Background: Elongator is an acetylase complex that regulates cell migration.
Results: DERP6 (ELP5) and C3ORF75 (ELP6) are characterized as Elongator subunits that control cell motility and tumorigenicity of melanoma cells. ELP5 ensures Elongator integrity by connecting ELP3 to ELP4.
Conclusion: ELP5 and ELP6 are new players for migration and tumorigenicity of transformed cells.
Significance: Elongator may be involved in both tumor initiation and progression.
Keywords: Cancer, Cell Invasion, Cell Migration, Histone Acetylase, Melanoma, Elongator, Tumorigenesis
Abstract
The Elongator complex is composed of 6 subunits (Elp1-Elp6) and promotes RNAPII transcript elongation through histone acetylation in the nucleus as well as tRNA modification in the cytoplasm. This acetyltransferase complex directly or indirectly regulates numerous biological processes ranging from exocytosis and resistance to heat shock in yeast to cell migration and neuronal differentiation in higher eukaryotes. The identity of human ELP1 through ELP4 has been reported but human ELP5 and ELP6 have remained uncharacterized. Here, we report that DERP6 (ELP5) and C3ORF75 (ELP6) encode these subunits of human Elongator. We further investigated the importance and function of these two subunits by a combination of biochemical analysis and cellular assays. Our results show that DERP6/ELP5 is required for the integrity of Elongator and directly connects ELP3 to ELP4. Importantly, the migration and tumorigenicity of melanoma-derived cells are significantly decreased upon Elongator depletion through ELP1 or ELP3. Strikingly, DERP6/ELP5 and C3ORF75/ELP6-depleted melanoma cells have similar defects, further supporting the idea that DERP6/ELP5 and C3ORF75/ELP6 are essential for Elongator function. Together, our data identify DERP6/ELP5 and C3ORF75/ELP6 as key players for migration, invasion and tumorigenicity of melanoma cells, as integral subunits of Elongator.
Introduction
The Elongator complex (Elp1-Elp6) was initially identified as a component of a hyper-phosphorylated RNA polymerase II (RNAPII)5 holoenzyme isolated from budding yeast chromatin and subsequently from human cells (1–4). Elp3, the catalytic subunit, harbors motifs found in the GNAT family of histone acetyltransferases (HATs) (5) and is essential for the ability of Elongator to acetylate histone H3, and to a lesser extent H4, in vitro (2, 5, 6). As a result, yeast elp3 mutation causes decreased histone H3 acetylation levels in chromatin in vivo (6, 7). These data, combined with studies describing the association of Elongator with nascent RNA emanating from elongating RNAPII along the coding region of several yeast genes (8) and with the preferential recruitment of Elongator to the transcribed regions of human genes (9–11), support a role for this complex in transcriptional elongation.
A substantial fraction of Elongator is cytoplasmic (2, 3, 13), raising the possibility that this complex performs additional molecular functions in the cell (14). In this context, genetic data in yeast demonstrated that Elongator is required for the presence of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups on uridines at the wobble position of some tRNAs (15–17). Although it is still unclear how Elongator precisely acts in the early step of the enzymatic cascade that ultimately generates these correctly modified tRNAs, this function is conserved, at least in Saccharomyces cerevisiae, Caenorhabditis elegans as well as in Arabidopsis thaliana (18, 19). Therefore, Elongator may act as a multitasking complex that regulates both transcriptional elongation through histone acetylation in the nucleus as well as translational fidelity through tRNA modifications in the cytoplasm (20). These data undoubtedly demonstrated that Elongator may not only acetylate nuclear histones but also additional and still poorly characterized substrates in the cytoplasm. In agreement with this hypothesis, recent studies indicated that Elongator promotes the acetylation of α-tubulin in both mice and C. elegans (21, 22) as well as of the ELKS family member Bruchpilot in Drosophila neurons (23).
Loss of function models for Elongator in distinct organisms revealed a variety of cellular processes that rely on this acetylase complex. Indeed, while Elongator yeast mutants (elp phenotypes) have slower growth adaptation, temperature sensitivity at 39 °C as well as exocytosis, telomeric gene silencing and DNA damage response defects (5, 24–26), Elongata mutants of A. thaliana showed impaired root growth due to decreased cell division rate (27). Moreover, deletion of Elp3 in Drosophila melanogaster resulted in larval lethality, at least because of an aberrant expression of stress response genes (28). Elongator deficiency is also lethal in mice (29) while migration and differentiation defects were observed in Elp3-depleted cortical neurons during embryogenesis (21). The consequence of impaired Elongator function in humans is exemplified by familial dysautonomia (FD), an autosomal recessive disease characterized by defects in the development and maintenance of autonomic and sensory system neurons (30, 31). FD is caused by a mutation in a splice site of the IKBKAP gene, which ultimately leads to decreased expression of ELP1, the scaffold protein that assembles Elongator, in a tissue-specific manner (32, 33). Interestingly, some ELP3 variants are associated with amyotrophic lateral sclerosis, a progressive motor neuron disease (34), thus strongly suggesting that the Elongator-dependent pathways may be deregulated in distinct neurological disorders (35).
We and others (11, 36–38) have reported that Elongator critically regulates cell migration, independently of the cell type studied. Indeed, primary or transformed cells depleted for Elongator systematically showed cell motility defects, suggesting that Elongator-dependent protein acetylation plays an important role in cell migration (11, 21). It is however still unclear whether and how other Elongator subunits beside ELP1 and ELP3 contribute to this process.
We report here the characterization of dermal papilla-derived protein 6 (DERP6) and C3ORF75 as human homologues of yeast Elp5 and Elp6, respectively. Moreover, we show that DERP6/ELP5 is essential for Elongator integrity as it connects ELP4 to ELP3. Importantly, ELP5- or ELP6-depleted melanoma-derived cells do not migrate properly and fail to efficiently generate colonies in soft agar, similar to what is observed in ELP1- and ELP3-depleted cells. Wild type DERP6/ELP5, but not a mutant that does not bind ELP1 or ELP3, restores motility in DERP6/ELP5-deficient cells. Taken together, our data identify the genes encoding ELP5 and ELP6 in human cells, define Elongator as a key actor for the tumorigenic potential and motility of melanoma cells, and provide further mechanistic insights into the function of DERP6/ELP5.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
ORFs encoding human ELP4 and DERP6/ELP5 were cloned into pIRESpuro (Clontech, Palo Alto, CA) with a FLAG tag at the C terminus. The pCMV-Myc ELP3, FLAG-ELP3, and FLAG-ELP1 expression constructs were previously described (21). DERP6/ELP5 mutants were generated by PCR and subcloned into the pcDNA3.1 expression vector (Invitrogen). Full-length DERP6/ELP5 was also subcloned into the pCMV-Myc expression construct (Clontech). Antibodies used were mouse anti-FLAG, mouse anti-α-tubulin (Sigma Aldrich), rabbit anti-Myc (Santa Cruz Biotechnologies), and rabbit anti-Histone H3 (Abcam, Cambridge, UK). The rabbit anti-Elp1, -Elp3, -Elp4, -Elp5, and Elp6 antibodies were previously described (39).
Cell Culture, Stable Cell Line Establishment
HEK293 cells were cultured as previously described (40). The mouse melanoma-derived B16-F10-luc-G5 Bioware cells (Caliper LifeSciences, Hopkinton, MA) were maintained in DMEM supplemented with 10% fetal bovine serum, 1% antibiotics, 1% l-glutamine, and G418. To generate HEK293 stably expressing ELP4- or DERP6/ELP5-FLAG proteins, cells were transfected with the relevant pIRESpuro construct and selected in 1 μg/ml puromycin (Sigma Aldrich). Cells were maintained in selecting media for 3 weeks and surviving cells used for experiment after transgene expression was checked.
Subcellular Fractionation
HEK293 cells were divided into cytoplasmic and nuclear fractions as previously described (41). Briefly, cells were trypsinized, washed in PBS, and lysed in cytoplasmic lysis buffer (10 mm Tris-HCl, pH 7.9, 340 mm sucrose, 3 mm CaCl2, 2 mm Mg(OAc)2, 0.1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, protease inhibitors). Nuclei were pelleted by centrifugation at 3,500 × g for 15 min, washed in cytoplasmic lysis without Nonidet P-40 and lysed in nuclear lysis buffer (20 mm Hepes pH 7.9, 250 mm KOAc, 1% SDS, 1 mm DTT, protease inhibitors). For ease of comparison, equal proportion of cytoplasmic or nuclear fractions was loaded on SDS-page gel.
Lentiviral Cell Infections
Control shRNA as well as shRNA Elp1, -Elp3, -Elp5, and -Elp6 lentiviral constructs were purchased from Sigma Aldrich. The corresponding sequences are listed in the supplemental Table S1. Lenti-X 293T cells (Clontech) were transfected with VSV-G, gag, and pol expressing constructs as well as with the vector (pLKO.1-puro) containing the shRNA sequence of interest. Infectious supernatants were collected 48–52 h post-transfection, and cleared by centrifugation. Polybrene was added (5 μg/ml), and the cleared supernatants were used to transduce B16-F10 or HEK293 cells. Infected cells were maintained in puromycin-containing media to produce stable knock down cells. The efficiency of the RNA interference was validated by either Western blot or qRT-PCR analysis.
Purification and Identification of Human Elongator Complex from Cytoplasmic Fractions
108 cells stably expressing ELP4-FLAG were lysed with cytoplasmic lysis buffer (10 mm Tris-HCl, pH 7.9, 340 mm sucrose, 3 mm CaCl2, 2 mm Mg(OAc)2, 0.1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, protease inhibitors). Nuclei were then pelleted by centrifugation at 3,500 × g for 15 min and discarded. The cytoplasmic fraction was further cleared by centrifugation at 20,000 × g for 30 min, and the supernatant was collected. For negative control purification, the same extracts were prepared from the same amount of untagged cells. The sample was then applied to M2-agarose beads (Sigma Aldrich) and incubated for 4 h at 4 °C. After binding, beads were washed extensively with washing buffer (20 mm Hepes pH 7.9, 250 mm KOAc, 1% Triton X-100, 10% glycerol, 3 mm EDTA, 1 mm DTT, protease inhibitors). Finally, proteins were eluted by using FLAG elution buffer (20 mm Hepes pH 7.9, 100 mm KOAc, 3 mm EDTA, 1 mm DTT, 200 μg/ml 3xFLAG peptide, protease inhibitors). Eluates were resolved by 4–12% bis-Tris gradient SDS-PAGE and analyzed by Sypro Ruby staining (Invitrogen). In-gel digestion of each band was then performed by addition of modified trypsin (Promega, Madison, WI) in 50 mm ammonium bicarbonate at 37 °C overnight and further analyzed by LC-Chip nano high performance liquid chromatography (HPLC) electrospray MS-MS using an XCT iontrap mass spectrometer (Agilent, Santa Clara, CA). The HPLC separations were performed on an RP C18 Zorbax column from Agilent. The mobile phase was a 60 min gradient mixture formed as follows: mixture A, water-acetonitrile-formic acid (97/3/0.1 [v/v/vol]); mixture B, acetonitrile-water-formic acid (90/10/0.1 [v/v/vol]). The flow rate was fixed at 300 nL/min. The collision energy was set automatically depending on the mass of the parent ion. Each MS full scan was followed by MS-MS scans of the first four most intense peaks detected in the prior MS scan. A list of peptide masses was subsequently introduced into the database for protein identification searches using MASCOT (Matrix Sciences). For size exclusion chromatography, samples were loaded on superdex 200 in buffer A (20 mm Hepes-KOH, 0.01% Nonidet P-40, 10% glycerol, 250 mm KOAc). 250 μl fractions were collected, and sizes were estimated by running protein size markers (Bio-Rad) in parallel.
Immunoprecipitations and Immunofluorescences
Immunoprecipitations involving ectopically expressed proteins were conducted as previously described (40). For immunostainings, HEK293 cells were washed twice in PBS, fixed in 4% PFA for 20 min at room temperature and washed three times in washing buffer (PBS, 0.1% Tween). Cells were subsequently permeabilized in PBS supplemented with 0.5% Triton X-100 for 15 min, incubated in a blocking buffer containing PBS, 0.3% Triton X-100 and 5% Donkey Serum for 30 min at room temperature, and then incubated with primary antibodies for 1 h. After three washes in the blocking buffer, cells were incubated with Alexa546- (Invitrogen) or FITC- (Jackson Immunoresearch Laboratories, West Grove, PA) conjugated secondary antibodies in the same buffer for 1 h at room temperature and subsequently washed three times. Cells were counterstained with To-Pro (Invitrogen), washed once and mounted in ProLong Gold antifade reagent (Invitrogen) between slide and coverslip. Images were acquired with a Leica TCS SP2 confocal (objective 63× with oil immersion).
Wound Healing, Soft Agar Colony, and Three-dimensional Cell Culture Assays
Wound healing assays were conducted as previously described (11). Briefly, 3 × 105 B16-F10 cells expressing shCTR, shElp1, -3, -5, or -6 were seeded in 24-well plates in triplicates. The next day, cells were treated with Mitomycin C (1 μg/ml) for 2 h before performing a linear scratch in the confluent cell monolayer. For each well, one picture was taken at time 0 (just after the scratch) and after 6 or 7 h. Cells were imaged using a Nikon Eclipse TS100 phase-contrast microscope equipped with a 10× objective. For soft agar assays, 2 × 103 B16-F10 cells expressing shCTRL, shElp1,-3, -5, or -6 lentiviral constructs were plated in 6-well plates in triplicates in culture media containing 0.4% agar on top of media containing 0.8% agar. Cells were incubated for 2 weeks and culture media was replaced every 2–3 days. At the end of the experiment, macroscopic colonies were stained with crystal violet 0.005% and scored with the ImageJ sofware. Three-dimensional cell culture assays were carried out mainly as described (42). Briefly, type I collagen from rat tail (a generous gift from A. Colige, Laboratory of Connective Tissues Biology, GIGA Cancer, University of Liege, Belgium) was diluted to a concentration of 2 mg/ml, and the pH was neutralized by adding 1:3 volume of DMEM. 800 μl aliquots were immediately added to 12-well culture plates and incubated at 37 °C until gelation occurred. A drop of collagen containing 2 × 105 control or Elp1-depleted cells was added on top of solidified collagen. After 30 min incubation at 37 °C, overlying collagen gels were generated. Wells were then filled with culture media and incubated for 3 weeks at 37 °C. The culture media was replaced every 2–3 days. Satellites colonies were stained using a solution of 0.2% methylene blue in 50% methanol and imaged with a Canon camera.
Statistical Analysis
Comparison between 2 groups was carried out by doing Student t-tests. Significance was reached when p < 0.1 *, p < 0.05 **, p < 0.01 ***.
RESULTS
Identification of DERP6 and C3ORF75 as Elongator Subunits
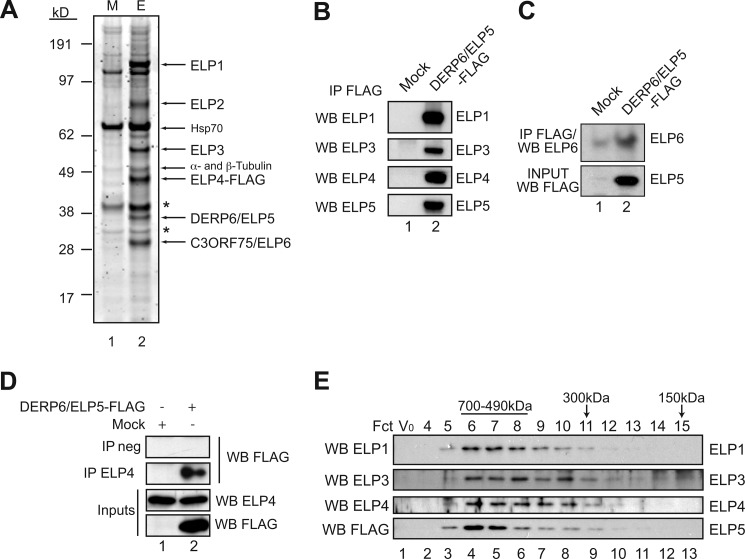
Human Elongator was previously purified from HeLa cells, which led to the identification of ELP1, ELP2, and ELP3 as subunits of the so-called “core” complex (2, 3). Elongator also exists as a larger six-subunit complex referred to as the “holo-complex” that has HAT activity and includes ELP4 as well as two uncharacterized polypeptides, namely p38 (ELP5) and p30 (ELP6) (3). These products are likely to be encoded by one of the several homologues of yeast elp5 and elp6 in human cells, but their identity has not been reported. To identify the p38 and p30 proteins, we generated HEK293 cells stably expressing FLAG-tagged ELP4, and used it as a bait to purify human Elongator (Fig. 1A). Eluted proteins were subjected to SDS-PAGE analysis and bands were identified by mass spectrometry (MS) (see supplemental Figs. S1 and S2) (43). They included ELP1, ELP2, ELP3, ELP4 as well as two products, namely DERP6 and C3ORF75, whose apparent molecular weights (38 and 30 kDa, respectively), matched that of the uncharacterized subunits of human Elongator (Fig. 1A, lane 2) (3). Sequence alignment confirmed that DERP6 and C3ORF75 correspond to human homologues of yeast Elp5 and Elp6, respectively (supplemental Figs. S3 and S4).
FIGURE 1.
Identification of DERP6 and C3ORF75 as Elongator subunits. A, Elongator was purified from the cytoplasmic fraction of ELP4-FLAG expressing cells. Equal amount of the M2 chromatography eluates from Mock (M) and ELP4-FLAG (E) were separated by 4–12% SDS-PAGE and stained with Sypro Ruby. Bands were extracted and analyzed by mass spectrometry. Arrows indicate the co-eluting Elongator subunits as well as Hsp70, α- and β-tubulin (* shows nonspecific bands). B, Western blot analysis of M2 eluates from control (Mock) or DERP6/ELP5-FLAG expressing cells. Proteins were detected with antibodies as shown on the left. C, Anti-ELP6 Western blot analysis on anti-FLAG immunoprecipitates from control or DERP6/ELP5-FLAG-expressing cells (top panel). At the bottom, anti-ELP5 Western blot performed on the crude cell extracts (input). D, endogenous ELP4 was immunoprecipitated from Mock or DERP6/ELP5-FLAG expressing cells and proteins detected with antibodies listed on the right. E, M2-purified Elongator complex from B was analyzed by size exclusion chromatography. Vo is the void volume fraction. Proteins were detected by Western blotting using the antibodies indicated on the left.
To further substantiate the notion that DERP6 and C3ORF75 are part of the human Elongator complex, we established a stable cell line expressing FLAG-tagged DERP6/ELP5 and applied the same purification protocol. DERP6/ELP5-FLAG co-immunoprecipitated endogenous ELP1, ELP3, ELP4 as well as C3ORF75/ELP6 (Fig. 1, B and C). Endogenous ELP4 immunoprecipitation also brought down DERP6/ELP5-FLAG (Fig. 1D). Moreover, ELP1, ELP3, ELP4 and DERP6/ELP5-FLAG co-eluted as a 600 kDa complex on gel filtration analysis, as previously described (1, 45) (Fig. 1E). We next performed immunofluorescence analysis to determine DERP6/ELP5 subcellular localization. Ectopically expressed ELP1 and ELP3 were mainly, but not exclusively, located in the cytoplasm, as previously described (Fig. 2A) (3, 21, 36, 46). ELP4 and DERP6/ELP5 were also mainly found in this cell compartment, showing that holo-Elongator subunits colocalize. In agreement with our immunofluorescence data, Western blot analysis performed on cytoplasmic versus nuclear fractions of HEK293 cells indicated that DERP6/ELP5-FLAG was mostly found in the cytoplasm (Fig. 2B, top panel, lane 1). About 14% of this protein was nevertheless detected in the nucleus of those cells (Fig. 2B, top panel, lane 2). Taken together, these data indicate that DERP6 and C3ORF75 are part of the Elongator complex, as human homologues of yeast Elp5 and Elp6. We therefore renamed these two proteins ELP5 and ELP6.
FIGURE 2.
ELP1, ELP3, ELP4, and ELP5 are mainly cytoplasmic. A, HEK293 cells were transfected with the indicated expression plasmids and immunofluorescence analysis were conducted using anti-Myc or -FLAG antibodies (left column). To-Pro (blue) was used to visualize the nuclei (middle and right columns). B, DERP6/ELP5-FLAG was transfected in HEK293 cells and cytoplasmic versus nuclear extracts (C and N, respectively) were isolated from the resulting cells. Those extracts were subjected to anti-FLAG, -α-tubulin, and -Histone H3 (cytoplasmic and nuclear markers, respectively), as indicated. A quantification of the cellular distribution of DERP6/ELP5 is illustrated as well.
Elongator-deficient Melanoma Cells Have Motility Defects
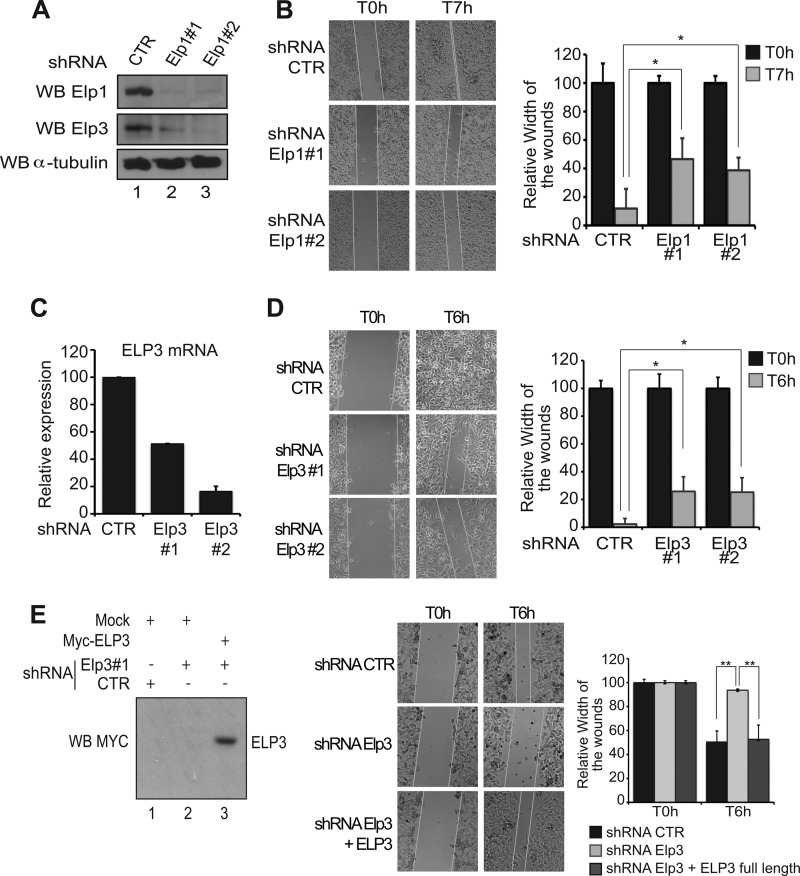
We previously reported that Elongator-depleted HeLa cells display migration defects, at least partly due to a failure to properly express genes coding for proteins involved in cell motility (11). To explore whether Elongator also regulates cell migration in melanomas, we first generated B16-F10 cells deficient for Elp1 through lentivirus based RNA interference using two distinct sequences targeting the Elp1 transcript, or a control sequence (“shRNA Elp1#1”, shRNA Elp1#2” and “shRNA control (CTR)”, respectively). Elp1 was efficiently depleted, as judged by Western blot analysis (Fig. 3A, top panel), which led to the destabilization of the Elp3 protein, as previously shown (11, 39) (Fig. 3A, middle panel). The migrating capacity of these cells was assessed by wound healing assay. Significant closure of the gaps in cell monolayers had occurred after 7 h in control B16-F10 cells, whereas a clear delay was observed in Elp1-depleted cells, independently of the sequence used to target the Elp1 transcript (Fig. 3B). We next generated Elp3-depleted B16-F10 cells using the same experimental approach (Fig. 3C) and subjected them to wound healing assays. Elp3 deficiency in B16-F10 cells caused a significant delay in wound healing as well (Fig. 3D). Importantly, ELP3 expression in these cells restored their migration potential (Fig. 3E). These data suggest that both Elp1 and Elp3 Elongator subunits control cell migration in melanoma-derived cells.
FIGURE 3.
Elp1 and Elp3 regulate cell motility of melanoma-derived B16-F10 cells. A, generation and characterization of Elp1-deficient B16-F10 cells. Anti-Elp1, -Elp3, and -α-tubulin (loading control) Western blot analysis were carried out using cell extracts from B16-F10 infected with lentiviral constructs delivering small hairpin RNAs targeting two distinct sequences of the Elp1 transcript, or a control sequence as a negative control (“shRNA Elp1#1,” “shRNA Elp1#2,” and “shRNA control (CTR),” respectively). B, migration of control (CTR) or Elp1-depleted (Elp1#1 or Elp1#2) melanoma-derived cells was measured by wound healing assay. Pictures were taken at the indicated times after the wound. A quantification of the data obtained is illustrated on the right. For each experimental condition, the width of the wound was set to 100% at time 0 and the width in other time points expressed relative to that. The figure shows the data from a representative experiment performed in triplicates (mean values + S.D.). C, generation and characterization of Elp3-depleted melanoma-derived cells. mRNA levels from B16-F10 cells infected with lentiviral constructs delivering small hairpin RNAs targeting two distinct sequences of the Elp3 transcript, or a control sequence (shRNA Elp3#1, shRNA Elp3#2, and shRNA CTR, respectively), were assessed by qRT-PCR. Elp3 mRNA levels in control B16-F10 cells were set to 100%, and mRNA levels in other experimental conditions are relative to that. The figure shows the data from a representative experiment performed in triplicates (mean values + S.D.). D, same as B, but using Elp3-depleted melanoma-derived generated in C. E, wound healing assays were conducted with shRNA control, shRNA Elp3 B16-F10 cells or with shRNA Elp3 B16-F10 cells transfected with full-length Myc-ELP3. A quantification of the data obtained is illustrated on the right. For each experimental condition, the width of the wound was set to 100% at time 0 and the width in other time points expressed relative to that. The figure shows the data from a representative experiment performed in triplicates (mean values + S.D.). On the left, anti-Myc Western blots were carried out with protein extracts from the indicated cells collected at the end of the wound healing assay. “Mock” denotes experimental conditions in which cells were transfected with a control plasmid.
Elongator Is Essential for Anchorage-independent Growth and Invasive Potential of Melanoma-derived Cells
Wound healing assays are used to assess the migrating capacity of any cell type, but this assay does not address the question of tumorigenesis. To determine whether Elongator regulates this latter process, we subsequently assessed the anchorage-independent growth of control-, Elp1-, or Elp3-depleted B16-F10 cells by counting colonies able to grow in soft agar. This cellular assay is widely used to study tumorigenic potential of cells. Whereas control B16-F10 cells efficiently generated colonies, Elp1 or Elp3 deficiency dramatically impaired the capacity of melanoma-derived cells to growth in an anchorage-independent manner (Fig. 4, A and B, respectively).
FIGURE 4.
Elongator affects tumorigenic potential of melanoma cells. A and B, ability of Elp1 (A) or Elp3 (B)-depleted B16-F10 cells to form colonies in soft agar was examined. The indicated cells were seeded in agar-containing media, as described under “Experimental Procedures,” and pictures were taken 2 weeks after seeding. The number of colonies observed in control B16-F10 cells was set to 100%, and the number of colonies obtained in other experimental conditions expressed relative to that. The figures show the data from a representative experiment performed in triplicates (mean values + S.D.). C, invasion of control or Elp1-depleted cells was evaluated by using a 3D cell culture system (described under “Experimental Procedures”). On the top, pictures were taken 3 weeks after seeding. At the bottom, the number of colonies obtained in control cells was set to 100%, and the number of colonies observed in other experimental conditions expressed relative to that. The figures show the data from a representative experiment performed in triplicates (mean values + S.D.).
To address the issue of tumor invasion and expansion through the extracellular matrix, we used three-dimensional (3D) cell culture systems (47). Briefly, this experimental approach is based on the growth of invasive cancer cells in collagen matrices, and the production of satellite colonies generated from the primary cancer cells. Consequently, key steps of the metastatic process are recapitulated including cell motility and invasion, expansion through a collagen matrix as well as cell survival at distant sites (42). We investigated to which extent satellite colony formation of melanoma-derived cells required Elongator. As expected, control B16-F10 cells generated multiple satellite colonies, a property that was dramatically affected in Elp1-depleted cells (Fig. 4C). Taken together, our data indicate that Elongator is crucial for transformation as well as for the invasive potential of melanoma-derived cells.
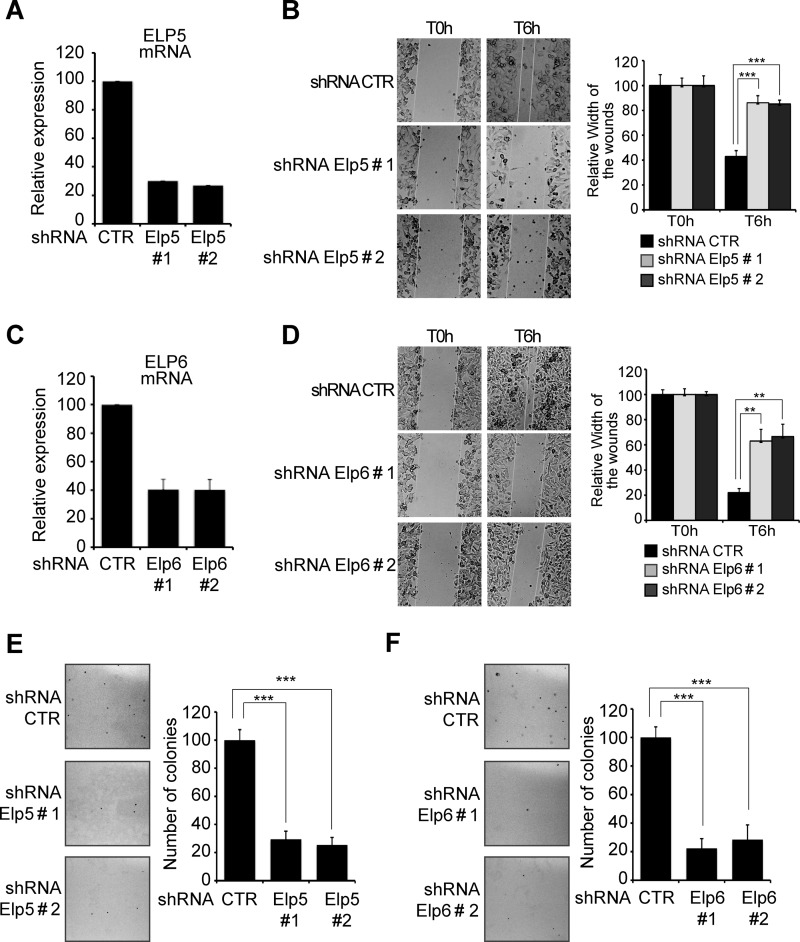
Elp5 and Elp6 Regulate Cell Migration and Tumorigenesis as Elongator Subunits
As ELP5 and ELP6 were identified as Elongator subunits, we next examined whether these proteins also regulate cell migration. Elp5 or Elp6 were efficiently depleted from B16-F10 cells, as judged by the dramatic decrease in their corresponding mRNA levels in cells infected with two distinct shRNA constructs (Fig. 5, A and C). Moreover, Elp5- or Elp6-depleted cells also showed cell motility defects, as evidenced by the significant delay in wound healing observed compared with control cells, independently of the shRNA sequence used to deplete them (Fig. 5, B and D, respectively T6h). Finally, Elp5- or Elp6-deficient B16-F10 cells also showed strong defects in their capacity to form colonies in soft agar (Fig. 5, E and F, respectively).
FIGURE 5.
Elp5 and Elp6 are essential for migration and tumorigenicity of melanoma-derived cells. A and C, Elp5 (A) or Elp6 (C) were depleted in B16-F10 cells by infection with two specific shRNAs (#1 and #2), as judged by qRT-PCR analysis. B and D, migration of control, Elp5 (B) or Elp6 (D)-depleted cells was assessed by wound healing assay. Pictures were taken at the indicated times. The quantification of the data is plotted. For each experimental condition, the width of the wound was set to 100% at time 0 and the width in other time points expressed relative to that. The figure shows the data from a representative experiment performed in triplicates (mean values + S.D.). E and F, number of colonies formed in soft agar was dramatically reduced in Elp5 (E) or Elp6 (F)-deficient versus control B16-F10 cells. The indicated cells were seeded in agar-containing media, as described under “Experimental Procedures.” Pictures were taken 2 weeks after seeding. The number of colonies observed in control B16-F10 cells was set to 100%, and the number of colonies obtained in Elp5 or Elp6-depleted cells expressed relative to that. The figure show the data from a representative experiment performed in triplicates (mean values + S.D.).
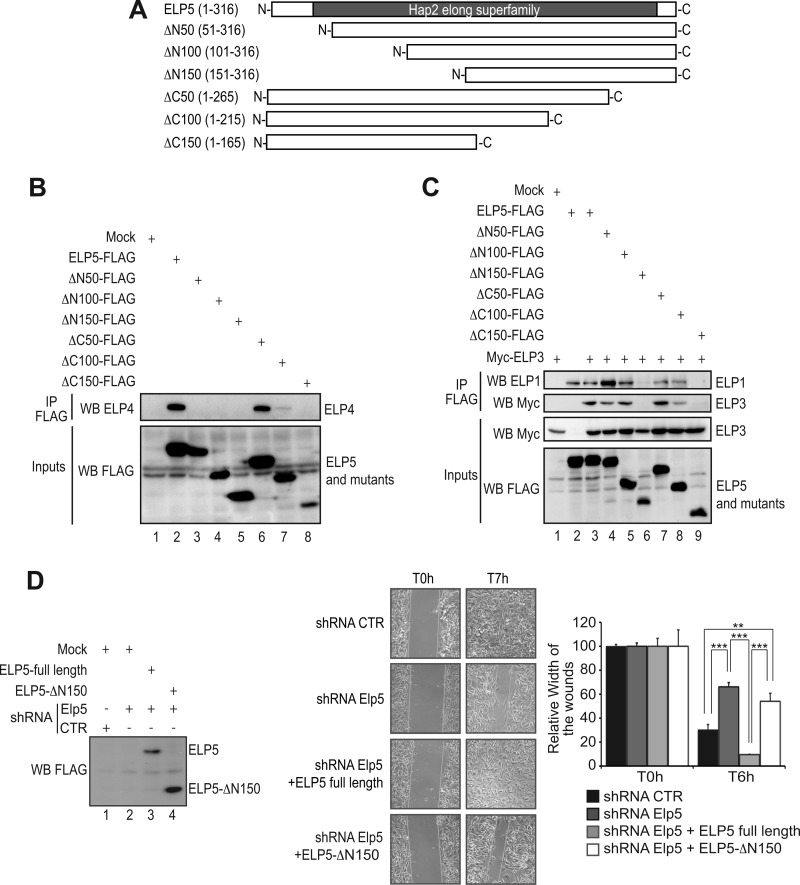
Having established the role of Elp5 and Elp6 in cell migration, we next explored the structure-function relationship of the Elp5 protein. We generated a variety of ELP5 mutants lacking N- or C-terminal amino acids (“ΔN50, ΔN100, ΔN150 as well as ΔC50, ΔC100, and ΔC150 ELP5”, respectively) (Fig. 6A). These mutants were subsequently tested for their ability to bind other Elongator subunits in co-immunoprecipitation experiments. While the ΔC50 mutant remained capable of binding endogenous ELP4, the ΔN50 ELP5 mutant failed to do so, suggesting that the first 50 amino acids are required for the interaction of ELP5 with ELP4 (Fig. 6B, top panel, compare lanes 2, 3, and 6, respectively). The ΔC100 ELP5 mutant also failed to efficiently bind ELP4, which might suggest that ELP5 harbors a second ELP4-interacting domain between amino acids 216 and 265 (Fig. 6B, top panel, compare lanes 2 and 7). Interestingly, despite its inability to bind ELP4, the ΔN50 ELP5 mutant still bound endogenous ELP1, or ectopically expressed ELP3, as did the ΔC100 mutant, suggesting that ELP5 binds ELP4 and ELP3 (or ELP1) through distinct domains (Fig. 6C, top and second panel from the top, lanes 4 and 8). Deletion of 150 N-terminal or C-terminal amino acids was required to disrupt the interaction of ELP5 with ELP3 and ELP1 (Fig. 6C, top panel and second panel from the top, lanes 6 and 9). Finally, as the ΔN150 ELP5 mutant failed to bind any of the tested Elongator subunits, we checked whether this product would efficiently restore cell migration when expressed in Elp5-depleted cells. Both full-length ELP5 and the ΔN150 mutant were detected by Western blot analysis using extracts collected at the end of the wound healing analysis (Fig. 6D, lanes 3 and 4). Interestingly, however, while full-length ELP5 completely restored the migration potential of these cells, the ΔN150 mutant failed to do so (Fig. 6D). Therefore, these results show that ELP5 regulates cell motility as a subunit of Elongator.
FIGURE 6.
Elp5 regulates cell migration of melanoma-derived cells as part of the Elongator complex. A, schematic representation of full-length ELP5 and mutants tested for interaction with ELP1, ELP3, and ELP4. The Hap2 elong superfamily domain (described in Ref. 12) is schematically illustrated. B, HEK293 cells were transfected with the indicated expression plasmids, and cell extracts were subjected to anti-FLAG immunoprecipitations followed by an anti-ELP4 Western blot (top panel). As inputs, FLAG expression constructs were detected in crude cell extracts by an anti-FLAG Western blot analysis (bottom panel). C, HEK293 cells were transfected with the indicated expression plasmids and cell extracts were subjected to anti-FLAG immunoprecipitations followed by anti-ELP1 or -Myc Western blots (top and second panel from the top). As inputs, FLAG- or Myc-expression constructs were detected in crude cell extracts by an anti-FLAG or anti-Myc Western blot analysis (bottom panel). D, wound healing assays were conducted with control, shRNA Elp5 B16-F10 cells or with shRNA Elp5 B16-F10 cells transfected with full-length ELP5 or with the ELP5 mutant lacking the first 150 N-terminal amino acids (“ELP5-ΔN150”). A quantification of the data obtained is illustrated on the right. For each experimental condition, the width of the wound was set to 100% at time 0 and the width in other time points expressed relative to that. The figure shows the data from a representative experiment performed in triplicates (mean values + S.D.). On the left, anti-FLAG Western blots were carried out with protein extracts from the indicated cells collected at the end of the wound healing assay. “Mock” denotes experimental conditions in which cells were transfected with a control plasmid.
ELP5 Connects ELP4 to ELP3
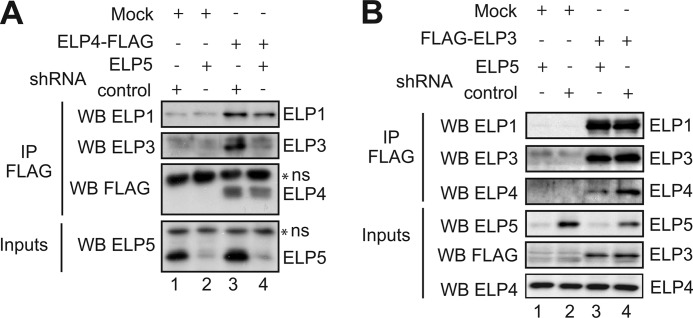
To learn more on the molecular mechanisms by which ELP5 is required for the activity of the Elongator complex, we experimentally assessed the interaction network of Elongator subunits in presence or absence of ELP5. As expected, endogenous ELP1 and ELP3 could be detected in an ELP4-FLAG pull down (Fig. 7A, top panel and second panel from the top, lanes 3). However, whereas ELP1 could efficiently bind ELP4 in ELP5-depleted HEK293 cells, ELP3-ELP4 interaction required ELP5, indicating that ELP5 connects ELP4 to ELP3 (Fig. 7A, top and second panel from the top, compare lanes 3 and 4). To validate these results, we immunoprecipitated FLAG-ELP3 from control or ELP5-depleted HEK 293 cells and observed that although endogenous ELP1 was detectable in the immunoprecipitates in both experimental conditions (Fig. 7B, top panel compare lanes 3 and 4), ELP4-ELP3 association was decreased in the absence of ELP5 (Fig. 7B, third panel from the top, compare lanes 3 and 4). Therefore, ELP5 is dispensable for the interaction between ELP3 and ELP1 but is required for optimal binding of ELP3 to ELP4.
FIGURE 7.
ELP5 connects ELP3 to ELP4. A and B, control or shRNA ELP5 HEK293 cells were transfected with a control plasmid (“Mock”) or a ELP4-FLAG (A) or FLAG-ELP3 (B) expression plasmid, as indicated. Cell extracts from the resulting cells were subjected to anti-FLAG immunoprecipitations followed by anti-ELP1 (A and B), -ELP3 (A and B), -FLAG (A), and -ELP4 (B) Western blots. As inputs, ELP5 (A and B), ELP4 (B) or FLAG-expressing constructs (B) were detected in crude cell extracts by Western blotting (bottom panels).
DISCUSSION
We report here the identification of two previously uncharacterized human Elongator subunits, namely DERP6/ELP5 and C3ORF75/ELP6. We show that both proteins control cell migration in melanoma-derived cells and provide evidences that ELP5 fulfills this function at least partly by connecting ELP3 to ELP4. Our results also point to a role of Elongator in regulating the growth of transformed cells in an anchorage-independent manner. Our data further demonstrate that ELP1-ELP6 act as integral Elongator subunits to control cell motility and tumorigenicity of transformed cells and provide additional insights into the structure-function of this acetylase complex.
Elongator is unique, as it includes two sub-complexes, namely the core-elongator (ELP1, -2, and -3) that has the acetyltransferase activity and the ELP456 subcomplex, which has recently been described as an hexameric RecA-like ATPase in yeast (48, 49). Only very recent studies actually started to explore the structure-function relationship of Elongator. In yeast, deletion of any of the (ELP) genes encoding the 6 subunits confers similar phenotypes, yet Elp2 deletion does not seem to impair Elongator integrity in vitro (6, 50). On the other hand, ELP3 is strongly destabilized in the absence of ELP1, thus defining this latter subunit as a crucial actor for assembling Elongator into a functional complex in yeast but also in human cells (11, 39). Beside a C-terminal HAT domain, yeast Elp3 also harbors an N-terminal iron-sulfur (FeS) cluster motif with no described catalytic activity but critical for the association of Elongator with accessory factors such as Kti11 and Kti12 (51).
Here, we show that ELP5 constitutively binds ELP1 and ELP3 and also connects ELP3 to ELP4. Our data suggest that this role underlies its ability to ensure Elongator integrity and function. We demonstrated that ELP5 bound ELP1/ELP3 through two distinct domains, namely a N-terminal motif spanning from amino acids 101 and 150 as well as a second motif from amino acids 166 to 216. It also binds ELP4 through amino acids 1 to 50 and also through amino acids 217 to 266 (Fig. 6, B and C). Interestingly, the very C-terminal region of ELP5 that is highly conserved throughout evolution is clearly located downstream of the ELP1/3-interacting domain while the motif required for binding to ELP4 only partially includes the most conserved residues. Indeed, the last 50 amino acids of ELP5 are dispensable for the interaction with ELP1/3 and 4, yet they include numerous conserved residues, which may indicate that ELP5 fulfills key functions that go beyond its capacity to connect Elongator subunits. A recent study performed in yeast defined amino acids E61 and D107 of Elp5 as critical residues for ATPase enzymatic activity (48). Surprisingly, these residues as well as their flanking sequences are poorly conserved throughout evolution (supplemental Fig. S3). Therefore, future work is needed to confirm whether this enzymatic activity can also be detected in human ELP5 and to examine to what extent this activity contributes to the capacity of ELP5 to promote cell migration.
Our report provides key insights into the biological functions of DERP6, a protein previously described as a positive regulator of the p53-dependent pathways (44). We previously showed that Elongator deficiency actually slightly stabilized p53 in colon cancer-derived HCT116 cells, but that it did not impair DNA damage-induced p53 phosphorylation (52). We also demonstrated that some p53 target genes were aberrantly expressed upon Elongator deficiency, which indicated that this acetylase complex protects cells from an inappropriate expression of p53-dependent genes rather than promoting p53 activation. Furthermore, we did not observe significant change in p53 levels upon Elp5 depletion (data not shown). Finally, a recent study in Drosophila pointed out for a role of Elongator as positive regulator of the insulin-receptor-TOR signaling, which ultimately control cell growth and cell death. Indeed, mutations in the poly gene (encoding the Elp6 homologue in Drosophila) led to increased apoptotic cell death in larvae (53). In support to this hypothesis, Elongator deficiency is linked to FD, a neurodevelopmental and neurodegenerative genetic disorder in which premature or excessive cell death is observed. Even if it is currently unclear whether the progression of this disease involves a deregulated p53 response, it provides enough evidence for Elongator acting as a negative, rather than a positive regulator of p53.
We show here that Elp1-, Elp3-, Elp5-, and Elp6-deficient melanoma-derived cells do not efficiently form colonies in soft agar. Therefore, Elongator is a protein complex that promotes cell growth and/or survival in an anchorage-independent. The molecular mechanisms by which Elongator is involved in this process are however not known. Based on the role of this acetylase complex in tumorigenesis but also in the migration of melanoma-derived cells, it is tempting to speculate that tumor initiation but also tumor progression may both rely on Elongator. The potential contribution of each subunit of this acetylase complex in both processes is currently under investigation.
Supplementary Material
Acknowledgments
We thank the GIGA Imaging facility for immunofluorescence analysis and to Charles Lambert, Audrey Hoffmann, and Alain Colige (GIGA Cancer, University of Liege, Belgium) for providing collagen.
This work was supported by grants from the F.N.R.S. the Belgian Federation against Cancer, the Concerted Research Action Program (11/16-01, University of Liege), the “Centre Anti-Cancéreux,” the “Leon Frédéricq” Foundation (Faculty of Medicine, University of Liege), the Inter-University Attraction Pole 6/12 (Federal Ministery of Science), and the King Baudouin Foundation (Brussels, Belgium). This work was also supported by a generous in-house grant from Cancer Research UK (to J. S.) and TELEVIE PhD fellowships (to M. G., A. L., and Z. J.).

This article contains supplemental Table S1 and Figs. S1–S4.
- RNAPII
- RNA polymerase II
- HAT
- histone acetyltransferase
- mcm5
- 5-methoxycarbonylmethyl
- ncm5
- 5-carbamoylmethyl
- FD
- familial dysautonomia
- DERP
- dermal papilla-derived protein.
REFERENCES
- 1. Otero G., Fellows J., Li Y., de Bizemont T., Dirac A. M., Gustafsson C. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118 [DOI] [PubMed] [Google Scholar]
- 2. Kim J. H., Lane W. S., Reinberg D. (2002) Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. U.S.A. 99, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hawkes N. A., Otero G., Winkler G. S., Marshall N., Dahmus M. E., Krappmann D., Scheidereit C., Thomas C. L., Schiavo G., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052 [DOI] [PubMed] [Google Scholar]
- 4. Krogan N. J., Greenblatt J. F. (2001) Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 8203–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wittschieben B. O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C. D., Tempst P., Svejstrup J. Q. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128 [DOI] [PubMed] [Google Scholar]
- 6. Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kristjuhan A., Walker J., Suka N., Grunstein M., Roberts D., Cairns B. R., Svejstrup J. Q. (2002) Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell 10, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilbert C., Kristjuhan A., Winkler G. S., Svejstrup J. Q. (2004) Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14, 457–464 [DOI] [PubMed] [Google Scholar]
- 9. Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 10. Kouskouti A., Talianidis I. (2005) Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Close P., Hawkes N., Cornez I., Creppe C., Lambert C. A., Rogister B., Siebenlist U., Merville M. P., Slaugenhaupt S. A., Bours V., Svejstrup J. Q., Chariot A. (2006) Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22, 521–531 [DOI] [PubMed] [Google Scholar]
- 12. Li Y., Takagi Y., Jiang Y., Tokunaga M., Erdjument-Bromage H., Tempst P., Kornberg R. D. (2001) A multiprotein complex that interacts with RNA polymerase II elongator. J. Biol. Chem. 276, 29628-29631 [DOI] [PubMed] [Google Scholar]
- 13. Holmberg C., Katz S., Lerdrup M., Herdegen T., Jäättelä M., Aronheim A., Kallunki T. (2002) A novel specific role for I κB kinase complex-associated protein in cytosolic stress signaling. J. Biol. Chem. 277, 31918–31928 [DOI] [PubMed] [Google Scholar]
- 14. Svejstrup J. Q. (2007) Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336 [DOI] [PubMed] [Google Scholar]
- 15. Huang B., Johansson M. J., Byström A. S. (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esberg A., Huang B., Johansson M. J., Byström A. S. (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148 [DOI] [PubMed] [Google Scholar]
- 17. Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D. A., Yoshida M., Hermand D. (2012) Translational control of cell division by elongator. Cell Rep. 1, 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehlgarten C., Jablonowski D., Wrackmeyer U., Tschitschmann S., Sondermann D., Jäger G., Gong Z., Byström A. S., Schaffrath R., Breunig K. D. (2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 76, 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen C., Tuck S., Byström A. S. (2009) Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS genetics 5, e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Versées W., De Groeve S., Van Lijsebettens M. (2010) Elongator, a conserved multitasking complex? Mol. Microbiol. 76, 1065–1069 [DOI] [PubMed] [Google Scholar]
- 21. Creppe C., Malinouskaya L., Volvert M. L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S., Belachew S., Malgrange B., Chapelle J. P., Siebenlist U., Moonen G., Chariot A., Nguyen L. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of α-tubulin. Cell 136, 551–564 [DOI] [PubMed] [Google Scholar]
- 22. Solinger J. A., Paolinelli R., Klöss H., Scorza F. B., Marchesi S., Sauder U., Mitsushima D., Capuani F., Stärzenbaum S. R., Cassata G. (2010) The Caenorhabditis elegans Elongator complex regulates neuronal α-tubulin acetylation. PLoS genetics 6, e1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miskiewicz K., Jose L. E., Bento-Abreu A., Fislage M., Taes I., Kasprowicz J., Swerts J., Sigrist S., Versées W., Robberecht W., Verstreken P. (2011) ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron 72, 776–788 [DOI] [PubMed] [Google Scholar]
- 24. Rahl P. B., Chen C. Z., Collins R. N. (2005) Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17, 841–853 [DOI] [PubMed] [Google Scholar]
- 25. Chen C., Huang B., Eliasson M., Rydén P., Byström A. S. (2011) Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS genetics 7, e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Q., Fazly A. M., Zhou H., Huang S., Zhang Z., Stillman B. (2009) The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS genetics 5, e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelissen H., Fleury D., Bruno L., Robles P., De Veylder L., Traas J., Micol J. L., Van Montagu M., Inzé D., Van Lijsebettens M. (2005) The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. U.S.A. 102, 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker J., Kwon S. Y., Badenhorst P., East P., McNeill H., Svejstrup J. Q. (2011) Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 187, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Y. T., Hims M. M., Shetty R. S., Mull J., Liu L., Leyne M., Slaugenhaupt S. A. (2009) Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell Biol. 29, 736–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slaugenhaupt S. A., Gusella J. F. (2002) Familial dysautonomia. Curr. Opin. Genet. Dev. 12, 307–311 [DOI] [PubMed] [Google Scholar]
- 31. Gold-von Simson G., Axelrod F. B. (2006) Familial dysautonomia: update and recent advances. Curr. Probl. Pediatr Adolesc Health Care 36, 218–237 [DOI] [PubMed] [Google Scholar]
- 32. Anderson S. L., Coli R., Daly I. W., Kichula E. A., Rork M. J., Volpi S. A., Ekstein J., Rubin B. Y. (2001) Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68, 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slaugenhaupt S. A., Blumenfeld A., Gill S. P., Leyne M., Mull J., Cuajungco M. P., Liebert C. B., Chadwick B., Idelson M., Reznik L., Robbins C., Makalowska I., Brownstein M., Krappmann D., Scheidereit C., Maayan C., Axelrod F. B., Gusella J. F. (2001) Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simpson C. L., Lemmens R., Miskiewicz K., Broom W. J., Hansen V. K., van Vught P. W., Landers J. E., Sapp P., Van Den Bosch L., Knight J., Neale B. M., Turner M. R., Veldink J. H., Ophoff R. A., Tripathi V. B., Beleza A., Shah M. N., Proitsi P., Van Hoecke A., Carmeliet P., Horvitz H. R., Leigh P. N., Shaw C. E., van den Berg L. H., Sham P. C., Powell J. F., Verstreken P., Brown R. H., Jr., Robberecht W., Al-Chalabi A. (2009) Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nguyen L., Humbert S., Saudou F., Chariot A. (2010) Elongator-an emerging role in neurological disorders. Trends Mol. Med. 16, 1–6 [DOI] [PubMed] [Google Scholar]
- 36. Johansen L. D., Naumanen T., Knudsen A., Westerlund N., Gromova I., Junttila M., Nielsen C., Bøttzauw T., Tolkovsky A., Westermarck J., Coffey E. T., Jäättelä M., Kallunki T. (2008) IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 121, 854–864 [DOI] [PubMed] [Google Scholar]
- 37. Boone N., Loriod B., Bergon A., Sbai O., Formisano-Tréziny C., Gabert J., Khrestchatisky M., Nguyen C., Féron F., Axelrod F. B., Ibrahim E. C. (2010) Olfactory stem cells, a new cellular model for studying molecular mechanisms underlying familial dysautonomia. PLoS One 5, e15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee G., Papapetrou E. P., Kim H., Chambers S. M., Tomishima M. J., Fasano C. A., Ganat Y. M., Menon J., Shimizu F., Viale A., Tabar V., Sadelain M., Studer L. (2009) Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petrakis T. G., Wittschieben B. Ø., Svejstrup J. Q. (2004) Molecular architecture, structure-function relationship, and importance of the Elp3 subunit for the RNA binding of holo-elongator. J. Biol. Chem. 279, 32087–32092 [DOI] [PubMed] [Google Scholar]
- 40. Viatour P., Dejardin E., Warnier M., Lair F., Claudio E., Bureau F., Marine J. C., Merville M. P., Maurer U., Green D., Piette J., Siebenlist U., Bours V., Chariot A. (2004) GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol. Cell 16, 35–45 [DOI] [PubMed] [Google Scholar]
- 41. Close P., East P., Dirac-Svejstrup A. B., Hartmann H., Heron M., Maslen S., Chariot A., Söding J., Skehel M., Svejstrup J. Q. (2012) DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature 484, 386–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gobeil S., Zhu X., Doillon C. J., Green M. R. (2008) A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 22, 2932–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sebastiaan Winkler G., Lacomis L., Philip J., Erdjument-Bromage H., Svejstrup J. Q., Tempst P. (2002) Isolation and mass spectrometry of transcription factor complexes. Methods 26, 260–269 [DOI] [PubMed] [Google Scholar]
- 44. Yuan J., Tang W., Luo K., Chen X., Gu X., Wan B., Yu L. (2006) Cloning and characterization of the human gene DERP6, which activates transcriptional activities of p53. Molecular Biol. Rep. 33, 151–158 [DOI] [PubMed] [Google Scholar]
- 45. Winkler G. S., Petrakis T. G., Ethelberg S., Tokunaga M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2001) RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743–32749 [DOI] [PubMed] [Google Scholar]
- 46. Pokholok D. K., Hannett N. M., Young R. A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9, 799–809 [DOI] [PubMed] [Google Scholar]
- 47. Doillon C. J., Gagnon E., Paradis R., Koutsilieris M. (2004) Three-dimensional culture system as a model for studying cancer cell invasion capacity and anticancer drug sensitivity. Anticancer Res. 24, 2169–2177 [PubMed] [Google Scholar]
- 48. Glatt S., Létoquart J., Faux C., Taylor N. M., Séraphin B., Müller C. W. (2012) The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 19, 314–320 [DOI] [PubMed] [Google Scholar]
- 49. Lin Z., Zhao W., Diao W., Xie X., Wang Z., Zhang J., Shen Y., Long J. (2012) Crystal structure of elongator subcomplex elp4–6. J. Biol. Chem. 287, 21501–21508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fellows J., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2000) The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J. Biol. Chem. 275, 12896–12899 [DOI] [PubMed] [Google Scholar]
- 51. Greenwood C., Selth L. A., Dirac-Svejstrup A. B., Svejstrup J. Q. (2009) An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem. 284, 141–149 [DOI] [PubMed] [Google Scholar]
- 52. Cornez I., Creppe C., Gillard M., Hennuy B., Chapelle J. P., Dejardin E., Merville M. P., Close P., Chariot A. (2008) Deregulated expression of pro-survival and pro-apoptotic p53-dependent genes upon Elongator deficiency in colon cancer cells. Biochem. Pharmacol. 75, 2122–2134 [DOI] [PubMed] [Google Scholar]
- 53. Bolukbasi E., Vass S., Cobbe N., Nelson B., Simossis V., Dunbar D. R., Heck M. M. (2012) Drosophila poly suggests a novel role for the Elongator complex in insulin receptor-target of rapamycin signalling. Open Biol. J. 2, 110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.