Background: The SIN3-HDAC is a multimeric histone deacetylase complex that regulates gene expression.
Results: FAM60A, a cell cycle-regulated protein, is a subunit of the SIN3-HDAC complex that influences the expression of certain SIN3-HDAC-regulated genes.
Conclusion: FAM60A is a novel subunit of SIN3-HDAC that regulates gene expression.
Significance: FAM60A, a novel component of SIN3-HDAC, is required for proper cyclin D1 expression.
Keywords: Cell Cycle, Cyclin D1, Epigenetics, Gene Regulation, Gene Transcription, Histone Deacetylase, FAM60A, HDAC1, SIN3A
Abstract
The SIN3A-HDAC complex deacetylates histones thereby repressing gene transcription. Here we describe family with sequence similarity 60A (FAM60A), a cell cycle-regulated protein that binds to the SIN3-HDAC complex. FAM60A expression peaks during G1 and S phases of the cell cycle in U2OS cells, in a manner similar to the G1 regulator cyclin D1, which is a known target of SIN3-HDAC. In this light we found that FAM60A binds to SIN3-HDAC-regulated promoters such as cyclin D1 in G1 and S phases. Cells depleted of FAM60A show increased histone acetylation at the cyclin D1 promoter and elevated levels of cyclin D1 mRNA and protein. Furthermore, depletion of FAM60A altered the periodic association of HDAC1 with the cyclin D1 promoter, increased cyclin D1 expression at all cell cycle phases, and caused premature S phase entry. The data in this study introduce FAM60A as a novel regulator of SIN3-HDAC function and gene expression.
Introduction
Acetylation of histones on lysines is a major mechanism for modulating chromatin conformation. Histone acetylation promotes a relaxed, transcriptionally active chromatin state whereas deacetylation catalyzed by histone deacetylases (HDACs)3 favor a silent, inactive state (1). Mammalian class I HDACs 1 and 2 form the catalytic core of several multisubunit co-repressor complexes including Mi-2/NURD, Co-REST, and SIN3-HDAC (2). The assembly of the SIN3-HDAC complex is coordinated by the SIN3A and SIN3B scaffold proteins. The SIN3 proteins cannot bind to DNA directly, and instead they are targeted to gene promoters by sequence-specific repressors (3, 4). Disruption of mouse Sin3a causes embryonic lethality indicative of important cell functions (5), and mouse Sin3a and Sin3b bind to a multitude of gene promoters (6). Furthermore, Sin3a deletion causes deregulation of genes involved in a broad range of biological processes (5). For example, SIN3-HDAC represses expression of cyclin D1 (5, 7).
Cyclins were so named because of their periodic cell cycle-dependent expression pattern. Cyclin D1, cyclin D2, and cyclin D3 are closely related proteins that bind to and activate CDK4 and CDK6 in G1 phase (8). Activation of CDK4/CDK6 leads to phosphorylation of the retinoblastoma protein in a manner that relieves the inhibitory effects which retinoblastoma protein exerts on E2F family transcription factors that control entry to S phase (9). Consequently, overexpression of cyclin D1 accelerates G1 phase in rodent fibroblasts causing premature S phase entry (10). This may explain the observations showing that cyclin D1 is an oncogene that is activated by chromosomal translocation or overexpressed in many types of human cancers (8, 11, 12). Based on these data, it is not surprising that cellular cyclin D1 levels are tightly controlled under normal circumstances, and multiple control mechanisms have been reported. For example, cyclin D1 protein levels are restrained by ubiquitin-dependent degradation (13). In this study we describe FAM60A, a cell cycle-regulated subunit of SIN3-HDAC, that influences gene expression.
EXPERIMENTAL PROCEDURES
Additional procedures are provided in the supplemental Materials and Methods.
Cell Lysis and Immunoprecipitation
Cells were lysed in a 50 mm Tris-HCl (pH 7.4) buffer containing 0.27 m sucrose, 150 mm NaCl, 1% (v/v) Triton X-100, 0.5% Nonidet P-40, and 0.1% (v/v) 2-mercaptoethanol and supplemented with protease inhibitors (Roche Applied Science). Where indicated, lysis buffer was supplemented with benzonase (50 units/ml; Novagen) and protein extracts incubated for 30 min at 4 °C. Immunoprecipitations were carried out in lysis buffer for 2 h at 4 °C with constant agitation.
Purification of GFP-FAM60A
HEK293 cells stably transfected with a tetracycline-inducible form of GFP-FAM60A, or untransfected parent cells, were incubated in the presence of 1 μg/ml tetracycline overnight. Approximately 400 mg of cell extracts were precleared with 500 μl of protein G-Sepharose and then incubated with 150 μl of GFP-Trap beads for 2 h at 4 °C. Beads were washed five times in lysis buffer and twice with a buffer containing 50 mm Tris (pH 7.4) and 0.27 m sucrose. GFP-Trap beads were boiled in SDS-PAGE sample buffer (Invitrogen) containing 5% (v/v) 2-mercaptoethanol and resolved in a 4–12% SDS-PAGE gradient gel (Nupage, Invitrogen). The gel was fixed and stained with colloidal Coomassie, and the gel lanes were cut into slices. Proteins present in each band were identified by tryptic digest followed by mass fingerprinting, as described before (14).
Measurement of Deacetylase Activity in Immunoprecipitates
HEK293 cells were transfected with the relevant siRNA for 24 h, harvested, and lysed as described above. Approximately 1.25 mg of protein lysate was incubated with 20 μl of protein G-Sepharose containing 1 μg of the relevant antibodies for 2 h at 4 °C. After extensive washing with lysis buffer, beads were washed twice in 1× histone deacetylase assay buffer (50 mm Tris-HCl (pH 8.0), 137 mm sodium chloride, 2.7 mm potassium chloride, 1 mm magnesium chloride). Deacetylase activity in the immunoprecipitates was measured using the Color de LysTM HDAC Colorimetric Assay/Drug Discovery Kit (Enzo Life Sciences) following the manufacturer's instructions. Essentially beads were incubated for 1 h at 37 °C in assay buffer containing 1 mm Color de LysTM substrate, reactions were stopped by adding 50 μl of developer solution for 10 min at 37 °C, and absorption at 405 nm was measured on a 96-well plate reader. When indicated, beads were incubated with 1 μm trichostatin A for 5 min prior to the addition of the histone deacetylase activity reaction mixture.
RESULTS
FAM60A Interacts with the SIN3-HDAC Complex
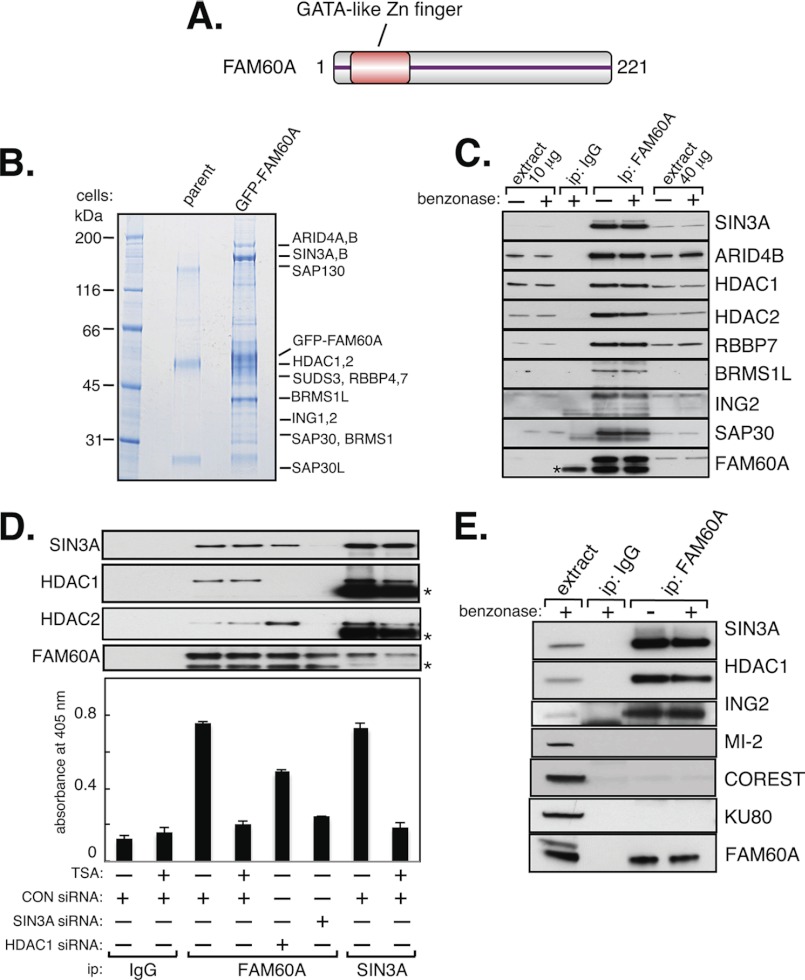
We, and others, identified the uncharacterized human protein FAM60A in a screen for substrates of the DNA damage-responsive kinase ATM4 (15). FAM60A is a small nuclear protein found from flies to humans, which has a conserved GATA-like zinc finger (Fig. 1A and supplemental Fig. S1) (16), suggestive of DNA binding and/or a role in transcriptional regulation. To gain clues to its function, we searched for FAM60A-interacting proteins.
FIGURE 1.
FAM60A is a component of the SIN3-HDAC complex. A, schematic diagram of human FAM60A. The GATA-like zinc finger is shaded in red. B, stable expression and purification of GFP-FAM60A are shown. GFP immunoprecipitates from HEK293 cells expressing GFP-FAM60A, or from parent cells, were subjected to SDS-PAGE. Proteins in the gel lane containing FAM60A-associated proteins were digested with trypsin before mass fingerprinting. C, endogenous FAM60A interacts with the SIN3-HDAC complex in HEK293 cells. Anti-GFP antibodies (IgG) were used as control. Where indicated, immunoprecipitates (ip) were treated with benzonase (50 units/ml). Whole cell extract (10 or 40 μg of protein) was used as input material. Asterisk indicates IgG light chain. D, SIN3A and FAM60A were immunoprecipitated from extracts of cells transfected with the indicated siRNAs. Precipitates were probed with the antibodies indicated (upper panels) or tested for HDAC activity (bottom panel) using a colorimetric assay as described under “Experimental Procedures.” Anti-GFP antibodies (IgG) were used as control. Asterisks indicate IgG chains. E, endogenous FAM60A does not co-immunoprecipitate with the MI-2 or Co-REST complexes. Whole cell extract (20 μg of protein) was used as input material. Anti-GFP antibodies (IgG) were used as control.
To this end, HEK293 cells were stably transfected with a tetracycline-inducible form of FAM60A with an N-terminal GFP tag, or with GFP only. Mass fingerprinting of proteins that co-immunoprecipitated with GFP-tagged FAM60A, but not GFP, revealed most of the subunits of the SIN3-HDAC complex (supplemental Table S1 and Fig. 1B), and we confirmed that endogenous FAM60A interacts with SIN3-HDAC using an antibody raised against recombinant human FAM60A (Fig. 1C). Consistent with these data, we found that FAM60A immunoprecipitates had HDAC activity that was comparable with the activity detected in SIN3A immunoprecipitates (Fig. 1D). The HDAC activity in FAM60A and SIN3 precipitates was inhibited by trichostatin A, an inhibitor of class I HDACs (Fig. 1D) (17). Furthermore, the HDAC activity in FAM60A precipitates was reduced by transfecting cells with siRNAs specific for SIN3A or HDAC1 (Fig. 1D). The effect of depleting HDAC1 was less pronounced than depletion of SIN3A because the SIN3-HDAC complex has two catalytic subunits: HDAC1 and HDAC2. FAM60A did not interact with other HDAC complexes which share HDACs 1 and 2 as catalytic subunits, such as Co-REST or Mi2/NURD (Fig. 1E) (2). Treatment of immunoprecipitates with recombinant benzonase, a deoxyribonuclease from Serratia macescens, did not affect the interaction of FAM60A with SIN3-HDAC. This excludes the possibility the interaction is mediated by DNA (Fig. 1C and E). Therefore, FAM60A appears to associate physically with the SIN3-HDAC complex.
FAM60A Associates with SIN3-HDAC-regulated Promoters and Restrains Cyclin D1 Levels
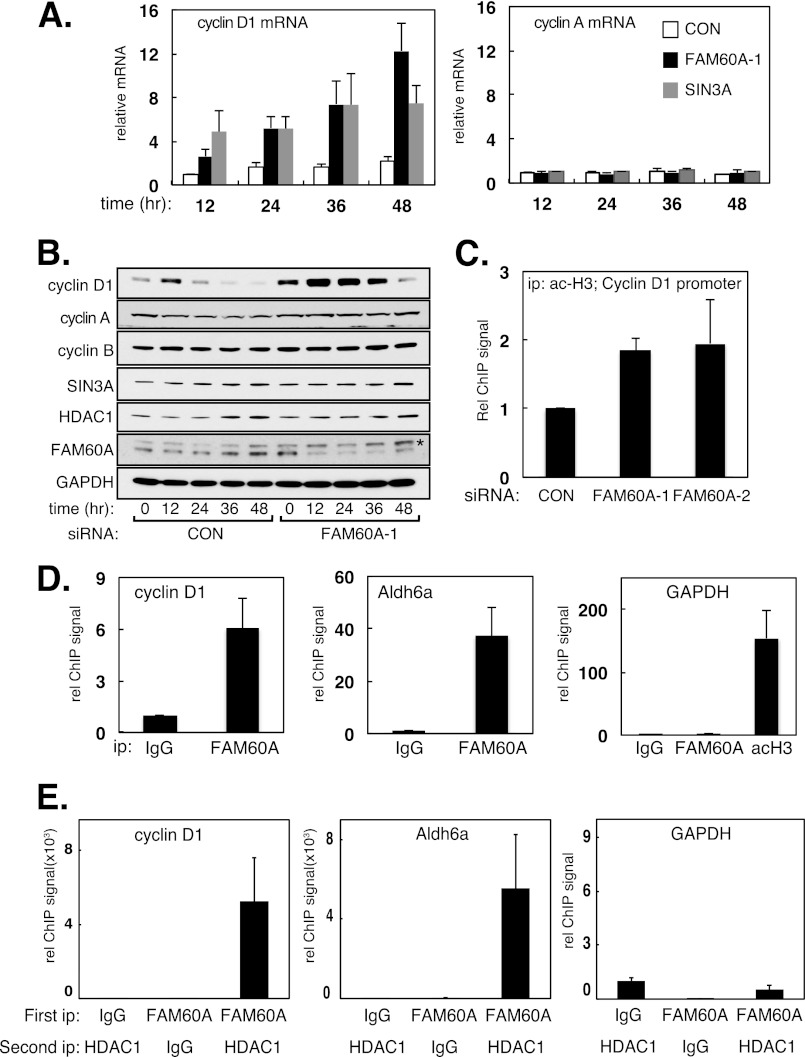
We next tested whether FAM60A affects the expression of SIN3-HDAC-regulated genes, and we focused on cyclin D1, a well characterized target of this repressor complex. Treatment of HeLa cells with siRNAs specific for FAM60A or SIN3A caused an increase in the steady-state levels of cyclin D1, but not cyclin A, mRNA and protein (Figs. 2A and B). A slight increase in histone H3 acetylation at the cyclin D1 promoter was also observed (Fig. 2C). U2OS cells depleted of FAM60A also showed increased levels of cyclin D1 mRNA (supplemental Fig. S2A). Further evidence that FAM60A regulates gene expression was obtained in experiments with cells harboring a luciferase reporter cassette under the control of the cyclin D1 promoter. As shown in supplemental Fig. S2B, overexpression of FLAG-tagged versions of SIN3A, HDAC1, or SIN3A inhibited luciferase expression from the cyclin D1 reporter cassette, whereas trichostatin A caused a 2-fold increase in reporter expression. These data indicate that FAM60A has the capacity to influence gene expression.
FIGURE 2.
FAM60A associates with SIN3-HDAC-regulated promoters and influences the expression of cyclin D1 mRNA and protein. A and B, depletion of FAM60A causes elevated levels of cyclin D1. HeLa cells were treated with the relevant siRNAs for the times indicated. Cyclin D1 and cyclin A2 mRNA levels were measured by quantitative PCR (A), and extracts were probed with the antibodies indicated (B). Asterisk indicates a nonspecific protein band. C, depletion of FAM60A increases H3 acetylation at the cyclin D1 promoter in U2OS cells tested by ChIP (acetyl-Lys9/Lys14 H3). Data are represented as means ± S.E. (error bars) of triplicate experiments. D, extracts of U2OS cells were subjected to ChIP analysis with the indicated antibodies. Anti-GST antibodies were used as control (IgG). Quantitative PCR was carried out on DNA extracted from the precipitates with the indicated primers. E, FAM60A co-occupies the cyclin D1 and ALDH6A promoters with HDAC1. Extracts of U2OS cells were subjected to re-ChIP analysis with the indicated antibodies. Anti-GST antibodies were used as control (IgG). Quantitative PCR was carried out on DNA extracted from the precipitates with the indicated primers.
We wished to test whether the effects of FAM60A depletion on cyclin D1 mRNA (Fig. 2A) were direct. We reasoned that if FAM60A affects expression of SIN3-HDAC-regulated genes directly, it should associate with the relevant promoters. To test this possibility, we carried out chromatin immunoprecipitation (ChIP) analysis. As shown in Fig. 2D, endogenous FAM60A associates with the cyclin D1 promoter and with the ALDH6A promoter, both of which are regulated by SIN3-HDAC (5, 6). In contrast, no association of FAM60A was observed with the GAPDH promoter. ChIP analysis with anti-GFP antibodies showed that a GFP-tagged form of FAM60A bound to the cyclin D1 promoter. However, mutations in pairs of conserved cysteines (Cys16/Cys19 or Cys53/Cys56; supplemental Fig. S1A) in the GATA-like zinc finger of FAM60A severely reduced binding of GFP-FAM60A to the cyclin D1 promoter (supplemental Fig. S2C). However, these mutations do not affect the ability of FAM60A to co-immunoprecipitate with components of the SIN3-HDAC complex (Fig. S2D), suggesting that the effect of the FAM60A zinc finger mutations on promoter binding is not due to dissociation from SIN3-HDAC.
To test whether the pool of FAM60A that associates with the gene promoters is part of the SIN3-HDAC complex, we carried out sequential ChIP or “re-ChIP” experiments. FAM60A immunoprecipitated from U2OS cells was eluted from the precipitates, and the eluate was subjected to immunoprecipitation with anti-HDAC1 antibodies. DNA was then extracted from the HDAC1 precipitates and subjected to quantitative PCR analysis. In this way, we found that FAM60A co-occupies the cyclin D1 and ALDH6A promoters with HDAC1 (Fig. 2E), which is consistent with the observation that FAM60A co-immunoprecipitates with SIN3-HDAC (Fig. 1). Taken together, the data in this section show that FAM60A co-occupies at least two gene promoters with SIN3-HDAC and restrains levels of cyclin D1 mRNA.
FAM60A Promotes Stable Retention of HDAC1 at the SIN3-regulated Promoters
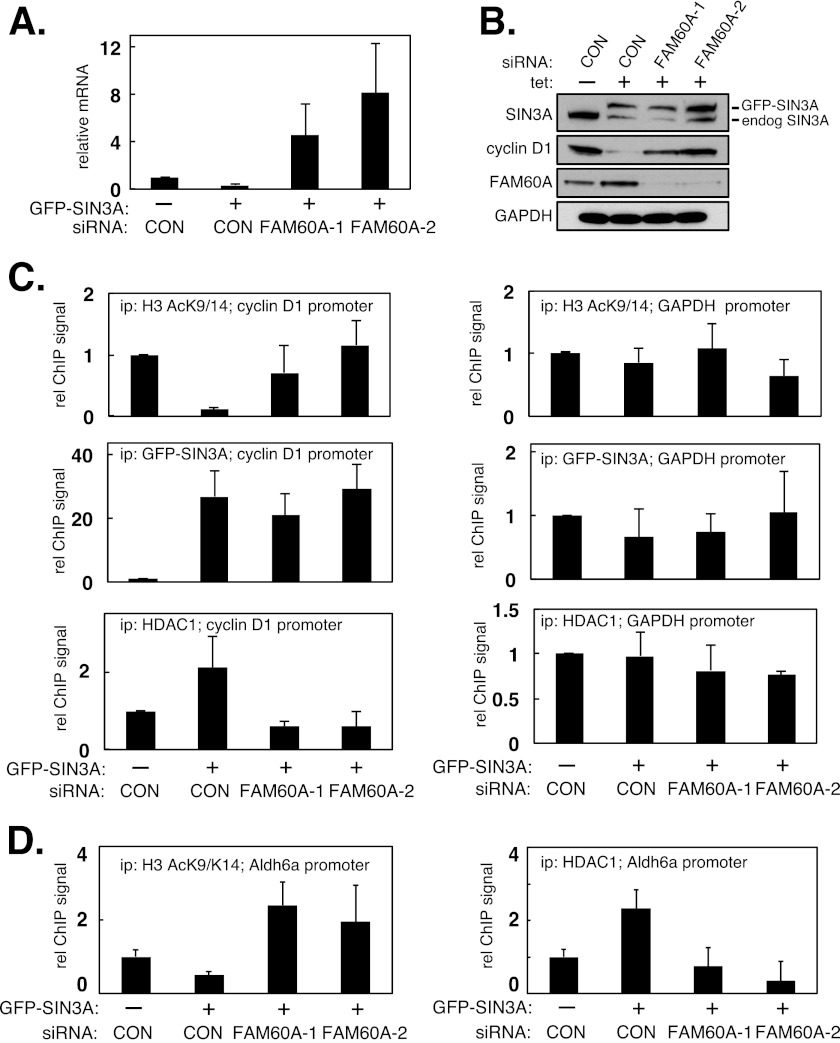
The data above show that FAM60A regulates the levels of cyclin D1 mRNA, but they do not address the underlying mechanisms. We reasoned that FAM60A might influence the association of one or more components of the SIN3-HDAC complex with the cyclin D1 promoter. It was not possible to examine promoter association of endogenous SIN3A (or SIN3B) because none of the commercially available SIN3 antibodies worked in ChIP experiments (data not shown). Instead, we stably overexpressed GFP-SIN3A in U2OS cells. Overexpression of SIN3A results in a decrease on the levels of cyclin D1 mRNA by increasing the levels of the holo-SIN3-HDAC complex at the cyclin D1 promoter. We used this system to test whether FAM60A is required for retention of HDAC1 at the cyclin D1 promoter, and the GFP tag on SIN3A allowed us to test SIN3A promoter binding.
U2OS cells were stably transfected with a tetracycline-inducible form of GFP-tagged SIN3A. Addition of tetracycline to these cells (but not to the parental cells; data not shown) elicited a marked decrease in the levels of cyclin D1 mRNA and protein whereas GAPDH was unaffected (Fig. 3, A and B). However, two separate FAM60A-specific siRNAs severely blunted the decrease in cyclin D1 caused by GFP-SIN3A induction (Fig. 3, A and B). ChIP experiments revealed that GFP-SIN3A induction caused increased HDAC1 association with the cyclin D1 promoter and a concomitant decrease in H3 Lys9/Lys14 acetylation. These effects were dampened by FAM60A-specific siRNAs (Fig. 3C). In these experiments, although there was less HDAC1 bound to the cyclin D1 promoter when FAM60A was depleted, the binding of GFP-SIN3A was unchanged (Fig. 3C). GFP-SIN3A induction also caused increased HDAC1 association with the ALDH6A promoter and a concomitant decrease in local H3 Lys9/Lys14 acetylation. These effects were again dampened by FAM60A-specific siRNAs (Fig. 3D), but GFP-SIN3A association with the ALDH6A promoter was unaffected (supplemental Fig. S3A). GFP-SIN3A overexpression had no effect on H3 acetylation of, or HDAC1 binding to, the GAPDH promoter (Fig. 3C), or the TGF-β promoter that is repressed by the MI-2 complex that shares HDAC1 as a catalytic subunit (supplemental Fig. S3B) (19). Taken together, these data show that FAM60A promotes histone deacetylation by influencing the stable retention of HDAC1 at SIN3-HDAC-regulated gene promoters, at least in U2OS cells.
FIGURE 3.
FAM60A promotes the retention of HDAC1 but not SIN3 at the cyclin D1 promoter in asynchronously growing cells. A and B, overexpression of SIN3A represses cyclin D1. U2OS cells stably expressing GFP-tagged SIN3A in a tetracycline-inducible manner were transfected with two separate FAM60A siRNAs or with a control (GFP-specific) siRNA. Cells were incubated, or not, with tetracycline. Cyclin D1 mRNA levels were quantitated by quantitative PCR (A), and cell extracts were subjected to Western blotting with the indicated antibodies (B). Data are represented as means ± S.E. (error bars) of triplicate experiments. C, extracts of cells from A were subjected to ChIP analysis with the indicated antibodies. Quantitative PCR was carried out on DNA extracted from the precipitates (ip) with primers specific for the cyclin D1 promoter (left panels) or the GAPDH promoter (right panels). D, extracts of cells from A were subjected to ChIP analysis with the indicated antibodies. Quantitative PCR was carried out on DNA extracted from the precipitates with primers specific for the ALDH6A promoter.
FAM60A Expression and Association with the Cyclin D1 Promoter Fluctuate during the Cell Cycle
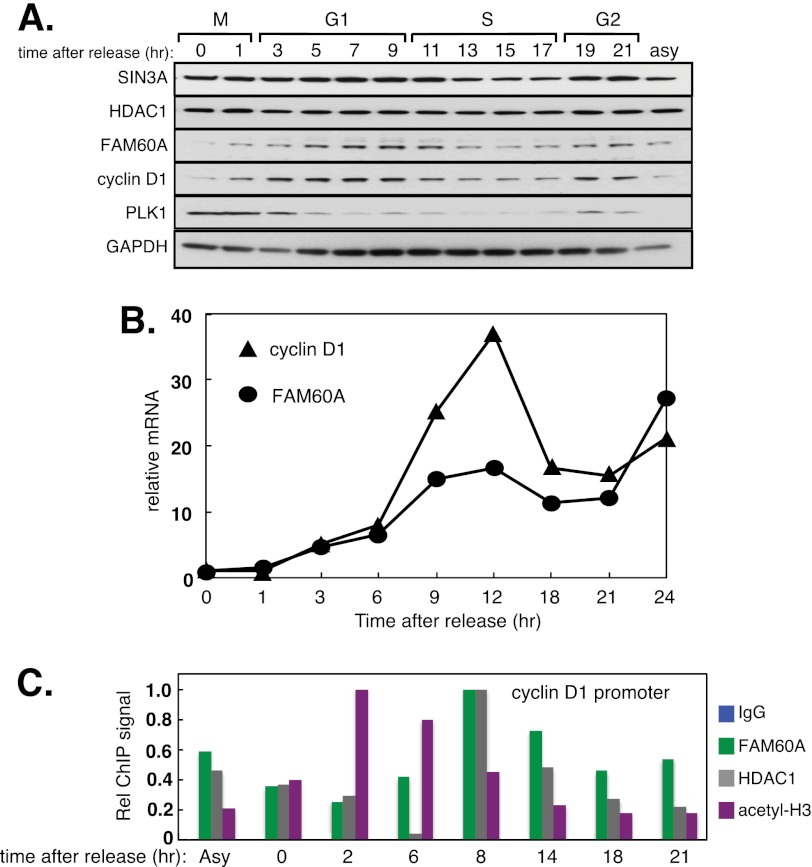
We noticed that FAM60A protein fluctuates in a cell cycle-dependent manner. U2OS cells were released from a double thymidine block into medium containing nocodazole to cause mitotic arrest. Cells were then released from mitotic arrest so that they proceeded synchronously through a cell cycle (supplemental Fig. S4). We found that FAM60A protein is almost undetectable in mitosis, but levels rise as cells exit mitosis, peak in late G1 phase before declining in late S phase (Fig. 4A). At least in U2OS cells, the fluctuations in FAM60A protein during the cell cycle are similar to cyclin D1 protein, although the FAM60A peak seems to lag very slightly behind the cyclin D1 peak (Fig. 4A). Furthermore, FAM60A mRNA showed a pattern of fluctuation similar to that of cyclin D1 mRNA (Fig. 4B).
FIGURE 4.
FAM60A expression and association with the cyclin D1 promoter fluctuate during the cell cycle. A, cell cycle regulation of FAM60A. U2OS cells were synchronized in mitosis by release from a double thymidine block into nocodazole for 20 h. The cells were then released from mitotic arrest and lysed at the times indicated. Extracts were subjected to Western blotting with the antibodies indicated. Asy, asynchronously growing cells. B, U2OS cells arrested in mitosis and released as in A. Levels of FAM60A and cyclin D1 mRNAs were measured by quantitative PCR. C, amount of FAM60A at the cyclin D1 promoter fluctuates during the cell cycle. Extracts of cells from A were subjected to ChIP analysis with the indicated antibodies. Quantitative PCR was carried out on DNA extracted from the precipitates with primers specific for the cyclin D1 promoter. Acetyl-H3 antibodies recognize Lys9/Lys14-acetylated H3.
We next tested whether FAM60A binding to the cyclin D1 promoter fluctuates in accordance with FAM60A protein levels during the cell cycle. U2OS cells proceeding synchronously through the cell cycle were lysed, and extracts were subjected to ChIP analysis with antibodies against FAM60A, HDAC1, and anti-acetyl H3 (Lys9/Lys14). As shown in Fig. 4C, the amount of FAM60A bound to the cyclin D1 promoter increased as cells exited mitosis, reaching a maximum around 8 h after release when cells were in late G1. The level of H3 acetylation at Lys9/Lys14 at the cyclin D1 promoter fell as FAM60A occupancy of the cyclin D1 promoter reached a maximum (Fig. 4C). These data showed that FAM60A fluctuates during the cell cycle.
FAM60A Promotes Retention of HDAC1 with the Cyclin D1 Promoter in G1 and S Phases
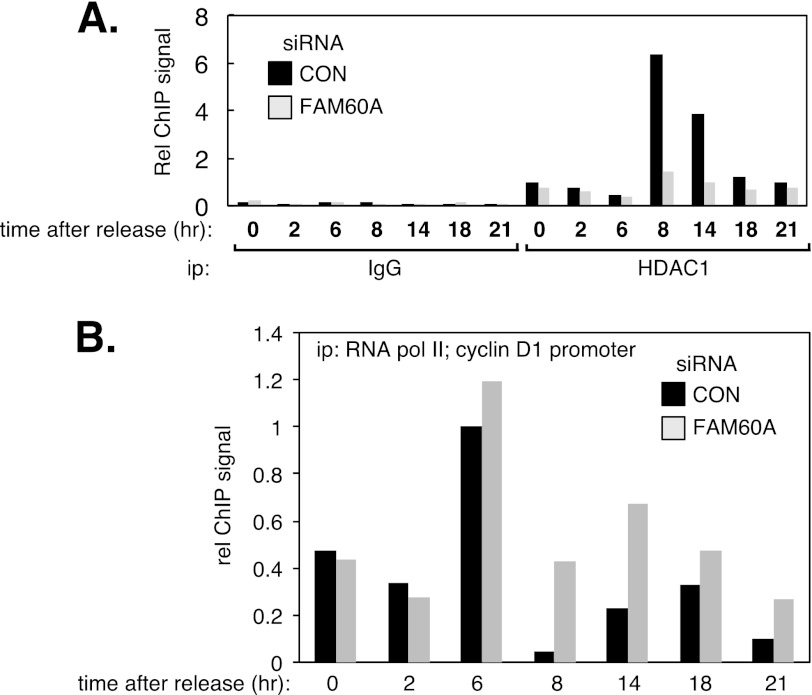
The data presented above showed that FAM60A promotes the association of HDAC1 with the cyclin D1 promoter in asynchronous cells (Fig. 3C). We next tested whether FAM60A influences the fluctuating association of HDAC1 with the cyclin D1 promoter throughout the cell cycle. To this end, U2OS cells that were treated with FAM60A-specific siRNA or control siRNA were released from a double thymidine block into medium containing the CDK1 inhibitor RO-3306, which arrests cells in G2 phase (20). Cells were then released from mitotic arrest so that they proceeded synchronously through a cell cycle. As shown in Fig. 5A, HDAC1 binding to the cyclin D1 promoter peaked at approximately 8 h after release from mitotic arrest (in late G1) in cells treated with control siRNA and declined subsequently. In contrast the binding of HDAC1 to the cyclin D1 promoter was markedly reduced in cells depleted of FAM60A (Fig. 5A) even though HDAC1 levels are constant throughout the cell cycle (Fig. 4A). Lower HDAC1 levels at the cyclin D1 promoter should lead to increased acetylation and increased association of RNA polymerase II. In this light we observed that association of RNA polymerase II with the cyclin D1 promoter decreases dramatically between 6 and 8 h after release of U2OS cells from nocodazole (Fig. 5B), at the same time that FAM60A and HDAC1 association with this promoter reached a maximum (Fig. 4C). In cells depleted of FAM60A, however, the levels of RNA polymerase II associated with the cyclin D1 promoter at these times is greater than in cell treated with control siRNA. Taken together, these data show that FAM60A promotes the association of HDAC1 with the cyclin D1 promoter during G1 and S phases of the cell cycle in U2OS cells.
FIGURE 5.
FAM60A influences cell cycle-dependent association of HDAC1 with the cyclin D1 promoter. A, U2OS cells treated with control siRNA or FAM60A-specific siRNA and synchronized in mitosis by release from a double thymidine block into RO-3306 for 20 h (supplemental Fig. S4). The cells were then released from mitotic arrest and lysed at the times indicated. Extracts were subjected to ChIP analysis with nonspecific antibodies (GST; IgG) or with antibodies against HDAC1. Quantitative PCR was carried out on DNA extracted from the precipitates with primers specific for the cyclin D1 promoter. B, same as A except that ChIP analysis was carried out with antibodies raised against the C terminus of the large subunit of RNA polymerase II.
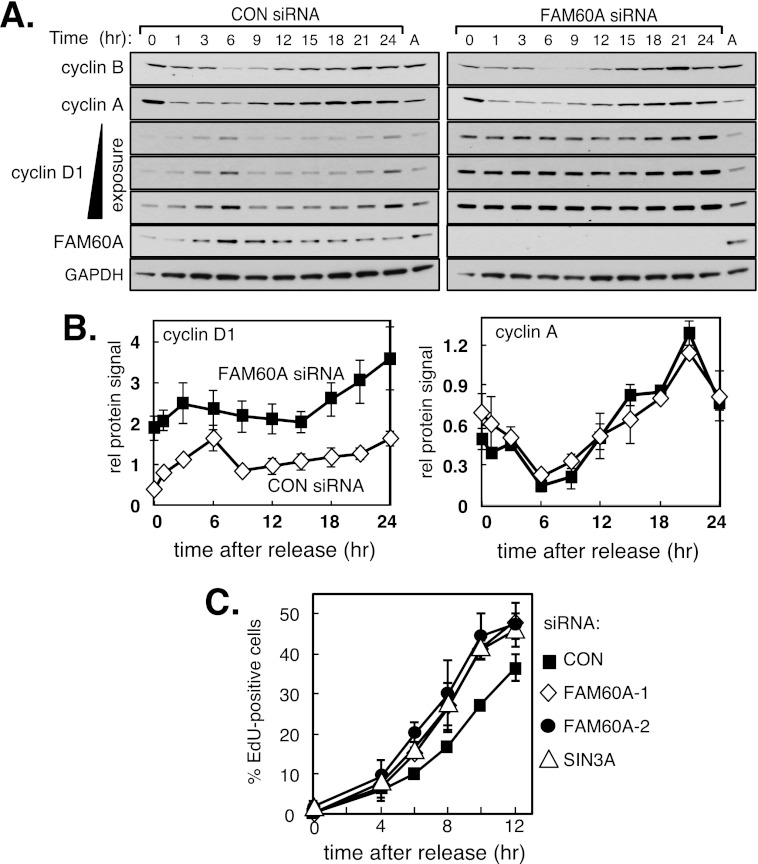
Because the pattern of FAM60A expression is similar to cyclin D1 expression throughout the cell cycle in U2OS cells, we tested whether cyclin D1 periodicity is affected by FAM60A depletion in these cells. To this end, U2OS cells depleted of FAM60A were released from mitotic arrest so that they proceeded synchronously though the cell cycle. As shown in Fig. 6, A and B, cyclin D1 periodicity was largely intact in cells depleted of FAM60A, but the levels of cyclin D1 protein were significantly higher at all cycle stages than in control cells. In contrast, neither the abundance nor periodicity of cyclins A and B was affected by FAM60A depletion (Fig. 6, A and B). These data indicate that FAM60A is required for the proper periodicity of cyclin D1 and for restraining levels of this protein.
FIGURE 6.
FAM60A controls cyclin D1 levels during the cell cycle and influences the timing of the G1–S transition. A, FAM60A depletion from U2OS cells hardly affects cyclin D1 periodicity but caused increased cyclin D1 levels at all cell cycle stages. U2OS transiently transfected with the FAM60A-1 siRNA or a control (GFP) siRNA were synchronized in mitosis by release from a double thymidine block into nocodazole for 20 h. The cells were then released from mitotic arrest and lysed at the times indicated. Extracts were subjected to Western blotting with the antibodies indicated. Lane A, asynchronously growing cells. B, quantitation of cyclin D1 and cyclin A2 protein levels in A was done using ImageJ (National Institutes of Health). C, depletion of FAM60A accelerates the G1–S transition. U2OS cells treated with the siRNAs indicated and arrested in mitosis were released from mitotic arrest into medium containing EdU, and samples taken at the times indicated were tested for EdU incorporation into DNA by FACS analysis. Data in B and C are represented as means ± S.E. (error bars) of triplicate experiments.
Elevated cyclin D1 levels cause accelerated G1–S phase transition and premature entry to S phase (8, 10, 21), and so we tested whether FAM60A depletion had similar effects. U2OS cells were synchronized in mitosis by a double thymidine block followed by release into nocodazole for 20 h. Cells were then released from mitotic arrest, and S phase entry was monitored by incorporation of EdU into replicating DNA. As shown in Fig. 6C, siRNAs specific for FAM60A decrease the time taken for cells to enter S phase. Six hours after release, ∼10% of cells treated with control siRNA had incorporated EdU, in contrast with ∼17–20% of cells treated with siRNAs specific for FAM60A or SIN3A (Fig. 6C). Therefore FAM60A is essential for preventing premature S phase entry potentially because of deregulated expression of cyclin D1.
DISCUSSION
In this study we showed that FAM60A associates physically with the SIN3-HDAC complex and co-occupies at least two promoters with SIN3-HDAC. Depletion of FAM60A causes increased levels of cyclin D1 mRNA and protein, possibly because FAM60A promotes the association of HDAC1 with promoters regulated by SIN3-HDAC. The molecular mechanism whereby FAM60A promotes HDAC1 retention at the promoters promoter is not yet clear, but we hypothesize that in the context of the holo-SIN3-HDAC complex FAM60A favors a conformation optimal for HDAC1 retention. This may involve binding of FAM60A not only to SIN3-HDAC but also to promoter DNA through the conserved GATA-like zinc finger.
FAM60A expression fluctuates during the cell cycle. Furthermore, the occupancy of the cyclin D1 promoter by FAM60A and HDAC1 mirrors the fluctuation in FAM60A levels. Moreover, depletion of FAM60A from U2OS cells reduces the level of HDAC1 associated with the cyclin D1 promoter during the cell cycle. These observations, together with the finding that FAM60A expression mirrors cyclin D1 expression in U2OS cells, prompted us to speculate that repression of the cyclin D1 promoter by FAM60A-SIN3-HDAC might contribute to the cyclical expression of cyclin D1. However, depletion of FAM60A from U2OS cells leaves cyclin D1 periodicity largely intact. Nonetheless, cyclin D1 levels are significantly higher at all cell cycle stages when FAM60A is depleted. We conclude that FAM60A restrains cyclin D1 expression, and even though its expression mirrors cyclin D1 there must be other mechanisms that are responsible for the cell cycle periodicity of cyclin D1 mRNA and protein. Multiple mechanisms can be envisaged such as control of mRNA stability and control of protein stability (13).
The increase in cyclin D1 protein levels in FAM60A-depleted cells is reminiscent of the overexpression of cyclin D1 in cancers (8, 11, 18). Cyclin D1 overexpression is sometimes caused by gene amplification or increased protein stability (13). However, there are many tumors in which cyclin D1 is elevated without gene amplification (12), and it will be interesting to test whether mutations in FAM60A could be responsible in some cancers.
We, and others, identified FAM60A as a substrate for the ATM kinase in cells exposed to genotoxins4 (15). Despite not having been able to obtain any evidence that FAM60A influences cell cycle checkpoints, DNA repair, or genome stability it remains possible that FAM60A influences histone acetylation at sites of DNA damage, but this will require further investigation.
Supplementary Material
Acknowledgments
We thank James Hastie, Hilary MacLauchlan, and the Antibody Production Team at Division of Signal Transduction Therapy, University of Dundee, for help with antibody production; Dave Campbell, Sanjay Kothiya, and Nick Morrice for help with mass spectrometry; the DNA Sequencing Service at CLS, University of Dundee; and Tom Owen-Hughes for critically reading the manuscript.
This work was supported by the Medical Research Council (to C. C. P. and J. R.), the Division of Signal Transduction Therapy, University of Dundee (to I.M.M., T.M., and J. R.), Cancer Research UK, and the Wellcome Trust (to S. R.).

This article contains supplemental Figs. S1–S4, Table S1, and Materials and Methods.
I. M. Muñoz and J. Rouse, unpublished data.
- HDAC
- histone deacetylase.
REFERENCES
- 1. Bannister A. J., Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang X. J., Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silverstein R. A., Ekwall K. (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 4. Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dannenberg J. H., David G., Zhong S., van der Torre J., Wong W. H., Depinho R. A. (2005) mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 19, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W. G., Jr., Kluger Y., Dynlacht B. D. (2008) A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rampalli S., Pavithra L., Bhatt A., Kundu T. K., Chattopadhyay S. (2005) Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Musgrove E. A., Caldon C. E., Barraclough J., Stone A., Sutherland R. L. (2011) Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572 [DOI] [PubMed] [Google Scholar]
- 9. Chen H. Z., Tsai S. Y., Leone G. (2009) Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. (1993) Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7, 1559–1571 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y. M., Sicinski P. (2006) Targeting cyclins and cyclin-dependent kinases in cancer: lessons from mice, hopes for therapeutic applications in human. Cell Cycle 5, 2110–2114 [DOI] [PubMed] [Google Scholar]
- 12. Han E. K., Lim J. T., Arber N., Rubin M. A., Xing W. Q., Weinstein I. B. (1998) Cyclin D1 expression in human prostate carcinoma cell lines and primary tumors. Prostate 35, 95–101 [DOI] [PubMed] [Google Scholar]
- 13. Shan J., Zhao W., Gu W. (2009) Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 36, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duro E., Lundin C., Ask K., Sanchez-Pulido L., MacArtney T. J., Toth R., Ponting C. P., Groth A., Helleday T., Rouse J. (2010) Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 40, 632–644 [DOI] [PubMed] [Google Scholar]
- 15. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 16. Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P. (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70 [DOI] [PubMed] [Google Scholar]
- 17. Yoshida M., Horinouchi S., Beppu T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17, 423–430 [DOI] [PubMed] [Google Scholar]
- 18. Bartkova J., Lukas J., Strauss M., Bartek J. (1995) Cyclin D1 oncoprotein aberrantly accumulates in malignancies of diverse histogenesis. Oncogene 10, 775–778 [PubMed] [Google Scholar]
- 19. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 20. Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C., Chen L. (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A 103, 10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Resnitzky D. (1997) Ectopic expression of cyclin D1 but not cyclin E induces anchorage-independent cell cycle progression. Mol. Cell. Biol. 17, 5640–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.