with dye pairs suitable for FRET experiments. The structural data provided by our complex structures between BTN3A1 and the 20.1 and 103.2 scFvs were key in this experimental design; the strategy is outlined in Fig. 6A, as are the possible conformations that would result in FRET signal or quenching of FRET signal. In the first strategy, we co-labeled soluble BTN3A with equimolar amounts of Atto532 and Cy5, which given equal labeling efficiencies should result in ∼50% of the protein co-labeled with the two dye pairs. Dimer 1 would give strong FRET signal, because the dye pair would be within the estimated 65 Å R0 distance for 50% FRET transfer efficiency (see methods for R0 calculation) (28). Dimer 2 would locate the donor and acceptor ∼80 Å apart. We also capitalized on the ability of the 20.1 scFv to disrupt Dimer 2 formation in this strategy. In our second strategy, we labeled BTN3A with Atto532 and the 103.2 scFv with Cy5. In this scenario, FRET would occur in Dimer 2, but not in Dimer 1, and would be quenched upon the addition of the 20.1 scFv (because it would disrupt Dimer 2). In both experimental strategies, FRET signal was measured in situations consistent with Dimer 1: strong signal was detected in the co-labeled BTN3A1 (Fig. 6) and co-labeled BTN3A1 incubated with 20.1 scFv, but no FRET signal was detected in the second scenario, where 103.2 was labeled with donor and BTN3A with the acceptor. These results establish that Dimer 1 is the predominant BTN3A dimer species in solution
FIGURE 6.
FRET investigation of BTN3A1 dimers. A, cartoon schematic presenting FRET strategy and experimental set-up. Upper panel, BTN3A1 was labeled with both donor (Atto532) and acceptor (Cy5) dyes. FRET efficiency was determined before and after the addition of the 20.1 scFv allowing for direct investigation of Dimer 1 in solution. Lower panel, BTN3A1 was labeled with donor and 103.2 scFv with acceptor (designated as 103.2*) to investigate the presence of the Dimer 2 in solution. FRET efficiency was determined as above. B, background subtracted fluorescence measurements after excitation of the donor. Dually labeled BTN3A1 measurements are shown in red, BTN3A1-103.2 inter-protein measurements are show in purple. Measurements after the addition of 20.1 are shown in light red and light purple, colored respective to the initial condition. C, calculated FRET efficiency for each scheme as determined from 665 to 750 nm for each.
Model for 20.1 and 103.2 Antibody Binding to BTN3A Molecules on the Cell Surface
Our structural and FRET data combined provide an excellent model by which to understand the binding of the 20.1 and 103.2 antibodies to BTN3A and how that might modulate the global conformation of BTN3A molecules on the cell surface. Our FRET data strongly favor BTN3A molecules in the Dimer 1 conformation, with little evidence that Dimer 2 exists in soluble BTN3A. It is therefore likely that BTN3A exists on the cell surface as Dimer 1. Both antibodies can bind the BTN3A Dimer 1 conformation; however our structural data suggest that they bind with different stoichiometries.
For the 20.1 antibody, the orientation observed for the 20.1scFv on the BTN3A monomer would make it improbable for one bivalent antibody molecule to bind one BTN3A dimer as visualized through superposition of Fab models onto the docking position of the 20.1 scFvs. The distance between the two C termini of these Fabs is ∼215 Å, a distance that could not be spanned by the linkers adjoining two Fabs to one Fc molecule. Even fully extended, these linkers span only ∼100 Å. Therefore our structural model provides a strong argument for the stoichiometry of the 20.1 full-length antibody binding to the BTN3A dimer as 1:2, with one bivalent antibody binding two separate BTN3A dimers, leading to a multimerization of BTN3A dimers on the cell surface (supplemental Fig. S3, left panel). Furthermore, the 20.1 antibody binds on the side of the BTN3A molecule, such that the top of the BTN3A dimer is accessible for other receptor binding. In contrast, the positioning of the 103.2 scFv on BTN3A would be consistent with a 1:1 stoichiometry of full-length antibody to BTN3A dimer, because modeling of Fab onto the scFv of our complex model positions the C terminus of each Fab within a reasonable distance of ∼70 Å to be connected by one Fc molecule (supplemental Fig. S3, right panel). Therefore, our model suggests a bivalent association of the agonist 20.1 antibody with two BTN3A dimers, leading to multimerization of BTN3A in the context of membrane-bound BTN3A on the cell surface. In contrast, our model suggests that the antagonist 103.2 antibody can bind BTN3A dimers either monovalently or bivalently.
DISCUSSION
The molecular basis by which PiP stimulates Vγ9Vδ2 T cells has puzzled immunologists for decades; the recent discovery of antibodies that modulate Vγ9Vδ2 activity in vitro has been the first significant step forward in defining the molecular components involved in this activation process. We have focused our molecular studies on structural determination of their targets: the BTN3A molecules, and provide the first structures of this subclass of the B7 superfamily. Furthermore we determined the molecular basis of how the agonist (20.1) and antagonist (103.2) antibodies interact with the BTN3A extracellular domains. Our crystal structures of the BTN3A isoforms reveal remarkable homology to myelin-oligodendrocyte glycoprotein and PD-L1, and comparisons within the BTN3A isoforms also demonstrate that these molecules have intrinsic flexibility between their membrane proximal Ig-C domains and their N-terminal Ig-V domains. However, these structures alone do not explain how these molecules could be involved in PiP-mediated Vγ9Vδ2 activation. Key to this characterization has been the two BTN3A specific antibodies that either mimic PiP-induced activation (20.1) or antagonize this effect (103.2) (13). The antigen-independent, antibody-mediated stimulation of Vγ9Vδ2 via the BTN3A molecules argues against up-regulation of BTN3A expression as the primary mechanism for Vγ9Vδ2 activation. Instead, our current model focuses on modification of the extracellular domains by the 20.1 antibody in such a way that mimics PiP activation as the switch from nonstimulatory BTN3A to that of stimulatory.
Our structural and biophysical data establish that the extracellular domains of BTN3A exist as dimers in solution and are thus likely preformed dimers on the cell surface. Two dimer forms were noted in our structures; both have appreciable BSAs to their interfaces that suggested both could exist in solution; however, our FRET data performed on our soluble extracellular domains of BTN3A in solution strongly favor the Dimer 1 conformation. Our structural data of the complexes between the 20.1 and 103.2 antibodies address how binding of antibody leads to changes in the organization of BTN3A molecules on the cell surface. Not only do the antibodies bind with different affinities (20.1 binds more weakly than 103.2), but their valency of binding likely differs as well. 20.1 appears capable of only cross-linking BTN3A dimers on the cell surface, whereas 103.2 can both cross-link and bind monovalently to a BTN3A dimer. It is likely that these structural and biophysical differences contribute to the different functional outputs of these two antibodies. Clues to the importance of the multimerization ability of these antibodies lie in the decreased potency of our scFv versions when used in functional assays. That the 103.2 scFv does not inhibit function in the monovalent, scFv state, suggests that it does not sterically block association of proteins that normally engage BTN3A during activation or that the bulk of a full-length antibody is required for steric hindrance.
How might this model be applied to PiP-mediated activation? BTN3A1 is the only isoform of the three BTN3A molecules that promotes Vγ9Vδ2 activation upon the addition of PiP. Removal of the BTN3A1 B30.2 intracellular domain of BTN3A1 inhibits PiP-mediated activation yet does not effect 20.1 Ab-mediated activation (13). Therefore, we speculate that the B30.2 intracellular domain plays a direct role in PiP-mediated activation and that addition of the 20.1 Ab modifies the extracellular domains of BTN3A molecules, mimicking the effect of PiP. From our crystal structures, MALS and FRET data, we propose that Dimer 1 is the primary dimer state on the cell surface (although we cannot rule out that Dimer 2 can exist). Dimer 1 would position the two homodimeric B30.2 intracellular domains in close proximity; therefore modifications to these intracellular domains may cause shifts that translate out to the extracellular domains. We observed a shift in the Dimer 1 interface in the complex structure with the agonist 20.1 scFv, demonstrating the rotational flexibility of this interface. We therefore propose a model whereby high concentrations of PiP can induce modification of the B30.2 domains, either through direct binding or recruitment of adaptor molecules, and thus translate a rotational shift to the extracellular domains. This “activated” Dimer 1 conformation is mimicked by a conformation induced and/or stabilized by the 20.1 antibody. “Inside-out” signaling such as this is seen in other receptor/ligand systems such as integrin signaling, whereby modification of the intracellular domains of integrins causes significant conformation changes in the extracellular domains that transforms a “low affinity” integrin to “high affinity” for ligand binding (29). Therefore our molecular studies provide a model by which to understand how antibody modification of the BTN3A extracellular domains may induce Vγ9Vδ2 TCR engagement and to further study the role of the B30.2 intracellular domain in PiP-mediated activation.
Supplementary Material
Acknowledgments
We thank the staff of the Advanced Proton Source at GM/CA-CAT (Beamline 23ID) and LS-CAT (Beamline21ID) for use of and assistance with x-ray beamlines and Ruslan Sanishvili and Joseph Brunzelle in particular for help and advice during data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI073922. This work was also supported by and the Kinship Foundation Searle Scholars Award (to E. J. A.).

This article contains supplemental Table S1 and S2 and Figs. S1–S4.
- TCR
- T cell receptor
- BTN3A
- butyrophilin-3
- PiP
- phosphoisoprenoid
- RMSD
- root mean square deviation
- CDR
- complementary determining region
- MALS
- multiangle light scattering
- PD-L1
- programmed cell death 1 ligand 1
- BSA
- buried surface area.
REFERENCES
- 1. Bonneville M., O'Brien R. L., Born W. K. (2010) γδ T cell effector functions. A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 [DOI] [PubMed] [Google Scholar]
- 2. Hayday A. C. (2009) γδ T cells and the lymphoid stress-surveillance response. Immunity 31, 184–196 [DOI] [PubMed] [Google Scholar]
- 3. Bonneville M., Scotet E. (2006) Human Vγ9Vδ2 T cells. Promising new leads for immunotherapy of infections and tumors. Curr. Opin. Immunol 18, 539–546 [DOI] [PubMed] [Google Scholar]
- 4. Morita C. T., Jin C., Sarikonda G., Wang H. (2007) Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vγ2Vδ2 T cells. Discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol. Rev. 215, 59–76 [DOI] [PubMed] [Google Scholar]
- 5. Kabelitz D. (2010) Human γδ T lymphocytes for immunotherapeutic strategies against cancer. F1000 Med. Rep. 2, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bukowski J. F., Morita C. T., Tanaka Y., Bloom B. R., Brenner M. B., Band H. (1995) Vγ2Vδ2 TCR-dependent recognition of non-peptide antigens and Daudi cells analyzed by TCR gene transfer. J. Immunol. 154, 998–1006 [PubMed] [Google Scholar]
- 7. Wang H., Fang Z., Morita C. T. (2010) Vγ2Vδ2 T cell receptor recognition of prenyl pyrophosphates is dependent on all CDRs. J. Immunol. 184, 6209–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allison T. J., Winter C. C., Fournié J. J., Bonneville M., Garboczi D. N. (2001) Structure of a human γδ T-cell antigen receptor. Nature 411, 820–824 [DOI] [PubMed] [Google Scholar]
- 9. Morita C. T., Beckman E. M., Bukowski J. F., Tanaka Y., Band H., Bloom B. R., Golan D. E., Brenner M. B. (1995) Direct presentation of nonpeptide prenyl pyrophosphate antigens to human γδ T cells. Immunity 3, 495–507 [DOI] [PubMed] [Google Scholar]
- 10. Wei H., Huang D., Lai X., Chen M., Zhong W., Wang R., Chen Z. W. (2008) Definition of APC presentation of phosphoantigen (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate to Vγ2Vδ2 TCR. J. Immunol. 181, 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lang F., Peyrat M. A., Constant P., Davodeau F., David-Ameline J., Poquet Y., Vié H., Fournié J. J., Bonneville M. (1995) Early activation of human Vγ9Vδ2 T cell broad cytotoxicity and TNF production by nonpeptidic mycobacterial ligands. J. Immunol. 154, 5986–5994 [PubMed] [Google Scholar]
- 12. Wang H., Lee H. K., Bukowski J. F., Li H., Mariuzza R. A., Chen Z. W., Nam K. H., Morita C. T. (2003) Conservation of nonpeptide antigen recognition by rhesus monkey Vγ2Vδ2 T cells. J. Immunol. 170, 3696–3706 [DOI] [PubMed] [Google Scholar]
- 13. Harly C., Guillaume Y., Nedellec S., Mönkkönen H., Mönkkönen J., Kuball J., Adams E. J., Netzer S., Déchanet-Merville J., Léger A., Breathnach R., Olive D., Bonneville M., Scotet E. (2012) Key implication of CD277/Butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T cell subset. Blood, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greenwald R. J., Freeman G. J., Sharpe A. H. (2005) The B7 family revisited. Annu. Rev. Immunol. 23, 515–548 [DOI] [PubMed] [Google Scholar]
- 15. Compte E., Pontarotti P., Collette Y., Lopez M., Olive D. (2004) Frontline. Characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur. J. Immunol. 34, 2089–2099 [DOI] [PubMed] [Google Scholar]
- 16. Arnett H. A., Escobar S. S., Viney J. L. (2009) Regulation of costimulation in the era of butyrophilins. Cytokine 46, 370–375 [DOI] [PubMed] [Google Scholar]
- 17. Yamashiro H., Yoshizaki S., Tadaki T., Egawa K., Seo N. (2010) Stimulation of human butyrophilin 3 molecules results in negative regulation of cellular immunity. J. Leukocyte Biol. 88, 757–767 [DOI] [PubMed] [Google Scholar]
- 18. Yamazaki T., Goya I., Graf D., Craig S., Martin-Orozco N., Dong C. (2010) A butyrophilin family member critically inhibits T cell activation. J. Immunol. 185, 5907–5914 [DOI] [PubMed] [Google Scholar]
- 19. Smith I. A., Knezevic B. R., Ammann J. U., Rhodes D. A., Aw D., Palmer D. B., Mather I. H., Trowsdale J. (2010) BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J. Immunol. 184, 3514–3525 [DOI] [PubMed] [Google Scholar]
- 20. Ruddy D. A., Kronmal G. S., Lee V. K., Mintier G. A., Quintana L., Domingo R., Jr., Meyer N. C., Irrinki A., McClelland E. E., Fullan A., Mapa F. A., Moore T., Thomas W., Loeb D. B., Harmon C., Tsuchihashi Z., Wolff R. K., Schatzman R. C., Feder J. N. (1997) A 1.1-Mb transcript map of the hereditary hemochromatosis locus. Genome Res. 7, 441–456 [DOI] [PubMed] [Google Scholar]
- 21. Tazi-Ahnini R., Henry J., Offer C., Bouissou-Bouchouata C., Mather I. H., Pontarotti P. (1997) Cloning, localization, and structure of new members of the butyrophilin gene family in the juxta-telomeric region of the major histocompatibility complex. Immunogenetics 47, 55–63 [DOI] [PubMed] [Google Scholar]
- 22. Rhodes D. A., Stammers M., Malcherek G., Beck S., Trowsdale J. (2001) The cluster of BTN genes in the extended major histocompatibility complex. Genomics 71, 351–362 [DOI] [PubMed] [Google Scholar]
- 23. Ikemizu S., Gilbert R. J., Fennelly J. A., Collins A. V., Harlos K., Jones E. Y., Stuart D. I., Davis S. J. (2000) Structure and dimerization of a soluble form of B7-1. Immunity 12, 51–60 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X., Schwartz J. C., Almo S. C., Nathenson S. G. (2003) Crystal structure of the receptor-binding domain of human B7-2. Insights into organization and signaling. Proc. Natl. Acad. Sci. U.S.A. 100, 2586–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y., Liu P., Gao F., Cheng H., Qi J., Gao G. F. (2010) A dimeric structure of PD-L1. Functional units or evolutionary relics? Protein Cell 1, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin D. Y., Tanaka Y., Iwasaki M., Gittis A. G., Su H. P., Mikami B., Okazaki T., Honjo T., Minato N., Garboczi D. N. (2008) The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clements C. S., Reid H. H., Beddoe T., Tynan F. E., Perugini M. A., Johns T. G., Bernard C. C., Rossjohn J. (2003) The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 100, 11059–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clegg R. M. (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211, 353–388 [DOI] [PubMed] [Google Scholar]
- 29. Luo B. H., Carman C. V., Springer T. A. (2007) Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. Holden H. M., Ito M., Hartshorne D. J., Rayment I. (1992) X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution. J. Mol. Biol. 227, 840–851 [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. DeBartolo J., Colubri A., Jha A. K., Fitzgerald J. E., Freed K. F., Sosnick T. R. (2009) Mimicking the folding pathway to improve homology-free protein structure prediction. Proc. Natl. Acad. Sci. U.S.A. 106, 3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haddadian E. J., Gong H., Jha A. K., Yang X., Debartolo J., Hinshaw J. R., Rice P. A., Sosnick T. R., Freed K. F. (2011) Automated real-space refinement of protein structures using a realistic backbone move set. Biophys. J. 101, 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugita M., Moody D. B., Jackman R. M., Grant E. P., Rosat J. P., Behar S. M., Peters P. J., Porcelli S. A., Brenner M. B. (1998) CD1. A new paradigm for antigen presentation and T cell activation. Clin. Immunol. Immunopathol. 87, 8–14 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z., Raifu M., Howard M., Smith L., Hansen D., Goldsby R., Ratner D. (2000) Universal PCR amplification of mouse immunoglobulin gene variable regions. The design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J. Immunol. Methods 233, 167–177 [DOI] [PubMed] [Google Scholar]
- 39. Maynard J., Adams E. J., Krogsgaard M., Petersson K., Liu C. W., Garcia K. C. (2005) High level bacterial secretion of single-chain αβ T-cell receptors. J. Immunol. Methods 306, 51–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.