Background: Head and neck cancer contains cancer stem cells (CSCs) characterized by a high level of CD44 expression.
Results: We discovered a new hyaluronan (HA)-CD44-mediated Oct4, Sox2, and Nanog pathway that regulates miR-302 production in CSCs.
Conclusion: HA-CD44v3-activated Oct4-Sox2-Nanog signaling and miR-302 play a pivotal role in CSC functions.
Significance: This information should provide new drug targets to treat head and neck cancer.
Keywords: Cancer Stem Cells, Extracellular Matrix, Head and Neck Cancer, MicroRNA, Signal Transduction, CD44, Differentiation, Hyaluronan, Self-renewal
Abstract
Human head and neck squamous cell carcinoma (HNSCC) is a highly malignant cancer associated with major morbidity and mortality. In this study, we determined that human HNSCC-derived HSC-3 cells contain a subpopulation of cancer stem cells (CSCs) characterized by high levels of CD44v3 and aldehyde dehydrogenase-1 (ALDH1) expression. These tumor cells also express several stem cell markers (the transcription factors Oct4, Sox2, and Nanog) and display the hallmark CSC properties of self-renewal/clonal formation and the ability to generate heterogeneous cell populations. Importantly, hyaluronan (HA) stimulates the CD44v3 (an HA receptor) interaction with Oct4-Sox2-Nanog leading to both a complex formation and the nuclear translocation of three CSC transcription factors. Further analysis reveals that microRNA-302 (miR-302) is controlled by an upstream promoter containing Oct4-Sox2-Nanog-binding sites, whereas chromatin immunoprecipitation (ChIP) assays demonstrate that stimulation of miR-302 expression by HA-CD44 is Oct4-Sox2-Nanog-dependent in HNSCC-specific CSCs. This process results in suppression of several epigenetic regulators (AOF1/AOF2 and DNMT1) and the up-regulation of several survival proteins (cIAP-1, cIAP-2, and XIAP) leading to self-renewal, clonal formation, and cisplatin resistance. These CSCs were transfected with a specific anti-miR-302 inhibitor to silence miR-302 expression and block its target functions. Our results demonstrate that the anti-miR-302 inhibitor not only enhances the expression of AOF1/AOF2 and DNMT1 but also abrogates the production of cIAP-1, cIAP-2, and XIAP and HA-CD44v3-mediated cancer stem cell functions. Taken together, these findings strongly support the contention that the HA-induced CD44v3 interaction with Oct4-Sox2-Nanog signaling plays a pivotal role in miR-302 production leading to AOF1/AOF2/DNMT1 down-regulation and survival of protein activation. All of these events are critically important for the acquisition of cancer stem cell properties, including self-renewal, clonal formation, and chemotherapy resistance in HA-CD44v3-activated head and neck cancer.
Introduction
Human head and neck squamous cell carcinoma (HNSCC)2 is the sixth most common cancer worldwide (1). The 3-year survival rate for patients with advanced-stage HNSCC and treatment with standard therapy is only 30–50% (1, 2). Resistance to standard therapy continues to be a limiting factor in the treatment of HNSCC. Nearly 40–60% of HNSCC patients subsequently develop recurrences or distant metastases (1, 2). Thus, there is currently a great need to clarify the key mechanisms of tumor initiation and progression underlying the clinical behavior of HNSCC.
Cisplatin is one of the most common chemotherapies used to treat head and neck cancer today. The ability of this drug to induce tumor cell death is often counteracted by the presence of anti-apoptotic regulators leading to chemoresistance (3–6). Several lines of evidence point toward the IAP family (e.g. cIAP-1, cIAP-2, and XIAP) playing a role in oncogenesis via their effective suppression of apoptosis (7). The mode of action of IAPs in suppressing apoptosis appears to be through direct inhibition of caspases and pro-caspases (primarily caspase 3 and 7) (7). IAPs also support chemoresistance directly by preventing tumor cell death induced by anticancer agents (8). Although certain anti-apoptotic proteins (e.g. Bcl-xL) have been shown to participate in anti-apoptosis and chemoresistance in HA-CD44-activated tumor cells (9), the involvement of IAPs in HA-CD44-mediated HNSCC cell survival and chemoresistance (6) only recently started to receive some attention.
Accumulating evidence has indicated that most tumors contain a small population of cells that persistently initiate tumor growth and promote tumor progression. These “tumor-initiating cells” are also called “cancer stem cells” (CSCs) because they share several hallmarks of normal stem cells (10, 11). For example, CSCs undergo self-renewal, maintain quiescence, display multipotentiality, and express survival protein/anti-apoptosis proteins (10, 11). Another well known property of CSCs is their ability to expand stem cell population by undergoing cell proliferation/survival and/or clone formation and differentiation (10, 11). CSCs also display chemotherapy resistance, thereby causing a relapse of the cancers (12).
A number of studies have identified specific molecules expressed in hyaluronans that correlate with both stem cell properties and tumor cell behaviors. Among such molecules is CD44, which is a multifunctional transmembrane glycoprotein expressed in many cells and tissues, including HNSCC cells and various carcinoma tissues (13). CD44 is commonly expressed in various isoforms generated by alternative mRNA splicing of variant exons inserted into an extracellular membrane-proximal site (14). CD44 is expressed in both normal and CSCs and has been suggested to be an important stem cell marker (13, 15).
HNSCC tumors appear to contain a subpopulation of CSCs characterized by a high level of CD44 expression (13). Purified CSCs overexpressing CD44 are capable of generating phenotypically distinct cells resulting in heterogeneous tumors in immunodeficient mice (13). These findings clearly indicate that CSCs overexpressing CD44 display the hallmark stem cell properties of self-renewal and the ability to generate heterogeneous cell populations. In fact, CD44 is considered to be one of the important cell surface markers for both normal stem cells and CSCs (13, 15). Most importantly, the expression of certain CD44 variant isoforms (in particular, CD44v3) is closely associated with head and neck cancer progression (16–18). The question of whether CD44v3 is expressed in CSCs of HNSCC remains unanswered.
Hyaluronan (HA) is a major component in the extracellular matrix of most mammalian tissues. HA is a nonsulfated, unbranched glycosaminoglycan consisting of repeating disaccharide units, d-glucuronic acid, and N-acetyl-d-glucosamine (19, 20). It is synthesized by specific HA synthases (20, 21) and digested into various smaller sized molecules by various hyaluronidases (22). HA is enriched in many types of tumors (23, 24) and has been found to be enriched in stem cell niches (25, 26). The unique HA-enriched microenvironment appears to be involved in both self-renewal and differentiation of normal human stem cells (25, 26). All CD44 isoforms contain an HA-binding site in their extracellular domain and thereby serve as a major cell surface receptor for HA (27). The fact that both CD44 and HA are overexpressed at tumor attachment sites and that HA binding to CD44 stimulates a variety of tumor cell-specific functions and tumor progression (5, 28, 29) suggests that the HA-CD44 interaction is a critical requirement for tumor progression.
The stem cell marker, Nanog, is an important transcription factor involved in the self-renewal and maintenance of pluripotency in the inner cell mass of mammalian embryos and embryonic stem (ES) cells (30, 31). Nanog signaling is regulated by interactions among various pluripotent stem cell regulators (e.g. Rex1, Sox2, and Oct3/4) that together control the expression of a set of target genes required for ES cell pluripotency (32–34). These findings confirm the essential role of Nanog in regulating a variety of essential cellular functions. Recently, several tumor cell types have been shown to express various embryonic stem cell markers, including Nanog (6, 35, 36). The Nanog family of proteins has also been found to act as growth-promoting regulators in various different tumor cells (37). Our recent work indicates that HA binding to epithelial tumor cells promotes Nanog protein association with CD44, followed by Nanog activation and the expression of pluripotent stem cell regulators (35). Most importantly, both HA-CD44 signaling and Nanog function appear to be involved in tumor cell growth, anti-apoptosis, cell survival, and chemoresistance in CD44-expressing HNSCC cells (6, 35, 36). These findings indicate that Nanog plays an important role in HA-CD44-mediated oncogenesis. Whether Nanog participates in HNSCC-specific CSC signaling and functions remains to be determined.
The SOX (short for Sex determining Region Y-box2) gene family encodes a group of transcription factors that are characterized by a highly conserved “high mobility group” domain (38). These genes play an important role in stem cell functions, organogenesis, and animal development (39, 40). SOX proteins, including SOX2, bind to specific DNA sequences (C(T/A)TTG(T/A)(T/A)) by means of their high mobility group domains as transcription factors in regulating their target gene expression (38). Because SOX2 by itself lacks strong affinity for DNA binding, it relies on forming a complex with other transcription factors (38). Recently, a number of studies have linked SOX2 overexpression with human cancers (41). However, information regarding the role of SOX2 in cancer development is very limited.
Oct4 (encoded by POU5f, also known as Oct3 or Oct3/4) belongs to the POU transcription factor family, in which the members regulate their target gene expression by binding to an octameric sequence motif containing the AGTCAAAT consensus sequence (42, 62). The Oct4 protein contains three functional domains, including POU (pit-Oct-Unc), DNA binding domain, an N-terminal transactivation domain, and a C-terminal cell type-specific transactivation domain (42, 62). Oct4 was first identified as a stem cell-specific and germ line-specific transcription factor in mice (43). In humans, Oct4 is the product of the OTF3 gene and has been shown to maintain “stemness” in pluripotent stem cells (44). Functionally, Oct4 is essential for early embryonic development and functions as a master regulator of the initiation and maintenance of pluripotent cells during embryonic development. It also interacts with other embryonic regulators, such as Nanog and Sox2, to oversee a vast regulatory network that maintains pluripotency and inhibits differentiation (45, 61).
Accumulating evidence indicates that noncoding microRNAs (miRNAs, ∼22-nucleotides) are involved in both cancer development and multidrug resistance (46, 47). The biological functions of certain miRNAs also appear to be involved in the regulation of self-renewal and/or differentiation (45, 48, 49). For example, miR-302 encodes a cluster of eight miRNAs that are expressed specifically in pluripotent embryonic stem cells (ESCs) and/or embryonic carcinoma cells (48, 49). A recent study indicates that the promoter of the miR-302 cluster was occupied by key ESC-specific transcriptional factors, including Oct4, Sox2, and Nanog in mouse and human cells (50). Most importantly, the miR-302 family is involved in maintaining stemness of human embryonal carcinoma cells (51) and reprogramming human skin cancer cells to a pluripotent ESC-like state (52). These findings strongly suggest that miR-302 plays an important role in the pluripotency of ESCs and embryonic carcinoma cells.
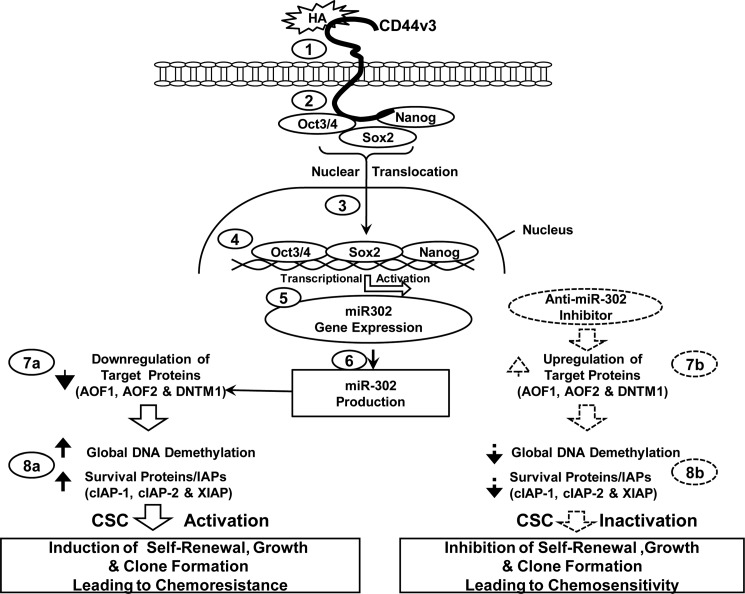
In this study we identified a unique subset of highly tumorigenic HNSCC cells that express high levels of CD44v3 and aldehyde dehydrogenase-1 (ALDH1) (designated as CD44v3highALDH1high cells). We also discovered a new HA-CD44v3-mediated signaling pathway that regulates stem cell marker (e.g. Oct4, Sox2, and Nanog)-associated miR-302 production. These events result in the following: (i) the reduction of epigenetic regulators (AOF1/AOF2 and DNMT1) resulting in DNA demethylation; and (ii) overexpression of several survival proteins leading to self-renewal, clonal formation, and cisplatin resistance. Inhibition of miR-302 expression/function not only results in AOF1/AOF2/DNMT1 up-regulation and DNA demethylation down-regulation, but it also causes a reduction of cIAP-1/cIAP-2 and XIAP expression and inhibition of CSC functions (e.g. self-renewal, clonal formation, and cisplatin resistance). These findings suggest the existence of a novel linkage between Oct4-Sox2-Nanog signaling and miR-302 expression that regulates cancer stem cell functions in HA-CD44v3-activated head and neck cancer.
MATERIALS AND METHODS
Cell Culture
Tumor-derived HSC-3 cell line (from human squamous carcinoma cells of mouth) was kindly provided from Dr. Randy Kramer (University of California, San Francisco). Cells were grown in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum.
Antibodies and Reagents
Monoclonal rat anti-CD44 antibody (clone, 020; isotype, IgG2b; obtained from CMB-TECH, Inc., San Francisco) recognizes a determinant of the HA-binding region common to CD44 and its principal variant isoforms such as CD44v3. This rat anti-CD44 was routinely used for HA-related blocking experiments, and the rabbit anti-CD44v3 antibody was used to identify v3-coated structure. Rabbit anti-CD44v3 antibody and goat anti-Sox2 antibody were obtained from EMD Chemicals (Gibbstown, NJ) and R & D Systems (Minneapolis, MN), respectively. Mouse anti-Oct4 antibody was obtained from Sigma, and rabbit anti-Oct4 antibody, goat anti-Nanog antibody, rabbit anti-AOF2 antibody, and goat anti-actin antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-cIAP2 antibody and mouse anti-XIAP antibody were purchased from BD Biosciences. Other immunoreagents such as rabbit anti-AOF1 antibody, rabbit anti-cIAP1 antibody, mouse anti-Nanog antibody, rabbit anti-Nanog antibody, mouse anti-Sox2 antibody, rabbit anti-DNMT1 antibody, rabbit anti-cytokeratin 17 antibody, rabbit anti-cytokeratin 19 antibody, FITC and Texas Red-conjugated goat anti-rabbit antibody, rabbit anti-goat antibody, and goat anti-mouse antibody were from Abcam (Cambridge, MA). Texas Red-conjugated control antibodies (normal rat/rabbit/goat IgG) were prepared using Texas Red sulfonyl chloride conjugation procedures (Pierce). Cisplatin was obtained from Sigma.
Previous studies indicated that HA fragments with a molecular mass of ∼500,000 daltons appear to promote CD44-mediated oncogenic signaling and cisplatin resistance in HNSCC cells (6, 35, 36). To investigate whether these HA fragments play a role in regulating stem cell marker signaling and cancer stem cell function, we decided to prepare HA fragments (∼500,000-dalton polymers) for our study. Briefly, intact high molecular weight (∼3,000,000-dalton polymers) healon HA polymers (500 mg) (obtained from Pharmacia & Upjohn Co. (Kalamazoo, MI)) was dissolved in 50 ml of 0.1 m acetate buffer (pH 5.4) containing 0.15 m NaCl and digested with 20,000 units of bovine testicular hyaluronidase (PH20) (Wyeth Laboratories Inc., Philadelphia) at 37 °C. Ten-milliliter aliquots were removed after 2-, 4-, 6-, 8-, and 24-h intervals, and the reaction was terminated by adding trichloroacetic acid at 10% final concentration (v/v). After incubating at 4 °C for at least 4 h, any precipitate was removed by centrifugation at 2,500 × g for 30 min. The supernatants were then pooled, dialyzed extensively against distilled water, recentrifuged, and freeze-dried. The HA fragments were then dissolved in 10 ml of 0.1 m acetic acid and applied to gel filtration chromatography using a Sephacryl S1000 column (GE Healthcare). Each fraction was collected and assayed for HA content and the size of the HA fragments (∼500,000-dalton polymers).
The purity of HA fragments (∼500,000-dalton polymers) used in our experiments was collected and further verified by anion exchange HPLC followed by protein and endotoxin analyses using BCA protein assay kit (Pierce) and an in vitro Limulus amebocyte lysate (LAL) assay (Cambrex Bio Science Walkersville Inc., Walkersville, MD), respectively. No protein or endotoxin contamination was detected in these HA preparations. HA fragments (∼500,000-dalton polymers) were then analyzed by using 4% polyacrylamide gradient gel electrophoresis followed by Alcian blue 8GX and silver staining. Select-HATM LowLadder (in the range of ∼27,000–495,000 daltons) obtained from Hyalose (Oklahoma City, OK) was used as HA standards as described previously (71).
Sorting Tumor-derived HSC-3 Cell Populations by Multicolor Fluorescence-activated Cell Sorter (FACS)
The identification of aldehyde dehydrogenase 1 (ALDH1) activity from tumor-derived HSC-3 cells was conducted using the ALDEFLUOR kit (StemCell Technologies, Durham, NC). Specifically, tumor cells were suspended in ALDEFLUOR assay buffer containing ALDH1 substrate (BODIPY-aminoacetaldehyde, 1 mol/liter per 1 × 106 cells) and incubated for 30 min at 37 °C. As a negative control, HSC-3 cells were treated with a specific ALDH1 inhibitor, 50 mmol/liter diethylaminobenzaldehyde (DEAB) (50 mmol/liter).
Next, for labeling the cell surface marker, tumor-derived HSC-3 cells were suspended in 100 μl of ALDEFLUOR buffer followed by incubating with 20 μl of allophycocyanin (APC)-labeled anti-CD44v3 antibody (recognizing the v3-specific domain of CD44) or APC-labeled normal mouse IgG (as a control) (BD Biosciences) for 15 min at 4 °C. For FACS sorting, tumor cells were incubated in PBS buffer followed by FACS (BD FACS Aria llu, BD Biosciences) sorting using dual wavelength analysis. Subsequently, CD44v3highALDH1high, CD44v3lowALDH1low, and unsorted (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting) tumor cell populations were collected and used for various experiments described in this study.
Tumorigenicity Assay
Nonobese, diabetic/severe combined immunodeficient (NOD/SCID) immunocompromised mice (5-week-old female mice) were purchased from Charles River Laboratories International, Inc. (Wilmington, MA), and maintained in micro-isolator cages. Specifically, these NOD/SCID were injected subcutaneously with sorted CD44v3highALDH1high cells (or CD44v3lowALDH1low cells) or unsorted HSC-3 cells (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting) ranging from 50, 500, to 5,000 cells. These mice were then monitored twice weekly for palpable tumor formation and euthanized 4 or 8 weeks after transplantation to assess tumor formation. Tumors were measured using a Vernier caliper, weighed, and photographed. A portion of the subcutaneous tumors was collected. Some tumors were cut into small fragments with sterile scissors and minced with a sterile scalpel, rinsed with Hanks' balanced salt solution containing 2% heat-inactivated calf serum (Invitrogen), and centrifuged for 5 min at 1,000 rpm. The resulting tissue specimen was placed in a solution of DMEM/F-12 containing 300 units/ml collagenase and 100 units/ml hyaluronidase (StemCell Technologies, Durham, NC). The mixture was incubated at 37 °C to dissociate cells. The digestion was terminated with the addition of FBS, and the cells were filtered through a 40-μm nylon sieve. The cells were washed twice with Hanks' balanced salt solution plus 2% heat-inactivated calf serum for FACS as described above. Some tumors were also fixed in 10% formaldehyde and embedded in paraffin for H&E staining to assess tumor pathology and CD44v3/stem cell marker expression.
Quantitative PCR (Q-PCR)
Total RNA was isolated from CD44v3highALDH1high cells or unsorted cells (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting) using Tripure isolation reagent kits (Roche Applied Science). First-stranded cDNAs were synthesized from RNA using Superscript first-strand synthesis system (Invitrogen). Gene expression was quantified using probe-based SYBR Green PCR master mix kits, ABI PRISM 7900HT sequence detection system, and SDS software (Applied Biosystems, Foster City, CA). A cycle threshold (minimal PCR cycles required for generating a fluorescent signal exceeding a preset threshold) was determined for each gene of interest and normalized to a cycle threshold for a housekeeping gene (36B4) determined in parallel. The 36B4 is a human acidic ribosomal phosphoprotein P0 whose expression was not changed in tumor cells. The Q-PCR primers used for detecting gene expression of Oct4, Sox2, and Nanog were as follows: specifically, two Oct4-specific primers (the sense primer 5′-GGTATTCAGCCAAACGACCA-3′ and the antisense primer 5′-CACACTCGGACCACATCCTT-3′); two Sox2-specific primers (the sense primer 5′-GACAGTTACGCGCACATGAA-3′ and the antisense primer 5′-TAGGTCTGCGAGCTGGTCAT-3′); and two Nanog-specific primers (the sense primer 5′-GTGATTTGTGGGCCTGAAGA-3′ and the antisense primer 5′-ACACAGCTGGGTGGAAGAGA-3′) were used. Finally, for detecting 36B4 gene expression, two 36B4-specific primers (the sense primer 5′-GCGACCTGGAAGTCCAACTAC-3′ and the antisense primer 5′-ATCTGCTGCATCTGCTTGG-3′) were used.
Transfection with Anti-miR-302a and Anti-miR-302b Inhibitors
CD44v3highALDH1high cells were transfected with anti-miRTM targeting miR-302 (anti-miR-302a and anti-miR-302b inhibitors) (Ambion, catalogue numbers AM12557–AM17000 and AM10081–AM17000) and its corresponding negative control (Ambion, catalogue number 17010) (Ambion, Foster City, CA) using Lipofectamine 2000 reagent (Invitrogen) for 24 h. Cells were then treated with HA or without HA in various experiments as described below. The final concentrations of anti-miR-302a and anti-miR302b inhibitors and miRNA-negative control used in various experiments were 30 nmol/liter.
Immunoprecipitation and Immunoblotting Techniques
First, CD44v3highALDH1high cells (untreated or treated with HA (50 μg/ml) or pretreated with anti-CD44 antibody plus HA (50 μg/ml) for various time intervals (e.g. 0, 5, 15, or 60 min)) were solubilized in a lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 20 mm MgCl2, 0.5% Nonidet P-40, 0.2 mm Na3VO4, 0.2 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 5 μg/ml aprotinin) (6, 9, 35, 36).
Immunoprecipitation of these Nonidet P-40 solubilized CD44v3highALDH1high cells was then carried out using rabbit anti-CD44v3 antibody followed by goat anti-rabbit IgG-beads. Subsequently, the immunoprecipitated materials were immunoblotted with rabbit anti-Oct4 antibody (2 μg/ml), goat anti-Sox2 antibody (2 μg/ml), or goat anti-Nanog antibody (2 μg/ml), respectively. In addition, anti-Oct4 antibody (or anti-Sox2 antibody or anti-Nanog antibody)-mediated immunoprecipitation was performed followed by immunoblotting with anti-CD44v3 antibody, respectively.
The nuclear fraction (using the extraction kit from Active Motif (Carlsbad, CA)) of CD44v3highALDH1high cells (untreated or treated with HA (50 μg/ml) or pretreated with anti-CD44 antibody plus HA (50 μg/ml) for various time intervals (e.g. 0, 5, 15, or 60 min)) was also immunoblotted with various immunoreagents (e.g. anti-Oct4 antibody (2 μg/ml), anti-Sox2 antibody (2 μg/ml), anti-Nanog antibody (2 μg/ml), or anti-lamin A/C antibody (a nucleus-specific marker) (2 μg/ml), respectively). Nuclear extract was also used for goat anti-Nanog antibody-mediated immunoprecipitation followed by rabbit anti-Oct4-mediated or goat anti-Sox2-mediated immunoblot, respectively.
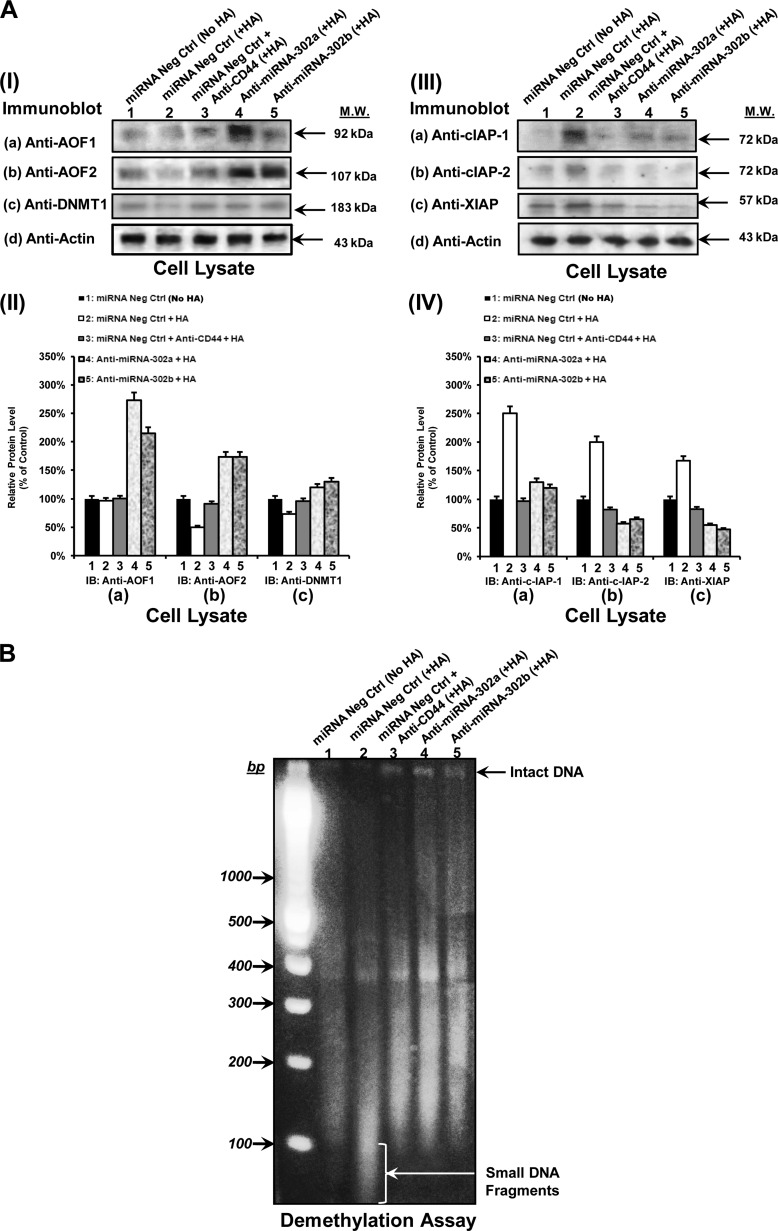
In some cases, Nonidet P-40-solubilized cell lysate of CD44v3highALDH1high cells (transfected with Oct4 siRNA or Sox2 siRNA, Nanog siRNA, or siRNA with scrambled sequences; or anti-miR-302a/anti-miR302b inhibitor or miRNA-negative control and treated with HA (50 μg/ml) for 24 h (or pretreated with anti-CD44 antibody plus HA or no HA) was processed for immunoblotting using various immunoreagents (e.g. anti-AOF1 (2 μg/ml), anti-AOF2 (2 μg/ml), anti-DNMT1 (2 μg/ml), or mouse anti-cIAP-1 (2 μg/ml) or mouse anti-cIAP-2 (2 μg/ml) and anti-XIAP (2 μg/ml) or goat anti-actin (2 μg/ml) (as a loading control), respectively). We routinely repeated our immunoblot analyses at least 4–5 times to verify the reproducibility and accuracy of our results.
Chromatin Immunoprecipitation (ChIP) Assay
To examine whether the Oct4-Sox2-Nanog complex directly interacts with the promoter region of miR-302 cluster, ChIP assays were performed in CD44v3highALDH1high cells (pretreated with normal rat IgG or anti-CD44 antibody or transfected with Oct4 siRNA or Sox2 siRNA or Nanog siRNA or siRNA with scrambled sequences, followed by 1 h of HA treatment (50 μg/ml) or no HA treatment) using a kit (EZ ChIP) from Millipore Corp. according to the manufacturer's instructions. Cross-linked chromatin lysates were sonicated and diluted with ChIP sonication buffer plus protease inhibitors, divided, and incubated with IgG isotype control, anti-Oct4 antibody, anti-Sox2 antibody, or anti-Nanog antibody at 4 °C overnight and then precipitated with protein G-agarose. Cross-linking was reversed by overnight 65 °C incubation; DNA fragments were then extracted with a PCR purification kit and analyzed by PCR or Q-PCR using the following primer pairs specific for the miR-302 cluster promoter containing the Oct4-Sox2-Nanog-binding sites: forward primer, 5′-CCCGTGGAAGCAATCTATTTATTT-3′, and reverse primer, 5′-TGTGTTTCTATCTGGAGGAACTCTGT-3′ on an agarose gel.
RNase Protection Assay Analysis of Mature miRNAs
Expression of miRNAs was qualitatively analyzed by RNase protection assay. For RNase protection assay, enriched small RNA isolated from CD44v3highALDH1high cells (transfected with Oct4 siRNA, Sox2 siRNA, Nanog siRNA, scrambled sequence siRNA, anti-miR-302a/miR-302b inhibitors, or miRNA-negative control in the presence or absence HA (or pretreated with anti-CD44 antibody plus HA) for various time intervals (e.g. 0, 5, 10, 15, and 30 min or 2 h) at 37 °C) was enriched and purified using the mirVana miRNA isolation kit (Ambion). RNA concentrations were verified by measuring absorbance (A260) on the NanoDrop spectrophotometer ND-1000 (NanoDrop). The mirVana miRNA probe construction kit (Ambion) was used to synthesize the 32P-labeled miR-302a or miR-302b antisense probe and miR-191 probe loading control.
Clinical Tumor Samples
IRB approval was obtained from our institution's Committee on Human Research. The clinical tissue specimens were portions of primary tumors of patients undergoing surgical treatment of HNSCC from multiple primary sites of the upper aerodigestive tract at a University of California at San Francisco-affiliated Veterans Affairs Medical Center from September 1, 2001, to June 30, 2006. The presence of ≥80% cancer cells in the procured samples was confirmed by a clinical pathologist as described previously (16).
Immunohistochemistry and Immunofluorescence Staining
The tumor tissue specimens (obtained from CD44v3highALDH1high cell-induced tumors from NOD/SCID mice or human HNSCC patients) were fixed in 4% formaldehyde in 0.1 m phosphate buffer for 24 h at 4 °C and embedded in paraffin. Five-μm-thick tissue sections were placed on positively charged glass slides (Fisher). Immunohistochemical stains were performed using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's protocol. Rabbit anti-Oct4 antibody, goat anti-Sox2 antibody, goat anti-Nanog antibody, or rabbit anti-CD44v3 antibody (dilution factor from 1:100 up to 1:1,000 determined by titration) were applied to tissue sections and incubated overnight at 4 °C. Secondary biotinylated antibody and streptavidin-HRP conjugate complex were applied for 60 and 30 min, respectively. After washing in buffer, the chromogen diaminobenzidine was applied for 5 min followed by a counterstain with Mayer's hematoxylin. Negative controls included substituting the primary antisera with preimmune sera from the same species, omitting the primary antibody, or using an antibody of irrelevant specificity.
For the detection of miR-302, we also performed in situ hybridization of miR-302 using digoxigenin (DIG)-labeled miR-302a and miR-302b LNA probe (and scrambled probe) (miRCURY LNATM detection probes, Exiqon, Inc., Woburn, MA) and tumor samples (obtained from CD44v3highALDH1high cell-induced tumors from NOD/SCID mice or human HNSCC patients) as well as normal mucosa from a group of healthy volunteers. First, 5-μm tissue sections (on coverslips) were treated with 5 μg/ml proteinase K (New England Biolabs) for 30 min followed by 4% formaldehyde (in 0.1 m phosphate buffer) treatment for 10 min. Tissue sections were then incubated with either miR-302a or miR-302b LNA probes 5′-labeled with DIG in 20 nm hybridization buffer in a hybridization chamber for 18 h at 51 °C. Subsequently, tissue sections were washed and incubated with an anti-DIG-AP (alkaline phosphatase)-FAB fragment at room temperature for 4 h. The final visualization was then performed with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Pierce).
Semiquantitative Analyses of the Immunohistochemical Staining and in Situ Hybridization
Semi-quantitative analyses of both CD44v3/stem cell marker protein expression and miRNA-302 production were performed on 10 mouse tumors and 66 oral cavity carcinoma primary tumors. Regions of tissue analysis for immunohistochemistry were confirmed to represent squamous cell carcinoma on H&E staining performed and examined by a mouse tumor biologist and a clinical oral pathologist. Specimens were then stained with a panel of antibodies (e.g. anti-Oct4, anti-Sox2, anti-Nanog, and anti-CD44v3) or performed with miR-302a/302b in situ as described above. Tumor specimens were reviewed by two blinded observers and grouped according to the following system. No staining was defined as rare or no tumor cells stained. Weak staining was defined as few tumor cells (<50%) stained. Strong staining was defined as majority of tumor cells stained.
Statistical Analysis
The statistical analysis was performed by an independent biostatistician. From the experimental results of the in vitro assays, a means ± S.E. were calculated for each experimental group. Student's t test with the resultant p value representing a two-sided test of statistical significance was used. Significance was set at 95% (p = 0.05).
The expression of various markers (e.g. CD44v3, Oct4, Sox2, Nanog, miR-302a, miR-302b) in primary tumors was analyzed statistically with respect to various demographic and clinical outcome parameters using Fisher's exact testing. The patient parameters examined included gender, age, presence of tobacco abuse, primary tumor T status, or grade of tumor differentiation.
Double Immunofluorescence Staining and Confocal Microscopic Analyses
CD44v3highALDH1high cells (untreated or treated with 5 min (or 15 min) of HA (50 μg/ml) or pretreated with anti-CD44 antibody plus 5 min (or 15 min) of HA (50 μg/ml)) were fixed with 2% formaldehyde. Subsequently, these cells were rendered permeable by ethanol treatment followed by incubation with rabbit anti-CD44v3 antibody followed by Texas Red-conjugated goat anti-rabbit IgG and mouse anti-Oct4 antibody (or mouse anti-Nanog antibody or mouse anti-Sox2 antibody) followed by FITC-conjugated goat anti-mouse IgG. In some cases, samples were incubated with rabbit anti-Nanog antibody followed by Texas Red-conjugated goat anti-rabbit IgG and mouse anti-Oct4 antibody followed by FITC-conjugated goat anti-mouse IgG. Samples were also labeled with mouse anti-Sox2 antibody followed by Texas Red-conjugated goat anti-mouse IgG and rabbit anti-Nanog antibody followed by FITC-conjugated goat anti-rabbit IgG. DAPI staining was used as a marker for nucleus. To detect nonspecific antibody, CD44v3highALDH1high cells were also incubated with FITC or Texas Red-conjugated normal IgG, respectively. No labeling was observed in control samples. These fluorescence-labeled samples were then examined with a confocal laser scanning microscope (Axiovert 200; Carl Zeiss Meditec, Inc., Jena, Germany).
In some cases, some fluorescence-labeled cells were scanned using a 0.2-μm/section XY and Z-axis step size to generate three-dimensional data sets extending from the cell surface through the cytoplasm and nucleus into the cell-substrate attachment sites. All images were recorded as 12-bit, 512 × 512 images. Three-dimensional data sets were then reconstructed using LSM Image Examiner (Carl Zeiss Meditec, GmbH). For each sample, a minimum of three three-dimensional data sets were collected.
Global DNA Demethylation Assays
Genomic DNAs were isolated from CD44v3highALDH1high cells (treated with anti-miR-302a/anti-miR302b inhibitor or miRNA-negative control followed by followed by HA (50 μg/ml) addition for 24 h at 37 °C (or anti-CD44 antibody plus HA or no HA addition)) using a DNA isolation kit (Roche Applied Science). Isolated DNAs (1 μg/per sample) were then digested with a CCGG-cutting restriction enzyme HpaII and analyzed by 1% agarose gel electrophoresis to determine genome-wide demethylation.
Spheroid Formation and Self-renewal Assays
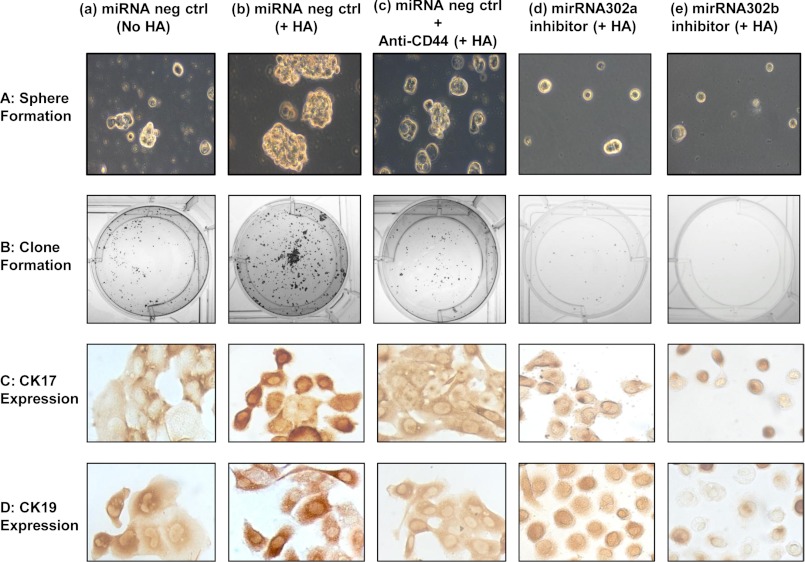
Sphere formation was induced by suspending CD44v3highALDH1high cells (treated with anti-miR-302a/anti-miR302b inhibitor or miRNA-negative control) in 1:1 Matrigel/basal medium in a total of volume of 100 μl. Samples (5 × 104cells) were then plated around the rims of wells in a 12-well plate and allowed to solidify at 37 °C for 10 min before 1 ml of basal medium (with B27 plus 20 ng/ml EGF, 10 ng/ml FGF, and 4 μg/ml insulin) containing HA (50 μg/ml) (or anti-CD44 antibody plus HA (50 μg/ml) or no HA) was added. Medium was replenished every 3 days. Ten days after plating, spheres (tight, spherical, nonadherent masses >40 μm in diameter) per well were counted, and at least 100 spheres per group were measured. The number of spheres containing CD44v3highALDH1high cells in each well were expressed as sphere-forming units. The number of sphere-forming units per well was counted in triplicate wells for each condition.
To recover CD44v3highALDH1high cells from the spheres, Matrigel-containing wells were treated with 1 mg/ml Dispase solution (Invitrogen). Spheres were then digested with trypsin and 0.05% EDTA. CD44v3highALDH1high cells dissociated from spheres were counted by hemocytometer and replated to generate spheres of next generation. Serial passage of individual spheres was regularly performed to verify self-renewal capability of cells associated with the spheres. Measurement of growth for CD44v3highALDH1high cells (dissociated from spheres) was also performed by incubating these cells in serum-free RPMI 1640 medium for 3 weeks using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based growth assay.
Clone Formation and Differentiation Assays
To analyze the clone formation properties, CD44v3highALDH1high cells (treated with anti-miR-302a/anti-miR302b inhibitor or miRNA-negative control) dissociated from spheres (preincubated with HA (50 μg/ml) or anti-CD44 antibody plus HA (50 μg/ml) or no HA as described above) were inoculated to 6-well plates at a density of 200 cells/well in 6-well plates and cultured with RPMI 1640 complete culture for ∼7–10 days. After most cell clones expanded to >50–100 cells, they were fixed with methanol followed by staining with crystal violet. The clone formation efficiency was expressed as the ratio of the clone number to the planted cell number. In some cases, colonies were fixed with 2% paraformaldehyde. Subsequently, these cells were rendered permeable by ethanol treatment followed by immunoperoxidase labeling of differentiation markers (e.g. cytokeratin 17 and cytokeratin 19) using anti-cytokeratin 17 antibody and anti-cytokeratin 19 antibody, respectively. In some cases, the clone formation capacity of various cell populations (e.g. CD44v3highALDH1high cells, CD44v3lowALDH1low cells, and unsorted cells) was also analyzed according to the procedures described above.
Tumor Cell Growth and Apoptosis Assays
To analyze tumor cell growth properties, sphere-derived CD44v3highALDH1high cells (pretreated with anti-miR-302a or anti-302b inhibitor or miRNA-negative control) (5 × 103 cells/well) were incubated in basal medium (with B27 plus 20 ng/ml EGF, 10 ng/ml FGF and 4 μg/ml insulin) containing HA (50 μg/ml) (or anti-CD44 antibody plus HA (50 μg/ml) or no HA). Medium was replenished every 3 days. Twenty one days after plating, the number of cell growth was then counted under a microscope at low magnification.
In some cases, these sphere-derived CD44v3highALDH1high cells (pretreated with anti-miR-302a or anti-302b inhibitor or miRNA-negative control) were also incubated with various concentrations of cisplatin (4 × 10−9 to 1.75 × 10−5 m) in the presence or absence of HA (50 μg/ml). After 24 h of incubation at 37 °C, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based growth assays were analyzed as described previously (6, 35, 36). The percentage of absorbance relative to untreated controls (i.e. cells treated with neither HA nor chemotherapeutic drugs) was plotted as a linear function of drug concentration. The 50% inhibitory concentration (IC50) was identified as a concentration of drug required to achieve a 50% growth inhibition relative to untreated controls. For apoptosis assay, FITC-conjugated annexin V (for measuring apoptotic cells) using apoptosis detection kit (Calbiochem) was used according to the manufacturer's protocol (6).
RESULTS
Isolation of a Highly Tumorigenic Cell Population from Tumor-derived HNSCC (HSC-3 Cells) Using Multicolor Fluorescence-activated Cell Sorter (FACS)
CSCs are those cancer cells that exhibit stem cell properties, especially the ability to undergo self-renewal/growth and clone formation (differentiation) into multiple cell types (10, 11). CSCs are also very tumorigenic and capable of forming tumors with high efficiencies (13). CD44 expression has been identified as one of the key CSC surface markers for head and neck cancer cells (13–15). Our previous data indicated that the level of CD44v3 isoform expression (e.g. CD44v3) increases as the histological grade of each of the HNSCC progresses (16–18). Aldehyde dehydrogenase-1 (ALDH1), a detoxifying enzyme responsible for the oxidation of intracellular aldehydes, is also considered to be a common marker for both normal and malignant human HNSCC CSCs (53, 54). Overexpression of both CD44v3 and ALDH1 appears to be closely associated with HNSCC development and progression.
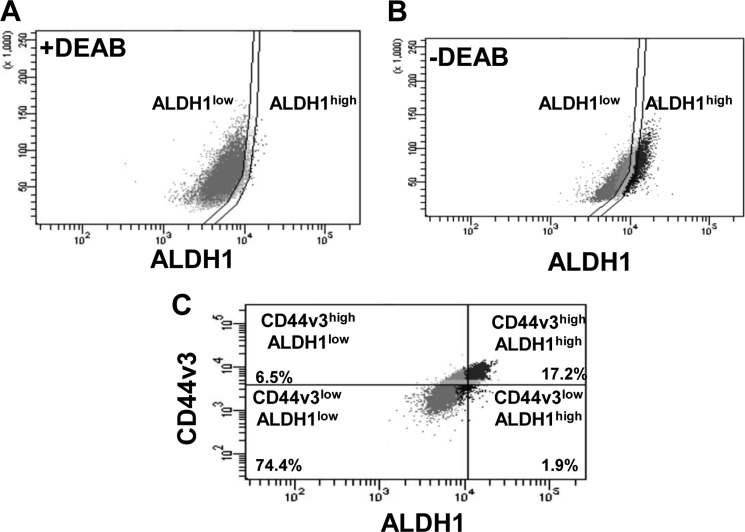
In this study we isolated CD44v3highALDH1high cell populations from tumor-derived human HNSCC cells (HSC-3) using both the ALDEFLUOR kit (to measure an ALDH1 enzymatic activity) (Fig. 1, A and B) and APC-labeled anti-CD44v3 antibody (recognizing the v3-specific domain of CD44) followed by FACS analysis as indicated in Fig. 1C. As a control for the ALDH1 activity measurement, an aliquot of cells was pretreated with 50 mmol/liter DEAB, which is a specific ALDH1 inhibitor followed by adding ALDEFLUOR as shown in Fig. 1A. Using flow cytometry (FACS) analyses of tumor cells stained with ALDEFLUOR and APC-anti-CD44v3 antibody, we identified four tumor cell populations, including CD44v3highALDH1high (Fig. 1C, top right quad), CD44v3highALDH1low (Fig. 1C, top left quad), CD44v3lowALDH1high (Fig. 1C, bottom right quad), and CD44v3lowALDH11ow (Fig. 1C, bottom left quad). CD44v3highALDH1high and CD44v3lowALDH11ow represented 17.2 and 74.4% of total tumor cell population, respectively, as the two major cell populations. CD44v3highALDH1low (Fig. 1C, representing 6.5%) and CD44v3lowALDH1high (Fig. 1C, representing 1.9%) are present as two minor tumor cell populations. Consequently, we employed a FACS to isolate the two major live tumor cell populations, CD44v3highALDH1high cells and CD44v3lowALDH11ow cells.
FIGURE 1.
Isolation of a highly tumorigenic cell population from tumor-derived HNSCC (HSC-3 cells) using multicolor FACS. Tumor-derived human HNSCC (HSC-3 cells) were incubated with both ALDEFLUOR kit (to measure an ALDH1 enzymatic activity) (A and B) and APC-labeled anti-CD44v3 antibody (recognizing the v3-specific domain of CD44) followed by FACS and cell sorting (C). A, flow cytometry analyses of tumor-derived HSC-3 cells treated with ALDEFLUOR (to measure an ALDH1 enzymatic activity) in the presence of DEAB (a specific ALDH1 inhibitor) as a control (noted that most of the tumor cell population shows a low level of ALDH1 activity (designated as ALDH1low cells)). B, flow cytometry analyses of tumor-derived HSC-3 cells treated with ALDEFLUOR (to measure an ALDH1 enzymatic activity) in the absence of DEAB (Noted that tumor cell population showing both a low level of ALDH1 activity (designated as ALDH1low cells) and a high level of ALDH1 activity (designated as ALDH1high cells)). C, flow cytometry analyses of tumor cell populations, including CD44v3highALDH1high (top right quad, 17.2%), CD44v3highALDH1low (top left quad, 6.5%), CD44v3lowALDH1high (bottom right quad, 1.9%), or CD44v3lowALDH11ow (bottom left quad, 74.4%). Live tumor cell sorting was then done using a FACS to isolate CD44v3highALDH1high cells or CD44v3lowALDH11ow for the study.
Previous studies reported that tumor cells with CD44highALDH1high (but to a much lesser extent CD44lowALDH11ow) phenotypes were found to display the typical CSC properties by forming tumors starting with a low number of tumor cells injected into animals (54). Therefore, we decided to evaluate the tumor-forming ability of sorted CD44v3highALDH1high cells and CD44v3lowALDH11ow cells (also unsorted cells as controls) using nonobese diabetic/severe combined immunodeficient (NOD/SCID) immunocompromised mice. Specifically, 5-week-old female NOD/SCID were injected subcutaneously with sorted CD44v3highALDH1high cells or CD44v3lowALDH11ow or unsorted HSC-3 cells ranging from 50, 500, to 5,000 cells. Our results showed that CD44v3highALDH1high (but not CD44v3lowALDH11ow cells or unsorted cells) cells were capable of forming tumors with high efficiency in NOD/SCID mice injected with as few as 50 CD44v3highALDH1high cells (Table 1). As a control, we routinely treated unsorted cells with APC-labeled anti-CD44v3 antibody and ALDEFLUOR reagent. Our data indicate that 5,000 unsorted cells (but not 500 or 50 unsorted cells) induce a very low number of tumors (1/16) within 6 weeks after being injected subcutaneously into NOD/SCID mice (Table 1). The small number of tumor formation (using unsorted cells) may be due to the reduced number of tumorigenic CD44v3highALDH1high cells present in the study or the interference (i.e. necrosis or cell death) from other cell populations such as CD44v3lowALDH1low cells in the tumorigenic assay. These findings indicate that CD44v3highALDH1high cells (to a lesser extent unsorted cells but not CD44v3lowALDH11ow cells) are very tumorigenic.
TABLE 1.
Analyses of tumor formation by CD44v3highALDHhigh cells, CD44v3lowALDHlow cells or unsorted cells subcutaneously injected into NOD/SCID mice
| Cell populations | Tumor formationa |
||
|---|---|---|---|
| 5,000 cells injected (6 weeks) | 500 cells injected (6 weeks) | 50 cells injected (6 weeks) | |
| CD44v3highALDHhigh cells | 16/16 | 14/16 | 10/16 |
| CD44v3lowALDHlow cells | 0/16 | 0/16 | 0/16 |
| Unsorted cells | 1/16 | 0/16 | 0/16 |
a For the tumor cell injection, each mouse was subcutaneously inoculated with CD44v3highALDHhigh cells or CD44v3lowALDHlow cells or unsorted cells with 5,000, 500, or 50 cells as described under “Materials and Methods.” The values expressed in the table represent the number of animals that developed tumors/total number of animals used in the study. The tumor formation assay was performed on at least five different experiments with a standard deviation less than ±5%.
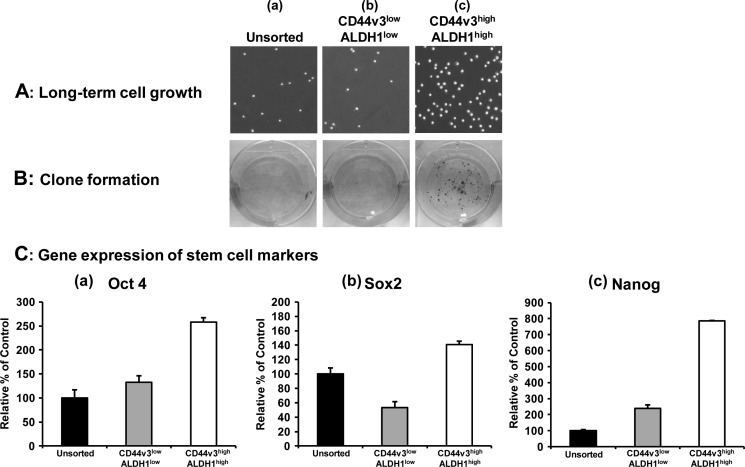
Further analysis indicated that CD44v3highALDH1high cells are also capable of growing in serum-free medium (containing EGF and bFGF) for 21 days (Fig. 2A, panel c). In contrast, both unsorted cells and CD44v3lowALDH11ow cells showed little cell growth under this serum-free medium condition (Fig. 2, A, panels b and c). To further test the ability of these sorted cells (CD44v3highALDH1high cells and CD44v3lowALDH11ow cells) versus unsorted cells to undergo clone formation, we conducted clone formation assays by incubating the tumor cells in serum-containing medium. Our data clearly indicated that CD44v3highALDH1high cells display a significantly higher clone formation capacity (Fig. 2B, panel c) compared with CD44v3lowALDH11ow cells or unsorted cells (Fig. 2B, panels a and b). These findings strongly suggest that CD44v3highALDH1high cells express cancer stem cell properties by exhibiting tumorigenicity, self-renewal/growth, and clone formation. However, the oncogenic mechanism that contributes to the highly tumorigenic CSC-like properties of CD44v3highALDH1high cells was unknown and therefore is the focus of this investigation.
FIGURE 2.
Characterization of CD44v3highALDH1high cells, CD44v3lowALDH1low cells, or unsorted cells. A, long term tumor cell growth was measured by incubating unsorted cells (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting) (panel a), CD44v3lowALDH1low cells (panel b), or CD44v3highALDH1high cells (panel c) in serum-free RPMI 1640 medium (containing EGF and bFGF) for 3 weeks (21 days) as described under “Materials and Methods.” B, clone formation was measured by incubating unsorted cells (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting) (panel a), CD44v3highALDH1high cells, CD44v3lowALDH1low cells (panel b), or CD44v3highALDH1high cells (panel c) in serum-containing RPMI 1640 medium for 10 days followed by crystal violet staining as described under “Materials and Methods.” C, detection of stem cell marker expression in CD44v3highALDH1high cells, CD44v3lowALDH1low cells, or unsorted cells (pretreated with ALDEFLUOR and APC-anti-CD44v3 antibody without cell sorting). The expression of stem cell markers such as Oct4, Sox2, and Nanog was measured using Oct4-Sox2-Nanog-specific primers followed by Q-PCR analyses according to the procedures described under “Materials and Methods.” Total RNA isolated from CD44v3highALDH1high cells, CD44v3lowALDH1low cells, or unsorted cells was reverse-transcribed and subjected to Q-PCR using Oct4-specific (panel a) or Sox2-specific (panel b) or Nanog-specific primer pairs to measure Oct4 gene expression (panel a), Sox2 gene expression (panel b), or Nanog gene expression, respectively. (Relative mRNA expression levels of Oct4, Sox2, and Nanog in CD44v3highALDH1high cells, CD44v3lowALDH1low cells, or unsorted cells were calculated after normalization with 36B4 mRNA levels as determined by Q-PCR.) The values expressed in this figure represent an average of triplicate determinations of three experiments with a standard deviation less than ±5%.
Detection of Stem Cell Marker Expression in Tumorigenic CD44v3highALDH1high Cells and HNSCC Tumors
Stem cell markers such as Oct4, Sox2, and Nanog are known to form a self-organized core of transcription factors that maintain pluripotency and self-renewal of human embryonic stem cells (45). In this study, we found that the gene expression of Oct4, Sox2, and Nanog was significantly increased in CD44v3highALDH1high cells as compared with CD44v3lowALDH1low cells or unsorted cells (Fig. 2C, panels a–c). The fact that a low level of stem cell marker expression was detected in CD44v3lowALDH1low cells suggests that some unknown stem cells are present in the CD44v3lowALDH1low cell subpopulation. The biochemical and cellular nature of this population awaits future investigation.
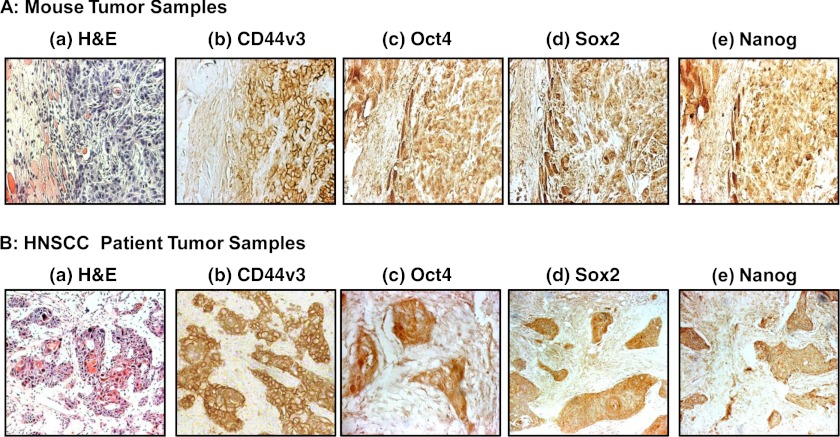
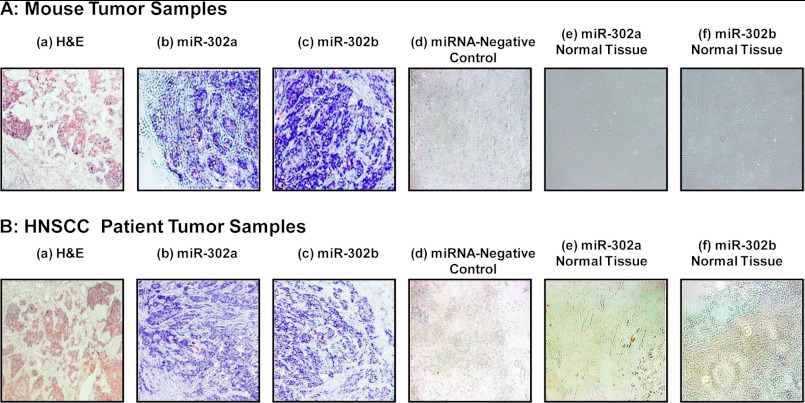
Our immunostaining data indicate that overexpression of the CD44v3, Oct4, Sox2, and Nanog proteins can be detected in all 10 CD44v3highALDH1high cell-induced tumor tissue samples (Fig. 3A). The expression of CD44v3 and three stem cell markers such as Oct4, Sox2, and Nanog was also analyzed in human HNSCC samples statistically with respect to various demographic and clinical outcome parameters. The patient parameters examined included gender, age, presence of tobacco abuse, primary tumor T status, or grade of tumor differentiation. Table 2 summarizes the expression of these four proteins in primary HNSCC tumors. Although no association of CD44v3 or three stem cell markers was found with respect to sex, age, presence of tobacco abuse, or grade of tumor differentiation, the expression of CD44v3, Oct4, Sox2, and Nanog expression was associated with advanced human HNSCC samples. Specifically, 50 of 66 such samples show CD44v3 overexpression (Fig. 3, A, panel b, and B, panel b; Table 2); 40 of 66 samples show Oct4 overexpression (Fig. 3, A, panel c, and B, panel c); 38 of 66 samples show Sox2 overexpression (Fig. 3, A, panel d, and B, panel d); and 45 our of 66 samples show Nanog overexpression (Fig. 3, A, panel e, and B, panel e; Table 2). These immunostaining results are statistically significant with a p < 0.02 (for CD44v3 staining), p = 0.01 (for Oct4 staining), p = 0.02 (for Sox2 staining), and p < 0.001 (for Nanog staining) as compared with CD44v3-Oct4-Sox2-Nanog detected in early (T1-T2) primary tumors (n = 44) from HNSCC patient samples. The staining patterns also reveal that CD44v3 is located in the plasma membrane of the tumor cells (Fig. 3, A, panel b, and B, panel b; Table 2). However, all three stem cell markers (e.g. Oct4, Sox2, and Nanog) appear to be accumulated inside the nucleus and/or in the cytoplasm of the tumor cells (Fig. 3, A, panels c–e, and B, panels c–e; Table 2). These findings clearly demonstrated a strong correlation between overexpression of CD44v3-Oct4-Sox2-Nanog and head and neck cancer progression.
FIGURE 3.
Detection of CD44v3, Oct4, Sox2, and Nanog expression in CD44v3highALDHhigh cell-induced mouse tumors and HNSCC patient samples. A panel of immunoreagents, including anti-CD44v3 antibody, Oct4 antibody, Sox2 antibody, and Nanog antibody, was used to examine the expression of CD44v3 and stem cell markers (e.g. Oct4, Sox2, and Nanog) in CD44v3highALDHhigh cell-induced mouse tumors (A) and HNSCC patient tumors (B). A, H&E staining (panel a) and immunoperoxidase staining of CD44v3 (panel b), Oct4 (panel c), Sox2 (panel d), and Nanog (panel e) using CD44v3highALDHhigh cell-induced mouse tumor samples. B, H&E staining (panel a) and immunoperoxidase staining of CD44v3 (panel b), Oct 4 (panel c), Sox2 (panel d), and Nanog (panel e) using HNSCC patient tumor samples.
TABLE 2.
Detection of CD44v3, Oct4, Nanog, Sox2, and miR-302 expression in early (T1-T2) (n = 44) versus advanced (T3-T4) (n = 66 patients) primary tumors from HNSCC patients
| Tumor markers | Weak staining (T1-T2) tumors | Strong staining (T1-T2) tumors | Weak staining (T3-T4) tumors | Strong staining (T3-T4) tumors |
|---|---|---|---|---|
| CD44v3 | 30 (68.18%) | 14 (31.82%) | 16 (24.24%) | 50 (75.76%)a |
| Oct4 | 38 (86.36%) | 6 (13.64%) | 26 (39.40%) | 40 (60.60%)b |
| Sox2 | 35 (79.55%) | 9 (20.45%) | 29 (43.94%) | 37 (56.06%)c |
| Nanog | 34 (77.27%) | 10 (22.73%) | 21 (31.82%) | 45 (68.18%)d |
| miR-302a | 36 (81.82%) | 8 (18.18%) | 25 (37.88%) | 41 (62.12%)e |
| miR-302b | 37 (84.10%) | 7 (15.90%) | 29 (43.94%) | 37 (56.06%)f |
a The procedures for detecting CD44v3, Oct4, Sox2, and Nanog expression (using semiquantitative analyses of the immunohistochemical staining) and miR-302a/miR-302b (using in situ hybridization) were described under “Materials and Methods” (weak staining, <50% tumor cells showing staining; strong staining: ≥50% tumor cells showing staining). Fisher exact testing was also used to determine whether the expression of various markers (CD44v3, Oct4, Sox2, Nanog, miR-302a and miR-302b) was significantly different in the advanced (T3-T4) primary tumors (n = 66) as compared with early (T1-T2) primary tumors (n = 44) from HNSCC patient samples. p < 0.02.
b p = 0.01.
c p = 0.02.
d p < 0.001.
e p = 0.03.
f p = 0.04.
Hyaluronan (HA)-induced Oct4, Sox2, and Nanog Complex Formation in Tumorigenic CD44v3highALDH1high Cells
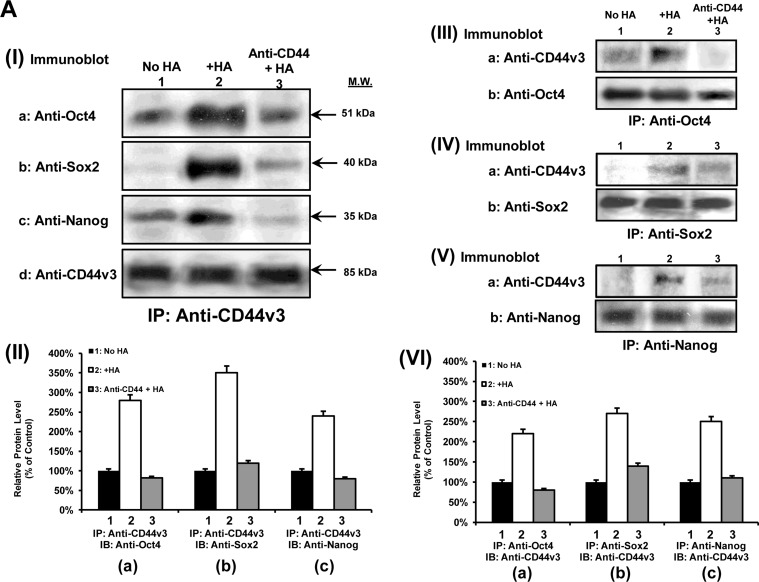
Oncogenic signaling induced by HA-CD44v3 has been previously implicated in head and neck cancer progression (16–18). However, the cellular and molecular mechanisms by which HA-CD44 signaling occurs in the tumorigenic CD44v3highALDH1high cell population are not known. In this study, we have first focused on the question of whether there is a physical linkage between CD44v3 and the three stem cell markers (e.g. Oct4, Sox2, and Nanog) in CD44v3highALDH1high cells. To this end, we performed an anti-CD44v3-mediated immunoprecipitation followed by anti-Oct4, anti-Sox2, anti-Nanog immunoblot, or anti-CD44v3 immunoblot, respectively. The results revealed that these stem cell markers are physically associated with CD44v3 as early as 5 min following HA addition to CD44v3highALDH1high cells (Fig. 4, A, panel I, a–d, lane 2, and A, panel II, a–c). In contrast, a very low level of Oct4 (or Sox2 or Nanog) was present in the anti-CD44v3-immunoprecipitated materials (reblotted with anti-CD44v3) in cells not treated with HA (Fig. 4, A, panel I, a–d, lane 1, and A, panel II, a–c) or in cells pretreated with anti-CD44 antibody (an HA-binding blocking agent) followed by a 5-min HA treatment (Fig. 4, A, panel I, a–d, lane 3, A, panel II, a–c). These findings establish that CD44 and three stem cell markers (Oct4, Sox2, and Nanog) are closely associated and that the treatment of CD44v3highALDH1high cells with HA promotes a complex formation between CD44v3 and all three stem cell markers.
FIGURE 4.
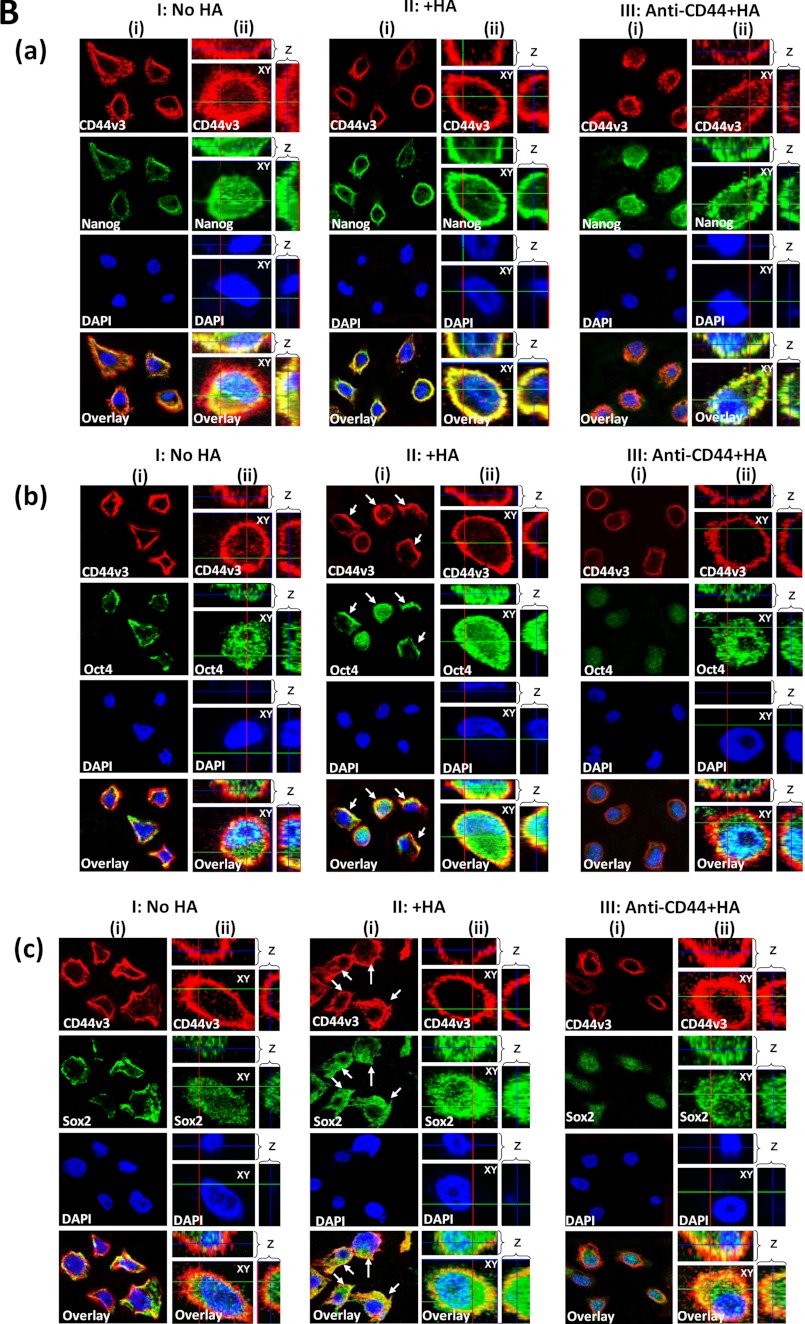
Detection of CD44v3 association with Oct4, Sox2, and Nanog in CD44v3highALDHhigh cells. A, panel I, detection of CD44v3 association with Oct4, Sox2, and Nanog by anti-CD44v3-mediated immunoprecipitation followed by immunoblotting with anti-Oct4 antibody (a), anti-Sox2 antibody (b), or anti-Nanog antibody (c) or reblotting with anti-CD44v3 (d) as a loading control using CD44v3highALDHhigh cells treated with no HA (lane 1) or with HA (50 μg/ml) (lane 2) for 5 min or pretreated with anti-CD44 antibody for 1 h followed by 5 min HA (50 μg/ml) addition (lane 3). A, panel II, quantification analysis of the protein levels of CD44v3-associated Oct4 (a), Sox2 (b), and Nanog (c) in CD44v3highALDHhigh cells with various treatments as shown in A, panel I. The relative protein level represents the ratio of Oct4 and total CD44v3 (the loading control) (a), the ratio of Sox2 and total CD44v3 (the loading control) (b), and the ratio of Nanog and total CD44v3 (the loading control) (c) as determined by densitometry, and these numbers were then normalized to untreated (no HA treatment) cell value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of five experiments with a standard deviation less than ±5%. A, panel III, detection of Oct4 association with CD44v3 by anti-Oct4-mediated immunoprecipitation followed by immunoblotting with anti-CD44v3 antibody (a) or reblotting with anti-Oct4 (b) as a loading control using CD44v3highALDHhigh cells treated with no HA (lane 1) or with HA (50 μg/ml) for 5 min (lane 2) or pretreated with anti-CD44 antibody for 1 h followed by 5 min of HA (50 μg/ml) addition (lane 3). A, panel IV, detection of Sox2 association with CD44v3 by anti-Sox2-mediated immunoprecipitation followed by immunoblotting with anti-CD44v3 antibody (a) or reblotting with anti-Sox2 (b) as a loading control using CD44v3highALDHhigh cells treated with no HA (lane 1) or with HA (50 μg/ml) for 5 min (lane 2) or pretreated with anti-CD44 antibody for 1 h followed by 5 min of HA (50 μg/ml) addition (lane 3). A, panel V, detection of Nanog association with CD44v3 by anti-Nanog-mediated immunoprecipitation followed by immunoblotting with anti-CD44v3 antibody (a) or reblotting with anti-Nanog (b) as a loading control using CD44v3highALDHhigh cells treated with no HA (lane 1) or with HA (50 μg/ml) for 5 min (lane 2) or pretreated with anti-CD44 antibody for 1 h followed by 5 min of HA (50 μg/ml) addition (lane 3). A, panel VI, quantification analysis of the protein levels of Oct4-associated CD44v3 (a), Sox2-associated CD44v3 (b), and Nanog-associated CD44v3 (c) in CD44v3highALDHhigh cells with various treatments as shown in A, panel III–V. The relative protein level represents the ratio of CD44v3 and total Oct4 (the loading control) (a as shown in A, panel III), the ratio of CD44v3 and total Sox2 (the loading control) (b as shown in A, panel IV), and the ratio of CD44v3 and total Nanog (the loading control) (c as shown in A, panel V) determined by densitometry, and these numbers were then normalized to untreated (no HA treatment) cell value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation less than ±5%. B, a, double immunofluorescence staining analysis of CD44v3/Nanog co-localization by Texas Red-conjugated anti-CD44v3 (red) and FITC-conjugated anti-Nanog (green) or nuclear marker, DAPI (blue) using CD44v3highALDHhigh cells treated with no HA (panel I, (i)), with HA (50 μg/ml) for 5 min (panel II, (i)), or pretreated with anti-CD44 antibody for 1 h followed by 5 min of HA (50 μg/ml) addition (panel III, (i)). Panels I, (ii), II, (ii), and III, (ii), represent three-dimensional reconstruction of images from fluorescence-labeled cells showing cellular distribution of CD44v3 and Nanog as described in B, a from a stack of confocal XY and Z-series images (0.2 μm/section in side views). An overlay image represents a combination of three colors (red, green, and blue). (Note: Nanog is closely associated with CD44v3-containing plasma membrane in all HA-treated cells.) B, b, double immunofluorescence staining analysis of CD44v3-Oct4 co-localization by Texas Red-conjugated anti-CD44v3 (red) and FITC-conjugated anti-Oct4 (green) or nuclear marker DAPI (blue) using CD44v3highALDHhigh cells treated with no HA (panel I, (i)) or with HA (50 μg/ml) for 5 min (panel II, (i)) or pretreated with anti-CD44 antibody for 1 h followed by 5 min of HA (50 μg/ml) addition (panel III, (i)). Panels I, (ii), II, (ii), and III, (ii), represent three-dimensional reconstruction of images from fluorescence-labeled cells showing cellular distribution of CD44v3 and Oct4 as described in B, b, from a stack of confocal XY and Z series images (0.2 μm/section in side views). An overlay image represents a combination of three colors (red, green, and blue). (Arrows indicate that Oct4 is closely associated with CD44v3-containing plasma membrane region in some HA-treated cells.) B, c, double immunofluorescence staining analysis of CD44v3/Sox2 co-localization by Texas Red-conjugated anti-CD44v3 (red) and FITC-conjugated anti-Sox2 (green) or nuclear marker DAPI (blue) using CD44v3highALDHhigh cells treated with no HA (panel I, (i)) or with HA (50 μg/ml) for 5 min (panel II, (i)) or pretreated with anti-CD44 antibody for 1 h followed by 5 min HA (50 μg/ml) addition (panel III, (i)). Panels I, (ii), II, (ii), and panel III, (ii), represent three-dimensional reconstruction of images from fluorescence-labeled cells showing cellular distribution of CD44v3 and Sox2 as described in B, c, from a stack of confocal XY and Z series images (0.2 μm/section in side views). An overlay image represents a combination of three colors (red, green, and blue). (Arrows indicate that Sox2 is closely associated with CD44v3-containing plasma membrane region in some HA-treated cells.)
To further establish the association of CD44v3 with certain stem cell markers (e.g. Oct4, Sox2, and Nanog), we also performed immunoprecipitation using anti-Oct4/anti-Sox2/anti-Nanog antibody followed by anti-CD44v3-mediated immunoblotting (Fig. 4, A, panel III, a and b, A, panel IV, a and b, A, panel V, a and b, and A, panel VI, a–,c). Our findings indicate that CD44v3 and three stem cell markers (Oct4, Sox2, and Nanog) are physically linked and that there is a significant increase in the recruitment of all three stem cell markers into the CD44v3-associated complex following a 5-min incubation of CD44v3highALDH1high cells with HA.
In addition, we performed double immunofluorescence staining using Texas Red-conjugated anti-CD44v3 and FITC-conjugated anti-Nanog/anti-Oct4/anti-Sox2 in CD44v3highALDH1high cells treated with HA (Fig. 4B, a–c, panel II, (i) and (ii)). Our results indicate that all three stem cell markers (in particular, Nanog (Fig. 4B, a, panel II, (i) and (ii)) and to a lesser extent Oct4 and Sox2 (Fig. 4, B, b, panel II, (i) and (ii), and B, c, panel II, (i) and (ii)) become localized to CD44v3-containing plasma membrane following a 5-min incubation of CD44v3highALDH1high cells with HA. In contrast, these three proteins are randomly distributed in the cytosol of these cells in the absence of HA treatment (Fig. 4B, a–c, panel I, (i) and (ii)). The fact that pretreatment of these cells with anti-CD44 antibody reduces HA-mediated Nanog-Oct4-Sox2 association with CD44v3 (Fig. 4B, a–c, panel III, (i) and (ii)) suggests that Oct4-Sox2-Nanog membrane localization with CD44v3 induced by HA in CD44v3highALDH1high cells is CD44-dependent.
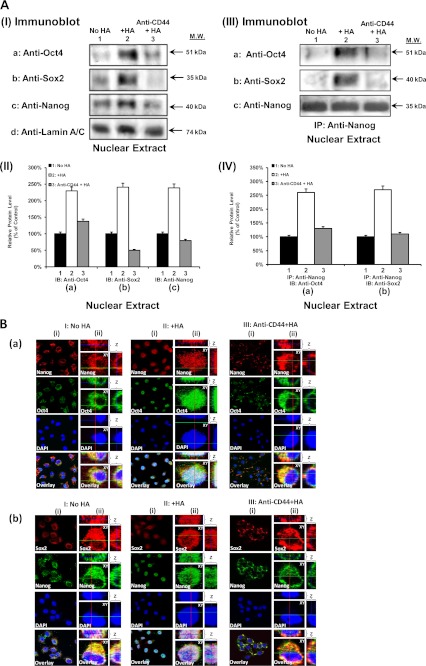
Moreover, we observed that a 15–20-min HA treatment of CD44v3highALDH1high cells stimulates a significant amount of Oct4-Sox2-Nanog accumulation (Fig. 5, A, panel I, a–d, lane 2, and A, panel II, a–c) and complex formation (Fig. 5, A, panel III, a–d, lane 2, A, panel IV, a and b) in the nucleus. In contrast, only a low level of Oct4-Sox2-Nanog accumulation and complexes is present in the nucleus of CD44v3highALDH1high cells, either pretreated with anti-CD44 antibody plus HA (Fig. 5, A, panel I, a–d, lane 3; A, panel II, a–c; A, panel III, a–d, lane 3, and A, panel IV, a and b) or without any HA treatment (Fig. 5, A, panel I, a–d, lane 1; A, panel II, a–c; A, panel III, a–d, lane 1, and A, panel IV, a and b). Using double immunofluorescence staining and confocal microscopic analyses, we also confirmed that three stem cell markers (e.g. Oct4, Nanog, and Sox2) are randomly distributed in the cytosol of CD44v3highALDH1high cells in the absence of HA (Fig. 5B, a and b, panel I, (i) and (ii)). Very low levels of these three molecules are detected in the nucleus (indicated by DAPI nuclear staining) in these cells without any HA treatment (Fig. 5B, a and b, panel I, (i) and (ii)). However, 15–20 min after HA treatment, Nanog together with Sox2 and Oct4 are co-translocated into the nucleus (indicated by DAPI nuclear staining) (Fig. 5B, a and b, panel II, (i) and (ii)). Pretreatment of cells with anti-CD44 antibody followed by HA addition effectively blocks nuclear translocation of Nanog, Oct4, and Sox2 complexes in these CD44v3highALDH1high cells (Fig. 5B, a and b, panel III, (i) and (ii)). These observations indicate that HA promotes Oct4-Sox2-Nanog accumulation and complex formation in the nucleus of CD44v3highALDH1high cells in a CD44-dependent manner.
FIGURE 5.
Analyses of HA-CD44-induced Oct4, Sox2, and Nanog accumulation and complex formation in the nuclear fraction. A, panel I, nuclear extract isolated from CD44v3highALDHhigh cells (treated with no HA (lane 1) or with HA (50 μg/ml) for 15 min (lane 2) or pretreated with anti-CD44 antibody for 1 h followed by 15 min of HA (50 μg/ml) addition (lane 3)) were immunoblotted with anti-Oct4 antibody (a) or anti-Sox2 antibody (b) or anti-Nanog antibody (c) or anti-lamin A/C antibody (d) (as a loading control), respectively. A, panel II, quantification analysis of the protein levels of Oct4 (a), Sox2 (b), and Nanog (c) in the nuclear extract of CD44v3highALDHhigh cells with various treatments as shown in A, panel I. The relative protein level represents the ratio of Oct4 and lamin A/C (the loading control) (a), the ratio of Sox2 and lamin A/C (the loading control) (b), and the ratio of Nanog and lamin A/C (the loading control) (c) determined by densitometry, and these numbers were then normalized to untreated (no HA treatment) cell value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation less than ±5%. A, panel III, nuclear extract isolated from CD44v3highALDHhigh cells (treated with no HA (lane 1) or with HA (50 μg/ml) for 15 min (lane 2) or pretreated with anti-CD44 antibody for 1 h followed by 15 min of HA (50 μg/ml) addition (lane 3)) were used for anti-Nanog-mediated immunoprecipitation followed by immunoblotting with anti-Oct4 antibody (a) or anti-Sox2 (b) or reblotting with anti-Nanog (c) as a loading control, respectively. A, panel IV, quantification analysis of the protein levels of Nanog-associated Oct4 (a) and Nanog-associated Sox2 (b) in the nuclear extract of CD44v3highALDHhigh cells with various treatments as shown in A, panel III. The relative protein level represents the ratio of Oct4 and Nanog (the loading control) (a) and the ratio of Sox2 and Nanog (the loading control) (b) determined by densitometry, and these numbers were then normalized to untreated (no HA treatment) cell value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation of less than ±5%. B, a, double immunofluorescence staining analysis of Nanog-Oct4 co-localization by Texas Red-labeled anti-Nanog (red) and FITC-labeled anti-Oct4 (green) or nuclear marker, DAPI (blue), using CD44v3highALDHhigh cells treated with no HA (panel I, (i)) or with HA (50 μg/ml) for 15 min (panel II, (i)) or pretreated with anti-CD44 antibody for 1 h followed by 15 min of HA (50 μg/ml) addition (panel III, (i)). Panels I, (ii), II, (ii), and III, (ii), represent three-dimensional reconstruction of images from fluorescence-labeled cells showing cellular distribution of Nanog and Oct4 as described in B, a, from a stack of confocal XY and Z series images (0.2 μm/section in side views). An overlay image represents a combination of three colors (red, green, and blue). (Note: Nanog and Oct4 are co-localized in the nucleus in all HA-treated cells but not in cells pretreated with anti-CD44 plus HA or treated with no HA.) B, b, double immunofluorescence staining analysis of Sox2/Nanog co-localization by Texas Red-labeled anti-Sox2 (red) and FITC-labeled anti-Nanog (green) or nuclear marker, DAPI (blue), using CD44v3highALDHhigh cells treated with no HA (panel I, (i)) or with HA (50 μg/ml) for 15 min (panel II, (i)) or pretreated with anti-CD44 antibody for 1 h followed by 15 min of HA (50 μg/ml) addition (panel III, (i)). Panels I, (ii), II, (ii), and III, (ii), represent three-dimensional reconstruction of images from fluorescence-labeled cells showing cellular distribution of Sox2 and Nanog as described in B, b, from a stack of confocal XY and Z series images (0.2 μm/section in side views). An overlay image represents a combination of three colors (red, green, and blue). (Note: Sox2 and Nanog are co-localized in the nucleus in all HA-treated cells but not in cells pretreated with anti-CD44 plus HA or treated with no HA.)
Furthermore, by stepping the objective through the depth of the fluorescence-labeled cells, both XY and Z series collection of optical sections were obtained and projected to re-construct the three-dimensional images. After rotating the image, the side view (Z-section) of a three-dimensional cell was then reconstructed using LSM Image Examiner (Carl Zeiss, GmbH). Our data indicate that the three stem cell markers (Nanog-Oct4-Sox2) are randomly distributed in the cytosol or beneath the plasma membrane, and very low levels of these three proteins are detected in the nucleus (indicated by DAPI nuclear staining) in CD44v3highALDH1high cells without any HA treatment (Fig. 4, B, a–c, panel I, (ii), and B, a and b, panel I, (ii)). Treatments of the cells with HA for 5 min result in the recruitment of Nanog-Oct4-Sox2 into CD44v3-associated plasma membrane (Fig. 4B, a–c, panel II, (ii)). When cells are treated with HA for 15–20 min, the Nanog-Oct4-Sox2 complexes are nuclearly translocated (Fig. 5B, a and b, panel II, (ii)). The fact that pretreatment of anti-CD44 antibody reduces HA-mediated stem cell marker plasma membrane localization and nuclear translocation (Fig. 4, B, a–c, panel III, (ii) and B, a and b, panel III, (ii)) strongly suggests that the cellular redistribution (plasma membrane versus nucleus) of Oct4-Sox2-Nanog induced by HA in CD44v3highALDH1high cells is CD44-dependent.
During the course of our double immunofluorescence staining analyses, we have consistently observed that the staining intensity of Nanog-Oct4-Sox2 in HA-treated cells is always higher than the control or anti-CD44 antibody-treated samples. It is possible that HA causes a large accumulation of these three proteins in the plasma membrane region (Fig. 4B, a–c, panel II, (ii)) and the nucleus of cells (Fig. 5B, a and b, panel II, (ii)) and also that HA may also induce conformational changes of these three stem cell markers during their plasma localization and nuclear translocation processes. The cellular redistribution of these three proteins induced by HA may lead to additional binding sites for anti-Nanog (or anti-Oct4 or anti-Sox2)-mediated immunofluorescence staining.
Our preliminary data also indicate that HA signaling-induced Nanog phosphorylation may be responsible for recruiting Oct4 and Sox2 complex formation and nuclear translocation. Detailed analysis of the Nanog-Oct4-Sox2 complex formation awaits further investigation.
Role of Oct4, Sox2, and Nanog in Regulating miR-302 Expression in HA-activated CD44v3highALDH1high Cells
In Vivo Binding of Oct4-Sox2-Nanog Complexes to the Promoter Region of the miR-302 in CD44v3highALDH1high Cells
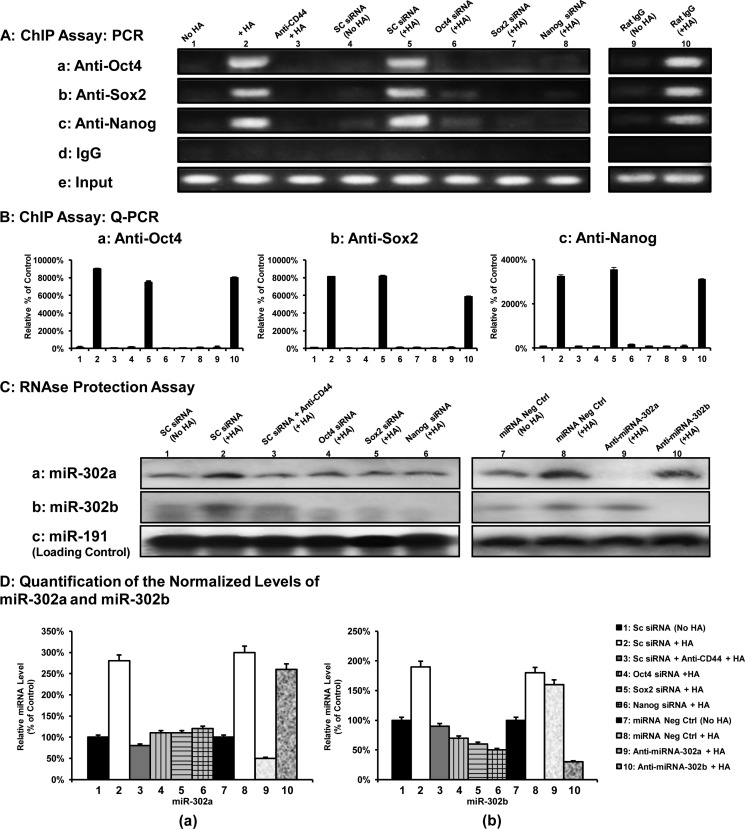
A previous study indicated that the gene encoding the stemness-related miR-302 cluster (e.g. miR-302a and miR-302b) is regulated by a promoter containing Oct4-Sox2-Nanog-binding sites (50). To examine whether the Oct4-Sox2-Nanog complex directly interacts with the promoter region of the miR-302 cluster, anti-Oct4-, anti-Sox2-, and anti-Nanog antibody-specific ChIP assays were performed with the CD44v3highALDH1high cells. As shown in Fig. 6, A and B, both PCR and Q-PCR results from anti-Oct4, anti-Sox2, and anti-Nanog-mediated precipitations from HA-treated CD44v3highALDH1high cells revealed a specific amplification product using a primer pair specific for the miR-302 cluster promoter region containing the Oct4-, Sox2-, and Nanog-binding sites (Fig. 6, A, panels a–c, lane 2, and B, panels a–c, bar 2). In contrast, significantly reduced amounts of Oct4, Sox2, or Nanog binding of the miR-302 cluster promoter region were found in cells treated with no HA (Fig. 6, A, panels a–c, lane 1, and B, panels a–c, bar 1) or pretreated with anti-CD44 antibody followed by HA addition (Fig. 6, A, panels a–c, lane 3; and B, panels a–c, bar 3). However, normal rat IgG does not appear to block HA-mediated Oct4-Sox2-Nanog binding to the miR-302 promoter (Fig. 6, A, panels a–c, lanes 9 and 10, and B, panels a–c, bars 9 and 10) These findings indicate that the recruitment of Oct4, Sox2, and Nanog into the miR-302 cluster promoter site is both HA- and CD44-dependent.
FIGURE 6.
Interaction between Oct4-Sox2-Nanog and the miR-302 cluster promoter in CD44v3highALDHhigh cells. A, ChIP assay was performed in CD44v3highALDHhigh cells using Oct4-Sox2-Nanog-binding site-containing miR-302 cluster promoter-specific primers and PCR analyses. Identical volumes from the final precipitated materials (anti-Oct4-mediated immunoprecipitated materials (panel a), anti-Sox2-mediated immunoprecipitated materials (panel b), anti-Nanog-mediated immunoprecipitated materials (panel c), IgG isotype control-precipitated materials (panel d), or total input materials (panel e)) were used for the PCRs (untreated cells (lane 1), cells treated with HA for 30 min (lane 2), cells pretreated with anti-CD44 antibody for 1 h plus 30 min of HA addition (lane 3), cells pretreated with scrambled (SC) siRNA with no HA (lane 4), cells pretreated with scrambled (SC) siRNA plus 30 min of HA addition (lane 5), cells pretreated with Oct4 siRNA plus 30 min of HA addition (lane 6), Sox2 siRNA plus 30 min of HA addition (lane 7), or Nanog siRNA plus 30 min of HA addition (lane 8), cells pretreated with normal rat IgG for 1 h without HA treatment (lane 9) and cells pretreated with normal IgG for 1 h plus 30 min of HA addition (lane 10)). B, ChIP assay was performed in CD44v3highALDHhigh cells using Oct4-Sox2-Nanog-binding site-containing miR-302 cluster promoter-specific primers and Q-PCR analyses. Identical volumes from the final precipitated materials (anti-Oct4-mediated immunoprecipitated materials (panel a), anti-Sox2-mediated immunoprecipitated materials (panel b), or anti-Nanog-mediated immunoprecipitated materials (panel c)) were used for the Q-PCRs (untreated cells (bar 1), cells treated with HA for 30 min (bar 2), cells pretreated with anti-CD44 antibody for 1 h plus 30 min of HA addition (bar 3), cells pretreated with scrambled (SC) siRNA with no HA (bar 4), cells pretreated with scrambled (SC) siRNA plus 30 min of HA addition (bar 5), cells pretreated with Oct4 siRNA plus 30 min of HA addition (bar 6), Sox2 siRNA plus 30 min of HA addition (bar 7), Nanog siRNA plus 30 min of HA addition (bar 8), cells pretreated with normal rat IgG for 1 h without HA treatment (bar 9), and cells pretreated with normal IgG for 1 h plus 30 min HA addition (bar 10). The values expressed in this figure represent an average of triplicate determinations of five experiments with a standard deviation less than ±5%. C, detection of HA-CD44-induced miR-302a and miR-302b production in CD44v3highALDHhigh cells. Detection of miR-302a and miR-302b in CD44v3highALDHhigh cells using RNase protection assay was described under “Materials and Methods.” Autoradiogram of miR-302a (panel a) and miR-302b (panel b) detected in CD44v3highALDHhigh cells incubated with scrambled (SC) siRNA (without HA (lane 1), with 2 h of HA treatment (lane 2), or pretreated with anti-CD44 antibody for 1 h followed by HA addition for 2 h (lane 3), incubated with Oct4 siRNA plus 2 h of HA treatment (lane 4) or Sox2 siRNA plus 2 h of HA treatment (lane 5) or Nanog siRNA plus 2 h of HA treatment (lane 6), incubated with miRNA-negative control (without HA (lane 7) or with 2 h of HA treatment (lane 8)), or incubated with an anti-miR-302a inhibitor plus 2 h of HA treatment (lane 9) or incubated with an anti-miR-302b inhibitor plus 2 h of HA treatment (lane 10). (Autoradiogram of miR-191 in each gel lane (panel c) was used as a loading control). D, quantification analysis of the relative levels of miR-302a and miR-302b production in CD44v3highALDHhigh cells with various treatments as shown in C. The relative miRNA level represents the ratio of miR-302a and miR-191 (the loading control) (panel a as shown in C) and the ratio of miR-302b and miR-191 (the loading control) (panel b as shown in C) determined by densitometry, and these numbers were then normalized to either scrambled siRNA (no HA)-treated cell value (bar 1, designated as 100%) for bars 1–6 or normalized to miRNA-negative control (no HA)-treated cell value (bar 7, designated as 100%) for bars 7–10. The values expressed in this figure represent an average of triplicate determinations of five experiments with a standard deviation less than ±5%.
To confirm the direct involvement of Oct4, Sox2, and Nanog in miR-302 cluster gene up-regulation, Oct4, Sox2, and Nanog were first down-regulated by Oct4 siRNA, Sox2 siRNA, or Nanog siRNA, respectively, followed by the miR-302 cluster promoter-specific ChIP assay as described above. The results revealed that transfection of CD44v3highALDH1high cells with Oct4 siRNAs (Fig. 6, A, panels a–c, lane 6, and B, panels a–c, bar 6), Sox2 siRNA (Fig. 6, A, panels a–c, lane 7, and B, panels a–c, bar 7), or Nanog siRNA (Fig. 6, A, panels a–c, lane 8, and B, panels a–c, bar 8) but not scrambled sequenced siRNA (Fig. 6, A, panels a–c, lane 4, versus lane 5, and B, panels a–c, bar 4 versus bar 5) effectively blocks HA-mediated Oct4-Sox2-Nanog binding to the miR-302 cluster promoter region in CD44v3highALDH1high cells. Identical amplification products were detected in the positive controls from total input chromatin (Fig. 6A, panel e, lanes 1–8). Moreover, no amplification was seen in samples that were precipitated with IgG isotype control (Fig. 6A, panel d, lanes 1–8). We observed similar results using quantitative RT-PCR (data not shown). Thus, these results verify that the Oct4-Sox2-Nanog complex binds directly (or forms as part of a complex) to the promoter region of miR-302 cluster in CD44v3highALDH1high cells following HA-CD44v3 activation.
Production of miR-302a and miR-302b in CD44v3highALDH1high Cells
The expression of a mature miR-302 cluster (e.g. miR-302a and miR-302b) has been shown to be involved in both normal stem cells and cancer stem cells (50, 51). To determine whether the level of miR-302 cluster (e.g. miR-302a or miR-302b) is increased following the binding of HA to CD44v3, we first isolated small RNAs followed by an RNase protection assay using the miRNA detection kit (Ambion). Our results indicate that the level of both miR-302a and miR-302b is increased in CD44v3highALDH1high cells treated with scrambled sequence siRNA plus HA (Fig. 6, C, panels a and b, lane 2, and D, panels a and b) compared with those cells without HA treatment (Fig. 6, C, panel a and b, lane 1, and D, panel a and b). We also noted that the increase of miR-302a/miR-302b expression was specifically a result of the interaction between HA and CD44v3 because pretreatment of CD44v3highALDH1high cells with anti-CD44 antibody plus HA addition significantly reduces miR-302a/miR-302b production (Fig. 6, C, panels a and b, lane 3, and D, panels a and b). Further analysis indicated that the CD44v3highALDH1high cells treated with Oct4 siRNA (Fig. 6, C, panels a and b, lane 4, and D, panels a and b), Sox2 siRNA (Fig. 6, C, panel a and b, lane 5, and D, panels a and b), or Nanog siRNA (Fig. 6, C, panels a and b, lane 6, and D, panels a and b) have significantly less HA-induced miR-302a/miR-302b expression. These findings support the contention that all three transcription factors, Oct4, Sox2, and Nanog, are required for miR-302a/miR-302b production in HA-activated CD44v3highALDH1high cells.
Moreover, we found that the expression of miR-302a and miR-302b can be up-regulated in cells treated with a miRNA-negative control upon addition of HA (Fig. 6, C, panels a and b, lanes 8 versus 7, and D, panels a and b). In contrast, the treatment of CD44v3highALDH1high cells with anti-miR-302a (in the presence of HA) results in an inhibition of miR-302a expression but not miR-302b expression (Fig. 6, C panels a and b, lane 9, and D, panels a and b). When CD44v3highALDH1high cells were treated with anti-miR-302b (in the presence of HA), the production of miR-302b but not miR-302a expression is significantly reduced (Fig. 6C, panels a and b, lane 10). We believe that anti-miR-302a and miR-302b inhibitors are specific, and these changes in miR-302a or miR-302b expression under various treatment conditions were not due to the variations of RNA extracted from each sample because there were very similar levels of the miR-191 control in all samples (Fig. 6, C, panel c, lanes 1–10, and D, panels a and b). Together, these findings strongly suggest that HA-CD44-activated Oct4, Sox2, and Nanog signaling plays an important role in the production of miR-302a and miR-302b in CD44v3highALDH1high cells.
Using in situ hybridization of DIG-labeled miR-302a and miR-30b LNA probe and anti-DIG-alkaline phosphatase (AP), we also observed overexpression of both miR-302a and miR-302b in tumor samples from CD44v3highALDH1high cell-derived mouse tumors (Fig. 7A) and clinical human HNSCC patient specimens (Fig. 7B). The expression levels of miR-302a and miR-302b were strongly elevated in all 10 CD44v3highALDH1high cell-induced tumor tissue samples (Fig. 7A, panels b and c). No miR-302a/miR-302b staining was detected in miRNA-negative control samples (Fig. 7A, panel d) or normal tissues (Fig. 7A, panels e and f).
FIGURE 7.
Detection of miR-302a/miR-302b expression in CD44v3highALDHhigh cell-induced mouse tumors (A) and HNSCC patient samples (B) using in situ hybridization of DIG-labeled miR-302a, miR-302b and miRNA-negative control LNA probe and anti-DIG-alkaline phosphatase (AP) staining. A, H&E staining of mouse tumors (panel a) and in situ hybridization of DIG-labeled miR-302a (panel b), miR-302b (panel c), and miRNA-negative control (panel d) LNA probe and anti-DIG-alkaline phosphatase (AP) staining using CD44v3highALDHhigh cell-induced mouse tumors (panels a–d) and normal tissues (panels e and f). B, H&E staining of HNSCC samples (panel a) and in situ hybridization of DIG-labeled miR-302a (panel b), miR-302b (panel c), and miRNA-negative control (panel d) LNA probe and anti-DIG-AP staining using human HNSCC samples (panels a–d) and normal tissues (panels e and f).
Our results also reveal that a high level expression of miR-302a (41 of 66 HNSCC samples) and miR-302b (37 of 66 HNSCC samples) is detected in the primary human tumors from HNSCC patients using in situ hybridization analyses (Fig. 7B, panels b and c); and this result was statistically significant (p = 0.03 (for miR-302a detection) and p = 0.04 (for miR-302b detection), respectively) as compared with miR-302a/miR-302b detected in early (T1-T2) primary tumors (n = 44) from HNSCC patient samples (Table 2). Both miR-302a and miR-302b appear to be accumulated inside the tumor cells. No miR-302a/miR-302b staining was detected in miRNA-negative control samples (Fig. 7B, panel d) or normal tissues (Fig. 7B, panels e and f). These observations further confirm that there is a close association between miR-302a/miR-302b overexpression and human HNSCC development.
Role of miR-302a/miR-302b in the Expression of Epigenetic Regulators and Survival Proteins in Tumorigenic CD44v3highALDH1high Cells
To further investigate the role of miR-302a and miR-302b in HNSCC, we placed a special focus on the downstream effector functions of these two miRNAs. Mammalian miRNAs act predominantly by binding to the 3′-untranslated region (UTR) of cognate mRNAs. Several epigenetic regulators such as lysine-specific histone demethylases (namely AOF1 and AOF2) have been identified as potential targets for the miR-302 cluster (55). Both AOF1 and AOF2 are required for stabilizing DNA (cytosine-5)-methyltransferase 1 (DNMT1) to maintain global DNA methylation (55). Previous studies showed that activation of the miR-302 cluster suppresses AOF1 and AOF2. This leads to degradation of DNMT1 (55). The deficiency of DNMT1 causes genomes to undergo demethylation resulting in co-expression of stem cell-specific genes (55). Here, we demonstrated that the expression of AOF1, AOF2, and DNMT1 was significantly reduced in HA-treated CD44v3highALDH1high cells transfected with a negative control miRNA (Fig. 8, A, panel I, a–c, lane 2, and A, panel II, a–c). Using the methylation site-sensitive enzyme (HpaII) digestion assay, we found that HpaII cleavage induced a significant loss of global CpG methylation identified by the increased presence of smaller DNA fragments, at a genome-wide scale in these cells (Fig. 8B, lane 2). In contrast, a basal level of AOF1/AOF2 and DNMT1 expression was detected in CD44v3highALDH1high cells transfected with negative control miRNA but not treated with HA (Fig. 8, A, panel I, a–c, lane 1, and A, panel II, a–c) or pretreated with anti-CD44 antibody followed by HA addition (Fig. 8, A, panel I, a–c, lane 3, and A, panel II, a–c). Under these conditions, incomplete cutting of DNA by HpaII was observed (reduced smaller DNA fragment formation) suggesting the presence of a lower level of DNA global demethylation (Fig. 8, B, lane 1, and B, lane 3). In addition, we found that down-regulation of miR-302a or miR-302b by treating CD44v3highALDH1high cells with an anti-miR-302a or miR-302b inhibitor promotes both AOF1/AOF2 and DNMT1 up-regulation (Fig. 8A, panel I, a–c, lanes 4 and 5, and Fig. 5A, panel II, a–c), which then leads to a decrease in DNA global demethylation (as evidenced the presence of smaller DNA fragments by HpaII digestion) in CD44v3highALDH1high cells treated with HA (Fig. 8B, lanes 4 and 5). These results indicate that both miR-302a and miR-302b are functionally coupled with the inhibition of AOF1/AOF2 and DNMT1 expression and DNA global demethylation. The HA-CD44-mediated miR-302 effects on DNA global demethylation may be closely linked to cancer stem cell-specific gene regulation and function.
FIGURE 8.
Analyses of HA-CD44-mediated AOF1, AOF2, DNMT1, cIAP-1, cIAP-2, and XIAP expression (A) and global DNA demethylation assay (B) in CD44v3highALDHhigh cells. A, panel I, detection of the expression of AOF1, AOF2, and DNMT1 by anti-AOF1-mediated immunoblotting (a), anti-AOF2-mediated immunoblotting (b), or anti-DNMT1-mediated immunoblotting (c) using cell lysate isolated from CD44v3highALDHhigh cells treated with miRNA-negative control (without HA (lane 1), with HA for 24 h (lane 2), or with anti-CD44 antibody plus HA for 24 h (lane 3)) or treated with anti-miR-302a inhibitor plus HA for 24 h (lane 4) or treated with anti-miR-302b inhibitor plus HA for 24 h (lane 5). The amount of actin detected by anti-actin-mediated immunoblot (d) in each gel lane was used as a loading control. A, panel II, quantification analysis of the protein levels of AOF1 (a), AOF2 (b), and DNMT1 (c) in CD44v3highALDHhigh cells with various treatments as shown in A, panel I. The relative protein level represents the ratio of AOF1 and total actin (the loading control) (a), the ratio of AOF2 and total actin (the loading control) (b), and the ratio of DNMT1 and total actin (the loading control) (c) determined by densitometry, and these numbers were then normalized to miRNA negative control (no HA treatment) value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation less than ±5%. A, panel III, detection of the expression of cIAP-1, cIAP-2, and XIAP by anti-cIAP-1-mediated immunoblotting (a), anti-cIAP-2-mediated immunoblotting (b), and anti-XIAP-mediated immunoblotting (c) using cell lysate isolated from CD44v3highALDHhigh cells treated with miRNA-negative control (without HA (lane 1), with HA for 24 h (lane 2), or with anti-CD44 antibody plus HA for 24 h (lane 3)) or treated with anti-miR-302a inhibitor plus HA for 24 h (lane 4) or treated with anti-miR-302b inhibitor plus HA for 24 h (lane 5). The amount of actin detected by anti-actin-mediated immunoblot (d) in each gel lane was used as a loading control. A, panel IV, quantification analysis of the protein levels of cIAP1 (a), cIAP2 (b), and XIAP (c) in CD44v3highALDHhigh cells with various treatments as shown in A, panel III. The relative protein level represents the ratio of cIAP1 and total actin (the loading control) (a), the ratio of cIAP2 and total actin (the loading control) (b), and the ratio of XIAP and total actin (the loading control) (c) as determined by densitometry, and these numbers were then normalized to miRNA negative control (no HA treatment) value (bar 1, designated as 100%). The values expressed in this figure represent an average of triplicate determinations of four experiments with a standard deviation less than ±5%. B, HpaII cleavage showing global DNA demethylation (formation of small DNA fragments) in CD44v3highALDHhigh cells treated with miRNA-negative control (without HA (lane 1), with HA for 24 h (lane 2), or with anti-CD44 antibody plus HA for 24 h (lane 3)), treated with anti-miR-302a inhibitor plus HA for 24 h (lane 4), or treated with anti-miR-302b inhibitor plus HA for 24 h (lane 5) (white arrow indicates small DNA fragments).
Involvement of HA-CD44-regulated miR-302a/miR-302b in CSC-like Properties and Chemoresistance in Tumorigenic CD44v3highALDH1high Cells
Self-renewal, proliferation, and differentiation are the essential stem cell properties that allow CSCs to generate both additional cancer stem cells and phenotypically diverse cancer cells with a limited proliferative potential (10, 11). To further investigate the role of HA-CD44-activated miR-302 cluster in regulating CD44v3highALDH1high cell-mediated cancer stem cell functions, we have conducted the following experiments.
Self-renewal and Proliferation
In vitro sphere formation assays using serum-free medium containing EGF and bFGF have been routinely used to evaluate stem cell activities (e.g. proliferation and self-renewal) in putative CSCs (56–58). In this study we assessed the ability of the tumorigenic CD44v3highALDH1high cells to grow in the sphere-forming culture by incubating them in a defined medium containing serum-free spheroid medium and the two mitogens, EGF and bFGF, in the presence or absence of HA. After 14 days of incubating these cells in serum-free medium, we observed that the HA-treated CD44v3highALDH1high cells form a large number of spheres, ranging from 50 to 100 cells per spheroid (Table 3). In contrast, a much reduced number of spheres were detected in those cells treated with anti-CD44 antibody plus HA or no HA (Table 3). Therefore, sphere formation with CD44v3highALDH1high cells appears to require HA binding to CD44.
TABLE 3.
Measurement of sphere formation, self-renewal/growth, and clone formation using CD44v3highALDHhigh cells
Effects of various agents on HA-mediated sphere formation are shown.
| Treatments | First generation sphere formationa | Second generation sphere formationa | Third generation sphere formationa |
|---|---|---|---|
| % control | % control | % control | |
| No HA treatment (control) | 100 ± 7 | 100 ± 5 | 100 ± 6 |
| HA treatment | 180 ± 10 | 209 ± 5 | 220 ± 9 |
| Anti-CD44 antibody + HA treatment | 80 ± 4 | 84 ± 4 | 90 ± 7 |
| miRNA-negative control-treated cells (no HA) | 95 ± 2 | 89 ± 5 | 102 ± 6 |
| miRNA-negative control-treated cells (+ HA) | 182 ± 9 | 200 ± 5 | 213 ± 13 |
| Anti-miR-302a inhibitor-treated cells (+ HA) | 57 ± 5 | 62 ± 5 | 65 ± 5 |
| Anti-miR-302b inhibitor-treated cells (+ HA) | 55 ± 4 | 60 ± 5 | 68 ± 7 |
a Sphere formation (using CD44v3highALDH1high cells from the serial passage of first, second, and third generation of spheres) was measured by sphere formation unit as described under “Materials and Methods.” The sphere formation in cells treated with no HA (Table IIIA-control) is designated as 100%.
To demonstrate CSC growth and self-renewal, individual spheres must be able to give rise to daughter spheres that retain same sphere-forming capacity. Our data indicate that CD44v3highALDH1high cells dissociated from spheres are capable of growing and self-renewing after multiple passages from spheres (Table 3). The ability of cell growth and self-renewal appears to be greatly enhanced in CD44v3highALDH1high cells treated with HA compared with those cells treated with anti-CD44 antibody plus HA or no HA (Table 3). Our data also indicated that CD44v3highALDH1high cells were capable of growing and self-renewing after dissociating from spheres followed by incubation in serum-free medium for 21 days (Table 4). The ability to undergo cell growth and self-renewal is greatly enhanced in CD44v3highALDH1high cells treated with HA compared with those cells treated with anti-CD44 antibody plus HA or no HA (Table 4). These observations indicate that HA-CD44 interaction promotes both cell-cell adhesion properties (as evidenced by high efficiency in sphere formation) and cell growth/self-renewal (demonstrated by serial passage of sphere-forming ability and long term tumor cell growth) of CD44v3highALDH1high cells.
TABLE 4.
Effects of various agents on HA-mediated tumor cell growth
| Treatments | Tumor cell growtha (cells dissociated from first generation spheres) | Tumor cell growtha (cells dissociated from second generation spheres) | Tumor cell growtha (cells dissociated from third generation spheres) |
|---|---|---|---|
| % control | % control | % control | |
| No HA treatment (control) | 100 ± 5 | 100 ± 5 | 100 ± 5 |
| HA treatment | 192 ± 10 | 204 ± 11 | 250 ± 16 |
| Anti-CD44 antibody + HA treatment | 84 ± 5 | 90 ± 4 | 93 ± 3 |
| miRNA-negative control-treated cells (no HA) | 92 ± 4 | 98 ± 3 | 95 ± 5 |
| miRNA-negative control-treated cells (+ HA) | 202 ± 15 | 200 ± 13 | 100 ± 16 |
| Anti-miR-302a inhibitor-treated cells (+ HA) | 52 ± 3 | 55 ± 5 | 53 ± 4 |
| Anti-miR-302b inhibitor-treated cells (+ HA) | 55 ± 2 | 66 ± 4 | 58 ± 5 |
a Measurement of growth for CD44v3highALDH1high cells (dissociated from spheres after a serial passage of first, second, and third generation of sphere formation) was performed by incubating these cells in serum-free RPMI 1640 medium for 3 weeks (21 days) using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based growth assay as described under “Materials and Methods.” Tumor cell growth in cells treated with no HA (Table IIIB-control) is designated as 100%.
Clone Formation and Differentiation
To further test the ability of these CD44v3highALDH1high cells (untreated or pretreated with miRNA-negative control followed by dissociation from spheres after a serial passage of 1st, 2nd, and 3rd generation of sphere formation) to undergo clone formation (Table 5) and differentiation, we conducted clone formation assays and analyzed differentiation marker expression (Fig. 9). Our data show that the level of clone formation efficiency and differentiation marker (cytokeratin 17 and cytokeratin 19) expression of HA-treated CD44v3highALDH1high cells dissociated from spheres appear to be significantly higher than those detected in CD44v3highALDH1high cells (pretreated with anti-CD44 antibody followed by HA addition or no HA) dissociated from spheres (Fig. 9 and Table 5). These observations suggest that CD44v3highALDH1high cells are capable of displaying cancer stem cell-like properties (e.g. sphere formation, cell growth/self-renewal, and clone formation/differentiation) in an HA-specific and CD44-dependent manner.
TABLE 5.
Effects of various agents on HA-mediated clone formation
| Treatments | Clone formationa (cells dissociated from first generation spheres) | Clone formationa (cells dissociated from second generation spheres | Clone formationa (cells dissociated from third generation spheres) |
|---|---|---|---|
| % control | % control | % control | |
| No HA treatment (control) | 100 ± 5 | 100 ± 4 | 100 ± 3 |
| HA treatment | 209 ± 12 | 250 ± 14 | 262 ± 15 |
| Anti-CD44 antibody + HA treatment | 72 ± 4 | 85 ± 3 | 93 ± 5 |
| miRNA-negative control-treated cells (no HA) | 98 ± 5 | 95 ± 5 | 102 ± 6 |
| miRNA-negative control-treated cells (+ HA) | 202 ± 10 | 218 ± 13 | 230 ± 16 |
| Anti-miR-302a inhibitor-treated cells (+ HA) | 62 ± 3 | 58 ± 5 | 63 ± 4 |
| Anti-miR-302b inhibitor-treated cells (+ HA) | 65 ± 2 | 60 ± 4 | 55 ± 5 |
a Clone formation was performed by incubating CD44v3highALDH1high cells (dissociated from spheres after a serial passage of first, second, and third generation of sphere formation) in serum-containing RPMI 1640 medium for 7–10 days and measured by the clone formation efficiency (the ratio of the clone number to the input CD44v3highALDHhigh cells (dissociated from spheres)) as described under “Materials and Methods.” Clone formation in cells treated with no HA is designated as 100%. The values expressed in this table represent an average of triplicate determinations of 4–5 experiments. All data represent mean ± S.E. (with n = 4) of the clone formation detected in each sample.
FIGURE 9.
Analyses of sphere formation (A) and clone formation and differentiation (B). A, sphere formation was induced by incubating CD44v3highALDHhigh cells treated with miRNA-negative control (without HA (panel a) or with HA for 10 days (panel b) or with anti-CD44 antibody plus HA for 10 days (panel c)), treated with anti-miR-302a inhibitor plus HA for 10 days (panel d), or treated with anti-miR-302b inhibitor plus HA for 10 days (panel e) in Matrigel/basal medium containing EGF, bFGF, and insulin. Ten days after cell plating, spheres with a diameter over 40 μm were photographed and counted as described under “Materials and Methods.” B–D, clone formation (B) and differentiation (C and D) were induced by incubating CD44v3highALDHhigh cells (dissociated from spheres treated with miRNA-negative control without HA (panel a), with HA for 10 days (panel b), with anti-CD44 antibody plus HA for 10 days (panel c), treated with anti-miR-302a inhibitor plus HA for 10 days (panel d), or treated with anti-miR-302b inhibitor plus HA for 10 days (panel e)) in RPMI 1640 complete culture containing 10% fetal bovine serum for ∼7–10 days. After most cell clones expand to >50–100 cells, they were fixed with methanol followed by staining with crystal violet to visualize clone formation (B) or processed for immunoperoxidase staining for differentiation markers (cytokeratin 17/CK17 (C) and cytokeratin 19/CK19 (D)), respectively, as described under “Materials and Methods.”
Moreover, our data indicate that down-regulation of miR-302a or miR-302b by transfecting CD44v3highALDH1high cells with either anti-miR-302a or miR-302b inhibitor (but not miRNA-negative control) effectively decreases HA-mediated sphere formation, self-renewal, and tumor cell growth (Fig. 9A and Tables 3 and 4) as well as clonal formation (Fig. 9B and Table 5) and differentiation marker expression (Fig. 9, C and D) in these cells. These observations strongly suggest both miR-302a and miR-302b are closely involved in HA-mediated CSC-like properties and functions.
Survival Protein Expression and Chemotherapy Resistance
Inhibitors of the apoptosis family of proteins (IAPs, e.g. cIAP-1, cIAP-2, and XIAP) are also frequently overexpressed by cancer stem cells. Importantly, up-regulation of IAPs increases cell survival by the binding of IAPs to caspases and suppressing apoptosis (7). In this study, we observed that IAP proteins (e.g. cIAP-1, cIAP-2, and XIAP) are up-regulated in CD44v3highALDH1high cells transfected with miRNA-negative control following HA treatment (Fig. 8, A, panel III, a–c, lane 2, and A, panel IV, a–c). In contrast, a reduced level of IAP protein (e.g. cIAP-1, cIAP-2, and XIAP) expression (Fig. 8, A, panel III, a–c, lanes 3 and 1, and A, panel IV, a–c) was detected in CD44v3highALDH1high cells (pretreated with anti-CD44 antibody followed by HA addition or no HA). Further analyses indicate that HA also enhances anti-apoptotic effects and decreases the ability of cisplatin to induce apoptosis and cell death of these CD44v3highALDH1high cells (Table 6) leading to the enhancement of chemoresistance (Table 6). In contrast, when CD44v3highALDH1high cells were treated with no HA or with anti-CD44 antibody, they appear to be more sensitive to cisplatin-induced apoptosis and cell death (Table 6). These findings suggest that HA-mediated cell survival contributes to both a decrease in apoptosis/cell death and an increase in cisplatin resistance in these CSC-like CD44v3highALDH1high cells.
TABLE 6.
Measurement of cisplatin-induced CD44v3highALDH1high cell apoptosis and growth inhibition
Effects of various signaling perturbation agents on cisplatin-induced apoptosis and cell growth inhibition in CD44v3highALDH1high cells.
| Treatments | Apoptotic cells (annexin V-positive cell/total cells × 100%)a |
Cisplatin-induced tumor cell growth inhibition IC50b | |
|---|---|---|---|
| No cisplatin | + Cisplatin | ||
| μm | |||
| Untreated cells (no HA) | 5 ± 0.7 | 36 ± 6.0 | 127.50 ± 6.4 |
| Untreated cells (+ HA) | 2 ± 0.5 | 12 ± 0.7 | 250.00 ± 18.0 |
| Anti-CD44 antibody-treated cells (+HA) | 7 ± 0.4 | 55 ± 1.5 | 93.75 ± 4.7 |
| miRNA-negative control-treated cells (no HA) | 4 ± 0.5 | 35 ± 1.5 | 125.00 ± 6.3 |
| miRNA negative control-treated cells (+ HA) | 2 ± 0.5 | 14 ± 0.9 | 250.00 ± 16.0 |
| Anti-miR-302a inhibitor-treated cells (+ HA) | 12 ± 0.6 | 58 ± 2.5 | 60.00 ± 3.0 |
| Anti-miR-302b inhibitor-treated cells (+ HA) | 17 ± 0.8 | 60 ± 6.5 | 62.50 ± 4.0 |
a Cells were designated apoptotic when displaying annexin V-positive staining. In each sample, at least 500 cells from five different fields were counted, with the percentage of cisplatin or SM-164 (2 h of treatment)-induced apoptotic cells calculated as annexin V-positive cells/total number of cells. The values are presented as the means ±S.D. All assays consisted of at least six replicates and were performed on at least 3–5 different experiments.
b Tumor cell growth inhibition (IC50) is designated as “the micromolar concentration of chemotherapeutic drug (e.g. cisplatin 24 h of treatment) that causes 50% inhibition of tumor cell growth” using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based growth assay as described under “Materials and Methods.” IC50 values are presented as the means ±S.D. All assays consisted of at least six replicates and were performed on at least 3–5 different experiments.
Furthermore, our data indicate that down-regulation of miR-302a or miR-302b by transfecting CD44v3highALDH1high cells with either anti-miR-302a or miR-302b inhibitor effectively reduces HA-mediated IAP protein (e.g. cIAP-1, cIAP-2, and XIAP) expression in CD44v3highALDH1high cells (Fig. 8, A, panel III, a–c, lanes 4 and 5, and A, panel IV, a–c). Consequently, the level of cisplatin-induced apoptosis and cell death of these cells is also significantly elevated. Taken together, we believe that the miR-302 cluster (e.g. miR-302a and miR-302b) plays a critical role in regulating HA-CD44-mediated chemotherapy resistance in these tumorigenic and CSC-like CD44v3highALDH1high cell populations from head and neck cancer.
Recently, a small molecule IAP protein inhibitor (bivalent SM-164) has been shown to concurrently target cIAP-1/cIAP-2 for degradation and antagonize XIAP leading to an enhancement of tumor cell apoptosis and tumor regression (59). In this study, we also found that chemosensitivity in CSC-like CD44v3highALDH1high cells increases with the newly developed IAP protein inhibitor SM164 (Table 7). These findings suggest that the use of a combination of IAP protein inhibitor (SM164) and cisplatin may significantly improve the efficacy of chemotherapeutic drug treatment by targeting CSCs in head and neck cancer.
TABLE 7.
Measurement of cisplatin-induced CD44v3highALDH1high cell apoptosis and growth inhibition
Effects of an IAP inhibitor (SM-164) on cisplatin-induced apoptosis and cell growth inhibition in CD44v3highALDH1high-expressing cells.
| Treatments | Apoptotic cells (annexin V-positive cell/total cells × 100%)a |
Cisplatin-induced tumor cell growth inhibition IC50b | |
|---|---|---|---|
| No cisplatin | + Cisplatin | ||
| μm | |||
| Vehicle control-treated cells (no HA) | 13 ± 0.2 | 42 ± 2.4 | 170.00 ± 8.2 |
| Vehicle control-treated cells (+ HA) | 6 ± 0.3 | 14 ± 0.5 | 235.00 ± 10.0 |
| SM-164-treated cells (no HA) | 12 ± 0.5 | 87 ± 2.6 | 70.50 ± 3.5 |
| SM-164-treated cells (+ HA) | 14 ± 0.4 | 68 ± 1.9 | 65.00 ± 3.0 |
DISCUSSION
HNSCC is a solid tumor malignancy that may involve the oral cavity, pharynx, larynx, and paranasal sinuses (1). Advanced HNSCC is an aggressive disease associated with major morbidity and mortality (1, 2). Therefore, there is currently a great need to clarify the mechanisms of tumor progression underlying the clinical behavior of HNSCC. In recent years, evidence has been accumulating to support the hypothesis that HNSCC tumors contain a small population of CSCs, which exhibit self-renewing capacities, chemoresistant properties, and the ability to differentiate into a heterogeneous tumor population leading to HNSCC progression (12).
A number of studies now indicate that tumor cells with high levels of CD44 expression appear to exhibit CSC properties in many cancers, including HNSCC (13). CD44 is a family of transmembrane receptors found on a number of different benign and malignant cells. The human CD44 gene contains 19 exons with up to 10 exons (primarily exons 6–14) being alternatively spliced to give rise to multiple variant CD44 (e.g. CD44v3, and CD44v6, etc.) isoforms, along with the standard CD44 (CD44s-no variable exons). These variants make up a class of CD44 receptors. It is now known that neither CD44s nor CD44v6 distinguishes normal from benign or malignant epithelia of head and neck (15). Consequently, the identification of other specific CD44 isoform-related CSC markers is needed.
In this regard, the role of CD44v3 isoform in HNSCC progression has been highlighted in studies that have identified an association of the v3-containing isoform with HNSCC growth, migration, and matrix metalloproteinase expression (16–18). We have demonstrated that transfection of the CD44v3 isoform into a nonexpressing HNSCC cell line results in significantly increased tumor cell migration (18). In addition, treatment of a CD44v3 isoform-expressing HNSCC cell line with anti-CD44v3 isoform antibody decreased in vitro proliferation and increased cisplatin sensitivity (17, 18). Using the same anti-CD44v3 isoform antibody, immunohistochemical tissue analysis revealed that the CD44v3 isoform is preferentially expressed in metastatic lymph nodes. Furthermore, CD44v3 isoform expression in primary tumors is significantly associated with advanced T status and positive lymph nodes (16). These findings suggest that the CD44v3 isoform is closely associated with HNSCC development and progression and may serve as a new CSC marker. Recently, an elevated level of aldehyde dehydrogenenase 1 (ALDH1) activity was also shown to identify CSC in HNSCC (53, 54).
In this study, we identified an HNSCC tumor cell population containing high levels of both CD44v3 and ALDH1 expression (CD44v3highALDH1high) (Fig. 1). Importantly, these CD44v3highALDH1high cells appear to exhibit CSC-like phenotypes and are highly tumorigenic for generating tumors starting with as few as 50 cells (Table 1). In breast cancer, a CD44highCD24low tumor population has also been shown to be highly tumorigenic (60). However, our preliminary data indicate that only a small portion (1.5–2.0%) of CD44v3highALDH1high cells also express CD24low. The question of whether the CD44v3highALDH1highCD24low cells also display high tumorigenic potential is currently under investigation in our laboratory. Although an enhanced level of ALDH1 activity is closely associated with CSC-like HNSCC cells, the role of ALDH1 in head and neck cancer has not been well established. In particular, HA effect on regulating ALDH1 activities in CD44v3highALDH1high CSC-like functions is not known. These questions will be addressed in our future studies. Our preliminary data also indicate that CD44v3lowALDH1low cells fail to interact with HA and display poor growth/survival properties. Therefore, comparison of HA effects on CD44v3lowALDH1low cell population versus CD44v3highALDH1high cell population cannot be performed at this time.
All stem cells are thought to exist in specialized microenvironments known as niches (25, 26). Components present in the niches can regulate stem cell behavior through direct binding to stem cell surface receptors or via indirect activation of paracrine signaling (25, 26). Extracellular matrix components, including HA, are known to be present in one of the stem cell niches (25, 26). Because CD44 is an HA receptor, it provides a physical linkage between matrix HA and various transcription factors that regulate tumor cell functions through distinct signaling pathways (5, 28, 29). In fact, both HA and CD44 have been shown to be involved in self-renewal and differentiation of human ESC (64, 65). In this study, we observed that the HA-CD44 interaction promotes sphere formation (cell-cell adhesion), self-renewal/growth, and clonal formation in a highly tumorigenic CD44v3highALDH1high tumor cell population (Table 2). These observations support the contention that HA signaling is directly involved in the regulation of CSC-like properties in CD44v3highALDH1high cells. The question of how these stemness properties are established in these tumor cell populations is addressed in this investigation.
One of the characteristics of CSCs is overexpression of stem cell-specific transcription factors such as Oct4, Sox2, and Nanog (61). Previous studies indicate that Oct4, Sox2, and Nanog often function in combinatorial complexes to regulate the expression of gene loci that are involved in self-renewal, proliferation, and differentiation (45, 61). These three molecules are also closely associated with higher histological grades and poorer clinical survival in head and neck cancer (6, 41, 63). In this study, we found that all three transcription factors (e.g. Oct4, Sox2, and Nanog) are overexpressed in CD44v3highALDH1high cells as compared with unsorted cells (Fig. 2). Moreover, a high level of Oct4, Sox2, and Nanog expression is detected in CD44v3highALDH1high cell-induced mouse tumors and in human HNSCC patient specimens (Fig. 3). These findings indicate that CD44v3highALDH1high cells are likely to be generating tumors through the stem cell processes of self-renewal, growth, and differentiation. These cells can persist in tumors as a distinct population, possibly causing chemotherapy resistance and tumor recurrence at a later time. The mechanism by which the regulatory network consisting of Oct4, Sox2, and Nanog contributes to the establishment of CSC-specific phenotypes in HNSCC is poorly understood at this time.
miRNAs are evolutionarily conserved and function as negative regulators of gene expression by inhibiting the translation of mRNAs that contain complementary target sites, referred to as the “seed region” (66). Studies have shown that miRNAs participate in a wide range of biological functions such as cellular proliferation, self-renewal, and differentiation (67). Computational predictions of miRNA targets suggest that up to 30% of human protein-coding genes may be regulated by miRNAs (68). This makes miRNAs one of the most abundant classes of post-transcriptional regulators of gene expression.
A number of studies indicate that more than 50% of microRNAs (small RNA molecules with ∼20–25 nucleotides) are located in cancer-associated genomic regions or fragile sites, suggesting that miRNAs may be closely associated with the pathogenesis of a variety of cancers, including HNSCC (46, 47). Our previous work has indicated that HA-CD44 promotes Nanog interaction with p68 and DROSHA leading to biosynthetic processing and production of microRNA-21 (miR-21) in breast tumor cells (36). Most recently, we have reported that Nanog binds to Stat-3 resulting in miR-21 up-regulation in HNSCC cells (6). These findings indicate that HA-CD44-mediated Nanog signaling is closely linked to the production of miRNAs during oncogenesis.
Previous genetic studies using mouse models revealed that transcription factors such as Oct4, Sox2, and Nanog have distinct roles but could use similar signaling pathways to maintain stemness functions during development (45, 61). Some studies have also indicated that Oct4, Sox2, and Nanog co-occupy the promoter of miR-302, which has been shown to target genes required for development and oncogenesis (50, 51). At the transcriptional level, Oct4, Sox2, and Nanog form a positive autoregulatory loop that is important for the maintenance of the undifferentiated state. At the post-translational level, miR-302 family is emerging as a key player in the control of cell proliferation and cell fate determination during differentiation (50, 51). Here, we provided new evidence that the HA-CD44v3 interaction activates Oct4, Sox2, and Nanog complex formation in CD44v3highALDH1high cells (Figs. 4 and 5). Furthermore, our results indicate that the miR-302 cluster is controlled by a promoter containing Oct4-Sox2-Nanog-binding sites in these cells, whereas ChIP assays demonstrate that stimulation of miR-302a and miR-302b production by HA is Oct4/Sox2 and Nanog complex-dependent in CD44v3highALDH1high cells (Fig. 6). Importantly, we have also detected overexpression of miR-302a and miR-302b in both mouse tumors induced by CD44v3highALDH1high cells and human HNSCC patient samples (Fig. 7). These findings clearly established the existence of a close association between miR-302 clusters (e.g. miR-302a and miR-302b) and HNSCC development.
Lysine-specific histone demethylases (namely AOF1 and AOF2) are epigenetic regulators known to be targeted by miR-302 (55). Both AOF1 and AOF2 repress gene transcription by demethylating histone 3 on lysine 4 (H3K4) (69, 70). AOF1-deficient mice fail to perform de novo DNA methylation imprint during oogenesis (69). AOF2 knock-out mice is embryonically lethal due to the loss of global DNA demethylation (70). DNA (cytosine-5)-methyltransferase 1 (e.g. DNMT1) plays an important role in the establishment and regulation of tissue-specific patterns of methylated cytosine residues and is involved in global DNA demethylation (70). A previous study indicated that suppression of AOF1 and AOF2 causes DNMT1 degradation leading to global demethylation during somatic cell reprogramming of human hair follicles cells into induced pluripotent stem cells (55). Thus, it is likely that inhibition of AOF1/AOF2 by HA-CD44-activated miR-302a or miR-302b (Fig. 8A) is also responsible for DNMT1 reduction, resulting in DNA demethylation as shown in CD44v3highALDH1high CSCs (Fig. 8B). This DNA demethylation process may subsequently activate the expression of self-renewal/survival-related proteins (e.g. c-IAP1, c-IAP2, and XIAP) (Fig. 8A) required for cancer stem cell-specific functions (e.g. self-renewal/growth, clonal formation, and survival/chemotherapy resistance (Tables 2 and 3–5)). Most importantly, an anti-miR-302a/miR-302b inhibitor is able to enhance the expression of AOF1/AOF2 and DNMT1 (Fig. 8A) and block HA-mediated CD44v3highALDH1high CSC-like functions (Table 2). Thus, we believe that this newly discovered HA-CD44-Oct4-Sox2-Nanog signaling pathway leading to miR-302a/miR-302b production/function is highly innovative and should provide important new drug targets to cause apoptosis in CSC-like CD44v3highALDH1high cells and to overcome chemotherapy resistance in head and neck cancer.
As outlined in Fig. 10, we propose that HA binding (step 1) promotes the formation of an Oct4, Sox2, and Nanog complex with CD44v3 (step 2). This Oct4-Sox2-Nanog signaling complex then translocates from the cytosol to the nucleus (Fig. 10, step 3) and interacts with the promoter region (containing Oct4-, Sox2-, and Nanog-binding sites) of the miR-302 cluster (Fig. 10, step 4) resulting in miR-302 cluster gene expression (step 5) and mature miR-302a and miR-302b production (step 6). The resultant miR-302a/miR-302b then functions to down-regulate the lysine-specific histone demethylases (namely AOF1 and AOF2) and DNA (cytosine-5)-methyltransferase 1 (DNMT1) (Fig. 10, step 7a) and induce global DNA demethylation (step 8a) leading to IAP (cIAP-1, cIAP-2, and XIAP) expression (step 8a), self-renewal, clonal formation, anti-apoptosis/survival, and chemoresistance in CSC-like CD44v3highALDH1high cells. In direct contrast, treatment of CD44v3highALDH1high cells with an anti-miR-302a/miR-302b inhibitor induces AOF1/AOF2 and DNMT1 up-regulation (Fig. 10, step 7b) resulting in a reduction of global DNA demethylation (step 8b). Subsequently, these changes result in the inhibition of both IAP (cIAP-1, cIAP-2, and XIAP) expression (Fig. 10, step 8b) and self-renewal/clonal formation, as well as stimulation of apoptosis and enhancement of chemosensitivity in CSC-like CD44v3highALDH1high cells. Taken together, these findings strongly suggest that targeting HA-CD44-mediated Oct4-Sox2-Nanog signaling pathways and miR-302 cluster function may provide important new drug targets to induce CSC apoptosis/death and overcome chemotherapy resistance in head and neck cancer.
FIGURE 10.
A proposed model for HA-CD44-mediated Oct4-Sox2-Nanog signaling in the regulation of miRNA-302 production and CSC functions in CD44v3highALDHhigh cells. HA binding (step 1) promotes CD44v3 association with Oct4, Sox2, and Nanog as a complex (step 2). This Oct4-Sox2-Nanog signaling complex then translocates from the cytosol to the nucleus (step 3) and interacts with the promoter region (containing Oct4-, Sox2-, and Nanog-binding sites) of the miR-302 cluster (step 4), resulting in miR-302 cluster gene expression (step 5) and mature miR-302a and miR-302b production (step 6). The resultant miR-302a/miR-302b then functions to down-regulate the lysine-specific histone demethylases (namely AOF1 and AOF2) and DNA (cytosine-5)-methyltransferase 1 (DNMT1) (step 7a) and induces global DNA demethylation (step 8a) leading to IAP (cIAP-1, cIAP-2, and XIAP) expression (step 8a), self-renewal, clonal formation, anti-apoptosis/survival, and chemoresistance in CSC-like CD44v3highALDH1high cells. In direct contrast, treatment of CD44v3highALDH1high cells with an anti-miR-302a/miR-302b inhibitor induces AOF1/AOF2 and DNMT1 up-regulation (step 7b) resulting in a reduction of global DNA demethylation (step 8b). Subsequently, these changes result in the inhibition of IAP (cIAP-1, cIAP-2, and XIAP) expression (step 8b) and self-renewal/clonal formation as well as stimulation of apoptosis and enhancement of chemosensitivity in CSC-like CD44v3highALDH1high cells.
Acknowledgments
We gratefully acknowledge the assistance of Drs. Gerard J. Bourguignon and Walter M. Holleran in the preparation and review of this manuscript. We also thank for Dr. Shaomeng Wang from the University of Michigan for providing us the IAP inhibitor SM164.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA66163 from USPHS. This work was also supported by Veterans Affairs Merit Review grants and a Department of Defense grant.
- HNSCC
- head and neck squamous cell carcinoma
- HA
- hyaluronan
- CSC
- cancer stem cell
- IAP
- inhibitor of apoptosis
- bFGF
- basic FGR
- Q-PCR
- quantitative PCR
- miRNA
- microRNA
- ESC
- embryonic stem cell
- DEAB
- diethylaminobenzaldehyde
- DIG
- digoxigenin
- APC
- allophycocyanin
- AP
- alkaline phosphatase.
REFERENCES
- 1. Parkin D. M., Bray F., Ferlay J., Pisani P. (2005) Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) Cancer statistics, 2008. CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 3. Nakamura M., Nakatani K., Uzawa K., Ono K., Uesugi H., Ogawara K. (2005) Establishment and characterization of a cisplatin-resistant oral squamous cell carcinoma cell line, H-1R. Oncol. Rep. 14, 1281–1286 [PubMed] [Google Scholar]
- 4. Torre C., Wang S. J., Xia W., Bourguignon L. Y. (2010) Reduction of hyaluronan-CD44-mediated growth, migration, and cisplatin resistance in head and neck cancer due to inhibition of Rho kinase and PI 3-kinase signaling. Arch. Otolaryngol. Head Neck Surg. 136, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S. J., Bourguignon L. Y. (2011) Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am. J. Pathol. 178, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourguignon L. Y., Earle C., Wong G., Spevak C. C., Krueger K. (2012) Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene 31, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunter A. M. (2007) The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis 12, 1543–1568 [DOI] [PubMed] [Google Scholar]
- 8. Gyrd-Hansen M., Meier P. (2010) IAPs. From caspase inhibitors to modulators of NF-κB, inflammation, and cancer. Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 9. Bourguignon L. Y., Xia W., Wong G. (2009) Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates β-catenin signaling and NFκB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J. Biol. Chem. 284, 2657–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dalerba P., Cho R. W., Clarke M. F. (2007) Cancer stem cells. Models and concepts. Annu. Rev. Med. 58, 267–284 [DOI] [PubMed] [Google Scholar]
- 11. Schulenburg A., Ulrich-Pur H., Thurnher D., Erovic B., Florian S., Sperr W. R., Kalhs P., Marian B., Wrba F., Zielinski C. C., Valent P. (2006) Neoplastic stem cells. A novel therapeutic target in clinical oncology. Cancer 107, 2512–2520 [DOI] [PubMed] [Google Scholar]
- 12. Al-Hajj M. (2007) Cancer stem cells and oncology therapeutics. Curr. Opin. Oncol. 19, 61–64 [DOI] [PubMed] [Google Scholar]
- 13. Prince M. E., Sivanandan R., Kaczorowski A., Wolf G. T., Kaplan M. J., Dalerba P., Weissman I. L., Clarke M. F., Ailles L. E. (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 104, 973–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Screaton G. R., Bell M. V., Jackson D. G., Cornelis F. B., Gerth U., Bell J. I. (1992) Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. U.S.A. 89, 12160–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mack B., Gires O. (2008) CD44s and CD44v6 expression in head and neck epithelia. PLoS One. 3, e3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S. J., Wong G., de Heer A. M., Xia W., Bourguignon L. Y. (2009) CD44 variant isoforms in head and neck squamous cell carcinoma progression. Laryngoscope 119, 1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franzmann E. J., Weed D. T., Civantos F. J., Goodwin W. J., Bourguignon L. Y. (2001) A novel CD44 v3 isoform is involved in head and neck squamous cell carcinoma progression. Otolaryngol. Head Neck Surg. 124, 426–432 [DOI] [PubMed] [Google Scholar]
- 18. Wang S. J., Wreesmann V. B., Bourguignon L. Y. (2007) Association of CD44 V3-containing isoforms with tumor cell growth, migration, matrix metalloproteinase expression, and lymph node metastasis in head and neck cancer. Head Neck 29, 550–558 [DOI] [PubMed] [Google Scholar]
- 19. Laurent T. C., Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 [PubMed] [Google Scholar]
- 20. Lee J. Y., Spicer A. P. (2000) Hyaluronan. A multifunctional, megadalton, stealth molecule. Curr. Opin. Cell Biol. 12, 581–586 [DOI] [PubMed] [Google Scholar]
- 21. Toole B. P. (2001) Hyaluronan in morphogenesis. Semin. Cell Dev. Biol. 12, 79–87 [DOI] [PubMed] [Google Scholar]
- 22. Stern R., Jedrzejas M. J. (2006) Hyaluronidases. Their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knudson W., Biswas C., Li X. Q., Nemec R. E., Toole B. P. (1989) The role and regulation of tumor-associated hyaluronan. CIBA Found Symp. 143, 150–159 [DOI] [PubMed] [Google Scholar]
- 24. Toole B. P., Wight T. N., Tammi M. I. (2002) Hyaluronan-cell interactions in cancer and vascular disease. J. Biol. Chem. 277, 4593–4596 [DOI] [PubMed] [Google Scholar]
- 25. Haylock D. N., Nilsson S. K. (2006) The role of hyaluronic acid in hemopoietic stem cell biology. Regen. Med. 1, 437–445 [DOI] [PubMed] [Google Scholar]
- 26. Astachov L., Vago R., Aviv M., Nevo Z. (2011) Hyaluronan and mesenchymal stem cells. From germ layer to cartilage and bone. Front. Biosci. 16, 261–276 [DOI] [PubMed] [Google Scholar]
- 27. Underhill C. (1992) CD44. The hyaluronan receptor. J. Cell Sci. 103, 293–298 [DOI] [PubMed] [Google Scholar]
- 28. Bourguignon L. Y. (2008) Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 18, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turley E. A., Noble P. W., Bourguignon L. Y. (2002) Signaling properties of hyaluronan receptors. J. Biol. Chem. 277, 4589–4592 [DOI] [PubMed] [Google Scholar]
- 30. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 31. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 32. Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- 34. Do J. T., Schöler H. R. (2006) Cell-cell fusion as a means to establish pluripotency. Ernst Schering Res. Found. Workshop 60, 35–45 [DOI] [PubMed] [Google Scholar]
- 35. Bourguignon L. Y., Peyrollier K., Xia W., Gilad E. (2008) Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J. Biol. Chem. 283, 17635–17651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bourguignon L. Y., Spevak C. C., Wong G., Xia W., Gilad E. (2009) Hyaluronan-CD44 interaction with protein kinase C(ϵ) promotes oncogenic signaling by the stem cell marker Nanog and the production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J. Biol. Chem. 284, 26533–26546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J., Wang X., Li M., Han J., Chen B., Wang B., Dai J. (2006) NANOGP8 is a retrogene expressed in cancers. FEBS J. 273, 1723–1730 [DOI] [PubMed] [Google Scholar]
- 38. Kamachi Y., Uchikawa M., Kondoh H. (2000) Pairing SOX off. With partners in the regulation of embryonic development. Trends Genet. 16, 182–187 [DOI] [PubMed] [Google Scholar]
- 39. Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gontan C., de Munck A., Vermeij M., Grosveld F., Tibboel D., Rottier R. (2008) Sox2 is important for two crucial processes in lung development. Branching morphogenesis and epithelial cell differentiation. Dev. Biol. 317, 296–309 [DOI] [PubMed] [Google Scholar]
- 41. Dong C., Wilhelm D., Koopman P. (2004) Sox genes and cancer. Genome Res. 105, 442–447 [DOI] [PubMed] [Google Scholar]
- 42. Pesce M., Schöler H. R. (2001) Oct-4. Gatekeeper in the beginnings of mammalian development. Stem Cells 19, 271–278 [DOI] [PubMed] [Google Scholar]
- 43. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 44. Wang X., Dai J. (2010) Concise review. Isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells 28, 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kashyap V., Rezende N. C., Scotland K. B., Shaffer S. M., Persson J. L., Gudas L. J., Mongan N. P. (2009) Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 18, 1093–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang S. S., Jiang W. W., Smith I., Poeta L. M., Begum S., Glazer C., Shan S., Westra W., Sidransky D., Califano J. A. (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer. 123, 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Houbaviy H. B., Murray M. F., Sharp P. A. (2003) Embryonic stem cell-specific microRNAs. Dev. Cell 5, 351–358 [DOI] [PubMed] [Google Scholar]
- 49. Suh M. R., Lee Y., Kim J. Y., Kim S. K., Moon S. H., Lee J. Y., Cha K. Y., Chung H. M., Yoon H. S., Moon S. Y., Kim V. N., Kim K. S. (2004) Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498 [DOI] [PubMed] [Google Scholar]
- 50. Card D. A., Hebbar P. B., Li L., Trotter K. W., Komatsu Y., Mishina Y., Archer T. K. (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 28, 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu H., Deng S., Zhao Z., Zhang H., Xiao J., Song W., Gao F., Guan Y. (2011) Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol. Biol. Rep. 38, 2155–2160 [DOI] [PubMed] [Google Scholar]
- 52. Lin S. L., Chang D. C., Chang-Lin S., Lin C. H., Wu D. T., Chen D. T., Ying S. Y. (2008) Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Y. C., Chen Y. W., Hsu H. S., Tseng L. M., Huang P. I., Lu K. H., Chen D. T., Tai L. K., Yung M. C., Chang S. C., Ku H. H., Chiou S. H., Lo W. L. (2009) Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem. Biophys. Res. Commun. 385, 307–313 [DOI] [PubMed] [Google Scholar]
- 54. Clay M. R., Tabor M., Owen J. H., Carey T. E., Bradford C. R., Wolf G. T., Wicha M. S., Prince M. E. (2010) Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck 32, 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin S. L., Chang D. C., Lin C. H., Ying S. Y., Leu D., Wu D. T. (2011) Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 39, 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reynolds B. A., Weiss S. (1996) Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 175, 1–13 [DOI] [PubMed] [Google Scholar]
- 57. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xin L., Lukacs R. U., Lawson D. A., Cheng D., Witte O. N. (2007) Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells 25, 2760–2769 [DOI] [PubMed] [Google Scholar]
- 59. Lu J., Bai L., Sun H., Nikolovska-Coleska Z., McEachern D., Qiu S., Miller R. S., Yi H., Shangary S., Sun Y., Meagher J. L., Stuckey J. A., Wang S. (2008) SM-164. A novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res. 68, 9384–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. (2007) let-7 regulates self- renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 61. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Herr W., Cleary M. A. (1995) The POU domain. Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Gene Dev. 9, 1679–1693 [DOI] [PubMed] [Google Scholar]
- 63. Wang Q., He W., Lu C., Wang Z., Wang J., Giercksky K. E., Nesland J. M., Suo Z. (2009) Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res. 29, 1233–1241 [PubMed] [Google Scholar]
- 64. Choudhary M., Zhang X., Stojkovic P., Hyslop L., Anyfantis G., Herbert M., Murdoch A. P., Stojkovic M., Lako M. (2007) Putative role of hyaluronan and its related genes, HAS2 and RHAMM, in human early preimplantation embryogenesis and embryonic stem cell characterization. Stem Cells 25, 3045–3057 [DOI] [PubMed] [Google Scholar]
- 65. Wheatley S. C., Isacke C. M. (1995) Induction of a hyaluronan receptor, CD44, during embryonal carcinoma and embryonic stem cell differentiation. Cell Adhes. Commun. 3, 217–230 [DOI] [PubMed] [Google Scholar]
- 66. Cowland J. B., Hother C., Grønbaek K. (2007) MicroRNAs and cancer. APMIS 115, 1090–1106 [DOI] [PubMed] [Google Scholar]
- 67. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs. MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 68. Sassen S., Miska E. A., Caldas C. (2008) MicroRNA. Implications for cancer. Virchows Arch. 452, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ciccone D. N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418 [DOI] [PubMed] [Google Scholar]
- 70. Wang J., Hevi S., Kurash J. K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 [DOI] [PubMed] [Google Scholar]
- 71. Bourguignon L. Y., Wong G., Earle C. A., Xia W. (2011) Interaction of low molecular weight hyaluronan with CD44 and toll-like receptors promotes the actin filament-associated protein 110-actin binding and MyD88-NFκB signaling leading to proinflammatory cytokine/chemokine production and breast tumor invasion. Cytoskeleton 68, 671–693 [DOI] [PMC free article] [PubMed] [Google Scholar]