Background: Porphyromonas gingivalis has low immunogenicity and synergizes with other periodontal pathogens, including Fusobacterium nucleatum.
Results: Porphyromonas gingivalis selectively represses the activation of the IL-1β-processing inflammasome by Fusobacterium nucleatum and inducers that are endocytosed.
Conclusion: Porphyromonas gingivalis suppresses inflammasome activity through a novel mechanism involving modulation of endocytosis.
Significance: Inflammasome suppression may contribute to periodontitis and other chronic diseases.
Keywords: Bacterial Pathogenesis, Bacterial Signal Transduction, Caspase, Endocytosis, Inflammation, Innate Immunity, Fusobacterium nucleatum, Inflammasome, Porphyromonas gingivalis
Abstract
The interleukin (IL)-1β-processing inflammasome has recently been identified as a target for pathogenic evasion of the inflammatory response by a number of bacteria and viruses. We postulated that the periodontal pathogen, Porphyromonas gingivalis may suppress the inflammasome as a mechanism for its low immunogenicity and pathogenic synergy with other, more highly immunogenic periodontal bacteria. Our results show that P. gingivalis lacks signaling capability for the activation of the inflammasome in mouse macrophages. Furthermore, P. gingivalis can suppress inflammasome activation by another periodontal bacterium, Fusobacterium nucleatum. This repression affects IL-1β processing, as well as other inflammasome-mediated processes, including IL-18 processing and cell death, in both human and mouse macrophages. F. nucleatum activates IL-1β processing through the Nlrp3 inflammasome; however, P. gingivalis repression is not mediated through reduced levels of inflammasome components. P. gingivalis can repress Nlrp3 inflammasome activation by Escherichia coli, and by danger-associated molecular patterns and pattern-associated molecular patterns that mediate activation through endocytosis. However, P. gingivalis does not suppress Nlrp3 inflammasome activation by ATP or nigericin. This suggests that P. gingivalis may preferentially suppress endocytic pathways toward inflammasome activation. To directly test whether P. gingivalis infection affects endocytosis, we assessed the uptake of fluorescent particles in the presence or absence of P. gingivalis. Our results show that P. gingivalis limits both the number of cells taking up beads and the number of beads taken up for bead-positive cells. These results provide a novel mechanism of pathogen-mediated inflammasome inhibition through the suppression of endocytosis.
Introduction
The innate immune inflammatory cytokine, interleukin (IL)-1β is critical in the host defense against infection, and consequently, pathogens have evolved an array of mechanisms for inhibiting its production (1). This includes the inhibition of the transcription of the pro-Il-1β precursor mRNA, the production of IL-1β decoy receptors, and most recently, the inhibition of the IL-1β processing inflammasome (see Ref. (2) for a review). The primary functions of the inflammasome are to cleave the precursor pro-IL-1β protein to its active secreted form and to promote caspase-dependent signaling pathways leading to cell death in response to foreign or intrinsic “danger signals,” including bacterial-encoded “pattern-associated molecular patterns” and host-derived “danger-associated molecular patterns.” In addition to its role in combating pathogenic infection, recent evidence suggests a role for the inflammasome in mediating host metabolic responses (3, 4). The dysregulation of inflammasome components has been linked to a number of inherited inflammatory and immune disorders, further underscoring its relevance in human disease (5).
Classically, the inflammasome is composed of a nucleotide-binding domain and leucine-rich repeat (NLR)4 family protein, the adaptor molecule PYCARD/ASC, and pro-caspase-1, which provides the core enzymatic activity of the complex. Each of these components has been postulated to serve as a target for pathogenic inhibition by specific bacteria and viruses. At least two pathogenic viruses have evolved strategies for suppressing the inflammasome by producing inactivating homologs of apoptosis-associated speck-like protein containing a CARD (ASC) (6, 7). Other viruses have been shown to mediate inflammasome inhibition through their production of serpin family proteins (8–10) and others (11) that can directly interact with caspase-1. Additionally, a viral NLR homolog was recently described that can repress inflammasome activation by Kaposi sarcoma-associated herpes virus (12). An alternative strategy for pathogenic inflammasome inhibition involves the modulation of the upstream signals that lead to its activation. Several bacteria minimize the inflammasome response through the production of Type III secretion system effector molecules that either regulate caspase-1 activity (13–15) or mask inflammasome detection (16). Staphylococcus aureus suppresses the inflammasome through enzymatic modification of its cell wall peptidoglycan (PGN) to make it resistant to lysosomal degradation following endocytosis (17). A variety of additional bacterial proteins suppress the inflammasome by unknown mechanisms (18, 19).
Porphyromonas gingivalis is one of the most common pathogens in chronic periodontitis, and its presence in the oral cavity is associated with a variety of related systemic diseases (20). In addition to its immunostimulatory properties, evidence is emerging to suggest that P. gingivalis has evolved several distinct mechanisms for evasion of the immune response that may contribute to its ability to perpetuate the chronic state of periodontal diseases. Like other oral pathogens, P. gingivalis produces proteases and toxins that directly attack host tissue, as well as lipopolysaccharide (LPS) and other microbial products that induce inflammation. However, cell wall extracts and purified cellular components from P. gingivalis have comparatively weak host immunostimulatory activity (21–24). Additionally, P. gingivalis lipid A has a unique structure that can either stimulate or antagonize Toll-like receptor 4 (TLR4) activation depending on environmental growth conditions and its phosphorylation and acylation states (25–29). Furthermore, P. gingivalis actively limits the immune response by producing proteases that degrade complement component C3 and IgG (30) and promotes intrinsic signaling that inhibits E-selectin expression and secretion of the neutrophil chemoattractant IL-8 (31, 32). Additionally, P. gingivalis confers resistance to apoptosis induced by pharmacologic agents (33–35). It is likely that these evasion mechanisms may explain the pathogenic synergism that is observed between P. gingivalis and other periodontal bacteria in animal models of infection (36–39).
Given the knowledge that P. gingivalis has evolved a unique set of mechanisms for subverting inflammatory activity, we hypothesized that P. gingivalis may function to suppress the inflammasome. Our results demonstrate that P. gingivalis suppresses inflammasome activation by other pathogenic bacteria, and that this suppression occurs by a novel mechanism involving the blockade of endocytosis.
EXPERIMENTAL PROCEDURES
Mouse Strains and Macrophage Culture
MyD88−/− mice were obtained from Dr. Shizuo Akira; Nlrp3−/− mice from Millenium Inc.; Asc−/− mice from Dr. Vishva Dixit at Genentech; and Casp1−/− mice from Dr. Richard Flavell, Yale University. All the mice were backcrossed for a minimum of nine generations to C57BL/6 mice. Bone marrow-derived macrophages were harvested from WT or gene-deletion mice and cultured in DMEM 10% fetal calf serum, M-CSF for 6–7 days. Cells were plated without M-CSF 16 h prior to infection. Peripheral blood macrophages were isolated on buffy coats from healthy donors (American Red Cross). Primary human macrophages were harvested by Ficoll-Hypaque gradients and seeded in 10 ml of RPMI 1640 medium containing 10% heat-inactivated FBS. Nonadherent cells were removed prior to infection.
Bacterial Strains and Bacterial Infection
P. gingivalis strain A7436 was isolated from a refractory case of periodontitis and has been described previously (40). P. gingivalis strain 381, F. nucleatum strain PK 1594, and Eschericha coli strain LF82 were obtained from American Type Culture Collection (Manassas, VA). P. gingivalis and F. nucleatum were cultured anaerobically, and E. coli aerobically until late exponential phase (0.8–1.2 optical density units at 660 nm). Aliquots were stored in media containing 20% glycerol at −80 °C and used within 3–4 months of preparation. Bacterial counts were confirmed to within 2–3-fold by replating of frozen cultures. Infections were performed by adding the desired multiplicity of infection (m.o.i.) of P. gingivalis either alone or together with F. nucleatum or E. coli to macrophages for 16–18 h. 10 μg/ml gentamycin was added to cell cultures 2 h following infection.
Treatment with Signal 1 and Signal 2 Inducers
To induce signal 1 activation in macrophages, 1 μg/ml Ultrapure E. coli LPS (InvivoGen) was added to macrophage cultures for 3 h prior to the addition of the signal 2 activator. Signal 2 activation was induced by the addition of 2 mm ATP (Sigma) or 20 μm nigericin (InvivoGen) for 30 min; 200 μg/ml monosodium urate (MSU) or 400 μg/ml alum crystals (InvivoGen) for 6 h; or 20 μg/ml PGN from S. aureus (InvivoGen) for 14–16 h. Inhibition of second signal activation by P. gingivalis was assessed by adding bacteria immediately prior to the second signal inducer.
ELISA Analysis
Supernatants were collected 18–24 h following infection unless otherwise indicated. Secreted cytokine levels were assessed using the human ELISA set for IL-1β (BD Biosciences) or the human ELISA kit for IL-18 (R&D Systems). Samples were assayed within linear range.
RNA Isolation and Real-time PCR
RNA was isolated using RNeasy kits (Qiagen). Real-time PCR was performed using TaqMan® Assays on Demand (Applied Biosystems). Values represent averages ±S.D. of biological triplicates. All values were standardized to 18 S rRNA expression.
Western Analysis
For assessment of secreted IL-1β levels, supernatants were clarified by centrifugation for 10 min and then boiled for 5 min in 1/3 volume of 3× SDS sample buffer (187.5 mm Tris, pH 6.8, 6% SDS, 30% glycerol, 150 mm DTT, 0.03% bromphenol blue). Cell lysates were prepared by washing cells in 1× PBS and then lysing for 20 min in ice-cold 1× lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100) supplemented with Complete EDTA-free Protease Inhibitor (Roche Applied Science). Lysates were centrifuged for 10 min and boiled for 5 min in 1/3 volume of 3× SDS sample buffer. Immunoblots were processed using 3ZD monoclonal antibody against Pro-IL-1 (Frederick National Laboratory for Cancer Research), H-153 against IL-1β (Santa Cruz Biotechnology), IMG-5028 against caspase-1 (Imgenex), AL177 against ASC (Enzo Life Sciences), Clone Cryo-2 against NLRP3 (Enzo Life Sciences), sc-1615 against actin (Santa Cruz Biotechnology), and MAB374 against GAPDH (Millipore).
Cell Death Measurement
ToxiLight® bioassays were performed according to the manufacturer's instructions (Lonza). Propidium iodide staining was performed as a measure of cell leakage upon death and Hoechst staining as a general nuclear stain as described previously (41).
Endocytosis Assays
FluoresbriteTN Carboxyl YG 2.0 Micron Microspheres latex beads (Polysciences, Inc.) were added to macrophages at a concentration of 10 beads/cell either alone or in combination with 100 moi P. gingivalis. 1 h following exposure, cells were washed three times with 1× PBS and then fixed with 4% paraformaldehyde. Cells and beads were visualized using a Zeiss 710 confocal microscope.
RESULTS
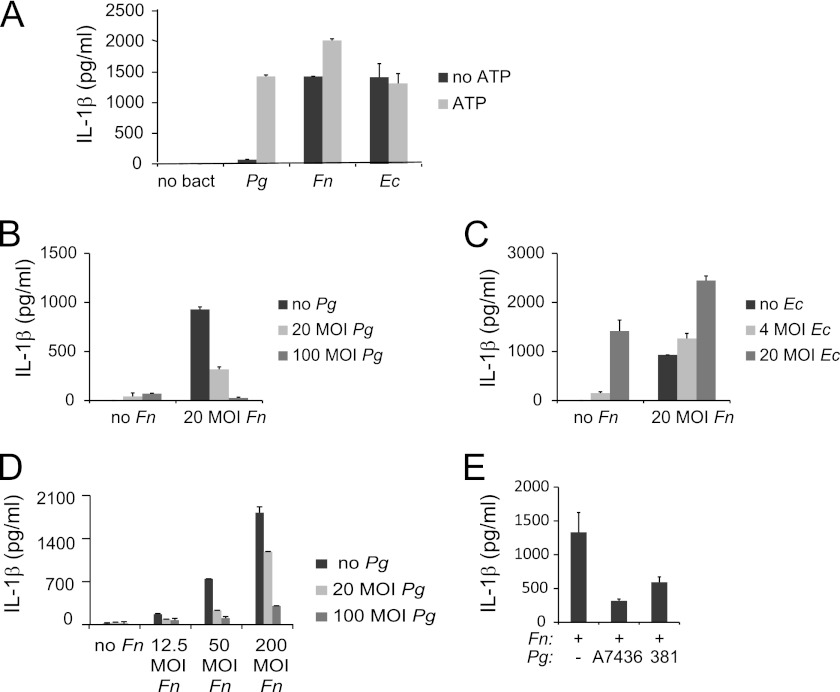
IL-1β is produced in many circumstances via a two-step process involving the activation of transcription and translation of pro-IL-1β (signal 1) followed by its proteolysis to an active secreted form by the inflammasome (signal 2). Most bacteria have the ability to provide both signals in macrophages, which are the most active physiologic producers of this potent inflammatory cytokine. By contrast, recent studies in epithelial cells suggest that P. gingivalis may lack the ability to produce a second, inflammasome-activating signal (21). To test whether P. gingivalis lacks “signal 2” activation capability in macrophages, we measured IL-1β production in bone marrow-derived mouse macrophages (BMDM) following infection. Our results showed that overnight infection with P. gingivalis was not sufficient for the stimulation of significant levels of IL-1β, but that IL-1β secretion could be induced by the subsequent addition of ATP, a well characterized second signal activator (42). This was in contrast to the high levels of IL-1β activation elicited in the absence of ATP for E. coli and for F. nucleatum, another periodontal pathogen that is commonly isolated from periodontal disease sites together with P. gingivalis (20) (Fig. 1A). This suggests that the lack of a signal 2 activation capability is a property of P. gingivalis that is not conserved among all bacteria.
FIGURE 1.
P. gingivalis lacks second signal activation ability for IL-1β production and can repress second signal activation by F. nucleatum. ELISAs of IL-1β in cell supernatants were performed following 12–16-h infection. A, mouse BMDM were infected with 20 m.o.i. P. gingivalis (Pg), F. nucleatum (Fn), or E. coli (Ec). Where indicated, cells were treated for 30 min with 2 mm ATP immediately prior to collection of supernatants. B, BMDM were infected with Fn and Pg alone or in combination as indicated. C, BMDM were infected with Fn and Ec alone or in combination as indicated. D, BMDM were treated with a range of doses of Fn and Pg. E, BMDM were treated with Fn and Pg strains A7436 or 381. Results represent the averages and standard deviation of duplicates and are representative of at least three independent experiments.
P. gingivalis has been shown to promote synergistic pathogenicity in polymicrobial infections with F. nucleatum (36–39). Given the emerging role of inflammasome repression as a mechanism for pathogenic stealth (2), we postulated that P. gingivalis may contribute to the synergistic effects of these two bacteria in vivo by repressing F. nucleatum-mediated activation of IL-1β processing. To assess this as a possible mechanism, IL-1β levels were measured following infection with 20 m.o.i. F. nucleatum and increasing doses of P. gingivalis. Results verified that P. gingivalis repressed IL-1β activation by F. nucleatum (Fig. 1B). This mechanism of P. gingivalis in repressing F. nucleatum is not a common feature shared with other bacteria, given that E. coli, when added to F. nucleatum, enhanced, rather than repressed IL-1β release (Fig. 1C). The repressive effect of P. gingivalis is dose dependent at a range of m.o.i. of F. nucleatum (Fig. 1D). Furthermore, this activity does not appear to be strain-specific, given that both the A7436 and 381 strains of P. gingivalis were able to repress F. nucleatum-mediated IL-1β activation (Fig. 1E).
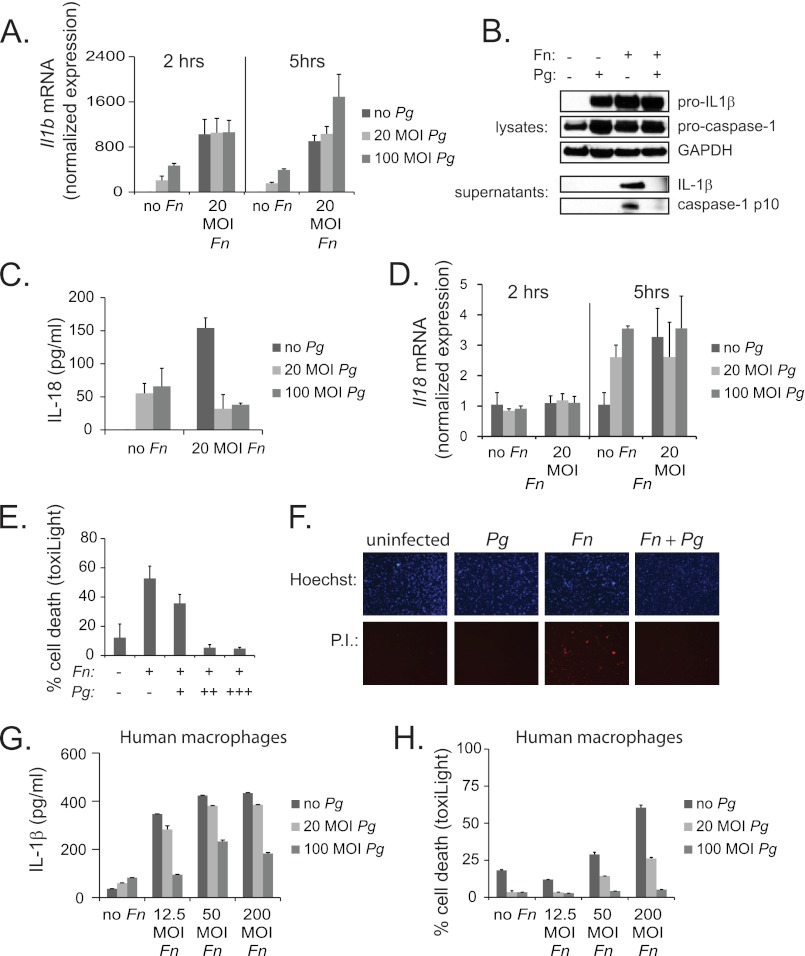
To distinguish the level at which P. gingivalis represses F. nucleatum-mediated IL-1β activation, RNA was isolated from macrophages following infection with F. nucleatum alone or in combination with P. gingivalis. Real-time PCR analysis of IL1b levels showed that this gene was transcriptionally induced by F. nucleatum, but that its induction was not repressed upon co-infection with P. gingivalis (Fig. 2A). These results indicate that P. gingivalis repression was not at the level of the activation of transcription (signal 1), implying a role in the IL-1β processing pathway. To determine whether P. gingivalis affects inflammasome activation by F. nucleatum (signal 2), levels of pro-IL-1β and pro-caspase-1 in cell lysates and levels of mature IL-1β and caspase-1 p10 in supernatants were examined by Western analysis. Whereas the pro-forms of each protein were expressed in F. nucleatum-infected cells both without and with P. gingivalis co-infection, the mature forms were only detected in cells infected with F. nucleatum alone (Fig. 2B). These results verify that P. gingivalis blocks IL-1β secretion at the level of inflammasome activation.
FIGURE 2.
Pg represses inflammasome activation by Fn. A, real-time PCR of Il1b mRNA in mouse BMDM was performed 2 or 5 h following infection with a range of doses of Fn and Pg. Expression is normalized to the expression of 18 S rRNA and standardized to 1 in uninfected cells. Results represent the average ±S.D. (error bars) of biological triplicates and are representative of three independent experiments. B, Western analysis is shown for pro-IL-1β and pro-caspase-1 protein in cell lysates and cleaved (activated) IL-1β and caspase-1 p10 in cell supernatants 14 h following infection with 20 m.o.i. Fn and 100 m.o.i. Pg. GAPDH is shown as a loading control. Results are representative of three independent experiments. C, ELISA of IL-18 expression in BMDM is shown following 14-h infection with Fn and Pg at indicated m.o.i. Results represent the averages ±S.D. of duplicates and are representative of at least three independent experiments. D, real-time PCR of Il18 mRNA in BMDM was performed 2 or 5 h following infection with a range of dose of Fn and Pg. Expression is normalized to the expression of 18 S rRNA and standardized to 1 in uninfected cells. Results represent the averages ±S.D. of biological triplicates and are representative of three independent experiments. E, percentage of cell death in BMDM was determined by ToxiLight® assay 16 h following infection with 20 m.o.i. Fn alone or together with (+) 12.5, (++) 50, or (+++) 200 m.o.i. Pg. Results are representative of three independent experiments. F, propidium iodide (P. I.) stain is shown as an indicator of cell death in BMDM 16 h following infection with 20 m.o.i. Fn and 100 m.o.i. Pg. Hoechst staining is shown as a control. Results are representative of two independent experiments. G, ELISA of IL-1β secretion in human macrophages is shown 14 h following infection with Fn and Pg at a range of m.o.i. Results represent the averages ±S.D. of duplicates and are representative of two independent experiments. H, percentage of cell death in human macrophages was determined by ToxiLight® assay 16 h following infection with Fn and Pg at a range of m.o.i.
The inflammasome is responsible for the processing and secretion of other inflammatory cytokines besides IL-1β, including IL-18, as well as for the promotion of cell death. Our results showed that, consistent with its role in inflammasome suppression, P. gingivalis repressed the secretion of IL-18 (Fig. 2C), and that this repression, like that of IL-1β, did not appear to be transcriptional (Fig. 2D). P. gingivalis also suppressed the induction of cell death by F. nucleatum as measured by ToxiLight® assay (Fig. 2E) or propidium iodide staining (Fig. 2F). These findings rule out the possibility that reduced cytokine secretion was caused by P. gingivalis-induced cell death in F. nucleatum- and P. gingivalis-infected cells and further support a role for P. gingivalis in inhibiting multiple signaling pathways downstream of the inflammasome. P. gingivalis was also able to repress IL-1β levels and cell death in human macrophages, demonstrating that these findings are not limited to murine cells (Fig. 2, G and H).
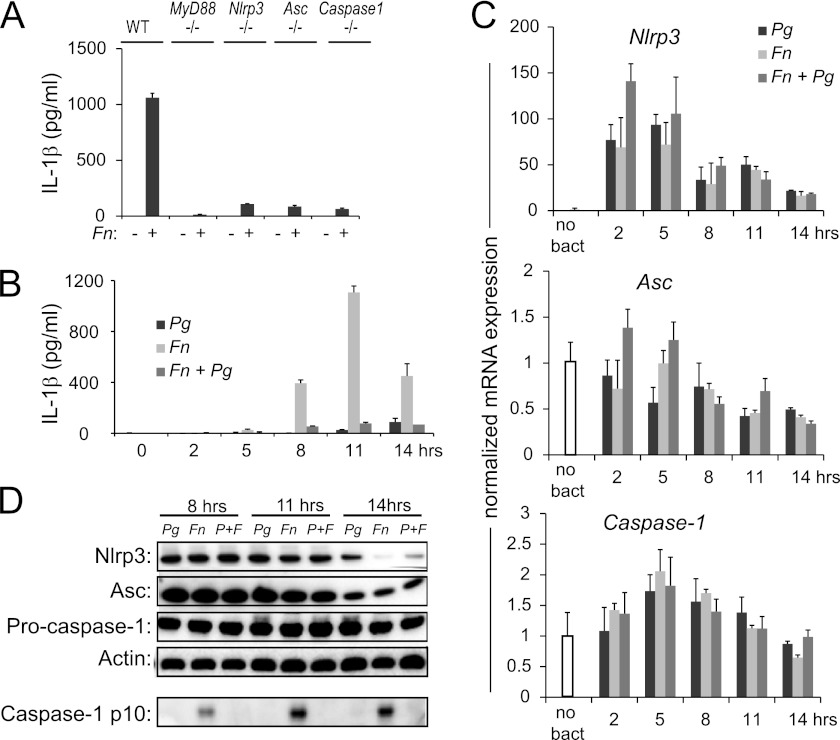
One potential mechanism of inflammasome repression involves the modulation of the expression of inflammasome components. The most commonly utilized pathway of inflammasome activation requires an initial activation through the MyD88 adaptor molecule, followed by the assembly of a protein complex composed of NLRP3, ASC, and caspase-1 (43). To determine whether MyD88 and these three inflammasome components are required for F. nucleatum activation, we infected macrophages from gene-deletion mice with F. nucleatum and measured IL-1β release. Our results showed that each of these genes was required for F. nucleatum-mediated IL-1β release (Fig. 3A). We next tested whether transcript levels of the three inflammasome components were regulated by F. nucleatum and P. gingivalis. RNA was isolated over a time course of infection that preceded and spanned the 11-h peak of IL-1β induction following F. nucleatum infection (Fig. 3B). Nlrp3 transcription was activated by both P. gingivalis and F. nucleatum, and levels persisted following infection with the two bacteria in combination at each time point (Fig. 3C, top panel). By contrast, levels of Asc and caspase-1 were not significantly modulated following infection with F. nucleatum either alone or in combination with P. gingivalis throughout the time course (Fig. 3C, bottom two panels). To determine whether repression of these inflammasome components occurs at the level of protein expression, Western blotting was performed 8, 11, or 14 h post-infection. Levels of Nlrp3, Asc, and pro-caspase-1 were similar in lysates from cells infected with F. nucleatum alone or in combination with P. gingivalis at each earlier time point. At the 14-h time point, levels of Nlrp3, Asc, and pro-caspase-1 in P. gingivalis-infected cells were similar or higher than F. nucleatum- and P. gingivalis+F. nucleatum-infected cells, though the processing of pro-caspase-1 to caspase-1 only occurred in F. nucleatum-infected cells and not the other groups. These findings clearly demonstrate that P. gingivalis does not suppress inflammasome activation by reducing the expression of the inflammasome components (Fig. 3D).
FIGURE 3.
Pg does not repress the inflammasome by ablating the expression of its protein components. A, ELISA of IL-1β secretion is shown for BMDM isolated from wild type (WT) and gene-deletion mice 16 h following infection with Fn. B, ELISA of IL-1β secretion in wild type mouse BMDM is shown following a time course of infection with 20 m.o.i. Fn and 100 m.o.i. Pg alone or in combination. C, real-time PCR of Nlrp3, Asc, and caspase-1 mRNA in BMDM following a time course of infection with 20 m.o.i. Fn and 100 m.o.i. Pg is shown. Expression is normalized to the expression of 18 S rRNA and standardized to 1 in uninfected cells. Results represent the average ±S.D. (error bars) of biological triplicates and are representative of three independent experiments. D, Western analysis was performed for Nlrp3, Asc, and caspase-1 protein levels in BMDM 8, 11, and 14 h following infection with 20 m.o.i. Fn and 100 m.o.i. Pg. Actin is shown as a loading control, and cleaved caspase-1 in cell supernatants is provided for reference. Results are representative of three independent experiments.
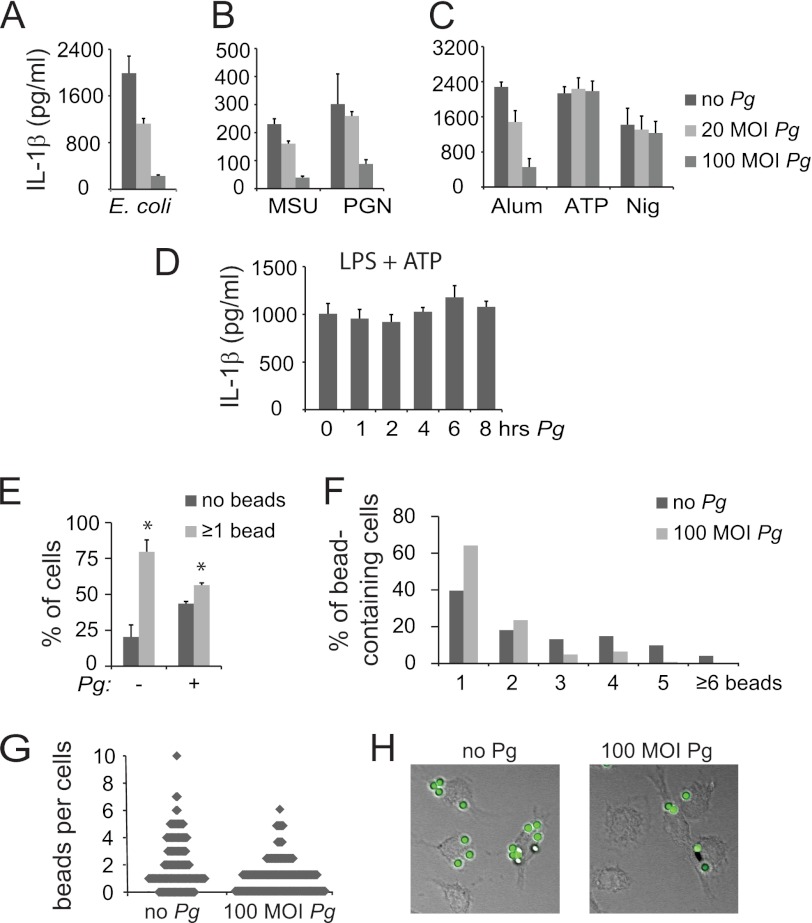
To determine whether P. gingivalis-mediated inhibition is limited to F. nucleatum activation of the inflammasome, we tested whether P. gingivalis inhibits E. coli as an alternate bacterial NLRP3 inflammasome inducer. Results showed that P. gingivalis also can inhibit E. coli-mediated IL-1β release (Fig. 4A). To further delineate the mechanism of P. gingivalis inflammasome inhibition, we tested the effect of P. gingivalis infection upon stimulation with LPS plus a panel of signal 2 inducers that are known to activate the NLRP3 inflammasome, including MSU, PGN, alum, ATP, and nigericin. Interestingly, P. gingivalis inhibited activation by MSU PGN, and alum but did not inhibit activation by ATP and nigericin under the same conditions of P. gingivalis addition (Fig. 4, B and C). One difference between ATP/nigericin and the other NLRP3 inflammasome inducers is the shortened time required for IL-1β stimulation. For inflammasome stimulation by ATP and nigericin, cells are treated with LPS for 3 h and then with ATP for 30 min. By contrast, to achieve optimal inflammasome activation, other inducers are applied for 6 h to overnight. To determine whether a critical time of P. gingivalis exposure is required for its inhibition of ATP-mediated inflammasome activation, P. gingivalis was applied at increasing times prior to the activation of the inflammasome by ATP. However, prolonged P. gingivalis exposure did not restore its ability to repress inflammasome activation, suggesting that its failure to repress ATP-mediated inflammasome activation is not explained by differential timing (Fig. 4D).
FIGURE 4.
Pg inflammasome repression is mediated through blockade of endocytosis. ELISA of mouse BMDM demonstrates that Pg can repress the activation of IL-1β by (A) E. coli; by (B) LPS plus MSU or PGN; and by (C) LPS plus alum, but not LPS plus ATP or nigericin (Nig). Results are representative of three independent experiments. D, IL-1β ELISA of BMDM treated with LPS plus ATP is shown. Pg was added over a time course preceding the ATP treatment. Representative of two independent experiments. E, percentages of BMDM uptaking beads following 2-h exposure are shown. Cells were pretreated with 100 m.o.i. Pg where indicated. F, numbers of beads in the bead-containing cell population for uninfected cells and cells infected with 100 m.o.i. Pg are shown. Numbers are expressed as a percentage of the total bead-containing cells. G, a diagrammatic representation of the number of beads per cell for uninfected cells and cells infected with 100 m.o.i. Pg is shown. Each cell is represented as a dot. H, a representative confocal microscopy image for bead uptake in uninfected cells and cells infected with 100 m.o.i. Pg is provided. The number of internalized beads was counted in >100 cells per each of three independent experiments.
An additional difference between ATP and nigericin and the other NLRP3 inflammasome inducers is the requirement for endocytosis to elicit IL-1β production. The inducers that were blocked by P. gingivalis all require endocytosis to activate the signal; however, ATP and nigericin are thought to activate the inflammasome directly (43). One possible explanation for the selective ability of P. gingivalis to block IL-1β production could be that P. gingivalis has the ability to regulate endocytosis. To determine whether this might be the case, we exposed macrophages to nonimmunogenic fluorescent latex beads and examined uptake by confocal microscopy 1 h following incubation. Our data showed that ∼80% of the uninfected cells took up one or more beads, with ∼20% of the cells having no bead uptake; however, cells that were infected with P. gingivalis prior to exposure to beads had a distribution of bead-positive and -negative cells closer to 50:50 (Fig. 4E). Furthermore, among the bead-positive cells, P. gingivalis-infected cells were more likely to contain one or two beads, whereas the uninfected cells were more likely to contain three or more beads (Fig. 4, F–H). These findings suggest that P. gingivalis infection causes reduction in levels of endocytosis.
DISCUSSION
P. gingivalis resides within periodontal tissue in combination with a wide variety of other pathogenic bacteria that can mediate a robust immune response, chief among which is F. nucleatum. In vivo studies have suggested that P. gingivalis and F. nucleatum can work synergistically to promote infection and pathogenesis (36–39). The mechanism for F. nucleatum-mediated synergy is postulated to involve its abundant production of adhesins that facilitate the co-aggregation of bacteria and attachment to host cells (44). However, the contribution of P. gingivalis to this synergy has remained largely unknown. We postulated that P. gingivalis may contribute to the synergy between the two bacteria through the evasion of F. nucleatum-mediated immune responses by inflammasome suppression. Inflammasome suppression is employed as a stealth mechanism by a variety of other pathogenic viruses and bacteria (2) and would be a likely avenue for promoting the chronic state of periodontitis through immune evasion. Furthermore, P. gingivalis has intrinsically low ability to stimulate immune responses (21–24), which could contribute to its stealth potential. Given these considerations, we sought to specify the effects of P. gingivalis on inflammasome activation in macrophages and to determine whether P. gingivalis has the ability to actively suppress inflammasome induction by F. nucleatum and other stimulatory agents.
Inflammasome activation is a two-step process, and generalized low stimulatory responses to P. gingivalis may be explained in part by our findings that P. gingivalis can induce the transcription of the IL-1β gene (signal 1) but that it lacks the ability to activate the processing of mature IL-1β (signal 2) in primary mouse macrophages. The ability of P. gingivalis to stimulate first signal activation without second signal activation is unique among pathogenic bacteria (Ref. 5 and Fig. 1). Additionally, this selective first signal activation could explain why P. gingivalis has the ability to stimulate IL-1β and cell death in monocytic cells (45–47), but not macrophages (Figs. 1 and 2). Immature monocytes are able to provide an endogenous signal that negates the requirement for second signal activation, but this endogenous second signal is lost upon maturation (48, 49).
Our results show that P. gingivalis can actively suppress the secretion of IL-1β induced by F. nucleatum. Given that co-infection with P. gingivalis blocks the F. nucleatum-mediated activation of caspase-1 without reducing F. nucleatum-induced Il-1β transcript levels, the inflammasome suppression by P. gingivalis occurs at the level of the second signal. These results are supported by the finding that P. gingivalis also can repress the secretion of IL-18, another cytokine that is known to be processed by the inflammasome, without affecting its transcript levels. Suppression is not explained by death induction by P. gingivalis. Instead, P. gingivalis is shown to suppress F. nucleatum-induced cell death. Cell death is an additional physiological process that is associated with inflammasome activation, and its suppression, therefore, is consistent with the notion that P. gingivalis IL-1β suppression is an inflammasome-mediated event. The induction of IL-1β and cell death also is suppressed in primary human macrophages, further supporting the findings that P. gingivalis acts at the level of the second signal. The suppression of the innate immune response by P. gingivalis in human macrophages could facilitate the maintenance of the chronic state of infection during periodontal diseases.
We propose a novel mechanism for P. gingivalis-mediated inflammasome inhibition through the suppression of endocytosis, as is indicated by the selectivity of P. gingivalis in its ability to inhibit Nlrp3 inflammasome inducers that are endocytosed. As a direct measure of effects on endocytosis, P. gingivalis infection was shown to modulate the levels of endocytosis of nonimmunogenic fluorescent beads. Additionally, our results rule out the possibility that P. gingivalis suppresses F. nucleatum-induced inflammasome activation through the reduction of endogenous levels of Nlrp3 or the other core components of its inflammasome. A recent study suggested that Nlrp3 is down-regulated by 33% in gingival fibroblast cultures colonized with a nine-species biofilm upon the inclusion of P. gingivalis (50). However, Il1b transcription was also reduced 35%, and it is therefore unclear whether reduced IL1b, rather than Nlrp3, may contribute to the reduced IL-1β secretion in that model. P. gingivalis can activate Nlrp3 transcription both in primary macrophages (Fig. 3) and in primary human gingival cells (51). Additionally, P. gingivalis does not significantly alter ASC or pro-caspase-1 levels. Therefore, the ability of P. gingivalis to suppress IL-1β in our model system is not likely to involve reduction in components of the core Nlrp3 inflammasome, but rather its ability to suppress endocytosis.
Studies to determine which components of F. nucleatum require phagocytosis to activate the inflammasome may help to further characterize mechanisms of P. gingivalis-mediated repression of endocytosis. There are multiple steps in the cellular pathway toward endocytosis, and it would be of interest to identify the step or steps that are affected. Given that P. gingivalis has been shown to cause actin cytoskeletal rearrangements (52), it is intriguing to postulate that P. gingivalis might suppress endocytosis by modulating the function of a core cytoskeletal protein. P. gingivalis has been shown to evade the endocytic pathway to lysosomes in favor of trafficking to autophagosomes (53), where it activates autophagy to provide a replicative niche (54); thus, there is a basis for its role in directing cellular trafficking.
An additional important future direction would include the identification of a P. gingivalis component responsible for inflammasome inhibition through the suppression of endocytosis. Interestingly, S. aureus produces an enzyme that suppresses its own inflammasome activation by making the cell wall PGN-resistant to degradation during phagocytosis (17). Although P. gingivalis produces abundant gingival proteases, a similar mechanism for enzymatic modulation of bacterial immunostimulatory components has not been determined. Our unpublished results suggest that P. gingivalis LPS is not sufficient for inflammasome inhibition, but that another secreted component may be involved.5 The identification and purification of this component could lead to the development of a therapeutic agent for the treatment of periodontitis, and considering that P. gingivalis can suppress the activation of the inflammasome by MSU and other danger-associated molecular patterns and pattern-associated molecular patterns, its therapeutic value could potentially extend to the treatment of gout and other inflammatory disorders.
This study was supported, in whole or in part, by National Institutes of Health Grants DE016326, AI029564, and AI057157 (to J. T.) and AI088255 (to J. A. D).
D. J. Taxman, unpublished data.
- NLR
- nucleotide-binding domain and leucine-rich repeat
- ASC
- apoptosis-associated speck-like protein containing a CARD
- PGN
- peptidoglycan
- BMDM
- bone marrow-derived mouse macrophages
- m.o.i.
- multiplicity of infection
- MSU
- monosodium urate.
REFERENCES
- 1. Dinarello C. A. (1998) Interleukin-1β, interleukin-18, and the interleukin-1β-converting enzyme. Ann. N.Y. Acad. Sci. 856, 1–11 [DOI] [PubMed] [Google Scholar]
- 2. Taxman D. J., Huang M. T., Ting J. P. (2010) Inflammasome inhibition as a pathogenic stealth mechanism. Cell Host Microbe 8, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M. T., Brickey W. J., Ting J. P. (2011) Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen H., Ting J. P., O'Neill L. A. (2012) A role for the NLRP3 inflammasome in metabolic diseases: did Warburg miss inflammation? Nat. Immunol. 13, 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davis B. K., Wen H., Ting J. P. (2011) The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnston J. B., Barrett J. W., Nazarian S. H., Goodwin M., Riccuto D., Wang G., McFadden G. (2005) A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity 23, 587–598 [DOI] [PubMed] [Google Scholar]
- 7. Dorfleutner A., Talbott S. J., Bryan N. B., Funya K. N., Rellick S. L., Reed J. C., Shi X., Rojanasakul Y., Flynn D. C., Stehlik C. (2007) A Shope fibroma virus pyrin-only protein modulates the host immune response. Virus Genes 35, 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ray C. A., Black R. A., Kronheim S. R., Greenstreet T. A., Sleath P. R., Salvesen G. S., Pickup D. J. (1992) Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1β-converting enzyme. Cell 69, 597–604 [DOI] [PubMed] [Google Scholar]
- 9. Kettle S., Alcamí A., Khanna A., Ehret R., Jassoy C., Smith G. L. (1997) Vaccinia virus serpin B13R (SPI-2) inhibits interleukin-1β-converting enzyme and protects virus-infected cells from TNF- and Fas-mediated apoptosis, but does not prevent IL-1β-induced fever. J. Gen. Virol. 78, 677–685 [DOI] [PubMed] [Google Scholar]
- 10. Petit F., Bertagnoli S., Gelfi J., Fassy F., Boucraut-Baralon C., Milon A. (1996) Characterization of a myxoma virus-encoded serpin-like protein with activity against interleukin-1β-converting enzyme. J. Virol. 70, 5860–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bump N. J., Hackett M., Hugunin M., Seshagiri S., Brady K., Chen P., Ferenz C., Franklin S., Ghayur T., Li P., Licari P., Mankovich J., Shi L., Greenberg A. H., Miller L. K., Wong W. W. (1995) Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 269, 1885–1888 [DOI] [PubMed] [Google Scholar]
- 12. Gregory S. M., Davis B. K., West J. A., Taxman D. J., Matsuzawa S., Reed J. C., Ting J. P., Damania B. (2011) Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331, 330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sutterwala F. S., Mijares L. A., Li L., Ogura Y., Kazmierczak B. I., Flavell R. A. (2007) Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galle M., Schotte P., Haegman M., Wullaert A., Yang H. J., Jin S., Beyaert R. (2008) The Pseudomonas aeruginosa type III secretion system plays a dual role in the regulation of caspase-1-mediated IL-1β maturation. J. Cell. Mol. Med. 12, 1767–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schotte P., Denecker G., Van Den Broeke A., Vandenabeele P., Cornelis G. R., Beyaert R. (2004) Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1β. J. Biol. Chem. 279, 25134–25142 [DOI] [PubMed] [Google Scholar]
- 16. Brodsky I. E., Palm N. W., Sadanand S., Ryndak M. B., Sutterwala F. S., Flavell R. A., Bliska J. B., Medzhitov R. (2010) A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 7, 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shimada T., Park B. G., Wolf A. J., Brikos C., Goodridge H. S., Becker C. A., Reyes C. N., Miao E. A., Aderem A., Götz F., Liu G. Y., Underhill D. M. (2010) Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1β secretion. Cell Host Microbe 7, 38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Master S. S., Rampini S. K., Davis A. S., Keller C., Ehlers S., Springer B., Timmins G. S., Sander P., Deretic V. (2008) Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe 3, 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Littmann M., Albiger B., Frentzen A., Normark S., Henriques-Normark B., Plant L. (2009) Streptococcus pneumoniae evades human dendritic cell surveillance by pneumolysin expression. EMBO Mol. Med. 1, 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darveau R. P. (2009) The oral microbial consortium's interaction with the periodontal innate defense system. DNA Cell Biol. 28, 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yilmaz O., Sater A. A., Yao L., Koutouzis T., Pettengill M., Ojcius D. M. (2010) ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 12, 188–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berezow A. B., Ernst R. K., Coats S. R., Braham P. H., Karimi-Naser L. M., Darveau R. P. (2009) The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 47, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peyret-Lacombe A., Brunel G., Watts M., Charveron M., Duplan H. (2009) TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine 46, 201–210 [DOI] [PubMed] [Google Scholar]
- 24. Okugawa T., Kaneko T., Yoshimura A., Silverman N., Hara Y. (2010) NOD1 and NOD2 mediate sensing of periodontal pathogens. J. Dent. Res. 89, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coats S. R., Jones J. W., Do C. T., Braham P. H., Bainbridge B. W., To T. T., Goodlett D. R., Ernst R. K., Darveau R. P. (2009) Human toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 11, 1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Q., Darveau R. P., Samaranayake L. P., Wang C. Y., Jin L. (2009) Differential modulation of human β-defensins expression in human gingival epithelia by Porphyromonas gingivalis lipopolysaccharide with tetra- and penta-acylated lipid A structures. Innate Immun. 15, 325–335 [DOI] [PubMed] [Google Scholar]
- 27. Reife R. A., Coats S. R., Al-Qutub M., Dixon D. M., Braham P. A., Billharz R. J., Howald W. N., Darveau R. P. (2006) Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 8, 857–868 [DOI] [PubMed] [Google Scholar]
- 28. Coats S. R., Do C. T., Karimi-Naser L. M., Braham P. H., Darveau R. P. (2007) Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol. 9, 1191–1202 [DOI] [PubMed] [Google Scholar]
- 29. Al-Qutub M. N., Braham P. H., Karimi-Naser L. M., Liu X., Genco C. A., Darveau R. P. (2006) Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 74, 4474–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cutler C. W., Kalmar J. R., Genco C. A. (1995) Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3, 45–51 [DOI] [PubMed] [Google Scholar]
- 31. Darveau R. P., Cunningham M. D., Bailey T., Seachord C., Ratcliffe K., Bainbridge B., Dietsch M., Page R. C., Aruffo A. (1995) Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect. Immun. 63, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darveau R. P., Belton C. M., Reife R. A., Lamont R. J. (1998) Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66, 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mao S., Park Y., Hasegawa Y., Tribble G. D., James C. E., Handfield M., Stavropoulos M. F., Yilmaz O., Lamont R. J. (2007) Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 9, 1997–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yilmaz O., Jungas T., Verbeke P., Ojcius D. M. (2004) Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 72, 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakhjiri S. F., Park Y., Yilmaz O., Chung W. O., Watanabe K., El-Sabaeny A., Park K., Lamont R. J. (2001) Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol. Lett. 200, 145–149 [DOI] [PubMed] [Google Scholar]
- 36. Feuille F., Ebersole J. L., Kesavalu L., Stepfen M. J., Holt S. C. (1996) Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 64, 2094–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Metzger Z., Lin Y. Y., Dimeo F., Ambrose W. W., Trope M., Arnold R. R. (2009) Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. J. Endod. 35, 86–94 [DOI] [PubMed] [Google Scholar]
- 38. Polak D., Wilensky A., Shapira L., Halabi A., Goldstein D., Weiss E. I., Houri-Haddad Y. (2009) Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. J. Clin. Periodontol. 36, 406–410 [DOI] [PubMed] [Google Scholar]
- 39. Ebersole J. L., Feuille F., Kesavalu L., Holt S. C. (1997) Host modulation of tissue destruction caused by periodontopathogens: effects on a mixed microbial infection composed of Porphyromonas gingivalis and Fusobacterium nucleatum. Microb. Pathog. 23, 23–32 [DOI] [PubMed] [Google Scholar]
- 40. Genco C. A., Cutler C. W., Kapczynski D., Maloney K., Arnold R. R. (1991) A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang M. T., Taxman D. J., Holley-Guthrie E. A., Moore C. B., Willingham S. B., Madden V., Parsons R. K., Featherstone G. L., Arnold R. R., O'Connor B. P., Ting J. P. (2009) Critical role of apoptotic speck protein containing a caspase recruitment domain (ASC) and NLRP3 in causing necrosis and ASC speck formation induced by Porphyromonas gingivalis in human cells. J. Immunol. 182, 2395–2404 [DOI] [PubMed] [Google Scholar]
- 42. Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 [DOI] [PubMed] [Google Scholar]
- 43. Jin C., Flavell R. A. Molecular mechanism of NLRP3 inflammasome activation. J. Clin. Immunol. 30, 628–631 [DOI] [PubMed] [Google Scholar]
- 44. Kolenbrander P. E., London J. (1993) Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175, 3247–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taxman D. J., Zhang J., Champagne C., Bergstralh D. T., Iocca H. A., Lich J. D., Ting J. P. (2006) Cutting edge: ASC mediates the induction of multiple cytokines by Porphyromonas gingivalis via caspase-1-dependent and -independent pathways. J. Immunol. 177, 4252–4256 [DOI] [PubMed] [Google Scholar]
- 46. Huang M. T., Mortensen B. L., Taxman D. J., Craven R. R., Taft-Benz S., Kijek T. M., Fuller J. R., Davis B. K., Allen I. C., Brickey W. J., Gris D., Wen H., Kawula T. H., Ting J. P. (2010) Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J. Immunol. 185, 5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taxman D. J., Holley-Guthrie E. A., Huang M. T., Moore C. B., Bergstralh D. T., Allen I. C., Lei Y., Gris D., Ting J. P. (2011) The NLR adaptor ASC/PYCARD regulates DUSP10, mitogen-activated protein kinase (MAPK), and chemokine induction independent of the inflammasome. J. Biol. Chem. 286, 19605–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Netea M. G., Nold-Petry C. A., Nold M. F., Joosten L. A., Opitz B., van der Meer J. H., van de Veerdonk F. L., Ferwerda G., Heinhuis B., Devesa I., Funk C. J., Mason R. J., Kullberg B. J., Rubartelli A., van der Meer J. W., Dinarello C. A. (2009) Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 113, 2324–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reig N., Jiang A., Couture R., Sutterwala F. S., Ogura Y., Flavell R. A., Mellman I., van der Goot F. G. (2008) Maturation modulates caspase-1-independent responses of dendritic cells to anthrax lethal toxin. Cell Microbiol. 10, 1190–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Belibasakis G. N., Guggenheim B., Bostanci N. (2012) Down-regulation of NLRP3 inflammasome in gingival fibroblasts by subgingival biofilms: involvement of Porphyromonas gingivalis. Innate Immun., in press [DOI] [PubMed] [Google Scholar]
- 51. Bostanci N., Emingil G., Saygan B., Turkoglu O., Atilla G., Curtis M. A., Belibasakis G. N. (2009) Expression and regulation of the NALP3 inflammasome complex in periodontal diseases. Clin. Exp. Immunol. 157, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hasegawa Y., Tribble G. D., Baker H. V., Mans J. J., Handfield M., Lamont R. J. (2008) Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 76, 2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dorn B. R., Dunn W. A., Jr., Progulske-Fox A. (2001) Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69, 5698–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bélanger M., Rodrigues P. H., Dunn W. A., Jr., Progulske-Fox A. (2006) Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy 2, 165–170 [DOI] [PubMed] [Google Scholar]