Background: Very short patch repair rectifies thymidine-guanine mismatches arising from spontaneous deamination of 5-methyl cytosine.
Results: The very short patch repair pathway was reconstituted using purified enzymes.
Conclusion: Patch length depends on the concentration of DNA ligase and may be regulated by the addition of MutS and MutL.
Significance: The results suggest roles for MutL and MutS in very short patch repair.
Keywords: DNA Damage, DNA Enzymes, DNA Methylation, DNA Mismatch Repair, DNA Polymerase, DNA Ligase, MutL, MutS, VSR Endonuclease
Abstract
The Escherichia coli very short patch (VSP) repair pathway corrects thymidine-guanine mismatches that result from spontaneous hydrolytic deamination damage of 5-methyl cytosine. The VSP repair pathway requires the Vsr endonuclease, DNA polymerase I, a DNA ligase, MutS, and MutL to function at peak efficiency. The biochemical roles of most of these proteins in the VSP repair pathway have been studied extensively. However, these proteins have not been studied together in the context of VSP repair in an in vitro system. Using purified components of the VSP repair system in a reconstitution reaction, we have begun to develop an understanding of the role played by each of these proteins in the VSP repair pathway and have gained insights into their interactions. In this report we demonstrate an in vitro reconstitution of the VSP repair pathway using a plasmid DNA substrate. Surprisingly, the repair track length can be modulated by the concentration of DNA ligase. We propose roles for MutL and MutS in coordination of this repair pathway.
Introduction
The repair of mismatched bases is of extraordinary importance in maintaining genomic integrity in both eukaryotic and prokaryotic organisms (1–3). In Escherichia coli, as is the case in other organisms, there are multiple endogenous mechanisms that can lead to a base-base mismatch including errors that result during DNA replication and the spontaneous deamination of DNA bases (4–6). Importantly, in every instance where a mismatch is generated, the repair machinery dedicated to correcting the error must be able to determine which DNA strand contains the correct base and which DNA strand contains the incorrect base, and subsequently repair the latter to maintain the integrity of the genome (reviewed in Ref. 7).
The occasional misincorporation of nucleotides by DNA polymerase III during DNA replication in E. coli is a well known source of base-base mismatches. The base mismatches that are missed by the proofreading exonuclease are repaired by the methyl-directed mismatch repair pathway (reviewed in Ref. 2 and 5), which discriminates between the parental and daughter DNA strands using the adenine methylation status of the DNA at nearby d(GATC) sequences (8). This pathway increases the overall fidelity of DNA replication by several orders of magnitude (9).
The spontaneous hydrolytic deamination of cytosine or adenine residues also leads to base pair mismatches, which are repaired by different pathways. The hydrolytic deamination of cytosine produces a uridine-guanine mismatch (10), whereas the hydrolytic deamination of adenosine produces a hypoxanthine-thymidine mismatch (11). The correct DNA strand is recognized in each case by virtue of the fact that uracil and hypoxanthine are not normal bases in DNA. The uridine-guanine mismatch is acted on by the uracil DNA glycosylase, producing an abasic site that is subsequently repaired by base excision repair proteins (12, 13). The hypoxanthine-thymidine mismatch recruits the hypoxanthine DNA glycosylase that removes the hypoxanthine base producing an abasic site (14, 15). This abasic site is then acted on by base excision repair enzymes to facilitate repair.
Hydrolytic deamination of 5-methyl cytosine (5-mC)3 presents a more challenging situation because the product of this event produces a T:G mismatch in which both bases are normal components of DNA. In addition, this deamination reaction is a common event in E. coli, as it is in all cells containing 5-mC, occurring at a rate two or three times higher than deamination of cytosine and more than sufficient to account for the mutational hot spots found associated with sites containing 5-mC (16). Because both guanine and thymidine are naturally occurring bases in E. coli, this mismatch is intractable to most repair pathways because strand discrimination is not possible. Presumably, this accounts for the high rate of mutation commonly associated with DNA sequences containing 5-mC (6, 17, 18). In addition, this T:G mismatch is refractory to the methyl-directed mismatch repair pathway during stationary phase because the double helix is fully methylated at the d(GATC) sequences that direct this repair process (19, 20). Therefore, this particular mismatch necessitates another DNA repair pathway.
In E. coli, the sequence 5′-CCWGG-3′/5′-CCWGG-3′ is methylated by the Dcm methylase at the second cytosine on each strand producing a 5-mC (21). The T:G mismatch generated by spontaneous deamination of the 5-mC is repaired in E. coli by a specific pathway that is able to discriminate between the erroneous thymidine and the correct guanine and selectively remove the incorrect thymidine (22). This repair mechanism is known as very short patch (VSP) repair reflecting the short (often less than 10 bases) patch used to restore the correct DNA sequence (23). It is important to note that this sequence is the only place in the E. coli chromosome that would produce a T:G mismatch caused by spontaneous hydrolytic deamination of 5-mC, because this sequence is the only place where the Dcm methyltransferase acts. Thus, a major function of VSP repair is to maintain 5′-C5meCWGG-3′ sequences within the genome (24).
As deduced from genetic assays, VSP repair requires the Vsr endonuclease, DNA polymerase I, DNA ligase I, MutL, and MutS to function at maximum efficiency (25–30). The gene product of vsr, the Vsr endonuclease, catalyzes the initiating step in VSP repair. The Vsr endonuclease recognizes the T:G mismatch in the appropriate sequence context and cleaves 5′ to the mismatched T, leaving a nick with a 3′-OH and a 5′-PO4 (22, 31–34). DNA polymerase I is thought to bind at the nick produced by Vsr cleavage and undergo nick translates, using its 5′ to 3′ exonuclease activity and its 5′ to 3′ polymerase activity, restoring the correct C:G base pair while adding a minimal number of additional nucleotides (27). This nick translation reaction leaves a ligatable nick that is presumably sealed by DNA ligase I to restore the integrity of the E. coli genome. The exact functions of MutL and MutS in the pathway remain unknown, although MutL has been suggested to stimulate the activity of the Vsr endonuclease (32, 35). Unlike methyl-directed mismatch repair, deletion of MutS or MutL does not abolish the activity of the VSP repair pathway. Rather, these deletions result in a significant decrease in the efficiency of repair. In a wild-type genetic background, VSP repair corrects a T:G mismatch in the 5′-CTWGG-3′/5′-C5meCWGG-3′ sequence with nearly 100% efficiency. However, in a mutL or a mutS mutant VSP corrects T:G mismatches in the appropriate sequence context with reduced efficiency (28–30). These data suggest that although MutL and MutS are not absolutely required for the function of the VSP repair pathway, they serve to augment or somehow regulate the pathway. Therefore, the minimal VSP repair pathway requires three proteins: the Vsr endonuclease, DNA polymerase I, and presumably DNA ligase I (23, 27, 36).
Here we report the reconstitution of the E. coli VSP repair reaction using purified proteins and a model plasmid DNA substrate. We demonstrate the requirement for the Vsr endonuclease, DNA polymerase I, and DNA ligase I in a minimal VSP repair reaction. In this reaction the length of the repair patch can be modulated by altering the DNA ligase concentration. Furthermore, we show that MutS and MutL are implicated in the regulation of VSP repair as demonstrated by their ability to reduce the length of the repair patch at a defined DNA ligase concentration.
MATERIALS AND METHODS
Cloning
The gene encoding the Vsr endonuclease (vsr) was amplified from E. coli GE1752 genomic DNA using the following primers: 5′-GGGAATTCCATATGGCCGACGTTCACGAT-3′ and 5′-TTCCGCTCGAGAGCGAGTAAATGAATCCC-3′ designed to introduce an NdeI site and an XhoI site on the N- and C-terminal ends of the gene, respectively. The amplified DNA fragment was digested to completion with NdeI and XhoI and inserted into the expression plasmid pTYB1 (New England Biolabs). The construct used for expression and purification of the Vsr endonuclease contained vsr in frame with the intein::CBD and was verified by sequencing.
The polA gene was amplified from GE1752 genomic DNA using the following primers: 5′-TTTTTCATATGGTTCAGATCCCC-3′ and 5′-TTTTTGGATCCTTAGTGCGCCTGATCC-3′ designed to introduce an NdeI site and a BamHI site on the N- and C-terminal ends of the gene, respectively. The amplified DNA fragment was inserted into the pET15b expression plasmid (Novagen) using the NdeI and BamHI restriction sites. This construct was verified by sequencing and used for expression and purification of DNA polymerase I.
The ligA gene was amplified from genomic GE1752 genomic DNA using the following primers: 5′-GGAATTCCATATGGAATCAATCGAACAACA-3′ and 5′-CGCGGATCCTCAGCTACCCAGCAAACGC-3′ designed to introduce an NdeI site and a BamHI site on the N-terminal and C-terminal ends of the gene, respectively. Cloning of the ligA gene was as detailed above for the polA gene. The construct was verified by sequencing.
The pUC19-VSR plasmid was constructed by digesting pUC19 DNA (New England Biolabs) with BamHI and EcoRI and inserting a duplex DNA fragment containing multiple Nt.BbvCI nicking sites. The duplex DNA fragment was prepared by annealing the following synthetic oligonucleotides: 5′-AATTCCTCAGCAATCCTCAGCCAGGCCTCAGCTGGCCTCAGCG-3′ and 5′-GATCCGCTGAGGCCAGCTGAGGCCTGGCTGAGGATTGCTGAGG-3′. The annealing reaction created compatible ends for insertion into the BamHI- and EcoRI-digested pUC19 vector. Clones containing the correct DNA insert were identified and verified by restriction digest. The supercoiled plasmid was purified using a CsCl/EtBr gradient.
Protein Purifications
BL21(λDE3) (F− ompT hsdSB(rB−mB−) gal dcm (λDE3)) cells harboring the pTYB1-Vsr plasmid were grown in 500 ml of LB medium containing 100 μg/ml ampicillin at 37 °C to an A600 of 0.6–0.8. The temperature was reduced to 16 °C, and protein expression was induced by the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. The cells were grown for 16 h at 16 °C, harvested by centrifugation, suspended in 5 ml of VSR lysis buffer (25 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.1 mm PMSF, 0.1 mm benzamidine, 500 mm NaCl, 10% (v/v) glycerol) and frozen for later use.
Cells in VSR lysis buffer were thawed at 4 °C. Lysozyme was added to a final concentration of 50 μg/ml, and the mixture was incubated at 4 °C for 60 min. Triton X-100 was added to a final concentration 0.1% (v/v), and the mixture was slowly heated to 20 °C with stirring. The mixture was briefly sonicated to reduce viscosity and centrifuged at 50,000 × g for 60 min to clarify the lysate. The supernatant was applied to a 1-ml chitin resin (New England Biolabs) column equilibrated with VSR lysis buffer and washed until the flow-through contained no detectable protein as measured by a Bradford protein assay. Five ml of VSR lysis buffer containing 50 mm DTT was applied and allowed to flow through the column. The column was then sealed and incubated at 4 °C for 48 h. Ten ml of VSR lysis buffer were used to elute the column, and 1-ml fractions were collected. Fractions containing Vsr were pooled and dialyzed twice against VSR Superdex buffer (250 mm NaCl, 25 mm Tris-HCl, pH 8.0, 1 mm EDTA, 2 mm DTT, 10% (v/v) glycerol). The dialyzed pool was concentrated to 300 μl using an Amicon Ultra 15 10,000 molecular weight cutoff (Millipore) centrifugal concentration device and applied to an equilibrated Superdex 200 gel filtration column (Amersham Biosciences). Seventy 0.5-ml fractions were collected, and fractions containing the Vsr endonuclease were pooled and dialyzed three times against VSR storage buffer (250 mm NaCl, 25 mm Tris-HCl, pH 8.0, 1 mm EDTA, 2 mm DTT, 50% (v/v) glycerol). Purified Vsr endonuclease was stored at −20 °C and was greater than 95% pure as judged by SDS-PAGE. Fresh protein preparations were made frequently to prevent possible artifacts resulting from degradation products.
BL21(λDE3) cells harboring the pET15b-PolA plasmid were grown in 500 ml of LB medium containing 100 μg/ml ampicillin at 37 °C to an A600 of 0.6. The production of DNA polymerase I was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm. The cells were incubated for an additional 4 h at 37 °C and harvested by centrifugation. The cells were suspended in 5 ml of Pol I lysis buffer (500 mm NaCl, 50 mm Tris-HCl, pH 7.5, 5 mm imidazole, 10% (v/v) glycerol) and frozen for later use.
The cells were thawed at 4 °C and lysed, and a cleared lysate was prepared as described above. The cleared lysate was applied to a 1-ml Talon metal affinity resin (BD Bioscience) column equilibrated with lysis buffer and washed as above. The column was eluted with 10 ml of Pol I lysis buffer containing 200 mm imidazole. Fractions containing Pol I were pooled and dialyzed twice against Pol I MonoQ buffer (40 mm NaCl, 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm DTT, 10% glycerol). The dialyzed pool was loaded onto a MonoQ HR (10/10) column (Amersham Biosciences) equilibrated with Pol I MonoQ buffer and washed until the flow-through contained no detectable protein. The column was eluted with a 160-ml linear NaCl gradient from 40 to 500 mm NaCl. DNA polymerase I eluted at ∼200 mm NaCl. Fractions containing DNA polymerase I were tested for endonuclease activity using supercoiled pUC19 DNA. Fractions that contained no detectable endonuclease activity were pooled and dialyzed three times against Pol I storage buffer (50 mm Tris-HCl, pH 7.5, 1 mm DTT, 0.5 mm EDTA, 50% (v/v) glycerol) and stored at −20 °C. DNA Polymerase I was determined to be greater than 95% pure as judged by SDS-PAGE.
BL21(λDE3) cells harboring pLysS (Novagen) and pET15b-LigA were grown at 37 °C in 500 ml of LB containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol to an A600 of 0.6–0.8. Expression of DNA ligase I was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm. The cells were incubated for an additional 4 h at 37 °C and harvested by centrifugation. The pellets were suspended in 5 ml of Ligase I lysis buffer (500 mm NaCl, 50 mm Tris-HCl, pH 7.5, 10% (v/v) glycerol) and frozen for later use.
The cells were thawed at 4 °C, lysed and a cleared lysate was prepared as described above. The lysate was applied to a 1-ml nickel-charged His bind resin (Novagen) column equilibrated in Ligase I lysis buffer and washed as above. The column was eluted using Ligase I lysis buffer containing 200 mm imidazole. Fractions containing Ligase I were pooled and dialyzed twice against Ligase I MonoQ buffer (50 mm NaCl, 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm DTT, 10% (v/v) glycerol). The dialyzed pool was loaded onto a MonoQ HR (10/10) column, washed, and eluted as described above for DNA polymerase I. Ligase I eluted at ∼250 mm NaCl. Relevant fractions were pooled and dialyzed twice against 100 volumes of Ligase I Superdex buffer (250 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1 mm DTT, 2 mm EDTA, 10% (v/v) glycerol). After dialysis the pool was concentrated to 300 μl using an Amicon Ultra 15 30,000 molecular weight cutoff (Millipore) centrifugal concentration device and loaded onto a Superdex 200 gel filtration column (Amersham Biosciences) equilibrated in Ligase I Superdex buffer. The column was eluted with 35 ml of Lig I Superdex buffer, and fractions (0.5 ml) were collected. Fractions containing DNA ligase I were tested for endonuclease activity using supercoiled pUC19 DNA. Fractions containing DNA ligase I and lacking detectable endonuclease activity were pooled and dialyzed into Ligase I storage buffer (10 mm Hepes-KOH, pH 7.4, 50 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 50% (v/v) glycerol). After dialysis DNA ligase I was stored at −20 °C. DNA ligase I was determined to be greater than 95% pure as measured by SDS-PAGE. MutS and MutL were expressed and purified as previously described (37, 38).
pUC19-VSR Substrate Construction
The pUC19-VSR plasmid (10 μg) was digested in a 50-μl reaction with 10 units of Nt.BbvCI at 37 °C for at least 10 h. After the addition of 250 μl of buffer PBI (supplied in Qiagen PCR clean kit), the reactions were heated to 80 °C for 20 min. The aliquots were immediately applied to a PCR clean column and centrifuged according to the protocol provided by the supplier. The resulting gapped pUC19-VSR DNA was eluted from the spin column in 30 μl of 10 mm Tris-HCl (pH 7.5). Three synthetic oligonucleotides were designed to be annealed into the gap created by the above procedure: 5′-TCAGCAATCCTCAGCTAGGCCTCAGCTGGCCTCAGCG-3′, 5′-TCAGCAATCCTCAGC5meCAGGCCTCAGCTGGCCTCAGCG-3′, and 5′-TCAGCAATCTTCAGCCAGGCCTCAGCTGGCCTCAGCG-3′. The first oligonucleotide creates a T:G mismatch within the sequence context 5′-CTAGG-3′/5′-CCTGG-3′ where the mismatch is underlined. The second oligonucleotide forms a homoduplex DNA and serves as a control. The third oligonucleotide creates a T:G mismatch (underlined) outside the 5′-CCWGG-3′/5′-CCWGG-3′ sequence context also serving as a control. Each oligonucleotide was purified on a 16% denaturing polyacrylamide gel prior to annealing and was phosphorylated using T4 polynucleotide kinase (New England Biolabs). The phosphorylated oligonucleotides were annealed into the gapped pUC19-VSR DNA as follows: annealing reaction mixtures (47 μl) contained 30 μl of gapped pUC19-VSR DNA, 30.6 pmol of phosphorylated oligonucleotide, 50 mm Tris-HCl (pH 7.5), 10 mm MgOAc, 5 mm dithiothreitol, 50 mm NaCl and were heated to 80 °C in a GeneAmp PCR System 2400 Thermocycler (Applied Biosystems). The temperature was subsequently reduced by 1 °C/min until the reaction temperature reached 16 °C. ATP (final concentration of 1 mm) and T4 DNA ligase (200 units; New England Biolabs) were added, and the reaction was incubated at 37 °C for 60 min. Covalently closed DNA was isolated using a CsCl/EtBr gradient or by gel extraction. The concentration of the substrate was measured spectrophotometrically at 260 nm. To produce a radiolabeled substrate the oligonucleotide was phosphorylated using T4 polynucleotide kinase in the presence of 40 μCi of [γ-32P]ATP.
Vsr Nicking Assays
Reaction mixtures (20 μl) contained 50 ng of the indicated DNA substrate (∼1.5 nm molecules), 6 mm MgCl2, 25 mm Tris-HCl (pH 7.5), 20 mm NaCl, 6.1 mm 2-mercaptoethanol, 50 μg/ml BSA and were preincubated at 37 °C for 2 min. The reactions were initiated by the addition of the indicated amount of the Vsr endonuclease and stopped after a 10-min incubation at 37 °C by the addition of 5 μl of stop solution (50 mm EDTA, 0.05% (v/v) bromphenol blue, 45% (v/v) glycerol). The products were resolved on a 1.4% 40 mm Tris-acetate, 1 mm EDTA (pH 8.3) (TAE)-buffered agarose gel containing 0.5 μg/ml ethidium bromide. The gel was UV-irradiated at 254 nm for 30 min to ensure equivalent staining of nicked and supercoiled DNA and restained using 0.5 μg/ml ethidium bromide for 30 min. The intensity of each species on the gel was quantified with Alpha Imager Software using the spot densitometry tool, and the fraction of the total DNA that was nicked was calculated.
VSR nicking reactions to determine location of nick site were as described above using 50 ng of pUC19-VSR heteroduplex [32P]DNA and were terminated by the addition of 180 μl of a solution containing 10 mm Tris-HCl (pH 7.5), 100 mm NaCl, 20 mm EDTA, and 162 μg/ml oyster glycogen. The Vsr endonuclease was removed by phenol/chloroform extraction followed by an ethanol precipitation. The pellets were suspended in 17 μl of 10 mm Tris-HCl (pH 7.5) followed by digestion with EcoRI + BamHI in an appropriate buffer. The digestion products were analyzed on a 16% denaturing polyacrylamide gel run at 30 W for 90 min.
DNA Polymerase I Repair Assays
Reaction mixtures (20 μl) contained 50 ng of pUC19-VSR heteroduplex DNA, 6 mm MgCl2, 25 mm Tris-HCl (pH 7.5), 20 mm NaCl, 6.1 mm 2-mercaptoethanol, 50 μg/ml BSA, 50 μm of each dNTP, and the indicated concentration of DNA polymerase I and were preincubated for 2 min at 37 °C. The reactions were initiated by the addition of the Vsr endonuclease at a final concentration of 52 nm. The reactions were incubated at 37 °C for 60 min and terminated as indicated above. The Vsr endonuclease and DNA polymerase I were removed by phenol/chloroform extraction followed by ethanol precipitation. Pellets were suspended in 17.5 μl of 10 mm Tris-HCl (pH 7.5) and digested with XcmI + AlwNI at 37 °C for 60 min. The reactions were stopped and analyzed using agarose gel electrophoresis as described above. The fraction of DNA repaired was calculated as the total DNA cleaved by both XcmI and AlwNI divided by the total DNA in each lane.
VSP Repair Patch Length Assays
Reaction mixtures (100 μl) contained 200 ng of pUC19-VSR heteroduplex DNA, 6 mm MgCl2, 25 mm Tris-HCl (pH 7.5), 20 mm NaCl, 6.1 mm 2-mercaptoethanol, 50 μg/ml BSA, 10 μm dATP, 0.5 μm dCTP, 10 μm dGTP, 10 μm dTTP, 2 μCi [α-32P]dCTP, 26 μm NAD+, 17.3 nm DNA polymerase I, and the indicated concentrations of DNA ligase I, MutS, and/or MutL and were preincubated at 37 °C for 2 min. Reactions that included MutS, MutL, or MutS and MutL also contained 1 mm ATP. The reactions were initiated by the addition of 50 nm Vsr endonuclease and allowed to proceed for 60 min. The reactions were stopped as indicated above, and the proteins were removed by phenol/chloroform extraction followed by ethanol precipitation. The pellets were suspended in 33 μl of 10 mm Tris-HCl (pH 7.5). Each suspended DNA pellet was split into three samples: one sample contained 16.5 μl and two samples contained 8.3 μl of the initial 33 μl. The first sample was prepared for agarose gel electrophoresis. The second sample was digested with AlwNI and XcmI in a final volume of 20 μl for 60 min at 37 °C. The third sample was digested with BfaI and PvuII at 37 °C for 90 min. 0.5 μl of TaqI was then added to the reaction, and the reaction was allowed to proceed at 65 °C for 60 min. The first and the second reactions were resolved on a 1.4% TAE-buffered agarose gel containing 0.5 μg/ml ethidium bromide. The gel was UV-irradiated and restained as described above and imaged using Alpha Imager Software. The gel was then soaked in 300 ml of 40% (v/v) methyl alcohol, 10% (v/v) acetic acid, and 3% (v/v) glycerol for 60 min, dried, and exposed to a phosphor storage screen followed by quantification using ImageQuant software. The third reaction was resolved on a 6% native polyacrylamide gel and stained with 0.5 μg/ml ethidium bromide. The gel was imaged using Alpha Imager Software and then exposed to a phosphor storage screen followed by quantification using ImageQuant software. Specific activity was calculated as the number of counts per cytosine per band as a function normalized to the specific activity of band 2 in the lane containing no ligase or band 2 in the lane containing no MutS and no MutL. The resulting specific activity was normalized by using the ethidium bromide-stained image to correct for small gel loading inequalities.
RESULTS
The components of the VSP repair pathway, as deduced from genetic studies, include the Vsr endonuclease, DNA polymerase I, DNA ligase I, MutL, and MutS (25). The current model for VSP repair postulates that the Vsr endonuclease incises the DNA backbone 5′ to the mismatched thymidine within the sequence context 5′-CTWGG-3′/5′-C5meCWGG-3′ producing a single strand break with a 3′-OH and a 5′-PO4 (34). DNA polymerase I then binds and removes the mismatched thymidine and several nucleotides beyond the mismatch via the 5′ → 3′ nick translation ability of the polymerase. DNA ligase I seals the nicked DNA to restore the integrity of the DNA strand, and genetic studies (27, 30, 39) suggest that the length of the repair track is short, perhaps 10 nucleotides or less. In addition, both MutL and MutS are utilized in this pathway, although they are not strictly required (27, 28, 40–42). We have completed a biochemical reconstitution of a minimal VSP repair reaction using purified VSR endonuclease, DNA polymerase I, and DNA ligase and have begun to investigate the role MutS and MutL in the context of a reconstituted VSP repair pathway.
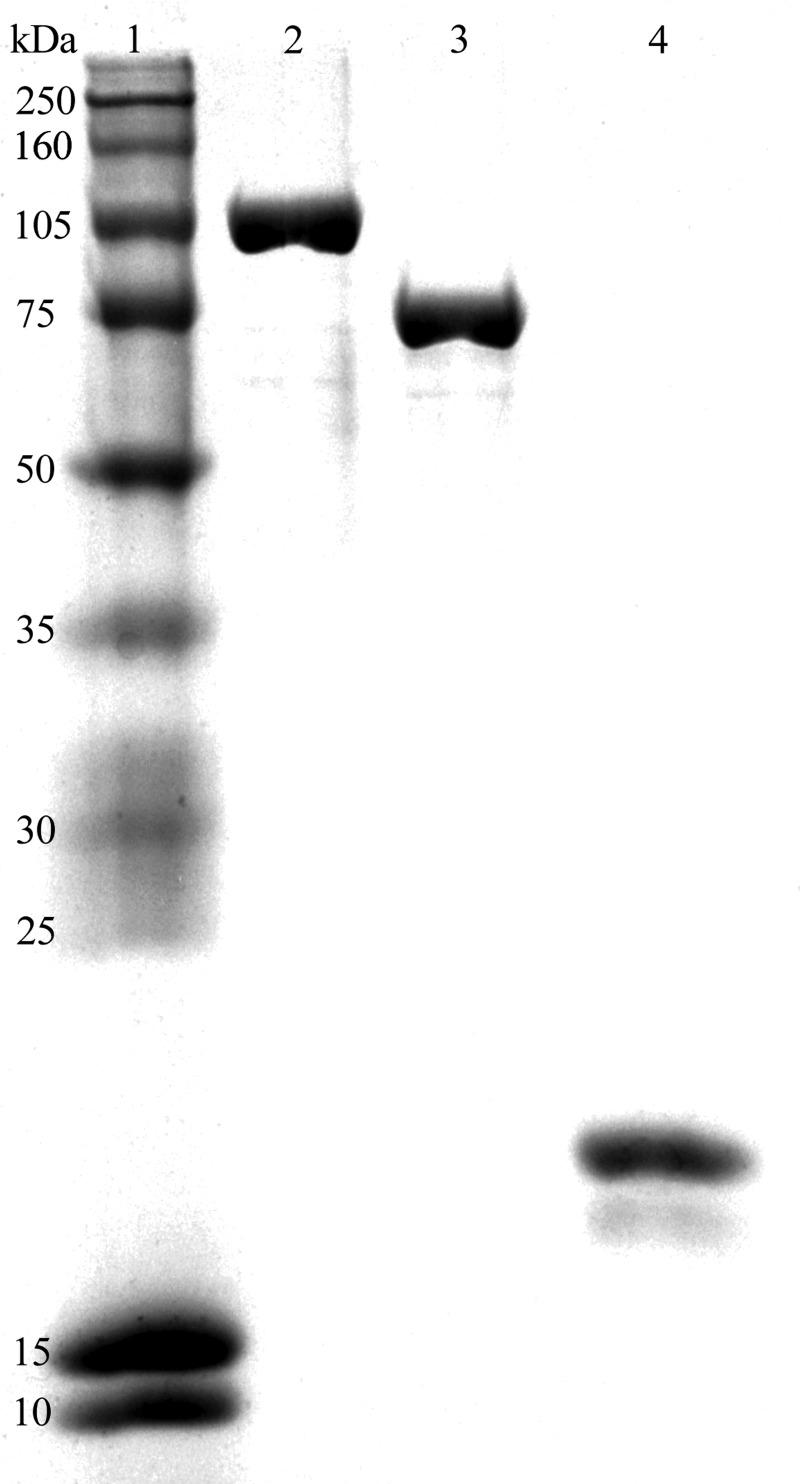
The Vsr endonuclease, DNA ligase I, and DNA polymerase I were purified as described under “Materials and Methods.” Each protein was judged to be greater than 95% homogeneous as evaluated by SDS-PAGE (Fig. 1). The visible protein product migrating below the full-length Vsr endonuclease is a well documented N-terminal degradation product of the Vsr endonuclease that partially reduces the activity of the protein, whereas the specificity of the nicking reaction is not altered (32, 33, 43). It is a minor contaminant and does not appear to increase with time. None of the purified proteins contained contaminating endonuclease activity.
FIGURE 1.
SDS-PAGE analysis of purified DNA polymerase I, DNA ligase I, and the Vsr endonuclease. Purified proteins (3 μg) were resolved on an 11% polyacrylamide gel run in the presence of SDS and stained with Coomassie Blue. Lane 1, molecular mass standards (size in kDa indicated on the left); lane 2, DNA polymerase I; lane 3, DNA ligase I; lane 4, the Vsr endonuclease.
The DNA substrate used for reconstitution of the VSP repair pathway (see Fig. 5) is a derivative of pUC19 and is fully Dcm- and Dam-methylated in vivo. A single T:G mismatch exists within the sequence context 5′-CTAGG-3′/5′-C5meCTGG-3′, making this covalently closed heteroduplex a substrate for the Vsr endonuclease. The T:G mismatch interrupts an XcmI restriction site rendering the substrate resistant to XcmI digestion. If the T:G mismatch is appropriately repaired to a C:G, the duplex becomes sensitive to digestion by XcmI. Repair in the opposite direction (i.e., to an T:A) leaves the site resistant to cleavage by the XcmI endonuclease. A similar substrate was constructed with no mismatch, and a substrate with a T:G mismatch outside the 5′CCAGG-3′/5′-CCTGG-3′ sequence context was also prepared.
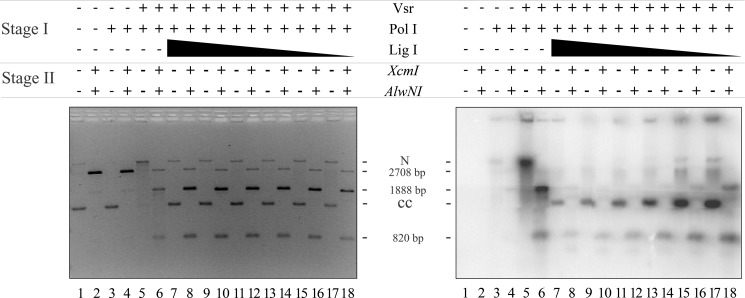
FIGURE 5.
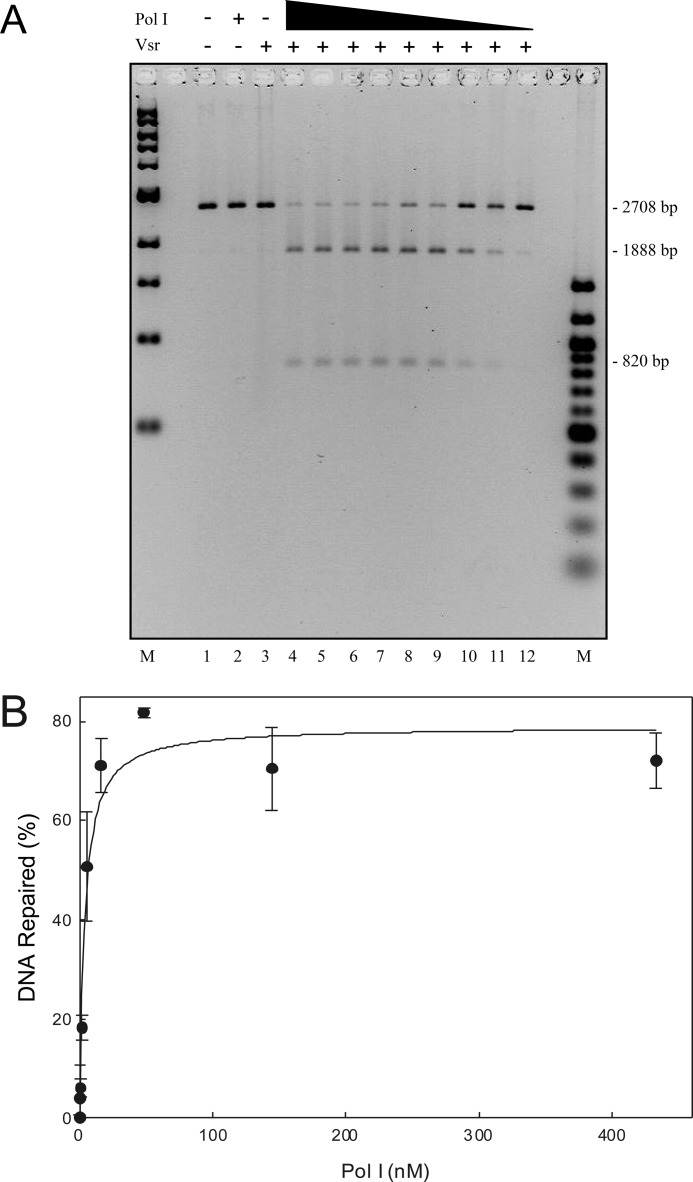
DNA ligase I seals the nick created by DNA polymerase I nick translation. Repair and ligation reactions were conducted as described under “Materials and Methods” using 200 ng of pUC19-VSR heteroduplex DNA (∼1.2 nm molecules), 10 μm dATP, dGTP, or dTTP, and 0.5 μm [α-32P]dCTP, 52 nm Vsr endonuclease, 17 nm DNA polymerase I and a titration of DNA ligase I from 1100 to 5 nm. After incubation for 60 min at 37 °C, the reactions were stopped, the proteins were removed by organic extraction, and the DNA was precipitated with EtOH. One-quarter of each reaction was digested with XcmI and AlwNI to confirm that repair had occurred (even-numbered lanes), and another quarter of the reaction remained untreated (odd-numbered lanes). Both the digested and undigested reactions were run on a 1.4% agarose gel containing 0.5 μg/ml ethidium bromide. The left panel shows a gel stained with EtBr (0.5 μg/ml), confirming that repair had occurred. The right panel shows an autoradiogram demonstrating the incorporation of [α-32P]dCMP and ligation to form covalently closed circles after repair. The positions of nicked DNA (N), covalently closed DNA (CC), and the expected products of the restriction digest (2708, 1888, and 820 bp) are shown.
The Vsr Endonuclease Reaction
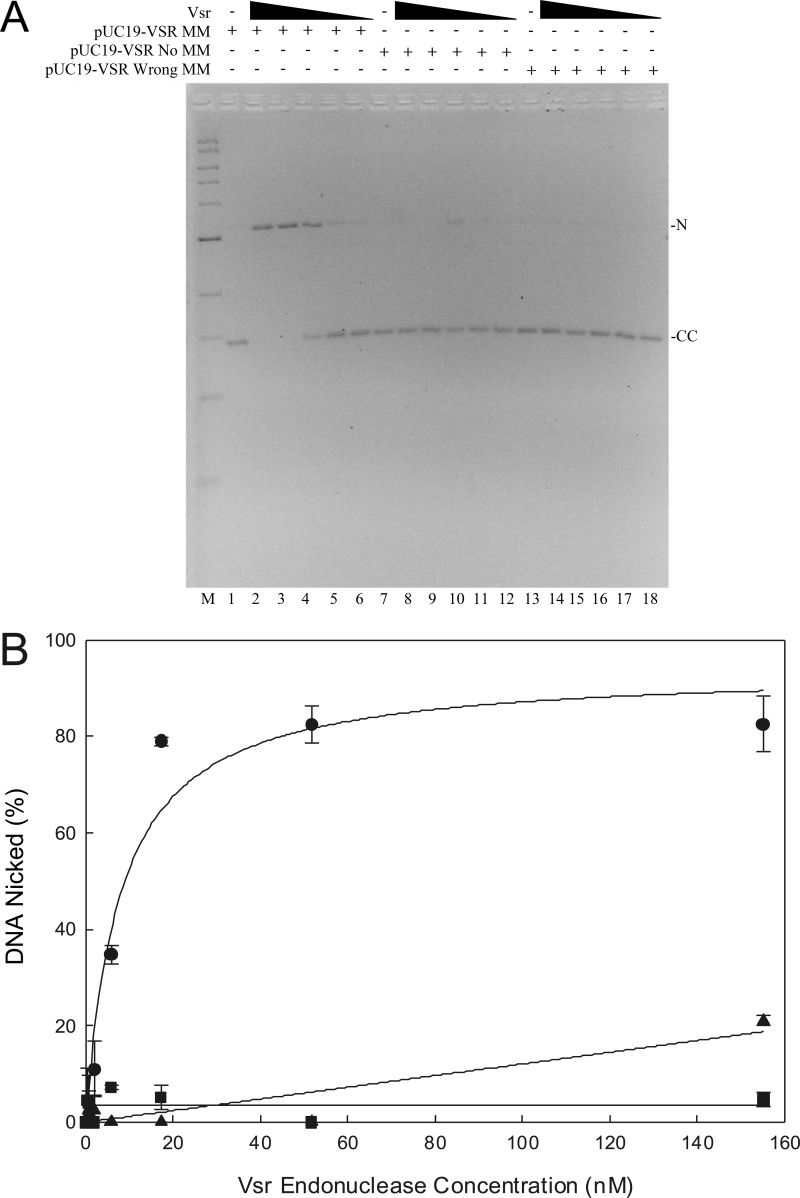
Previous reports (22, 31) have shown that the Vsr endonuclease introduces a nick 5′ to the mismatched thymidine within the sequence context 5′-CTWGG-3′/5′-C5meCWGG-3′. However, the Vsr nicking reaction has largely been studied using synthetic oligonucleotides. To demonstrate Vsr-catalyzed nicking on a plasmid substrate, the covalently closed circular heteroduplex described above was used (Fig. 2). As the Vsr endonuclease concentration increased, the fraction of the substrate converted to nicked DNA increased, demonstrating a dependence on the Vsr endonuclease for the conversion of covalently closed circular DNA to nicked circular DNA (Fig. 2A, lanes 2–6). At a concentration of 52 nm Vsr, essentially all of the substrate was converted to a nicked form. There is no evidence for the production of a linear DNA species.
FIGURE 2.
Vsr-catalyzed nicking of a covalently closed circular DNA substrate. Vsr-dependent nicking reactions were as described under “Materials and Methods.” A, the titration of the Vsr endonuclease was from 153 to 0 nm with each DNA substrate, and incubation was for 10 min at 37 °C. The reactions were stopped, and the products were resolved on a 1.4% agarose gel containing 0.5 μg/ml ethidium bromide. The positions of nicked (N) and covalently closed (CC) DNA are noted on the right. A representative gel is shown. B, quantification of Vsr nicking reaction using covalently closed DNA substrates. The fraction of nicked DNA was determined as indicated under “Materials and Methods.” The data represent the average of at least three experiments with error bars indicating the standard deviation about the mean. The plasmids used were covalently closed DNA containing a T:G mismatch the Vsr recognition sequence (●), covalently closed homoduplex DNA (■), and covalently closed DNA containing a T:G mismatch outside the Vsr recognition sequence (▴).
We performed the same experiment using either a homoduplex DNA substrate or a substrate that contained a T:G mismatch outside the 5′-CTWGG-3′/5′-CCWGG-3′ sequence context. Using the homoduplex substrate, we were unable to detect measurable nicking catalyzed by the Vsr endonuclease consistent with previous reports using oligonucleotide substrates (31) (Fig. 2A, lanes 8–12). We were, however, able to detect a small amount of nicking when using the substrate that contained a T:G mismatch outside the 5′-CTWGG-3′/5′-CCWGG-3′ sequence context (Fig. 2A, lanes 14–18). This result is consistent with previous reports indicating the ability of Vsr to recognize and cleave DNA, albeit much less efficiently, in sequences that are related to but not identical with the canonical Vsr recognition sequence (31, 33). Importantly, the fraction of the DNA substrate nicked at this alternate location is significantly reduced. Taken together, these results demonstrate that the preferred substrate for the Vsr endonuclease contains a T:G mismatch within the 5′-CTWGG-3′/5′-CCWGG-3′ sequence, and nicking of a covalently closed circular molecule is robust in vitro.
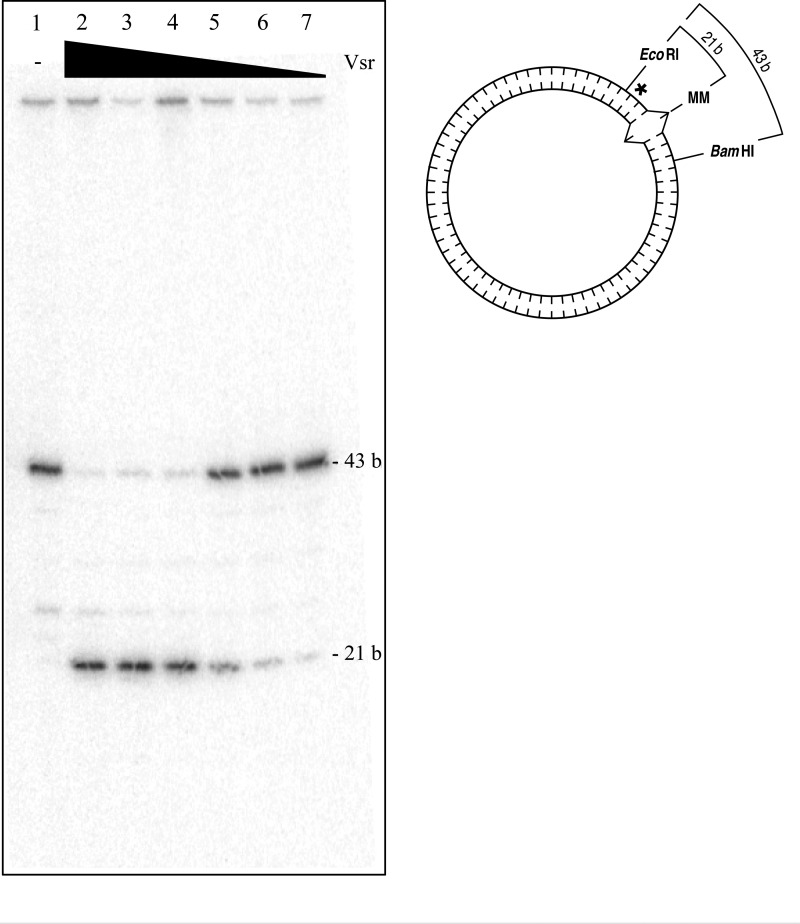
Although the experiment described above demonstrates that the Vsr endonuclease produces a nick in the pUC19-VSR heteroduplex substrate in a Vsr-dependent manner, the assay does not demonstrate that the nick is produced immediately 5′ to the mismatched thymidine. To precisely define the location of the nick produced by the Vsr endonuclease, a covalently closed pUC19-VSR heteroduplex substrate containing an internal 32P label was prepared as described under “Materials and Methods.” The [32P]DNA substrate was incubated with varying concentrations of the Vsr endonuclease and subsequently cleaved with EcoRI and BamHI to release the relevant DNA fragment for analysis by denaturing PAGE. The expected product of Vsr-catalyzed cleavage immediately 5′ to the mismatched thymidine is a DNA fragment 21 nucleotides in length. The 43-nucleotide DNA fragment represents the DNA species that was not nicked by Vsr. As seen in Fig. 3, nicking by the Vsr endonuclease resulted in the production of a unique product 21 nucleotides in length, indicating that Vsr cleaves the DNA immediately 5′ to the mismatched thymidine.
FIGURE 3.
The Vsr endonuclease incises DNA immediately 5′ to the mismatched thymidine. Vsr nicking reactions were as described under “Materials and Methods.” The Vsr endonuclease was incubated with covalently closed pUC19-VSR heteroduplex [32P]DNA (shown on the right; asterisk denotes location of 32P, and carats denote the G:T mismatch) for 10 min at 37 °C. The reactions were stopped, and the Vsr endonuclease was removed by organic extraction followed by EtOH precipitation. The circular DNA was incubated with EcoRI and BamHI, which flank the Vsr endonuclease recognition sequence as noted in the depiction of the substrate on the right. The resulting products were resolved on 16% denaturing polyacrylamide gel. Lane 1, no protein control; lane 2, 153 nm Vsr; lane 3, 51 nm Vsr; lane 4, 17 nm Vsr; lane 5, 5.7 nm Vsr; lane 6, 1.9 nm Vsr; lane 7, 0.6 nm Vsr.
The Vsr Endonuclease and DNA Polymerase I Are Able to Repair a T:G Mismatch in Vitro
A previous report (27) suggested that DNA polymerase I removed the mismatched thymidine by nick translation with the incorporation of the correct deoxycytosine. To test this in the context of an in vitro reconstitution, we incubated the pUC19-VSR heteroduplex substrate with 52 nm Vsr endonuclease (a concentration that will produce maximal nicking of the substrate) and increasing concentrations of DNA polymerase I (Fig. 4). The resulting product was digested simultaneously with XcmI to confirm repair and AlwNI to linearize the plasmid to increase quantification accuracy. If the substrate is repaired with 100% efficiency, then two products are expected on an agarose gel (1888 and 820 bp) representing cleavage by both XcmI and AlwNI. A single product at 2708 bp represents an unrepaired species cleaved only by AlwNI. As expected, DNA polymerase I is required for the repair of the mismatched T:G in the pUC19-VSR heteroduplex because in the absence of DNA polymerase I, there is no repair of the DNA substrate (i.e., not cleaved with XcmI; Fig. 4A, lane 3). Moreover, as the concentration of DNA polymerase I was increased, the fraction of the substrate repaired also increased. We conclude that DNA polymerase I binds the 3′-OH produced by VSR cleavage and extends the end by at least one nucleotide, replacing the mismatched T with a C and restoring the XcmI restriction site. In fact, DNA polymerase I catalyzes significant extension of the 3′-OH as shown below.
FIGURE 4.
DNA polymerase I and the Vsr endonuclease are sufficient to repair a T:G mismatch. The VSP repair reaction contained 50 ng of covalently closed heteroduplex substrate, 52 nm Vsr endonuclease, and a titration of DNA polymerase I from 467 to 0.2 nm as described under “Materials and Methods.” After incubation at 37 °C for 60 min, the reactions were stopped, DNA polymerase I and the Vsr endonuclease were removed by organic extraction, and the DNA was precipitated with EtOH. A, the DNA was digested with XcmI and AlwNI, and products were resolved on a 1.4% agarose gel containing 0.5 μg/ml ethidium bromide. Marker lanes are shown on both sides of the gel. B, experiments identical to the one shown in A were quantified as described under “Materials and Methods.” The data presented represent the averages of at least three experiments with error bars representing standard deviations from the mean. The fraction of DNA repaired was calculated as described under “Materials and Methods.”
DNA Ligase I Seals the Nick Produced by DNA Polymerase I-catalyzed Nick Translation
Initial experiments demonstrated that purified DNA ligase I was able to catalyze the ligation of a Vsr-nicked DNA substrate, albeit poorly, where the mismatched base had not been removed (data not shown). Because of this confounding ligation product, we measured ligation after the incorporation of [α-32P]dCMP by DNA polymerase I to ensure that any ligation product visible in the autoradiograph had undergone the complete VSP repair reaction. We incubated pUC19-VSR heteroduplex substrate, Vsr, DNA polymerase I (both proteins at concentrations sufficient to ensure a complete reaction; see Figs. 2 and 4) and a titration of DNA ligase I. After incubation at 37 °C, proteins were removed by organic extraction, the DNA was precipitated, and the products were divided into two aliquots. One aliquot was untreated to reveal covalently closed DNA, and the second aliquot was digested with XcmI and AlwNI to confirm repair and enhance quantification.
The result of this experiment, shown in Fig. 5, demonstrated that DNA ligase I was able to seal the nick generated as a result of nick translation by DNA polymerase I as evidenced by the formation of covalently closed product containing [α-32P]dCMP (Fig. 5, right panel, lanes 7, 9, 11, 13, 15, and 17). Additional evidence supporting this conclusion derives from the fact this product can be cleaved by XcmI and AlwNI to yield the expected products at 1888 and 820 bp (Fig. 5, left panel, lanes 8, 10, 12, 14, 16, and 18). A small amount of DNA migrates at the position of linear DNA (2708 bp), indicating cleavage with AlwN1 in the absence of DNA repair (i.e., resistant to XcmI). Thus, the product of the DNA polymerase I extension reaction is a ligatable nick consistent with nick translation catalyzed by DNA polymerase I.
Inspection of the autoradiograph in the right panel reveals an unexpected result. There was significantly more [α-32P]dCMP incorporated into the repaired DNA at low ligase concentrations than at high ligase concentrations. This is evident upon comparison of the covalently closed DNA product (Fig. 5, right panel, compare lanes 7 and 17), as well as comparison of the 820-bp DNA fragment produced after restriction endonuclease digestion (Fig. 5, right panel, compare lanes 8 and 18). We suspected that the increased incorporation of [α-32P]dCMP at low DNA ligase I concentrations might be due to multiple rounds of DNA polymerase I binding, synthesis, and dissociation prior to the ligation event because DNA polymerase I has a modest processivity (10–15 nucleotides/binding event) and would not be expected to synthesize a long repair patch in a single binding event on a nicked DNA template (44). At higher concentrations of DNA ligase I, there would be fewer rounds of synthesis and rebinding by DNA polymerase I. Effectively, there appeared to be a competition between DNA polymerase I and DNA ligase I for the nick generated by the DNA polymerase I nick translation event. Thus, at high concentrations of DNA ligase I there was a higher probability of sealing the nick prior to binding by another molecule of DNA polymerase I. At low concentrations of ligase I, there were multiple rounds of binding and synthesis by DNA polymerase I. If this is the case, then the length of the repair track should be much greater at low ligase I concentrations than at high ligase I concentrations.
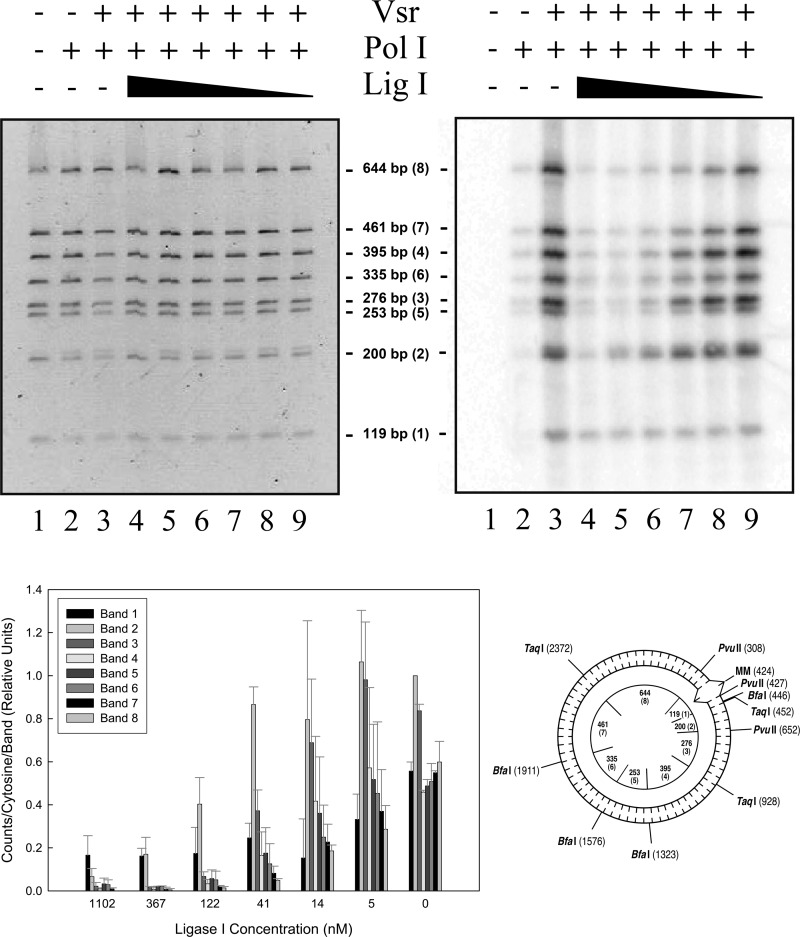
To test this hypothesis, we completed the reaction as detailed above with a modification that allowed the measurement of repair track length. Upon completion of the repair reaction at multiple ligase I concentrations, the DNA from each sample was analyzed in three separate reactions. The first reaction was resolved on an agarose gel to determine whether the DNA species was covalently closed. The second reaction was digested with AlwNI and XcmI to confirm that the DNA species was indeed repaired (data not shown). The third reaction was digested with BfaI, PvuII, and TaqI, which produces eight restriction fragments resolvable on a 6% polyacrylamide gel (Fig. 6). By quantifying the relative amount of radioactivity incorporated into each restriction fragment, it was possible to estimate the distance over which DNA polymerase I had incorporated nucleotides prior to sealing of the nick by DNA ligase I.
FIGURE 6.
The impact of DNA ligase I concentration on repair track length. Ligation and repair reactions were as described under “Materials and Methods” using 200 ng of covalently closed pUC19-VSR heteroduplex DNA (∼1.2 nm molecules), 10 μm dATP, 10 μm dGTP, 10 μm dTTP, 0.5 μm [α-32P]dCTP, 26 μm NAD+, 50 nm Vsr endonuclease, 17 nm DNA polymerase I, and DNA ligase I (Lig I) at the indicated concentrations. After incubation for 60 min at 37 °C, the reactions were stopped, the proteins were removed by organic extraction, and the DNA was precipitated with EtOH. Each reaction was digested with BfaI, PvuII, and TaqI, and the products were resolved on a 6% native polyacrylamide gel followed by staining with 0.5 μg/ml ethidium bromide. The upper left panel shows the stained polyacrylamide gel, and the upper right panel shows the autoradiogram. The lower left panel shows quantification of [32P]dCMP incorporated into each band normalized to band 2 in the lane containing no DNA ligase I. The data presented represent the averages of at least two experiments with error bars representing standard deviations about the mean. The pUC19-VSR heteroduplex plasmid DNA substrate is shown in the lower right panel. The T:G mismatch within the canonical Vsr endonuclease recognition sequence is denoted as MM. Relevant restriction sites and their positions are shown on the outside circle. Fragments are listed by size with the relative distance from the mismatch shown in parentheses. Restriction fragment sizes generated by cleavage with BfaI, PvuII, and TaqI are shown inside the circle. The agarose gel run to demonstrate repair of the DNA substrate, similar to that shown in Fig. 5, is not shown.
It is clear that the length of the synthetic repair track decreases significantly as a function of increasing DNA ligase I concentrations. In the absence of DNA ligase, there is significant incorporation into each of the DNA fragments produced by the restriction digest. Band 1 (119 bp) contains the site nicked by the Vsr endonuclease, and band 8 (644 bp) is located furthest from the nick site. As the DNA ligase I concentration was increased, the relative radioactive [α-32P]dCMP incorporated into band 8 decreased until it is essentially at background levels at 120 nm ligase I. In similar fashion, the relative radioactivity in other bands far removed from the nick site decreased as the DNA ligase I concentration was increased. Conversely, the relative incorporation in band 1 was highest at the highest DNA ligase I concentration. Taken together, this semiquantitative analysis of patch length strongly suggests that DNA polymerase I has the ability to synthesize a very long repair patch, presumably by loading on the DNA substrate multiple times, if DNA ligase I is not present at sufficient concentrations to seal the nick immediately after the dissociation of DNA polymerase I from the substrate. The reported processivity of DNA polymerase I is ∼15–20 nucleotides while nick translating (44). We presume that DNA polymerase I dissociates after each synthesis event, leaving a substrate suitable for DNA ligase I. If ligase I fails to seal the nick, then DNA polymerase I can bind again to extend the repair track. Thus, in this minimal in vitro reconstitution, it is possible to modulate the length of the repair track by varying the DNA ligase concentration.
MutS and MutL Coordinate the VSP Repair Reaction in Vitro
MutS and MutL are required for maximal efficiency of the VSP repair reaction in vivo; in the absence of either protein, VSP repair efficiency is significantly decreased (28–30). In addition, genetic evidence suggests that VSP repair tracks are quite short and seldom extend more than 10 nucleotides in length (30, 39). The data presented above clearly indicate that the repair track length in the reconstituted system lacking MutL and MutS extends well beyond the short repair tracks observed in vivo. Perhaps MutS and MutL coordinate the VSP repair system and act to reduce the length of the repair track. For example, if MutL and MutS were to increase the local concentration of the Vsr endonuclease, DNA polymerase I, and DNA ligase I at the site of repair, this would make DNA ligase I immediately available to seal the nick subsequent to DNA polymerase I-catalyzed nick translation and reduce the probability of loading of a molecule of DNA polymerase I a second time. This would have the overall effect of creating a shorter repair track. In addition, this would be consistent with previously reported results, indicating that MutL interacts with the Vsr endonuclease and stimulates the binding of this protein to its substrate (24, 32, 45, 46).
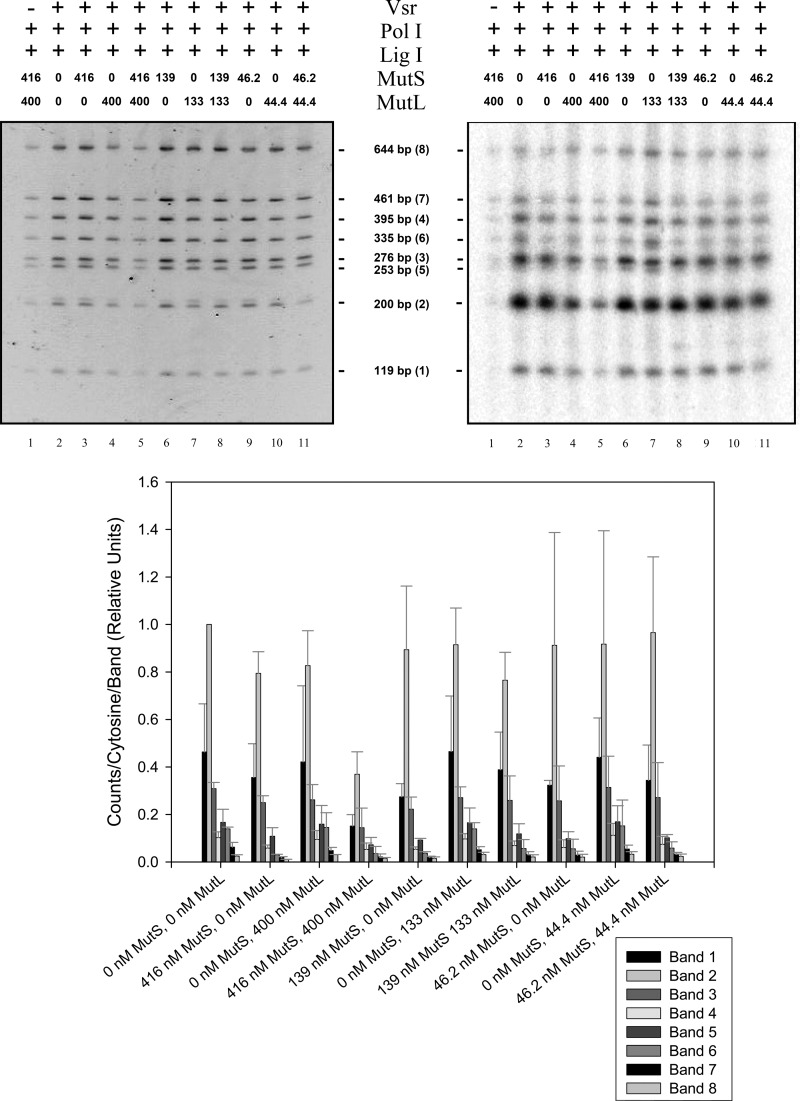
We tested the hypothesis that MutL and MutS may act to decrease the repair patch length by performing the repair track length assay detailed above using a fixed concentration of Vsr, DNA polymerase I, and DNA ligase I with multiple concentrations of MutS, MutL, or both MutS and MutL (Fig. 7). The results of this experiment showed a significantly decreased amount of radioactivity incorporated into the substrate molecule in the presence of both MutS and MutL and showed that the majority of radioactivity that was incorporated by DNA polymerase I was immediately adjacent to the mismatch site. In reactions containing 416 nm MutS and 400 nm MutL, it is clear that the length repair track is greatly reduced compared with reactions that contain no MutS or MutL proteins. This effect was dependent both MutS and MutL. The apparent track length in reactions containing one protein or the other was indistinguishable from control reactions lacking either protein. Moreover, the effect of MutL+MutS on track length was concentration-dependent. Reactions containing a 10-fold lower concentration of MutS and MutL showed a decrease in repair track length that was not as pronounced as the higher concentrations of MutS and MutL.
FIGURE 7.
MutS and MutL shorten VSP repair track lengths in vitro. Ligation and repair reactions were conducted as described in the legend for Fig. 5 with the addition of 1 mm ATP, a single concentration of DNA ligase I (4.5 nm), and the indicated concentration of MutS, MutL or both MutS and MutL. The reactions were processed and analyzed as described for Fig. 5. The upper left panel shows the stained polyacrylamide gel, and the upper right panel shows the autoradiogram. Fragments are listed by size with the relative distance from the mismatch shown in parentheses. The bottom panel shows quantification of [32P]dCMP incorporated into each band normalized to band 2 in the lane containing no MutS or MutL. The data presented represent the averages of at least two experiments with error bars representing standard deviations about the mean. The agarose gel run to demonstrate repair of the DNA substrate, similar to that shown in Fig. 5, is not shown.
DISCUSSION
The VSP repair reaction was reconstituted in vitro using purified Vsr endonuclease, DNA polymerase I, DNA ligase I, and a plasmid DNA substrate, verifying genetic experiments that established the basic biochemical requirements of the pathway. However, the length of the repair track in this reconstitution was much longer than observed in vivo and could be manipulated by altering the concentration of DNA ligase I. For this reason we describe this as the “minimal” VSP repair pathway using only the components of the system believed to be able to initiate and complete the repair of a T:G mismatch resulting from deamination of 5-mC within the sequence context 5′-CTWGG-3′/5′-CCWGG-3′.
The data presented in Figs. 2 and 3 clearly demonstrate the concentration dependence and specificity of the Vsr endonuclease on a covalently closed circular heteroduplex DNA molecule. Thus, the endonuclease reaction described here using a plasmid DNA substrate is consistent with that previously described for this protein using oligonucleotide DNA substrates (31, 33).
The reconstituted repair reaction was dependent on DNA polymerase I, as expected. Moreover, the appropriate nucleotide was incorporated as evidenced by the sensitivity of the site to XcmI cleavage after DNA synthesis. Efforts to reconstitute the repair reaction using T7 DNA polymerase were not successful (data not shown). This is likely because T7 DNA polymerase is not capable of strand displacement synthesis under these conditions (47) and therefore cannot act upon the nicked DNA substrate available after the Vsr reaction.
The repair reaction was also dependent on the addition of DNA ligase I, as expected. Because the product of the reconstituted reaction was a covalently closed molecule, DNA polymerase I must use its 5′ to 3′ exonuclease to remove the damaged DNA while using its 5′ to 3′ polymerase to synthesize new DNA. If repair synthesis was the result of stand displacement synthesis, the resulting product would contain a single-stranded DNA flap and would not be sealed by DNA ligase. Thus, the reconstitution reaction we have described using purified proteins effectively demonstrates that nick translation, as opposed to strand displacement, by DNA polymerase I is responsible for repair-resynthesis in the VSP pathway.
Preliminary experiments indicated that ligase I was capable of ligating the nick introduced by the Vsr endonuclease, which restored the integrity of the DNA strand but left the mismatch intact (data not shown). The ability of ligases to ligate ends containing a mismatch, albeit with reduced efficiency, has been previously reported (48). To ensure we were measuring a “repair” reaction, we included [α-32P]dCTP to monitor reactions that included the incorporation of dCMP to repair the T:G mismatch. Inspection of these data (Fig. 5) suggested that the length of a repair track was inversely proportional to DNA ligase I concentration. This interpretation was confirmed by measuring the length of repair tracks at multiple ligase I concentrations. At low concentrations of DNA ligase I, the length of the repair track was quite long, on the order of kilobase pairs in length, whereas at higher DNA ligase I concentrations, the length of the repair track was much shorter.
Because in vivo measurements have suggested that the VSP repair track lengths are generally less than 10 nucleotides in length (30, 39), we interrupted this result to signify that the minimal VSP repair event was uncoordinated and did not accurately represent the in vivo reaction. This result is perhaps not surprising because each of the three proteins added to the reaction is capable of acting independently on the substrate or the intermediate in the reaction pathway. Thus, once the substrate has been nicked by Vsr endonuclease, the nick can be sealed by DNA ligase I, leaving the mismatch in place, or the 3′-OH can be extended by DNA polymerase I, removing the mismatch and ultimately leaving a ligatable nick. In the absence of DNA ligase I or at low concentrations of this protein, DNA polymerase I can apparently bind and extend the 3′-OH multiple times, resulting in long repair tracks. We note that DNA polymerase I and ligase have been reported at roughly equal concentrations in the cell (56, 57).
In vivo VSP repair corrects a significant fraction of T:G mismatches in the sequence context 5′-CTWGG-3′/5′-C5meCWGG-3′ in the absence of MutS or MutL (29, 30). However, in the presence of MutS and MutL, the efficiency of VSP repair increases to nearly 100% (28). Therefore, MutS and MutL must have some role in the VSP repair pathway, such that these proteins increase the efficiency of VSP repair. Previous results (24, 32) have shown that MutL interacts with the Vsr endonuclease and apparently stimulates binding to the DNA substrate. We have confirmed this result that, in our hands, is only apparent at high concentrations of MutL (data not shown). However, this leaves the role of MutS unexplained, whereas the VSP-related repair phenotype of mutL and mutS mutants is essentially identical, suggesting that they function together in the VSP repair pathway.
MutL has a well documented role in coordinating the activities of several proteins involved in the methyl-directed mismatch repair pathway (2, 5). Recent work has suggested that the general function of MutL may be to coordinate a wide variety of repair activities in the cell (49, 50). MutS binds to mismatched base pairs and is known to bind the T:G mispair with high affinity (51, 52). Thus, it is easy to imagine MutS and MutL being recruited to the T:G mispair that results from deamination of 5-mC. We suggest that MutS and MutL serve to coordinate the VSP repair pathway in some way not yet understood. We have shown that the addition of MutS and MutL, in the presence of ATP, significantly decreased the length of the in vitro repair track at relatively low DNA ligase I concentrations (Fig. 7). Importantly, this required the addition of both MutS and MutL, although the addition of MutS alone may have a modest effect. This coordination might be similar to the role played by these two proteins in both methyl-directed mismatch repair and in the control of homeologous recombination.
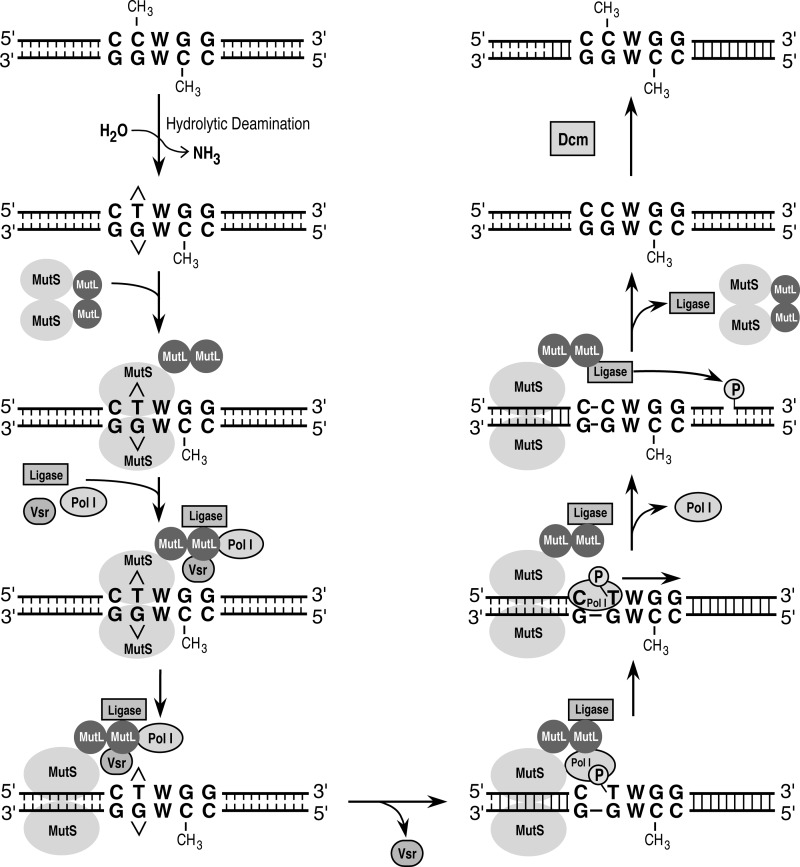
There are several ways in which the VSP repair reaction might be coordinated by MutS and MutL. MutL has been shown to stimulate the nicking reaction catalyzed by the Vsr endonuclease (24, 32, 35). Therefore, it is possible that MutL is able to increase the efficiency of VSP repair by stimulating the nicking reaction catalyzed by the Vsr endonuclease. MutL is also known to interact with the MutS protein when MutS is bound at a mismatched base pair (53, 54). These interactions and other potential interactions have led us to propose a model for VSP repair diagramed in Fig. 8. MutS binds to the T:G mismatch in the sequence 5′-CTWGG-3′/5′-C5meCWGG-3′ in a similar fashion to the way MutS binds to a mismatch in methyl-directed mismatch repair. MutL is then recruited to the mismatch-MutS complex. MutL subsequently recruits the Vsr endonuclease and coordinates its activity with that of DNA polymerase I and DNA ligase I, effectively increasing the local concentration of each of these proteins. The MutS-MutL complex moves away from the mismatch by some as yet undefined mechanism and allows the Vsr endonuclease to nick 5′ to the mismatched thymidine. The MutS-MutL-DNA polymerase I-DNA ligase I complex, still bound at a short distance from the nicked site, then loads DNA polymerase I at the nick generated by the Vsr endonuclease. DNA polymerase I then undergoes nick translates to form the repair-resynthesis track. This nick translation event repairs the T:G mismatch and produces a ligatable nick. The MutS-MutL-DNA ligase I complex then loads DNA ligase I at the newly formed nick created by DNA polymerase I, allowing it to seal the nick and restore the integrity of the DNA. In this model proteins are added sequentially by the MutS-MutL complex to coordinate the repair event. This coordination allows for the short patch length that has been observed in vivo. Potential protein-protein interactions or regulation of protein activities by MutS-MutL remain to be explored.
FIGURE 8.
A model for VSP repair. A T:G mismatch is generated by the spontaneous hydrolytic deamination of a 5-methyl cytosine residue. MutS and MutL are recruited to the T:G mismatched base, presumably through the interaction of MutS with the mismatch. MutS and MutL then recruit the Vsr endonuclease, DNA ligase I, and DNA polymerase I, effectively increasing the local concentration of each of the proteins. A physical interaction between MutL and the Vsr endonuclease has been demonstrated (24, 35); other protein-protein interactions, if they exist, are hypothetical. The Vsr endonuclease catalyzes the hydrolysis of a phosphodiester bond 5′ to the mismatched thymidine. DNA polymerase I is then loaded onto the nick and undergoes nick translates to repair the mismatch and synthesize a short repair patch. DNA ligase I seals the nick to restore the integrity of the DNA strand, and the Dcm methyltransferase methylates the appropriate cytosine on the newly synthesized repair patch.
In vivo and in vitro experiments have shown that the Vsr endonuclease and DNA polymerase I are essential for VSP repair (22, 27), and MutS and MutL are required for maximal efficiency of the VSP repair reaction (28, 29). However, it has yet to be shown that DNA ligase I is responsible for sealing the nick created by nick translation of DNA polymerase I. A second ligase has recently been identified: DNA ligase II (55), which could potentially have a role in VSP repair. This result remains to be explored further.
Acknowledgments
We thank Susan Whitfield for preparation of the artwork and the members of the Matson laboratory for suggestions and careful reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 33476 (to S. W. M.).
- 5-mC
- 5-methyl cytosine
- VSP
- very short patch
- Pol I
- polymerase I.
REFERENCES
- 1. Harfe B. D., Jinks-Robertson S. (2000) DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34, 359–399 [DOI] [PubMed] [Google Scholar]
- 2. Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) DNA mismatch repair. Functions and mechanisms. Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 3. Jiricny J. (2006) The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 4. Schofield M. J., Hsieh P. (2003) DNA mismatch repair. Molecular mechanisms and biological function. Annu. Rev. Microbiol. 57, 579–608 [DOI] [PubMed] [Google Scholar]
- 5. Kunkel T. A., Erie D. A. (2005) DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 6. Walsh C. P., Xu G. L. (2006) Cytosine methylation and DNA repair. Curr. Top. Microbiol. Immunol. 301, 283–315 [DOI] [PubMed] [Google Scholar]
- 7. Modrich P., Lahue R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65, 101–133 [DOI] [PubMed] [Google Scholar]
- 8. Au K. G., Welsh K., Modrich P. (1992) Initiation of methyl-directed mismatch repair. J. Biol. Chem. 267, 12142–12148 [PubMed] [Google Scholar]
- 9. Umar A., Kunkel T. A. (1996) DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur. J. Biochem. 238, 297–307 [DOI] [PubMed] [Google Scholar]
- 10. Lindahl T., Nyberg B. (1974) Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13, 3405–3410 [DOI] [PubMed] [Google Scholar]
- 11. Karran P., Lindahl T. (1980) Hypoxanthine in deoxyribonucleic acid. Generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry 19, 6005–6011 [DOI] [PubMed] [Google Scholar]
- 12. Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 71, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedberg E. C., Ganesan A. K., Minton K. (1975) N-Glycosidase activity in extracts of Bacillus subtilis and its inhibition after infection with bacteriophage PBS2. J. Virol. 16, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao M., Hatahet Z., Melamede R. J., Kow Y. W. (1994) Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 269, 16260–16268 [PubMed] [Google Scholar]
- 15. Yao M., Kow Y. W. (1995) Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 270, 28609–28616 [DOI] [PubMed] [Google Scholar]
- 16. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lutsenko E., Bhagwat A. S. (1999) Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat. Res. 437, 11–20 [DOI] [PubMed] [Google Scholar]
- 18. Duncan B. K., Miller J. H. (1980) Mutagenic deamination of cytosine residues in DNA. Nature 287, 560–561 [DOI] [PubMed] [Google Scholar]
- 19. Längle-Rouault F., Maenhaut-Michel G., Radman M. (1987) GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. EMBO J. 6, 1121–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welsh K. M., Lu A. L., Clark S., Modrich P. (1987) Isolation and characterization of the Escherichia coli mutH gene product. J. Biol. Chem. 262, 15624–15629 [PubMed] [Google Scholar]
- 21. Bhagwat A. S., Sohail A., Roberts R. J. (1986) Cloning and characterization of the dcm locus of Escherichia coli K-12. J. Bacteriol. 166, 751–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hennecke F., Kolmar H., Bründl K., Fritz H. J. (1991) The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature 353, 776–778 [DOI] [PubMed] [Google Scholar]
- 23. Lieb M. (1991) Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics 128, 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heinze R. J., Giron-Monzon L., Solovyova A., Elliot S. L., Geisler S., Cupples C. G., Connolly B. A., Friedhoff P. (2009) Physical and functional interactions between Escherichia coli MutL and the Vsr repair endonuclease. Nucleic Acids Res. 37, 4453–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lieb M., Bhagwat A. S. (1996) Very short patch repair. Reducing the cost of cytosine methylation. Mol. Microbiol. 20, 467–473 [DOI] [PubMed] [Google Scholar]
- 26. Sohail A., Lieb M., Dar M., Bhagwat A. S. (1990) A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J. Bacteriol. 172, 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dzidic S., Radman M. (1989) Genetic requirements for hyper-recombination by very short patch mismatch repair. Involvement of Escherichia coli DNA polymerase I. Mol. Gen. Genet. 217, 254–256 [DOI] [PubMed] [Google Scholar]
- 28. Zell R., Fritz H. (1987) DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine rersidues. EMBO J. 6, 1809–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lieb M. (1987) Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J. Bacteriol. 169, 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones M., Wagner R., Radman M. (1987) Mismatch repair and recombination in E. coli. Cell 50, 621–626 [DOI] [PubMed] [Google Scholar]
- 31. Gonzalez-Nicieza R., Turner D. P., Connolly B. A. (2001) DNA binding and cleavage selectivity of the Escherichia coli DNA G:T-mismatch endonuclease (Vsr protein). J. Mol. Biol. 310, 501–508 [DOI] [PubMed] [Google Scholar]
- 32. Monastiriakos S. K., Doiron K. M., Siponen M. I., Cupples C. G. (2004) Functional interactions between the MutL and Vsr proteins of Escherichia coli are dependent on the N-terminus of Vsr. DNA Repair 3, 639–647 [DOI] [PubMed] [Google Scholar]
- 33. Turner D. P., Connolly B. A. (2000) Interaction of the E. coli DNA G:T-mismatch endonuclease (Vsr protein) with oligonucleotides containing its target sequence. J. Mol. Biol. 304, 765–778 [DOI] [PubMed] [Google Scholar]
- 34. Elliott S. L., Brazier J., Cosstick R., Connolly B. A. (2005) Mechanism of the Escherichia coli DNA T:G-mismatch endonuclease (Vsr protein) probed with thiophosphate-containing oligodeoxynucleotides. J. Mol. Biol. 353, 692–703 [DOI] [PubMed] [Google Scholar]
- 35. Polosina Y. Y., Mui J., Pitsikas P., Cupples C. G. (2009) The Escherichia coli mismatch repair protein MutL recruits the Vsr and MutH endonucleases in response to DNA damage. J. Bacteriol. 191, 4041–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhagwat A. S., Sohail A., Lieb M. (1988) A new gene involved in mismatch correction in Escherichia coli. Gene 74, 155–156 [DOI] [PubMed] [Google Scholar]
- 37. Robertson A. B., Pattishall S. R., Gibbons E. A., Matson S. W. (2006) MutL-catalzyed ATP hydrolysis is required at a post-UvrD loading step in methyl-directed mismatch repair. J. Biol. Chem. 281, 19949–19959 [DOI] [PubMed] [Google Scholar]
- 38. Robertson A., Pattishall S. R., Matson S. W. (2006) The DNA binding activity of MutL is required for methyl-directed mismatch repair in Escherichia coli. J. Biol. Chem. 281, 8399–8408 [DOI] [PubMed] [Google Scholar]
- 39. Lieb M., Allen E., Read D. (1986) Very short patch mismatch repair in phage lambda. Repair sites and length of repair tracts. Genetics 114, 1041–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhagwat A. S., Lieb M. (2002) Cooperation and competition in mismatch repair. Very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol. Microbiol. 44, 1421–1428 [DOI] [PubMed] [Google Scholar]
- 41. Macintyre G., Doiron K. M., Cupples C. G. (1997) The Vsr endonuclease of Escherichia coli. An efficient DNA repair enzyme and a potent mutagen. J. Bacteriol. 179, 6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lieb M., Rehmat S. (1995) Very short patch repair of T:G mismatches in vivo. Importance of context and accessory proteins. J. Bacteriol. 177, 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsutakawa S. E., Muto T., Kawate T., Jingami H., Kunishima N., Ariyoshi M., Kohda D., Nakagawa M., Morikawa K. (1999) Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell 3, 621–628 [DOI] [PubMed] [Google Scholar]
- 44. Bambara R. A., Uyemura D., Choi T. (1978) On the processive mechanism of Escherichia coli DNA polymerase I. Quantitative assessment of processivity. J. Biol. Chem. 253, 413–423 [PubMed] [Google Scholar]
- 45. Mansour C. A., Doiron K. M., Cupples C. G. (2001) Characterization of functional interactions among the Escherichia coli mismatch repair proteins using a bacterial two-hybrid assay. Mutat. Res. 485, 331–338 [DOI] [PubMed] [Google Scholar]
- 46. Drotschmann K., Aronshtam A., Fritz H. J., Marinus M. G. (1998) The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 26, 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lechner R. L., Engler M. J., Richardson C. C. (1983) Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J. Biol. Chem. 258, 11174–11184 [PubMed] [Google Scholar]
- 48. Liu P., Burdzy A., Sowers L. C. (2004) DNA ligases ensure fidelity by interrogating minor groove contacts. Nucleic Acids Res. 32, 4503–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Polosina Y. Y., Cupples C. G. (2010) MutL. Conducting the cell's response to mismatched and misaligned DNA. Bioessays. 32, 51–59 [DOI] [PubMed] [Google Scholar]
- 50. Polosina Y. Y., Cupples C. G. (2010) Wot the 'L-Does MutL do? Mutat. Res. 705, 228–238 [DOI] [PubMed] [Google Scholar]
- 51. Jiricny J., Su S. S., Wood S. G., Modrich P. (1988) Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 16, 7843–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su S. S., Lahue R. S., Au K. G., Modrich P. (1988) Mispair specificity of methyl-directed DNA mismatch correction in vitro. J. Biol. Chem. 263, 6829–6835 [PubMed] [Google Scholar]
- 53. Grilley M., Welsh K. M., Su S. S., Modrich P. (1989) Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 264, 1000–1004 [PubMed] [Google Scholar]
- 54. Schofield M. J., Nayak S., Scott T. H., Du C., Hsieh P. (2001) Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 276, 28291–28299 [DOI] [PubMed] [Google Scholar]
- 55. Sriskanda V., Shuman S. (2001) A second NAD+-dependent DNA ligase (LigB) in Escherichia coli. Nucleic Acids Res. 29, 4930–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kornberg A., Baker T. A. (1992) in DNA Replication, W. H. Freeman & Co., New York [Google Scholar]
- 57. Modrich P., Anraku Y., Lehman I. R. (1973) Deoxyribonucleotide acid ligase. Isolation and physical characterization of the homogeneous enzyme from Escherichia coli. J. Biol. Chem. 248, 7495–7501 [PubMed] [Google Scholar]