Background: Aberrant expression of annexin A2 has been found in several cancers; however, its role in tumorigenesis is unknown.
Results: Silencing annexin A2 partly caused p53-regulated cell cycle arrest at the G2 phase by inactivating JNK/c-Jun signaling.
Conclusion: Annexin A2 promotes the cell cycle and cell proliferation in lung cancer in part by inhibiting p53.
Significance: This study provides new insight into the tumorigenic role of annexin A2.
Keywords: Cell Cycle, Jun N-terminal Kinase (JNK), Jun Transcription Factor, Lung Cancer, p53, Proliferation, shRNA, siRNA, Annexin A2, NSCLC
Abstract
Annexin A2 (ANXA2) overexpression is required for cancer cell proliferation; however, the molecular mechanisms underlying ANXA2-mediated regulation of the cell cycle are still unknown. ANXA2 is highly expressed in non-small cell lung cancer (NSCLC) and is positively correlated with a poor prognosis. NSCLC A549 cells lacking ANXA2 exhibited defects in tumor growth in vivo and in cell proliferation in vitro without cytotoxicity. ANXA2 knockdown induced cell cycle arrest at G2 phase. Unexpectedly, ANXA2 silencing increased the expression of p53 and its downstream genes, which resulted in p53-dependent and -independent G2 arrest. Aberrant JNK inactivation, which was observed in ANXA2-deficient cells, inhibited cell proliferation following G2 arrest. A lack of ANXA2 caused a loss of JNK-regulated c-Jun expression, resulting in an increase in p53 transcription. These results demonstrate a novel role for ANXA2 in NSCLC cell proliferation by facilitating the cell cycle partly through the regulation of p53 via JNK/c-Jun.
Introduction
Annexin A2 (ANXA2) is a 36-kDa protein that is also known as ANX II, p36, calpactin I, lipocortin II, chromobindin VIII, and placental anticoagulant protein IV. It contains three distinct functional regions, including the N terminus, the core domain, and the C terminus (1). The N terminus contains an S100/A10 (p11)-binding site (2); the core domain contains a phospholipid-binding site (3); and the C terminus contains heparin- and plasminogen-binding sites (4, 5). The core domain is composed of four repeats, each containing an α-helix domain for calcium binding. ANXA2 is regulated in a calcium-dependent manner and undergoes a conformational change that exposes a hydrophobic amino acid, thereby allowing heterotetramer formation with p11 (S100/A10). This complex has a high affinity for membrane phospholipid binding (6–8). In general, ANXA2 can be expressed as an extracellular complex with p11 and as an intracellular unbound protein and/or complexed with other partner proteins.
ANXA2 is overexpressed in brain tumors (9, 10), acute promyelocytic leukemia (APL)2 (11), glioma (12), gastric cancer (13), colorectal carcinoma (14), pancreatic cancer (15), breast cancer (16), Lewis lung carcinoma (17), hepatocellular carcinoma (18), and multiple myeloma (19). However, ANXA2 expression is decreased in prostate cancer (20), esophageal squamous cell carcinoma (21), osteosarcoma (22), and oral squamous cell carcinoma (23). In the tumorigenesis of APL and pancreatic cancers, overexpressed ANXA2 acts as a tissue plasminogen activator receptor; the activated plasminogen causes increases invasiveness of cancer cells through reconstitution of the extracellular matrix (11, 24). Additionally, ANXA2 overexpression in the breast cancer cell line MDA-MB231 correlates with its high invasiveness compared with that of MCF-7, a poorly invasive cancer cell line with lower ANXA2 expression (16). Although ANXA2 expression is related to metastasis and invasion, ANXA2 expression is limited in proliferating ductular adenocarcinoma (25). ANXA2 silencing in cervical cancer-derived HeLa cells and multiple myeloma-derived U266 and RPMI8226 cells inhibits cell division and proliferation by blocking DNA synthesis (19, 26). Nuclear ANXA2 acts as a primer recognition protein and is involved in DNA replication (27–29). In MDA-MB-435 cells, ANXA2-targeted siRNA reduces the levels of p11 and c-Myc and causes cell cycle arrest (30). ANXA2 has therefore been hypothesized to be involved in cell proliferation and cell cycle progression.
Lung cancer is the leading cause of cancer deaths worldwide (31). Non-small cell lung cancer (NSCLC), which includes adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, accounts for 80% of all lung cancers and is epithelial in origin (32). Despite advances in surgical therapies radiotherapies, and chemotherapies the 5-year lung cancer survival rate remains at less than 15% (33, 34). Early stage NSCLC can initially be radically treated in ∼30% of stage I patients (35) and 50% of those with stage II (36). However, more than 70% of stage III individuals (37) will relapse and die from the disease as a result of resistance to anticancer drugs. Unfortunately, the majority of patients with NSCLC are often diagnosed during the later stages of lung cancer and die within the first 18 months following diagnosis without successful treatment (34, 38). It is therefore essential to identify biomarkers that are able to predict survival or therapeutic benefit.
ANXA2 is up-regulated in the doxorubicin-selected, multidrug-resistant small cell lung cancer cell line H69AR (39). Intense ANXA2 immunoreactivity has been detected in lung adenocarcinoma and squamous cell carcinoma (40). However, the role of ANXA2 in the molecular tumorigenesis of lung cancer has not been determined. In this report, we investigated the potential for ANXA2 to regulate the cell cycle and cell proliferation in NSCLC. We also investigated the impact of ANXA2-mediated p53 expression.
EXPERIMENTAL PROCEDURES
Cell Cultures and Reagents
Human lung adenocarcinoma A549 (CCL-185, ATCC) and H1975 (CRL-5908, ATCC) cells, human large cell lung cancer H460 cells (HTB-177, ATCC), human cervical cancer SiHa cells (HTB-35, ATCC), human gastric cancer MKN45 cells (JCRB0254, RIKEN Cell Bank, Japan), human embryonic kidney 293FT cells (PTA-5077, ATCC), human cervical carcinoma CaSki cells (CRL-1550, ATCC), and human hepatoma Hep3B (HB-8064, ATCC), and Huh-7 (PTA-4583, ATCC) cells were cultured in Dulbecco's modified Eagle's medium or RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen). They were maintained at 37 °C in 5% CO2. Nocodazole, cyclohexamide (Sigma), MG132, U0126, LY294002 (Cayman Chemical Co., Ann Arbor, MI), SB203580, PD98059, and SP600125 (Cayman Chemical Co.) were dissolved in DMSO.
Tissue Collection
Paraffin-embedded tumor specimens (n = 51) were obtained from patients with lung cancer who resided in southern Taiwan. Patients were recruited at the National Cheng Kung University Hospital between 2005 and 2010. All patients signed a consent form. Clinical and pathological information was obtained from medical records and pathology reports. Disease staging was performed according to the TNM system of the American Joint Committee on Cancer/Union Internationale Contre le Cancer (41). The collection of tumor specimens and clinical and pathological information was reviewed and approved by the National Cheng Kung University Hospital Institutional Review Board (Tainan, Taiwan).
Animals and Xenograft Models
Six-week-old BALB/c nude mice progeny were purchased from the National Laboratory Animal Center (National Applied Research Laboratories, Taipei, Taiwan). The mice were fed standard laboratory chow and water ad libitum in the Laboratory Animal Center of National Cheng Kung University. They were raised and cared for in a pathogen-free environment according to the guidelines set by the National Science Council, Taiwan. The experimental protocol adhered to the rules of the Taiwan Animal Protection Act and was approved by the Laboratory Animal Care and Use Committee of National Cheng Kung University. For tumor model development, a suspension (1 × 106 cells/0.1 ml of PBS) of ANXA2-deficient A549 cells (shANXA2-A549) was subcutaneously injected into the right side of the dorsal flanks of six BALB/c nude mice, and their corresponding control cells (shLuc-A549) were injected into the left side of the same mice. We measured the tumor volume by caliper weekly for up to 4 weeks by the following formula: length (mm) × width2 (mm2)/2. After 30 days, we sacrificed the mice and obtained the tumor nodules. For each tumor, a portion was fixed in 4% buffered formaldehyde and processed for histological analysis, and another portion was frozen in liquid nitrogen and stored at −80 °C.
Western Blot Analysis
Cell extracts were separated by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA). After blocking, blots were developed with a series of primary antibodies against ANXA2 (BD Biosciences), p53, p21, growth arrest and DNA damage-inducible protein (GADD45A) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), Cdc2 (Cell Signaling Technology, Danvers, MA), cyclin B1 (MDBio, Inc., Taipei, Taiwan), anti-c-Jun (BD Biosciences), enhanced green fluorescent protein (Santa Cruz Biotechnology, Inc.), and β-actin (Sigma). After washing twice with PBS, blots were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Millipore) and developed using an ECL development kit (Pierce).
siRNA and Lentiviral-based shRNA Transfection
ANXA2 expression was silenced using commercial ANXA2 stealth siRNA oligonucleotide (Invitrogen, catalogue nos. 146996F05 and 146996F06) in A549 cells. The target sequences of Stealth siRNA oligonucleotide of ANXA2 were as follows: sense, 5′-AUCAGUUCAUAAUCAAUGACAGAGC-3′; antisense, 5′-GCUCUGUCAUUGAUUAUGAACUGAU-3′. A nonspecific scramble siRNA was the negative control. HtrA2 (catalogue no. 35615) was silenced using a commercial siRNA kit (Santa Cruz Biotechnology, Inc.). Transfection was performed by electroporation using a pipette-type microporator (Microporator system, Digital BioTechnology, Suwon, Korea). Non-targeting shRNA control vector (shLuc; TRCN0000072247) and shRNA constructs targeting human ANXA2 (shANXA2; TRCN0000056145 containing 5′-CGGGATGCTTTGAACATTGAA-3′), human p53 (shp53; TRCN0000003753 containing 5′-CGGCGCACAGAGGAAGAGAAT-3′), human GADD45A (TRCN0000062349 containing 5′-CGAATCCACATTCATCTCAAT-3′), and human cyclin-dependent kinase inhibitor 1A (CDKN1A) (TRCN0000287021 containing 5′-CGCTCTACATCTTCTGCCTTA-3′) were purchased from the National RNAi Core Facility (Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taipei, Taiwan). Lentivirus was prepared as described previously (42). Briefly, human TE671 cells were cotransfected with two helper plasmids, pCMVΔR8.91 and pMD.G, plus pLKO.1-puro-shRNA, using GeneJammer transfection reagent (Stratagene, La Jolla, CA). The transfected cells were incubated for 24 h, and then the medium was replaced with fresh medium. Cell supernatants containing the viral particles were harvested at 36, 48, 60, and 72 h after transfection. The supernatants were filtered using a 0.45-μm low protein-binding filter and concentrated by centrifugation at 20,000 × g at 4 °C for 3 h using a JA25.50 (Beckman Coulter Inc., Brea, CA) rotor. Supernatant from TE671 cells was used to infect A549 cells.
Colony Formation Assay
Colony formation was performed using a colony formation assay as described previously (43). A total of 1 × 103 A549 cells were plated into 100-mm Petri dishes. After incubation for 10 days, colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reagent (1 mg/ml), and the number of colonies was counted. After counting, the cells were dissolved in DMSO, and the absorbance at 570 nm was detected using a microplate reader (SpectraMax 340PC, Molecular Devices, Sunnyvale, CA). The data were analyzed using Softmax Pro software (Molecular Devices).
WST-8 Assay
To determine cell proliferation, a colorimetric assay (Cell Counting Kit 8; Dojindo Molecular Technologies, Kumamoto, Japan) was used according to the manufacturer's instructions. A microplate reader (SpectraMax 340PC) was used to measure the absorbance at 450 nm, and the data were analyzed using Softmax Pro software.
Lactate Dehydrogenase Assay
To evaluate cell damage, lactate dehydrogenase activity was assayed using a colorimetric assay (cytotoxicity detection kit; Roche Applied Science) according to the manufacturer's instructions. Aliquots of the culture medium were transferred to 96-well microplates. A microplate reader (Spectra MAX 340PC) was used to measure the absorbance at 620 nm with a reference wavelength of 450 nm, and the data were analyzed with Softmax Pro software.
Cell Cycle Analysis
The cell cycle and apoptosis was analyzed using nuclear propidium iodide (Sigma-Aldrich) plus RNase staining and then analyzed using flow cytometry (FACSCalibur, BD Biosciences) with excitation set at 488 nm and emission detected at the FL-2 channel (565–610 nm). For cell cycle analysis, the distribution of cells in different phases of the cell cycle was calculated using MetaMorph software (Molecular Devices, Downingtown, PA). For apoptotic analysis, the samples were analyzed using CellQuest Pro 4.0.2 software (BD Biosciences), and quantification was performed using WinMDI 2.8 software (Scripps Institute, La Jolla, CA). Apoptosis levels were reported as percentages of sub-G1. Further, the mitotic cells were analyzed by two-dimensional flow cytometry using mitotic marker MPM-2 and a DNA content marker (44).
Immunostaining
For tissue staining, the tumor specimens were fixed in 10% neutral formalin, embedded in paraffin, and cut into 5-μm-thick sections. The sections were deparaffinized, rehydrated, and subjected to heat-induced antigen retrieval by boiling for 10 min in 10 mm sodium citrate. Subsequently, the specimens were incubated with the primary antibodies against ANXA2, Ki-67, p53, or active caspase-3 (Santa Cruz Biotechnology, Inc.) (1:100) at 4 °C for 16 h, followed by Alexa Fluor 488/594-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR) and Alexa Fluor 488/594-conjugated goat anti-Rabbit IgG (Molecular Probes) staining. To quantify the histological changes, morphometric analysis of photomicrographs of the tumor sections was performed using ImageJ software (version 1.41o) from W. Rasband (National Institutes of Health, Bethesda, MD). The fluorescent intensities of each specimen were evaluated, and a relative intensity score less than the mean value of total tumors (n = 54) was considered as lower expression, whereas a score of more than the mean value was considered as high expression. All of the immunostaining results were reviewed and scored independently by two pathologists. For cell staining, the cells were fixed in 3.7% formaldehyde in PBS for 10 min. After washing twice with PBS, the cells were incubated with primary antibodies against ANXA2 (Abcam, Cambridge, MA), Ki-67, p53, phospho-ERK Thr-202/Tyr-204, phospho-Akt Ser-473, phospho-p38MAPK Thr-180/Tyr-182, and phospho-JNK Thr-183/Tyr-185 (Cell Signaling Technology) followed by Alexa Fluor 488/594-conjugated goat anti-rabbit IgG and Alexa Fluor 594-conjugated goat anti-mouse IgG (Molecular Probes) staining or MPM2 with or without PI for DNA content staining and incubated at 4 °C overnight. To observe nuclear staining, 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich)-stained cells were observed using a fluorescence microscope (BX51, Olympus, Tokyo, Japan). For flow cytometric analysis, the samples were acquired with FACSCalibur with excitation set at 488 and 633 nm; emission was detected using the FL-1 (515–545 nm) and FL-2 (565–610 nm) channels. The samples were analyzed using CellQuest Pro 4.0.2 software, and quantification was performed using WinMDI 2.8 software.
Microarray
Microarray analysis was performed on A549 cells harvested 2 days after siRNA electroporation. Cells were lysed in TRIzol reagent (Invitrogen), and total RNA was isolated from the aqueous phase according to the protocol of the manufacturer. The subsequent process was performed in the Microarray and Gene Expression Analysis Core Facility (National Research Progress for Genomic Medicine), where a HG-U133 Plus 2 microarray chip (Affymetrix) was hybridized. The data were analyzed using GeneSpring GX 11.5 (Agilent Technologies) and DAVID (45, 46). The data were initially normalized by robust multiarray average algorithms. Statistically significant data were selected by the one-way analysis of variance test (p < 0.05) between duplicate data for A549, scrambled A549 and siANXA2 A549 cell lines. The genes were filtered using volcano plots (with a cut-off value of 0.1), and genes up-regulated and down-regulated more than 1.5-fold were considered for gene list preparation. The gene lists were further processed using the DAVID Bioinformatics Resources 6.7 Web site to identify statistically overrepresented GO term/pathway associations. In this study, a GO term/pathway was considered significantly enriched if its p value was less than 0.05 adjusted for multiple hypothesis testing. The expression patterns of the genes found in selected overrepresented GO terms were plotted in heat maps using R with the gplots (47) package using Euclidean hierarchical clustering of these genes.
Real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the protocol of the manufacturer and was reverse-transcribed with the ReveretAidTM first strand cDNA synthesis kit (Fermentas, Vilnius, Lithuania). The cDNA was PCR-amplified with Taq polymerase (Fermentas). mRNA samples were analyzed with SYBR Green-based real-time quantitative RT-PCR (Applied Biosystems, Foster City, CA) with β-actin as the reference gene in each reaction. The p53 and β-actin primers, designed by the Primer 3 program (Steve Rozen and Helen Skaletsky, Whitehead Institute for Biomedical Research, Cambridge, MA), were as follows: p53, 5′-TTTGCGTGTGGAGTATTTGGATG-3′ (sense) and 5′-CCAGTGTGATGATGGTGAGG-3′ (antisense); GADD45A, 5′-GAGAGCAGAAGACCGAAAGC-3′ (sense) and 5′-TGGATCAGGGTGAAGTGGA-3′ (antisense); CDKN1A, 5′-AGGGGACAGCAGAGGAAGAC-3′ (sense) and 5′-GGCGTTTGGAGTGGTAGAAA-3′ (antisense); and β-actin, 5′-ACTGCCGCATCCTCTTCCTC-3′ (sense) and 5′-TGCCACAGGATTCCATACCC-3′ (antisense). The experiments were conducted in triplicate, and the results were expressed as 2(gene of interest of cycles − β-actin number of cycles).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using reagents commercially obtained from Millipore according to the protocol of the manufacturer with modifications. Briefly, the cells were cross-linked in 1% formaldehyde for 10 min at room temperature. After washing twice with PBS containing protease inhibitors, the cells were lysed in 200 μl of SDS-lysis buffer, and the chromatin/DNA was sonicated to around 500 bp. Immunoprecipitations were performed by incubating the sheared chromatin (2 × 106 cells) overnight at 4 °C with 5 μg of antibody followed by binding to 60 μl of salmon sperm DNA/protein G-Sepharose beads. The antibodies used in these experiments were anti-c-Jun (BD Biosciences) and anti-mouse normal IgG (Millipore). The immunoprecipitates were washed and eluted in a solution containing 1% SDS, 0.1 m NaHCO3 and incubated overnight at 65 °C with 20 μl of 5 m NaCl to reverse the cross-linking. The DNA was purified using phenol/chloroform/isoamyl alcohol, and each fragment was PCR-amplified using the following sets of primers: p53, 5′-GCTGAGAGCAAACGCAAAAG-3′ (sense) and 5′-GAAATGGAGTTGGGGAGGAG-3′ (antisense).
Statistical Analysis
Values are expressed as means ± S.D. Groups were compared using Student's two-tailed unpaired t test with Prism software (GraphPad Software, La Jolla, CA) or one-way analysis of variance followed by Dunnet's post hoc test, as appropriate. Statistical significance was set at p < 0.05. Cumulative disease-free survival curves for patients were plotted using the Kaplan-Meier method with commercially available statistical software (SPSS 18.0 for Windows; Systat Software, Inc., San Jose, CA).
RESULTS
ANXA2 Overexpression Predicts Poor Survival in Patients with NSCLC
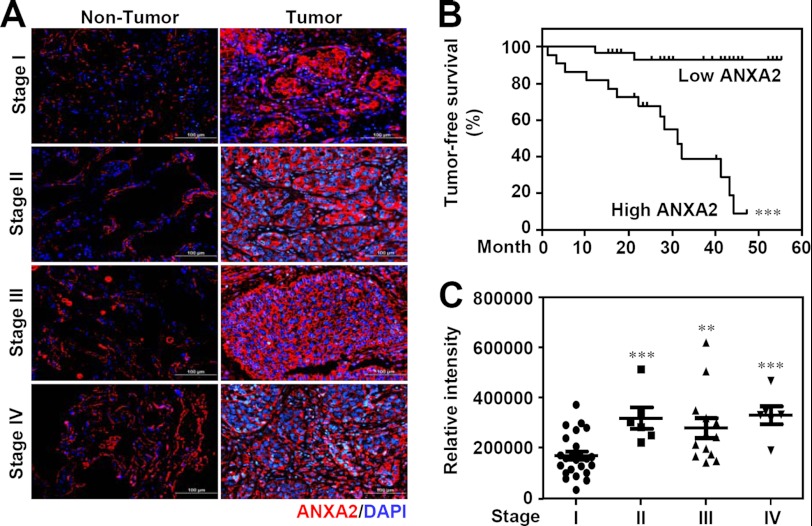
To determine whether ANXA2 expression is associated with lung cancer progression, immunohistochemistry was used to examine ANXA2 expression in surgical resections of NSCLC specimens from different clinical stages. We found that ANXA2 is overexpressed in tumors (n = 51) compared with adjacent non-tumor tissues (n = 12) (p < 0.05; Fig. 1A). Kaplan-Meier analysis showed that a high level of ANXA2 (compared with the mean; n = 23) was found in 45% of the tested tumor specimens (n = 51) and was associated with an increased mortality rate (p < 0.001; Fig. 1B) compared with patients with a low level of ANXA2 (compared with the mean) in tumors. Compared with stage I NSCLC patients (n = 26), ANXA2 was significantly increased in stages II (p < 0.001, n = 6), III (p < 0.01, n = 13), and IV (p < 0.001, n = 6) (Fig. 1C). The relationship between ANXA2 expression and various clinicopathological factors, including age (p = 0.110), sex (p = 0.429), histological type (p = 0.758), tumor stage (p = 0.006), tumor status (p = 0.064), lymph node status (p = 0.047), distal metastasis status (p = 0.045), tumor differentiation (p = 0.539), and recurrence status (p = 0.004), are summarized in Table 1. These results show that overexpression of ANXA2 results in a worse prognosis in NSCLC patients.
FIGURE 1.
ANXA2 expression in lung tumors and its relationship with survival in patients with NSCLC. A, representative immunohistochemical analysis of ANXA2 (red) in formalin-fixed, paraffin-embedded NSCLC tissues from non-tumor (left) and tumor (right) tissues from clinical stages I, II, III, and IV. DAPI (blue) was used for nuclear staining. Scale bar, 100 μm. B, Kaplan-Meier survival curves of tumor-free lung cancer patients with high ANXA2 (n = 26) or low ANXA2 (n = 28) levels, shown as relative intensities. The differences in the curves were assessed using a log-rank test. ***, p < 0.001. C, comparison of ANXA2 expression with clinical stage progression of NSCLC (stage I, n = 26; stage II, n = 7; stage III, n = 14; stage IV, n = 7). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
TABLE 1.
Relationship between ANXA2 expression and clinicopathologic factors in 51 lung cancer patients
| Characteristic | Low ANXA2 (n = 28) | High ANXA2 (n = 23) | p valuea |
|---|---|---|---|
| Age | 0.110 | ||
| Years (mean ± S.D.) | 65 ± 13 | 59 ± 10 | |
| Sex | 0.429 | ||
| Male | 11 | 11 | |
| Female | 17 | 12 | |
| Histological type | 0.758 | ||
| Adenocarcinoma | 22 | 16 | |
| Squamous cell carcinoma | 5 | 6 | |
| Large cell carcinoma | 1 | 1 | |
| Stageb | 0.006 | ||
| I | 20 | 6 | |
| II | 1 | 5 | |
| III | 6 | 7 | |
| IV | 1 | 5 | |
| Tumor status | 0.064 | ||
| T1-2 | 26 | 17 | |
| T3-4 | 2 | 6 | |
| Lymph node status | 0.047 | ||
| N0 | 22 | 12 | |
| N1–N3 | 6 | 11 | |
| Distal metastasis status | 0.045 | ||
| M0 | 27 | 18 | |
| M1 | 1 | 5 | |
| Tumor differentiation | 0.539 | ||
| Well | 9 | 5 | |
| Moderate | 12 | 8 | |
| Poor | 5 | 8 | |
| Unknown | 2 | 2 | |
| Recurrence status | 0.004 | ||
| No | 22 | 9 | |
| Yes | 6 | 14 | |
a p value for age was derived from a two-tailed Student's t test; other p values were derived with a two-tailed Pearson's χ2 test.
b The tumor stage, tumor status, lymph node status, distal metastasis status, and tumor differentiation were classified according to the international system for staging lung cancer.
ANXA2 Knockdown Causes Defects in A549 Cell-derived Tumor Growth in Vivo and A549 Cell Proliferation in Vitro
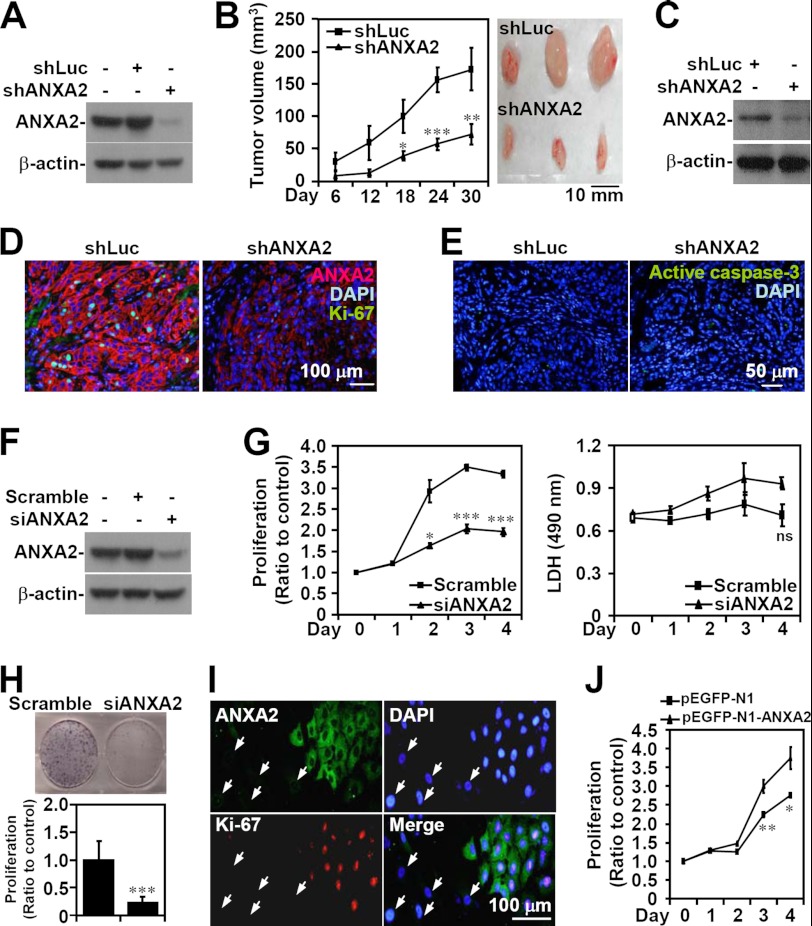
To determine the role of ANXA2 in lung tumorigenesis, lentivirus-based shRNA was used to knock down ANXA2 in A549 cells (shANXA2), as demonstrated by Western blot analysis (Fig. 2A). After subcutaneous injection of either shANXA2-transfected A549 cells or control luciferase shRNA (shLuc)-transfected A549 cells for 30 days, a marked decrease in tumor growth was present in the shANXA2 group (p < 0.05; Fig. 2B). Western blot analysis confirmed the lack of ANXA2 expression in shANXA2-transfected, A549 cell-derived tumor nodules (Fig. 2C). Immunostaining showed that the proliferation marker Ki-67 was considerably reduced in shANXA2-transfected, A549 cell-derived tumors (Fig. 2D); however, there was no detectable caspase-3 activation (Fig. 2E). These data suggest that ANXA2 knockdown restricts tumor growth in vivo without inducing apoptosis.
FIGURE 2.
ANXA2 controls cell growth, but not cell survival, in NSCLC. ANXA2 (ANXA2) was silenced in human NSCLC A549 cells by lentivirus-based short hairpin transfection (shANXA2). An shRNA targeting luciferase (shLuc) was used as a negative control. Western blot analysis was used to detect the expression of ANXA2 in transfected A549 cells (A) and tumors from BALB/c nude mice 30 days postinoculation with shLuc- or shANXA2-transfected cells (C). B, tumor growth was analyzed in xenografts with a subcutaneous injection of A549 cells transfected with shLuc or shANXA2 for the indicated time. Tumor volume was calculated (n = 5/group), and the representative morphology of the tumors was photographed (n = 3/group). D and E, sections of subcutaneous tumor were stained for ANXA2 (red), Ki-67 (green), active caspase-3 (green), and nuclei using DAPI (blue). F, small interfering RNA (siANXA2) was also transfected to silence ANXA2 in A549 cells. A non-targeting siRNA (scramble) was used as a negative control. Western blot analysis was used to detect the expression of ANXA2. G, after transfection for the indicated time, cell proliferation and cytotoxicity were determined by WST-8 and lactate dehydrogenase assays, respectively. The data are the means ± S.D. (error bars) of triplicate cultures. *, p < 0.05; ***, p < 0.001 compared with the control. ns, not significant. H, colony formation was used to measure the growth of siANXA2-transfected A549 cells 10 days post-transfection. The data are the means ± S.D. of triplicate cultures. ***, p < 0.001 compared with scramble. I, immunostaining followed by fluorescence microscopy was used to determine the expression of ANXA2 (green) and Ki-67 (red) in siANXA2-transfected A549 cells. The arrows indicate successfully transfected cells. DAPI (blue) was used for nuclear staining. One representative image of three individual experiments is shown. J, ANXA2 was overexpressed in A549 cells by pEGFP-N1-ANXA2 transfection. A WST-8 assay was used to measure the growth of transfected cells. pEGFP-N1 was used as the negative control. For Western blot analysis, β-actin was used as an internal control. One representative data set of three individual experiments is shown.
To further confirm these findings and to study the effects of ANXA2 silencing on lung cancer cell proliferation in vitro, we used small interfering RNA (siRNA) to transiently target ANXA2 (siANXA2) expression in A549 cells, as confirmed by Western blot analysis (Fig. 2F). A WST-8 assay showed that ANXA2 knockdown significantly (p < 0.05) inhibited A549 cell proliferation compared with the scrambled control (Fig. 2G). A lactate dehydrogenase activity assay showed that ANXA2 silencing did not cause cytotoxicity. A colony formation assay also confirmed that A549 cell growth was inhibited by ANXA2 silencing (p < 0.001; Fig. 2H). Immunostaining indicated that A549 cells transfected with siANXA2 showed less Ki-67 staining, suggesting a key role for ANXA2 in cell proliferation (Fig. 2I). To further investigate the role of ANXA2 in cell proliferation, a WST-8 assay showed that ANXA2 overexpression significantly (p < 0.01) promoted A549 cell proliferation compared with the scrambled control (Fig. 2J). In addition, we found that ANXA2 knockdown decreased EGF-enhanced A549 cell proliferation in vitro (data not shown). These results indicate that ANXA2 deficiency retards lung cancer cell proliferation in vitro without causing cytotoxicity.
ANXA2 Silencing Induces Cell Cycle Arrest at G2 Phase in a Partially p53-regulated Manner
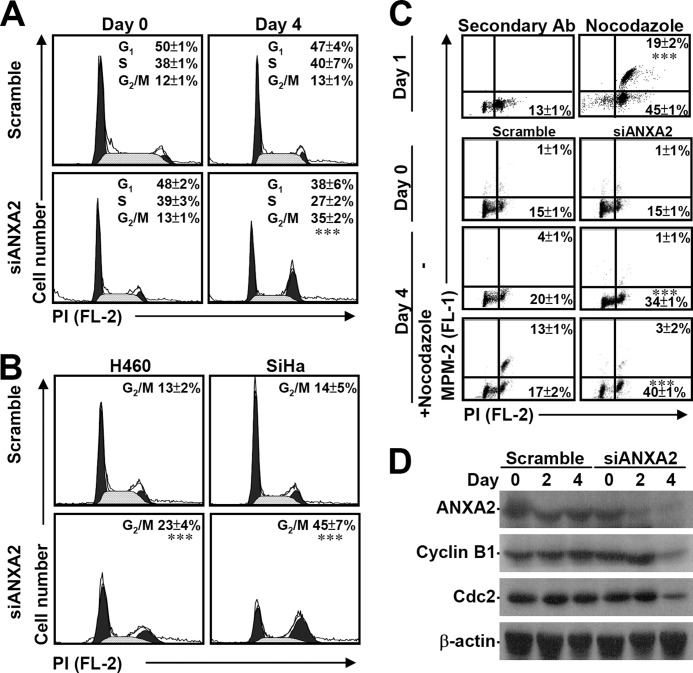
ANXA2 plays a role in tumor hyperproliferation (19, 26, 48); however, the mechanism underlying this control of cell proliferation is still unknown. A β-galactosidase assay demonstrated that although ANXA2 knockdown inhibited cell growth, it did not result in senescence in A549 cells (data not shown). PI staining followed by flow cytometric analysis demonstrated that the loss of ANXA2 significantly (p < 0.001) causes cell cycle arrest at G2/M phase (Fig. 3A). We also demonstrated that ANXA2 silencing generally and significantly (p < 0.001) caused cell cycle arrest at G2/M phase in human large cell lung cancer-derived H460 cells and human cervical cancer-derived SiHa cells (Fig. 3B). Combined immunostaining for MPM-2, an M phase marker protein, showed that ANXA2 silencing specifically and significantly (p < 0.001) caused cell cycle arrest at G2 phase (Fig. 3C). This finding is inconsistent with results obtained with nocodazole, which inhibits mitosis and cytokinesis by interfering with microtubule polymerization at M phase. Even with nocodazole treatment, knockdown of ANXA2 significantly caused G2 arrest. In a synchronized model using aphidicolin, ANXA2 knockdown also induced G2/M arrest (data not shown). Moreover, the expression of cyclin B1 and Cdc2, which are required for cells to enter M phase, was down-regulated under ANXA2-deficient conditions (Fig. 3D). These results indicate that ANXA2 knockdown leads to cell cycle arrest at G2 phase, not at M phase.
FIGURE 3.
ANXA2 knockdown causes G2 arrest. A small interfering RNA (siANXA2) was used to silence ANXA2 in A549, H460, and SiHa cells. A non-targeting siRNA (scramble) was used as a negative control. A and B, the cell cycle stages (G1, S, and G2/M) of siANXA2-transfected cells were determined using PI staining followed by flow cytometry 4 days post-transfection. One representative data set and the percentage of cells in the G2/M phase are shown as the means ± S.D. from three individual experiments with triplicate cultures. ***, p < 0.001 compared with scramble. C, the M phase cells were determined by analysis of DNA content (PI) and MPM2 staining 4 days post-transfection with or without nocodazole treatment given at 3 days post-transfection. Nocodazole treatment in A549 cells for 24 h was used as a positive control. The percentages of G2 and M phase (MPM2-positive) cells are shown as the means ± S.D. from three individual experiments with triplicate cultures. ***, p < 0.001 compared with scramble. D, Western blotting was used to determine the expression of ANXA2, cyclin B1, and Cdc2 for the indicated times. β-Actin was the internal control. The data shown are representative of three individual experiments.
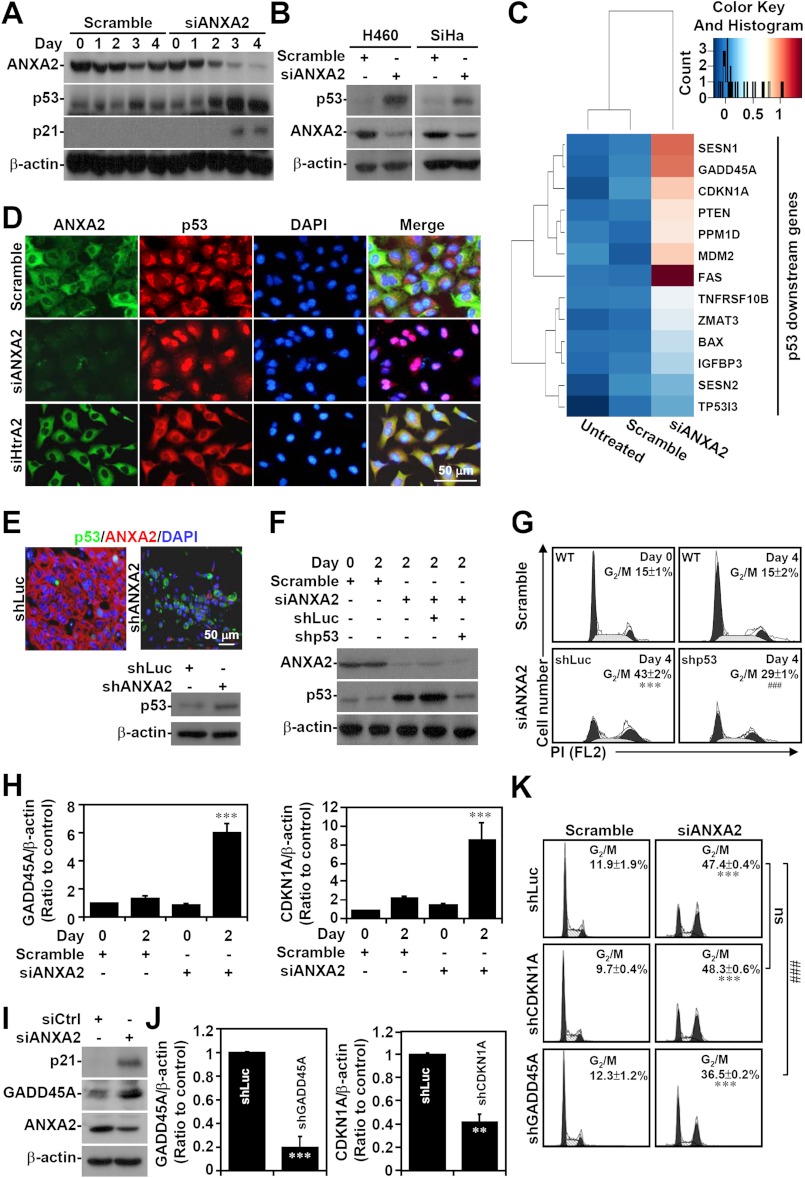
To determine the molecular mechanisms underlying the G2 arrest caused by ANXA2 silencing, we analyzed the involvement of the cyclin-dependent kinase inhibitor p53, which is a general regulator of G2 arrest (49). Remarkably, Western blot (Figs. 4, A and B) and cDNA microarray analyses (Fig. 4C) showed that p53 and its downstream genes, including p21, were induced in ANXA2-deficient A549, H460, and SiHa cells. Notably, ANXA2 knockdown specifically caused p53 translocation from the cytoplasm to the nucleus in A549 cells in vitro (Fig. 4D). Immunostaining and Western blotting confirmed that p53 was also induced in shANXA2-transfected, A549 cell-derived tumors (Fig. 4E). To further characterize whether p53 mediates G2 arrest, the expression of both p53 and ANXA2 was silenced in A549 cells (Fig. 4F). Using PI staining followed by flow cytometry, we found that simultaneous p53 and ANXA2 knockdown resulted in a decrease in total G2/M cells compared with ANXA2 knockdown alone (p < 0.001; Fig. 4G). Because p53 was induced, we next investigated the downstream cell cycle genes controlled by p53. Microarray and bioinformatics analyses showed increased expression of CDKN1A (p21) and GADD45A in ANXA2-silenced A549 cells. qRT-PCR (p < 0.001; Fig. 4H) and Western blot analysis (Fig. 4I) confirmed the increase in the mRNA and protein levels of these p53-regulated factors. Notably, knockdown of GADD45A, but not of CDKN1A, partially reversed the G2/M arrest induced by ANXA2 deficiency (p < 0.001; Fig. 4, J and K). These results demonstrate that ANXA2 knockdown induces G2 arrest in part through p53 and GADD45A.
FIGURE 4.
p53 is partially responsible for G2 arrest in ANXA2-deficient cells. Small interfering RNA (siANXA2) was used to silence ANXA2 in A549, H460, and SiHa cells. A non-targeting siRNA (scramble) was used as a negative control. A and B, Western blotting was used to determine the expression of ANXA2, p53, and p21 at the indicated times. C, the gene expression of A549 cells lacking ANXA2 (siANXA2) or the control (scramble) was analyzed by hierarchical clustering 2 days post-transfection. The data for transcripts with at least a 2-fold difference are shown in a heat map. Selected p53 downstream genes are labeled on the right. D and E, immunostaining followed by fluorescence microscopy was used to determine the expression of ANXA2 (green or red) and p53 (red or green) in scramble-, siANXA2-, and siHtrA2-transfected A549 cells 2 days post-transfection, as well as in shLuc- or shANXA2-transfected A549-derived tumor nodules 30 days postinoculation. DAPI (blue) was used for nuclear staining. Western blot analysis showed the expression of p53 in shLuc- or shANXA2-transfected A549-derived tumor nodules. One representative image obtained from one of three individual experiments is shown. F, p53 was silenced in ANXA2-deficient A549 cells with a lentivirus-based shRNA approach. shLuc was used as a negative control. Western blot analysis was used to detect the expression of ANXA2 and p53 2 days post-transfection. G, the cell cycle was analyzed using PI staining followed by flow cytometry 4 days post-transfection. One representative data set and the percentages of cells in the G2/M phase are shown as the means ± S.D. from three individual experiments with triplicate cultures. H, ANXA2 was silenced by siANXA2 in A549 cells. Real-time RT-PCR was used to determine the mRNA expression of GADD45A and CDKN1A in ANXA2-deficient A549 cells 2 days post-transfection. A non-targeting siRNA (scramble) was used as a negative control. The data are shown as the mean ± S.D. (error bars) of the -fold change relative to untreated cells in triplicate cultures. I, Western blotting showed the protein expression of ANXA2, CDKN1A (p21), and GADD45A in ANXA2-deficient A549 cells 2 days post-transfection. J, CDKN1A (p21) and GADD45A was silenced in ANXA2-deficient A549 cells with a lentivirus-based shRNA approach. shLuc was used as a negative control. Real-time RT-PCR was performed to determine the mRNA expression of GADD45A and CDKN1A. The data are shown as the means ± S.D. of the -fold change relative to untreated cells from triplicate cultures. K, the cell cycle was analyzed 4 days post-transfection by using PI staining followed by flow cytometry. One representative data set and the percentages of cells in the G2/M phase are shown as the means ± S.D. from three individual experiments with triplicate cultures. **, p < 0.01; ***, p < 0.001 compared with the scramble or shLuc control group. ###, p < 0.001 compared with the shLuc group. ns, not significant. For Western blot analysis, β-actin was used as an internal control. One representative data set obtained from three individual experiments is shown.
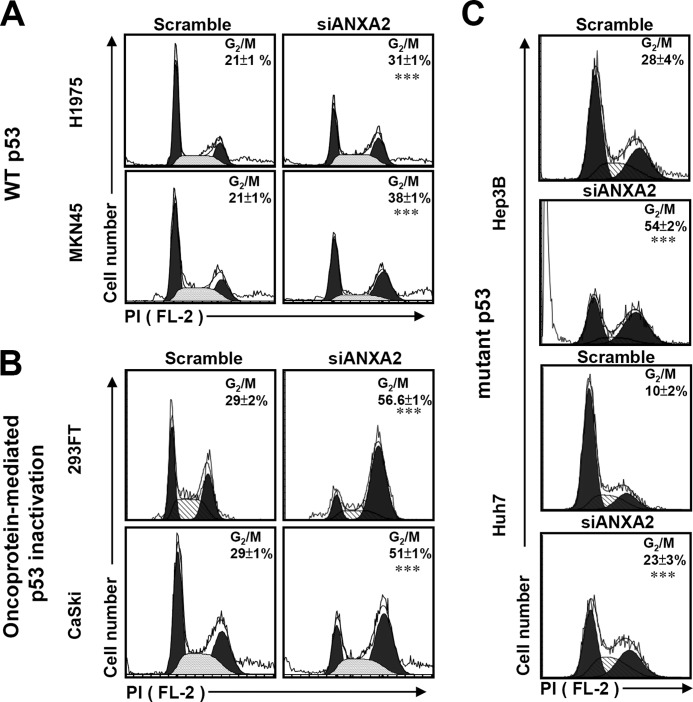
To further clarify the role of p53, we found that knockdown of ANXA2 caused G2/M arrest not only in p53 wild-type cells (H1975 and MKN45) (p < 0.001; Fig. 5A) but also in cells with oncoprotein-mediated p53 inactivation (293FT and CaSki) (p < 0.001; Fig. 5B) or a p53 mutation (Hep3B and Huh7) (p < 0.001; Fig. 5C). These data suggest that ANXA2 regulates the G2/M transition not only through the repression of p53 but also through a p53-independent pathway.
FIGURE 5.
Knockdown of ANXA2 causes G2/M arrest in p53-inactivated and p53 mutant cells. siANXA2 or scramble was transfected into either the wild-type (WT) p53 cell lines H1975 and MKN45 (A), the oncoprotein-mediated p53-inactivated cell lines 293FT and CaSki (B), or the mutant p53 cell lines Hep3B and Huh7 (C). The cell cycle stages were determined using PI staining followed by flow cytometric analysis, and the percentages of G2/M cells are shown as the means ± S.D. from three individual experiments with triplicate cultures. ***, p < 0.001 compared with the scramble group.
JNK Is Inactivated by ANXA2 Deficiency
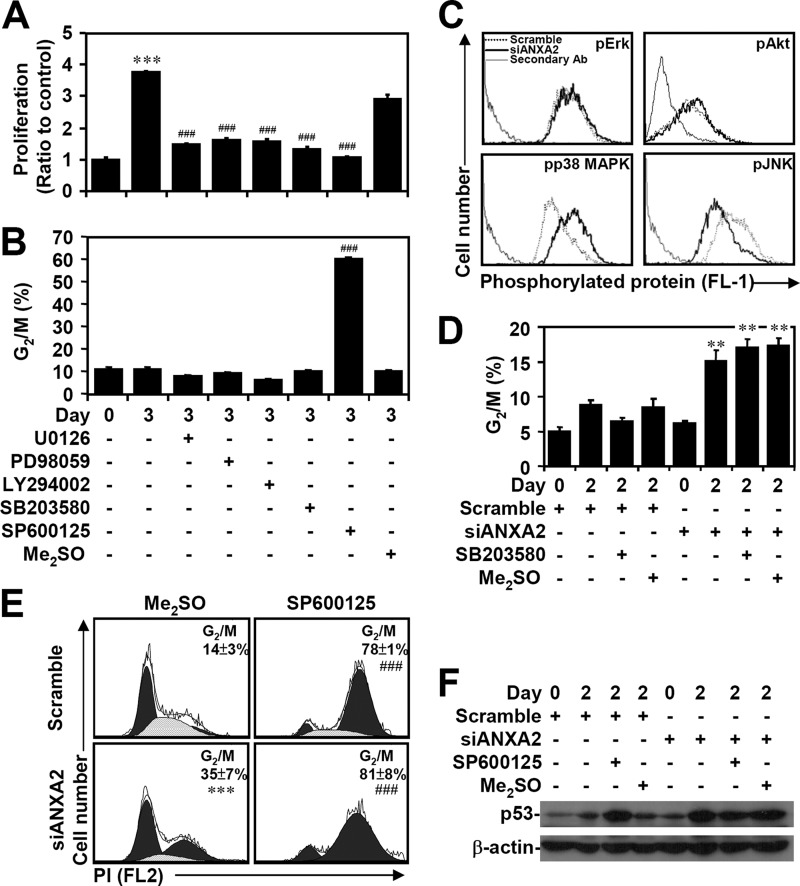
We next examined the possible mechanism of ANXA2-regulated p53 suppression. To address this issue, several known p53 expression pathways, including transcriptional regulation in particular, were investigated. To determine the role of MAPKs and Akt, which are involved in A549 lung cancer cell proliferation (50–52), we used the MEK inhibitors U0126 and PD98059, the PI3K inhibitor LY294002, the p38 MAPK inhibitor SB203580, and the JNK inhibitor SP600125. A WST-8 assay showed that all of these inhibitors retarded cell proliferation (p < 0.001; Fig. 6A). A cell cycle assay using PI staining followed by flow cytometric analysis demonstrated that only SP600125 treatment significantly (p < 0.001) caused G2/M arrest (Fig. 6B). MPM2 immunostaining further demonstrated that JNK inhibition caused cell cycle arrest at G2 phase (data not shown). Notably, immunostaining and flow cytometric analysis showed that ANXA2 silencing caused a decrease in the phosphorylation of JNK (Thr-183/Tyr-185) but not that of ERK (Thr-202/Tyr-204) or Akt (Ser-473). In contrast, ANXA2 silencing caused an increase in the phosphorylation of p38 MAPK (Thr-180/Tyr-182) (Fig. 6C). However, SB203580 treatment did not inhibit G2/M arrest in ANXA2-deficient A549 cells (Fig. 6D). Treatment with the JNK inhibitor SP600125 significantly (p < 0.001) caused cell cycle arrest at G2/M phase in ANXA2 knockdown cells compared with ANXA2 knockdown alone (Fig. 6E). However, p53 expression did not change significantly after SP600125 treatment (Fig. 6F). These results suggest that ANXA2 knockdown causes JNK inactivation-related G2/M arrest through an unknown mechanism.
FIGURE 6.
Inactivation of JNK results in G2 arrest caused by ANXA2 deficiency. A549 cells were treated with the indicated inhibitors for 3 days. WST-8 assays (A) and PI staining followed by flow cytometry (B) were used to evaluate cell proliferation and the cell cycle, respectively. The results are shown as the means ± S.D. (error bars) of the -fold change relative to untreated cells or to the percentages of G2/M cells obtained from three cultures. ***, p < 0.001 compared with the untreated group at day 0. ###, p < 0.001 compared with the untreated group at day 3. C, immunostaining was used to determine the expression of phospho-ERK Thr-202/Tyr-204 (pErk), phospho-Akt Ser-473 (pAkt), phospho-p38 MAPK Thr-180/Tyr-182 (pp38 MAPK), and phospho-JNK Thr-183/Tyr-185 (pJNK) in A549 cells with or without scramble or siANXA2 2 days post-transfection. One data set representative of three individual experiments is shown. D, ANXA2 was silenced in A549 cells using siANXA2. Twenty-four hours after transfection, cells were treated with the p38 MAPK inhibitor SB203580 for an additional 24 h. DMSO and scrambled siRNA were used as negative controls. The cell cycle stages were determined using PI staining followed by flow cytometric analysis, and the percentages of G2/M cells are shown as the means ± S.D. from three individual experiments with triplicate cultures. **, p < 0.01 compared with scramble. A549 cells were transfected with siANXA2, incubated for 24 h, and then treated with the JNK inhibitor SP600125 for another 24 h. DMSO and scrambled siRNA were used as negative controls. E, the cell cycle phase was determined using PI staining followed by flow cytometric analysis, and the percentages of G2/M cells are shown as the means ± S.D. from three individual experiments with triplicate cultures. ***, p < 0.001 compared with scramble. ###, p < 0.001 compared with the DMSO group. F, Western blot analysis of p53 expression. β-Actin was the internal control. The data shown are representative of three individual experiments.
ANXA2 Silencing Attenuates the JNK/c-Jun-regulated Suppression of p53 Transcription
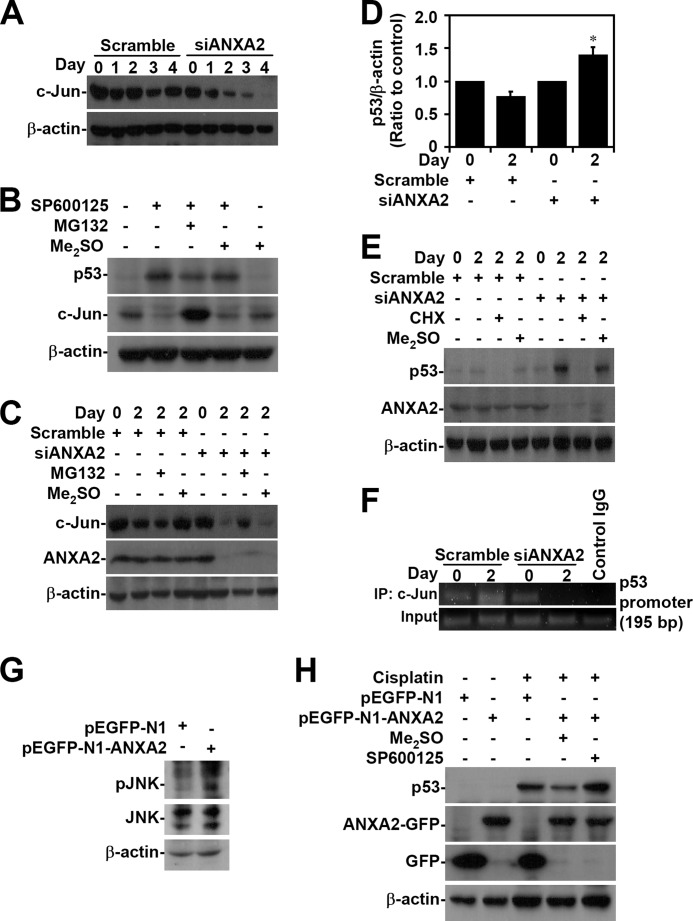
Whereas c-Jun acts as a repressor that negatively regulates p53 mRNA expression through promoter suppression (53, 54), JNK negatively regulates p53 through c-Jun stabilization (55, 56). Western blot analysis demonstrated that c-Jun was down-regulated when ANXA2 was silenced in A549 cells (Fig. 7A). In SP600125-treated A549 cells, JNK inhibition caused p53 induction accompanied by c-Jun degradation, which occurred via an MG132-sensitive, proteasome-mediated pathway (Fig. 7B). The loss of c-Jun caused by silencing of ANXA2 was restored after treatment with the proteasome inhibitor MG132 (Fig. 7C). We next evaluated whether ANXA2 regulates p53 expression transcriptionally and/or via post-translational modification. Real-time RT-PCR showed that ANXA2 silencing significantly (p < 0.05) increased the mRNA expression of p53 in A549 cells (Fig. 7D). Furthermore, cycloheximide treatment effectively inhibited p53 expression in ANXA2-deficient A549 cells (Fig. 7E), suggesting that ANXA2 transcriptionally regulates p53. For chromatin immunoprecipitation, we designed primers for the putative c-Jun binding site in the p53 promoter. The results showed that ANXA2 silencing caused the loss of c-Jun binding to the p53 promoter (Fig. 7F). Notably, overexpression of ANXA2 induced JNK activation (Fig. 7G). Furthermore, A549 cells overexpressing ANXA2 showed JNK-dependent resistance to cisplatin-induced p53 expression (Fig. 7H). These results indicate that ANXA2 negatively regulates p53 mRNA expression through c-Jun stabilization following JNK activation.
FIGURE 7.
ANXA2 deficiency causes JNK/c-Jun down-regulation, which causes the loss of transcriptional p53 suppression. A, ANXA2 was silenced with siANXA2 in A549 cells. Western blotting was used to determine the expression of c-Jun at the indicated times. A non-targeting siRNA (scramble) was used as a negative control. B, A549 cells were treated with the JNK inhibitor SP600125 (25 μm) in the absence or presence of the proteasome inhibitor MG132 (1.5 μm) for 24 h. Western blotting was used to determine the expression of p53 and c-Jun. C, ANXA2-deficient A549 cells were treated with MG132 for 48 h. Western blot analysis was used to detect the expression of c-Jun and ANXA2. DMSO and scrambled siRNA were used as negative controls. D, we performed real-time RT-PCR to determine the p53 mRNA expression in ANXA2-deficient A549 cells 2 days post-transfection. The data are shown as the mean -fold change ± S.D. (error bars) normalized to untreated cells grown in triplicate cultures. *, p < 0.05 compared with scramble. E, ANXA2-deficient A549 cells were treated with cycloheximide (CHX; 5 μg/ml) for 48 h. Western blot analysis was used to detect the expression of p53 and ANXA2. DMSO and the scrambled siRNA were used as negative controls. F, chromatin immunoprecipitation was used to determine the binding capacity of c-Jun to p53 promoter in ANXA2-deficient A549 cells. The enrichment of the p53 promoter in immunoprecipitated (IP) c-Jun and the input of c-Jun are shown. IgG was used as a negative control. One representative data set of three individual experiments is shown. G, ANXA2 was overexpressed in A549 cells by pEGFP-N1-ANXA2 transfection. Western blotting showed the expression of phospho-JNK Thr-183/Tyr-185 (pJNK) and JNK. H, pEGFP-N1-ANXA2-transfected A549 cells were treated with cisplatin for 24 h. Western blotting showed the expression of p53 and ANXA2. pEGFP-N1 was used as a negative control. For Western blot analysis, β-actin was the internal control. The data shown are representative of three individual experiments.
DISCUSSION
Aberrant ANXA2 expression has oncogenic effects in several tumor types. We provide evidence that in patients with NSCLC, a poor prognosis for survival is correlated with ANXA2 expression, and this observation is consistent with the results of ANXA2 tissue staining in lung cancer (40). However, the molecular significance of aberrant ANXA2 expression remained unclear. Based on our present results, ANXA2 facilitates cell cycle progression and cell proliferation in part by inhibiting p53 expression in NSCLC cells in vivo and in vitro. Through an unknown mechanism, ANXA2 maintains JNK/c-Jun signaling, which transcriptionally represses p53. Notably, NSCLC cells with aberrant ANXA2 expression inhibit p53-mediated tumor suppression. According to these findings on the tumorigenic role of ANXA2, we speculate that ANXA2 may be a good target for anticancer therapeutics.
ANXA2 knockdown leads to the inhibition of proliferation in multiple types of cancers, including multiple myeloma, prostate tumors, and breast cancer (19, 30, 57). Intratumoral administration of pDrive-shAnxA2-loaded polylactide-co-glycolide nanoparticles decreases xenograft prostate tumor growth in nude mice, demonstrating the oncogenic effects of ANXA2 (57). While our study was being completed, another study (58) reported that ANXA2 knockdown results in a blockade of proliferation and migration in A549 cells, which is consistent with our findings. However, the mechanism by which ANXA2 functions in the cell cycle and in cell proliferation is still unknown. Our findings show that ANXA2 silencing induces p53 expression. We therefore propose a role for p53 in ANXA2-regulated cancer cell proliferation and migration. ANXA2 expression is down-regulated in response to a number of growth-inhibiting conditions, including nutrient deprivation and anticancer drug treatment (59). In contrast, p53 is up-regulated under the same conditions (60, 61). Either ANXA2 knockdown or p53 overexpression induces G2 arrest and growth inhibition (19, 57). This observation raises the possibility that ANXA2 negatively regulates p53 and promotes tumorigenesis. However, the findings of Huang et al. show that p53 overexpression in Anip973 and AGZY-83a lung cancer cells down-regulates ANXA2 prior to apoptosis (62). Cells transfected with an ANXA2 siRNA proliferated more slowly than the control, which is consistent with our findings. Our findings and those of Huang et al. (62) suggest that the activation of p53 leads to the suppression of ANXA2, which in turn activates p53 to a higher level.
p53 up-regulation is classically regarded to be the result of p53 stabilization (63). In this study, p53 mRNA expression after ANXA2 silencing was demonstrated by real-time RT-PCR. Furthermore, p53 protein expression was reduced after cycloheximide treatment. Hence, we investigated the possibility that ANXA2 negatively regulates p53 expression at the transcriptional level. Of the transcription factors that regulate p53, c-Jun represses p53 mRNA expression (53, 64). Notably, c-Jun expression was reduced following JNK inactivation in ANXA2-deficient cells. A ChIP assay also demonstrated that ANXA2 facilitates JNK/c-Jun-mediated p53 repression, whereas ANXA2 silencing causes the loss of c-Jun binding to the p53 promoter as a result of JNK inactivation. Hence, it is possible that c-Jun loss is responsible for the p53 up-regulation in ANXA2-deficient cells, whereas c-Jun knockout causes p53 induction (53–56). We also show that ANXA2 silencing causes p53 nuclear translocation in vitro and induces the expression of genes downstream of p53, including p21, sestrin-1, sestrin-2, GADD45A, PTEN, Mg2+/Mn2+ 1D, MDM2, Fas, Bax, TNF receptor superfamily member 10b, zinc finger matrin-type 3, IGF binding protein 3, and Tp53 inhibitor 3. The mechanism for p53 activation following increased p53 expression in ANXA2-deficient cells requires further investigation.
Simultaneous knockdown of p53 and GADD45A, a downstream gene controlled by p53 (65), can partially reverse siANXA2-induced G2/M arrest in p53 wild-type cells. However, knockdown of ANXA2 also causes G2/M arrest in cells with oncoprotein-mediated p53 inactivation or a natural p53 mutation. Based on these findings, ANXA2 regulates the G2/M transition through p53-dependent, GADD45A-regulated, and p53-independent pathways. GADD45A is also induced by a p53-dependent pathway in response to arachidonic acid metabolites and ultraviolet stimulation, leading to cell cycle arrest (66–68). In addition to p53, the possible regulation of GADD45A by ANXA2 in response to DNA damage remains unclear. The cross-talk between p53, GADD45A, and other ANXA2-associated molecules for such signaling pathways involved in ANXA2-regulated cell cycle and proliferation needs further investigation.
JNK interacts with the δ domain of c-Jun (amino acids 30–57) through strong hydrophobic interactions and phosphorylates c-Jun at Ser-63 and Ser-73, which is sufficient to protect c-Jun from ubiquitination and degradation (69). JNK is often activated in NSCLC and promotes oncogenic transformation (51) because JNK negatively regulates p53 through c-Jun (55, 56). In this study, we show that ANXA2 silencing causes JNK dephosphorylation through an unknown mechanism. JNK is positively regulated by MKK4/7 following the activation of the upstream kinases MEKK1/2 and MLK1/2/4 (70, 71). Generation of aberrant reactive oxygen species causes the activation of apoptosis signaling kinase followed by apoptosis signaling kinase-mediated MKK4/7 and JNK activation (72–74). Notably, oxidative stress induces ANXA2 up-regulation and triggers ANXA2-related cell proliferation and metastasis in renal carcinoma cells (75). Inhibiting reactive oxygen species reduced endogenous activation of JNK in A549 cells; however, ANXA2 silencing did not affect reactive oxygen species generation (data not shown). It has been speculated that aberrant oxidative stress in NSCLC cells may cause ANXA2 up-regulation followed by JNK activation, whereas such oxidative stress has been involved in lung tumorigenesis (76).
Pyk2 is a redox-sensitive tyrosine kinase (77). Previous studies have shown that 64% of NSCLC patients have increased Pyk2 expression in cancers (78). Notably, overexpression of Pyk2 activates JNK (79), whereas Pyk2 is reported to be co-localized with ANXA2 and mediates downstream MEKK4 activation (80). Therefore, redox-sensitive Pyk2 may be a candidate to mediate ANXA2-assocated JNK activation. This hypothesis needs further investigation.
A549 cells are a K-Ras-mutated NSCLC line. Notably, K-Ras signaling could activate JNK through Rac/MAPKKKs/MKK4/7 signaling (81–83), and JNK is required for the oncogenic Ras-induced transformed phenotype, including morphological reversion and inhibition of anchorage-independent growth and low serum growth (84). Rac1 is a GTPase that belongs to the Ras superfamily of small GTP-binding proteins. Further, the complex of Rac1 and ANXA2 is localized to cadherin-mediated cell-cell contacts (85). It is possible that the loss of ANXA2 may retard Rac1 activation followed by JNK inactivation. However, our preliminary data showed that the activation of JNK in A549 cells is independent of Ras and Rac activity (data not shown). According to these possibilities, understanding the possible roles of ANXA2 in mediating reactive oxygen species/apoptosis signaling kinase- and/or Ras/Rac/MAPKKKs-regulated MKK4/7/JNK signaling is critical for future studies to explore oncogenic ANXA2.
As a partner for p11, ANXA2 maintains p11 stability by inhibiting polyubiquitination and degradation at its C terminus (86). Previous studies have shown that p11 is expressed mainly in regions with strong proliferating capacity in melanocytic lesions comprising melanocytic nevi and melanomas (87). Further, knockdown of p11 contributes to a marked inhibition of cell migration, invasiveness, and colony formation in A549 lung cancer cells (88), which is consistent with our unpublished data3 indicating that knockdown of p11 attenuated the proliferation rate of A549 cells. However, the mechanism of p11-regulated cell proliferation and the possible cross-talk with the ANXA2-p11 complex are unclear (89).
In conclusion, our results confirm that ANXA2 knockdown retards cell proliferation following cell cycle arrest. Mechanistically, ANXA2 loss not only induces p53 expression followed by p53-regulated G2 arrest in p53 wild-type cells but also causes p53-independent G2 arrest. Furthermore, JNK is down-regulated in ANXA2-deficient cells and is accompanied by a loss of c-Jun-mediated repression of the p53 promoter, which leads to an increase in p53 transcription. One of the challenges that have resulted from the wealth of knowledge of ANXA2-regulated JNK activity has been how to develop a strategy to manipulate ANXA2 while ANXA2 is overexpressed as an oncogenic factor for cell proliferation and p53 is repressed in NSCLC cells. Therefore, targeting ANXA2 could inhibit the growth of NSCLC.
Acknowledgments
We thank the Immunobiology Core, Research Center of Clinical Medicine, National Cheng Kung University Hospital, for providing services that included training, technical support, and assistance with experimental design and data analysis using the Flow Cytometry Core facilities. We thank Chia-Hung Liu (Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taiwan) for analyzing the array data using the DAVID software and for plotting the heat map. Computational analyses and data mining were performed using the system provided by the Bioinformatics Core at the National Cheng Kung University, supported by the National Science Council.
This work was supported by National Science Council, Taiwan, Grant 100-2320-B-006-009-MY3.
C.-Y. Wang, C.-L. Chen, Y.-L. Tseng, Y.-T. Fang, Y.-S. Lin, W.-C. Su, C.-C. Chen, K.-C. Chang, Y.-C. Wang, and C.-F. Lin, unpublished data.
- APL
- acute promyelocytic leukemia
- NSCLC
- non-small cell lung cancer
- PI
- propidium iodide.
REFERENCES
- 1. Hajjar K. A., Krishnan S. (1999) Annexin II. A mediator of the plasmin/plasminogen activator system. Trends Cardiovasc Med. 9, 128–138 [DOI] [PubMed] [Google Scholar]
- 2. Johnsson N., Marriott G., Weber K. (1988) p36, the major cytoplasmic substrate of Src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 7, 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camors E., Monceau V., Charlemagne D. (2005) Annexins and Ca2+ handling in the heart. Cardiovasc. Res. 65, 793–802 [DOI] [PubMed] [Google Scholar]
- 4. Hajjar K. A., Jacovina A. T., Chacko J. (1994) An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. J. Biol. Chem. 269, 21191–21197 [PubMed] [Google Scholar]
- 5. Kassam G., Manro A., Braat C. E., Louie P., Fitzpatrick S. L., Waisman D. M. (1997) Characterization of the heparin binding properties of annexin II tetramer. J. Biol. Chem. 272, 15093–15100 [DOI] [PubMed] [Google Scholar]
- 6. Gerke V., Moss S. E. (1997) Annexins and membrane dynamics. Biochim. Biophys. Acta 1357, 129–154 [DOI] [PubMed] [Google Scholar]
- 7. Waisman D. M. (1995) Annexin II tetramer. Structure and function. Mol. Cell Biochem. 149, 301–322 [DOI] [PubMed] [Google Scholar]
- 8. Raynal P., Pollard H. B. (1994) Annexins. The problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim. Biophys. Acta 1197, 63–93 [DOI] [PubMed] [Google Scholar]
- 9. Nygaard S. J., Haugland H. K., Kristoffersen E. K., Lund-Johansen M., Laerum O. D., Tysnes O. B. (1998) Expression of annexin II in glioma cell lines and in brain tumor biopsies. J. Neurooncol. 38, 11–18 [DOI] [PubMed] [Google Scholar]
- 10. Roseman B. J., Bollen A., Hsu J., Lamborn K., Israel M. A. (1994) Annexin II marks astrocytic brain tumors of high histologic grade. Oncol. Res. 6, 561–567 [PubMed] [Google Scholar]
- 11. Menell J. S., Cesarman G. M., Jacovina A. T., McLaughlin M. A., Lev E. A., Hajjar K. A. (1999) Annexin II and bleeding in acute promyelocytic leukemia. N. Engl. J. Med. 340, 994–1004 [DOI] [PubMed] [Google Scholar]
- 12. Mai J., Finley R. L., Jr., Waisman D. M., Sloane B. F. (2000) Human procathepsin B interacts with the annexin II tetramer on the surface of tumor cells. J. Biol. Chem. 275, 12806–12812 [DOI] [PubMed] [Google Scholar]
- 13. Emoto K., Sawada H., Yamada Y., Fujimoto H., Takahama Y., Ueno M., Takayama T., Uchida H., Kamada K., Naito A., Hirao S., Nakajima Y. (2001) Annexin II overexpression is correlated with poor prognosis in human gastric carcinoma. Anticancer Res. 21, 1339–1345 [PubMed] [Google Scholar]
- 14. Emoto K., Yamada Y., Sawada H., Fujimoto H., Ueno M., Takayama T., Kamada K., Naito A., Hirao S., Nakajima Y. (2001) Annexin II overexpression correlates with stromal tenascin-C overexpression. A prognostic marker in colorectal carcinoma. Cancer 92, 1419–1426 [DOI] [PubMed] [Google Scholar]
- 15. Esposito I., Penzel R., Chaib-Harrireche M., Barcena U., Bergmann F., Riedl S., Kayed H., Giese N., Kleeff J., Friess H., Schirmacher P. (2006) Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J. Pathol. 208, 673–685 [DOI] [PubMed] [Google Scholar]
- 16. Sharma M. R., Koltowski L., Ownbey R. T., Tuszynski G. P., Sharma M. C. (2006) Angiogenesis-associated protein annexin II in breast cancer. Selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp. Mol. Pathol. 81, 146–156 [DOI] [PubMed] [Google Scholar]
- 17. Sharma M. R., Rothman V., Tuszynski G. P., Sharma M. C. (2006) Antibody-directed targeting of angiostatin's receptor annexin II inhibits Lewis lung carcinoma tumor growth via blocking of plasminogen activation. Possible biochemical mechanism of angiostatin's action. Exp. Mol. Pathol. 81, 136–145 [DOI] [PubMed] [Google Scholar]
- 18. Mohammad H. S., Kurokohchi K., Yoneyama H., Tokuda M., Morishita A., Jian G., Shi L., Murota M., Tani J., Kato K., Miyoshi H., Deguchi A., Himoto T., Usuki H., Wakabayashi H., Izuishi K., Suzuki Y., Iwama H., Deguchi K., Uchida N., Sabet E. A., Arafa U. A., Hassan A. T., El-Sayed A. A., Masaki T. (2008) Annexin A2 expression and phosphorylation are up-regulated in hepatocellular carcinoma. Int. J. Oncol. 33, 1157–1163 [PubMed] [Google Scholar]
- 19. Bao H., Jiang M., Zhu M., Sheng F., Ruan J., Ruan C. (2009) Overexpression of Annexin II affects the proliferation, apoptosis, invasion, and production of proangiogenic factors in multiple myeloma. Int. J. Hematol. 90, 177–185 [DOI] [PubMed] [Google Scholar]
- 20. Liu J., Rothermund C. A., Ayala-Sanmartin J., Vishwanatha J. K. (2003) Nuclear annexin II negatively regulates growth of LNCaP cells and substitution of Ser-11 and -25 to Glu prevents nucleo-cytoplasmic shuttling of annexin II. BMC Biochem. 4, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X., Zhi H. Y., Zhang J., Wang X. Q., Zhou C. N., Wu M., Sun Y. T., Liu Z. H. (2003) [Expression of annexin II in human esophageal squamous cell carcinoma]. Zhonghua Zhong Liu Za Zhi 25, 353–355 [PubMed] [Google Scholar]
- 22. Gillette J. M., Chan D. C., Nielsen-Preiss S. M. (2004) Annexin 2 expression is reduced in human osteosarcoma metastases. J. Cell Biochem. 92, 820–832 [DOI] [PubMed] [Google Scholar]
- 23. Rodrigo J. P., Lequerica-Fernández P., Rosado P., Allonca E., Garcia-Pedrero J. M., de Vicente J. C. (2011) Clinical significance of annexin A2 down-regulation in oral squamous cell carcinoma. Head Neck 33, 1708–1714 [DOI] [PubMed] [Google Scholar]
- 24. Paciucci R., Torà M., Díaz V. M., Real F. X. (1998) The plasminogen activator system in pancreas cancer. Role of t-PA in the invasive potential in vitro. Oncogene 16, 625–633 [DOI] [PubMed] [Google Scholar]
- 25. Vishwanatha J. K., Chiang Y., Kumble K. D., Hollingsworth M. A., Pour P. M. (1993) Enhanced expression of annexin II in human pancreatic carcinoma cells and primary pancreatic cancers. Carcinogenesis 14, 2575–2579 [DOI] [PubMed] [Google Scholar]
- 26. Chiang Y., Rizzino A., Sibenaller Z. A., Wold M. S., Vishwanatha J. K. (1999) Specific down-regulation of annexin II expression in human cells interferes with cell proliferation. Mol. Cell Biochem. 199, 139–147 [DOI] [PubMed] [Google Scholar]
- 27. Kumble K. D., Iversen P. L., Vishwanatha J. K. (1992) The role of primer recognition proteins in DNA replication. Inhibition of cellular proliferation by antisense oligodeoxyribonucleotides. J. Cell Sci. 101, 35–41 [DOI] [PubMed] [Google Scholar]
- 28. Vishwanatha J. K., Jindal H. K., Davis R. G. (1992) The role of primer recognition proteins in DNA replication. Association with nuclear matrix in HeLa cells. J. Cell Sci. 101, 25–34 [DOI] [PubMed] [Google Scholar]
- 29. Vishwanatha J. K., Kumble S. (1993) Involvement of annexin II in DNA replication. Evidence from cell-free extracts of Xenopus eggs. J. Cell Sci. 105, 533–540 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Guo B., Zhang Y., Cao J., Chen T. (2010) Silencing of the annexin II gene down-regulates the levels of S100A10, c-Myc, and plasmin and inhibits breast cancer cell proliferation and invasion. Saudi Med. J. 31, 374–381 [PubMed] [Google Scholar]
- 31. Jemal A., Siegel R., Ward E., Murray T., Xu J., Smigal C., Thun M. J. (2006) Cancer statistics, 2006. CA Cancer J. Clin. 56, 106–130 [DOI] [PubMed] [Google Scholar]
- 32. Brambilla E., Travis W. D., Colby T. V., Corrin B., Shimosato Y. (2001) The new World Health Organization classification of lung tumors. Eur. Respir. J. 18, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 33. Sun S., Schiller J. H., Spinola M., Minna J. D. (2007) New molecularly targeted therapies for lung cancer. J. Clin. Invest. 117, 2740–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang T., Nelson R. A., Bogardus A., Grannis F. W., Jr. (2010) Five-year lung cancer survival. Which advanced stage nonsmall cell lung cancer patients attain long-term survival? Cancer 116, 1518–1525 [DOI] [PubMed] [Google Scholar]
- 35. Win T., Sharples L., Groves A. M., Ritchie A. J., Wells F. C., Laroche C. M. (2008) Predicting survival in potentially curable lung cancer patients. Lung 186, 97–102 [DOI] [PubMed] [Google Scholar]
- 36. Feigenberg S. J., Hanlon A. L., Langer C., Goldberg M., Nicolaou N., Millenson M., Coia L. R., Lanciano R., Movsas B. (2007) A phase II study of concurrent carboplatin and paclitaxel and thoracic radiotherapy for completely resected stage II and IIIA non-small cell lung cancer. J. Thorac. Oncol. 2, 287–292 [DOI] [PubMed] [Google Scholar]
- 37. Albain K. S., Swann R. S., Rusch V. W., Turrisi A. T., 3rd, Shepherd F. A., Smith C., Chen Y., Livingston R. B., Feins R. H., Gandara D. R., Fry W. A., Darling G., Johnson D. H., Green M. R., Miller R. C., Ley J., Sause W. T., Cox J. D. (2009) Radiotherapy plus chemotherapy with or without surgical resection for stage III non-small-cell lung cancer. A phase III randomized controlled trial. Lancet 374, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato M., Shames D. S., Gazdar A. F., Minna J. D. (2007) A translational view of the molecular pathogenesis of lung cancer. J. Thorac. Oncol. 2, 327–343 [DOI] [PubMed] [Google Scholar]
- 39. Cole S. P., Pinkoski M. J., Bhardwaj G., Deeley R. G. (1992) Elevated expression of annexin II (lipocortin II, p36) in a multidrug-resistant small cell lung cancer cell line. Br. J. Cancer 65, 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brichory F. M., Misek D. E., Yim A. M., Krause M. C., Giordano T. J., Beer D. G., Hanash S. M. (2001) An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 98, 9824–9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langfort R. (2010) [The new recommendation of the 7th edition of TNM classification for Lung Cancer in pathologic assesssment (pTNM)]. Pneumonol. Alergol. Pol. 78, 379–383 [PubMed] [Google Scholar]
- 42. Tsai C. C., Kai J. I., Huang W. C., Wang C. Y., Wang Y., Chen C. L., Fang Y. T., Lin Y. S., Anderson R., Chen S. H., Tsao C. W., Lin C. F. (2009) Glycogen synthase kinase-3β facilitates IFN-γ-induced STAT1 activation by regulating Src homology-2 domain-containing phosphatase 2. J. Immunol. 183, 856–864 [DOI] [PubMed] [Google Scholar]
- 43. Sasaki K., Tsuno N. H., Sunami E., Tsurita G., Kawai K., Okaji Y., Nishikawa T., Shuno Y., Hongo K., Hiyoshi M., Kaneko M., Kitayama J., Takahashi K., Nagawa H. (2010) Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on colon cancer cells. BMC Cancer 10, 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jascur T., Brickner H., Salles-Passador I., Barbier V., El Khissiin A., Smith B., Fotedar R., Fotedar A. (2005) Regulation of p21WAF1/CIP1 stability by WISp39, a Hsp90 binding TPR protein. Mol. Cell 17, 237–249 [DOI] [PubMed] [Google Scholar]
- 45. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID. Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 46. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 47. Warnes G. R., Bolker B., Lumley T. (2009) gplots: Various R programming tools for plotting data. R Package version 2.7.4 [Google Scholar]
- 48. Shiozawa Y., Havens A. M., Jung Y., Ziegler A. M., Pedersen E. A., Wang J., Wang J., Lu G., Roodman G. D., Loberg R. D., Pienta K. J., Taichman R. S. (2008) Annexin II/annexin II receptor axis regulates adhesion, migration, homing, and growth of prostate cancer. J. Cell Biochem. 105, 370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taylor W. R., Stark G. R. (2001) Regulation of the G2/M transition by p53. Oncogene 20, 1803–1815 [DOI] [PubMed] [Google Scholar]
- 50. Greenberg A. K., Basu S., Hu J., Yie T. A., Tchou-Wong K. M., Rom W. N., Lee T. C. (2002) Selective p38 activation in human non-small cell lung cancer. Am. J. Respir. Cell Mol. Biol. 26, 558–564 [DOI] [PubMed] [Google Scholar]
- 51. Khatlani T. S., Wislez M., Sun M., Srinivas H., Iwanaga K., Ma L., Hanna A. E., Liu D., Girard L., Kim Y. H., Pollack J. R., Minna J. D., Wistuba I. I., Kurie J. M. (2007) c-Jun N-terminal kinase is activated in non-small-cell lung cancer and promotes neoplastic transformation in human bronchial epithelial cells. Oncogene 26, 2658–2666 [DOI] [PubMed] [Google Scholar]
- 52. Hollander M. C., Maier C. R., Hobbs E. A., Ashmore A. R., Linnoila R. I., Dennis P. A. (2011) Akt1 deletion prevents lung tumorigenesis by mutant K-ras. Oncogene 30, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schreiber M., Kolbus A., Piu F., Szabowski A., Möhle-Steinlein U., Tian J., Karin M., Angel P., Wagner E. F. (1999) Control of cell cycle progression by c-Jun is p53-dependent. Genes Dev. 13, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stepniak E., Ricci R., Eferl R., Sumara G., Sumara I., Rath M., Hui L., Wagner E. F. (2006) c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 20, 2306–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eferl R., Ricci R., Kenner L., Zenz R., David J. P., Rath M., Wagner E. F. (2003) Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell 112, 181–192 [DOI] [PubMed] [Google Scholar]
- 56. Das M., Jiang F., Sluss H. K., Zhang C., Shokat K. M., Flavell R. A., Davis R. J. (2007) Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 15759–15764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Braden A. R., Kafka M. T., Cunningham L., Jones H., Vishwanatha J. K. (2009) Polymeric nanoparticles for sustained down-regulation of annexin A2 inhibit prostate tumor growth. J. Nanosci. Nanotechnol. 9, 2856–2865 [DOI] [PubMed] [Google Scholar]
- 58. Wang Y. X., Lv H., Li Z. X., Li C., Wu X. Y. (2012) Effect of shRNA-mediated down-regulation of annexin A2 on biological behavior of human lung adencarcinoma cells A549. Pathol. Oncol. Res. 18, 183–190 [DOI] [PubMed] [Google Scholar]
- 59. Wang C. Y., Lin Y. S., Su W. C., Chen C. L., Lin C. F. (2009) Glycogen synthase kinase-3 and Omi/HtrA2 induce Annexin A2 cleavage followed by cell cycle inhibition and apoptosis. Mol. Biol. Cell 20, 4153–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fritsche M., Haessler C., Brandner G. (1993) Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene 8, 307–318 [PubMed] [Google Scholar]
- 61. Shang L., Zhou H., Xia Y., Wang H., Gao G., Chen B., Liu Q., Shao C., Gong Y. (2009) Serum withdrawal up-regulates human SIRT1 gene expression in a p53-dependent manner. J. Cell Mol. Med. 13, 4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang Y., Jin Y., Yan C. H., Yu Y., Bai J., Chen F., Zhao Y. Z., Fu S. B. (2008) Involvement of Annexin A2 in p53-induced apoptosis in lung cancer. Mol. Cell Biochem. 309, 117–123 [DOI] [PubMed] [Google Scholar]
- 63. Kruse J. P., Gu W. (2009) Modes of p53 regulation. Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hollstein M., Hainaut P. (2010) Massively regulated genes. The example of TP53. J. Pathol. 220, 164–173 [DOI] [PubMed] [Google Scholar]
- 65. Kastan M. B., Zhan Q., el-Deiry W. S., Carrier F., Jacks T., Walsh W. V., Plunkett B. S., Vogelstein B., Fornace A. J., Jr. (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71, 587–597 [DOI] [PubMed] [Google Scholar]
- 66. Ohtani-Fujita N., Minami S., Mimaki S., Dao S., Sakai T. (1998) p53-Independent activation of the gadd45 promoter by Δ12-prostaglandin J2. Biochem. Biophys. Res. Commun. 251, 648–652 [DOI] [PubMed] [Google Scholar]
- 67. Pedeux R., Lefort K., Cuenin C., Cortes U., Kellner K., Doré J. F., Nakazawa H. (2002) Specific induction of gadd45 in human melanocytes and melanoma cells after UVB irradiation. Int. J. Cancer 98, 811–816 [DOI] [PubMed] [Google Scholar]
- 68. Ishida T., Suzuki T., Hirashima S., Mizutani K., Yoshida A., Ando I., Ikeda S., Adachi T., Hashimoto H. (2006) Benzimidazole inhibitors of hepatitis C virus NS5B polymerase. Identification of 2-[(4-diarylmethoxy)phenyl]-benzimidazole. Bioorg. Med. Chem. Lett. 16, 1859–1863 [DOI] [PubMed] [Google Scholar]
- 69. Treier M., Staszewski L. M., Bohmann D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell 78, 787–798 [DOI] [PubMed] [Google Scholar]
- 70. Wagner E. F., Nebreda A. R. (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 71. Huang P., Han J., Hui L. (2010) MAPK signaling in inflammation-associated cancer development. Protein Cell 1, 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Inanami O., Ohta T., Ito S., Kuwabara M. (1999) Elevation of intracellular calcium ions is essential for the H2O2-induced activation of SAPK/JNK but not for that of p38 and ERK in Chinese hamster V79 cells. Antioxid. Redox Signal. 1, 501–508 [DOI] [PubMed] [Google Scholar]
- 73. Liu S. L., Lin X., Shi D. Y., Cheng J., Wu C. Q., Zhang Y. D. (2002) Reactive oxygen species stimulated human hepatoma cell proliferation via cross-talk between PI3K/PKB and JNK signaling pathways. Arch. Biochem. Biophys. 406, 173–182 [DOI] [PubMed] [Google Scholar]
- 74. Matsukawa J., Matsuzawa A., Takeda K., Ichijo H. (2004) The ASK1-MAP kinase cascades in mammalian stress response. J. Biochem. 136, 261–265 [DOI] [PubMed] [Google Scholar]
- 75. Tanaka T., Akatsuka S., Ozeki M., Shirase T., Hiai H., Toyokuni S. (2004) Redox regulation of annexin 2 and its implications for oxidative stress-induced renal carcinogenesis and metastasis. Oncogene 23, 3980–3989 [DOI] [PubMed] [Google Scholar]
- 76. Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., Chandel N. S. (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 107, 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Frank G. D., Motley E. D., Inagami T., Eguchi S. (2000) PYK2/CAKβ represents a redox-sensitive tyrosine kinase in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 270, 761–765 [DOI] [PubMed] [Google Scholar]
- 78. Zhang S., Qiu X., Gu Y., Wang E. (2008) Up-regulation of proline-rich tyrosine kinase 2 in non-small cell lung cancer. Lung Cancer 62, 295–301 [DOI] [PubMed] [Google Scholar]
- 79. Tokiwa G., Dikic I., Lev S., Schlessinger J. (1996) Activation of Pyk2 by stress signals and coupling with JNK signaling pathway. Science 273, 792–794 [DOI] [PubMed] [Google Scholar]
- 80. Halfter U. M., Derbyshire Z. E., Vaillancourt R. R. (2005) Interferon-γ-dependent tyrosine phosphorylation of MEKK4 via Pyk2 is regulated by annexin II and SHP2 in keratinocytes. Biochem. J. 388, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Minden A., Lin A., Claret F. X., Abo A., Karin M. (1995) Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 82. Teramoto H., Coso O. A., Miyata H., Igishi T., Miki T., Gutkind J. S. (1996) Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J. Biol. Chem. 271, 27225–27228 [DOI] [PubMed] [Google Scholar]
- 83. Brumby A. M., Goulding K. R., Schlosser T., Loi S., Galea R., Khoo P., Bolden J. E., Aigaki T., Humbert P. O., Richardson H. E. (2011) Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster. A RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics 188, 105–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xiao L., Lang W. (2000) A dominant role for the c-Jun NH2-terminal kinase in oncogenic ras-induced morphologic transformation of human lung carcinoma cells. Cancer Res. 60, 400–408 [PubMed] [Google Scholar]
- 85. Hansen M. D., Ehrlich J. S., Nelson W. J. (2002) Molecular mechanism for orienting membrane and actin dynamics to nascent cell-cell contacts in epithelial cells. J. Biol. Chem. 277, 45371–45376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. He K. L., Deora A. B., Xiong H., Ling Q., Weksler B. B., Niesvizky R., Hajjar K. A. (2008) Endothelial cell annexin A2 regulates polyubiquitination and degradation of its binding partner S100A10/p11. J. Biol. Chem. 283, 19192–19200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Petersson S., Shubbar E., Enerbäck L., Enerbäck C. (2009) Expression patterns of S100 proteins in melanocytes and melanocytic lesions. Melanoma Res. 19, 215–225 [DOI] [PubMed] [Google Scholar]
- 88. Yang X., Popescu N. C., Zimonjic D. B. (2011) DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. 71, 2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. König J., Prenen J., Nilius B., Gerke V. (1998) The annexin II-p11 complex is involved in regulated exocytosis in bovine pulmonary artery endothelial cells. J. Biol. Chem. 273, 19679–19684 [DOI] [PubMed] [Google Scholar]