Background: Myeloid cell lines were generated from Npm1+/− mice to understand the role of NPM1 in MDS.
Results: Compromised myeloid gene expression in NPM1+/− cells was due to up-regulation of C/EBPαp30 and eIF4E.
Conclusion: Altered ratio of C/EBPαp42:C/EBPαp30 accounts for compromised gene expression.
Significance: Aberrant feed-forward pathways up-regulate eIF4E and C/EBPαp30 levels, thus contributing to the MDS phenotype associated with NPM1 deficiency.
Keywords: C/EBP Transcription Factor, eIF4E, Gene Expression, Gene Regulation, Leukocyte, C/EBPα, Myelodysplastic Syndrome, Myeloid Development, Nucleophosmin 1
Abstract
NPM1 is a ubiquitously expressed nucleolar phosphoprotein, the gene for which maps to chromosome 5q35 in close proximity to a commonly deleted region associated with (del)5q, a type of myelodysplastic syndrome (MDS). This region is also a frequent target of deletions in de novo and therapy-related MDS/acute myeloid leukemia. Previous studies have shown that Npm1+/− mice develop an MDS-like disease that transforms to acute myeloid leukemia over time. To better understand the mechanism by which NPM1 haploinsufficiency causes an MDS phenotype, we generated factor-dependent myeloid cell lines from the bone marrow of Npm1+/+ and Npm1+/− mice and demonstrated compromised neutrophil-specific gene expression in the MNPM1+/− cells. We attribute these observations to increased levels of the shorter, dominant negative leukemogenic isoform (p30) of CCAAT enhancer-binding protein α (C/EBPα). We show that this increase is caused, in part, by elevated levels of the activated translation initiation factor eIF4E, overexpression of which also increases translation of C/EBPαp30 in HEK293 cells. In a positive feedback loop, eIF4E expression is further elevated both at the mRNA and protein levels by C/EBPαp30 but not by the full-length C/EBPαp42. Re-expression of C/EBPαp42 or NPM1 but not C/EBPαp30 in MNPM1+/− cells partially rescues the myeloid phenotype. Our observations suggest that the aberrant feed-forward pathway that keeps eIF4E and C/EBPαp30 elevated in NPM1+/− cells contributes to the MDS phenotype associated with NPM1 deficiency.
Introduction
NPM1 is a conserved, ubiquitously expressed nucleolar phosphoprotein that belongs to the nucleoplasmin family of nuclear chaperones and resides on chromosome 5q35. Two NPM isoforms have been described in humans. The most abundant one encodes a 294-amino acid protein that carries an oligomerization domain at its N terminus, a bipartite nuclear localization signal, and a nucleolar localization signal in its C terminus (reviewed in Ref. 1). An important feature of NPM1 is its ability to shuttle rapidly between the nucleolus and the cytoplasm. This shuttling allows NPM1 to participate in diverse cellular processes including ribosome biogenesis, transport of preribosomal particles, maintenance of genomic stability by the control of cellular ploidy, DNA repair, and regulation of DNA transcription by controlling chromatin condensation and decondensation (reviewed in Ref. 2). Recently, NPM1 has been shown to be associated with centrosome duplication (3) and regulation of the p53 tumor suppressor (4). There is growing evidence that NPM1 may also act as a tumor suppressor (2). NPM1 is an essential gene, because germ line inactivation in mice leads to embryonic lethality at midgestation (embryonic day 11.5) due to severe anemia (5). NPM1 is overexpressed in some nonhematologic tumors and is deleted or involved with chromosomal translocations in several hematologic malignancies (reviewed in Ref. 1). The region on chromosome 5q harboring NPM1 is a frequent target of deletions in de novo and therapy-related MDS (6). Mutations in NPM1 occur in over 50% of acute myeloid leukemia (AML)3 patients presenting with normal karyotype. The mutant NPM1 protein termed NPMc+ is mislocalized to the cytoplasm, an event that may contribute to its role in leukemogenesis (7, 8). Npm1+/− mice develop a hematological syndrome very similar to that observed in MDS patients (5) and subsequently develop myeloid and lymphoid malignancies, with a preponderance of the former (9). Because one copy of the NPM1 gene is often deleted in MDS with (del) 5q, we hypothesized that haploinsufficiency of NPM1 may contribute to this form of MDS. We further postulated that myeloid abnormalities observed in NPM1 deficiency may reflect an aberrant NPM-1-C/EBPα axis, given that C/EBPα is a master regulator of the myeloid development program (10). C/EBPα is the founding member of a family of basic region/leucine zipper (bZip) transcription factors (reviewed in Ref. 11). It is expressed at high levels throughout myeloid differentiation and has been shown to bind to the promoters of multiple myeloid-specific genes at different stages of myeloid maturation, including the primary (myeloperoxidase, defensins, and neutrophil elastase) and secondary granule (lactoferrin, MMP8, MMP9, and TC1) protein genes. C/EBPα is a single exon gene, but it is expressed as two isoforms that arise from alternate translation start sites that give rise to a full-length C/EBPαp42 and a truncated dominant negative C/EBPαp30 isoform (12). Translational control of C/EBPα isoform expression is orchestrated by a conserved upstream ORF in the 5′-UTR. This region is thought to be responsive to the activities of the translation initiation factors eIF4E and eIF2 (reviewed in Ref. 13) such that an increase in the activity of eIF2 or eIF4E results in an increase in expression of the shorter p30 isoform (reviewed in Ref. 12; supplemental Fig. S1 for details).
Several groups have reported mutations in the C/EBPα gene in a subset of patients with AML presenting with normal karyotypes (reviewed in Ref. 14). These mutations can be broadly classified into two main categories. The first includes in-frame mutations clustered in the highly conserved C terminus of the C/EBPα protein. The second category involves frameshift mutations at the N terminus of C/EBPα, resulting in the premature termination of the full-length C/EBPαp42 isoform while keeping the truncated C/EBPα p30 protein intact (15). The remaining C/EBPαp42 is thought to be rendered inactive by the dominant negative activity of the p30 isoform by an unknown mechanism. Thus, changes in the expression ratio of the two C/EBPα isoforms play a role in cell fate (reviewed in Ref. 14; Ref. 16 and references therein).
In this paper we show that NPM1 haploinsufficiency is associated with increased expression of the shorter p30 isoform of C/EBPα compared with the full-length p42 isoform in a cell line derived from the bone marrow of Npm1+/− mice (MNPM1+/− cells). We show that this ratio change is associated with an increase in phosphorylated levels of the rate-limiting translation initiation factor eIF4E. eIF4E forms the mRNA 7-methyl guanosine cap (m7-Gppp)-binding component of the translation initiation complex eIF4F (reviewed in Ref. 13). Approximately 30% of malignancies are associated with increased activity of eIF4E (reviewed in Ref. 17). Increased levels of active eIF4E in the MNPM1+/− cells are initially likely because of an increase in expression of the oncogene c-Myc, which regulates eIF4E at the transcriptional level (18). eIF4E then increases C/EBPαp30 protein levels by a previously documented mechanism involving ribosome stalling (reviewed in Ref. 19; supplemental Fig. S1). Subsequently, in a positive feedback loop, increased levels of C/EBPαp30 but not C/EBPαp42 transcriptionally increase levels of eIF4E. Finally, we demonstrate that re-expression of either C/EBPαp42 or NPM1 but not C/EBPαp30 in MNPM1+/− cells rescues the myeloid phenotype. Our data therefore suggest that haploinsufficiency of NPM1 modulates neutrophil-specific gene expression by altering the activity of the master myeloid regulator C/EBPα. The activation of an aberrant feed-forward mechanism that keeps levels of both eIF4E/eIF4E-P and C/EBPαp30 elevated likely contributes to the molecular landscape of MDS associated with NPM1 deficiency.
EXPERIMENTAL PROCEDURES
Generation of Factor-dependent Cell Lines from Npm1+/− and Npm1+/+ Bone Marrow
Factor-dependent cell lines were generated from the bone marrow of wild type and NPM1 haploinsufficient mice using methodology previously described (20). Briefly, Npm1+/− mice and wild type littermates were injected with 5-fluorouracil (100 mg/kg). Whole bone marrow was harvested 3–5 days later and cultured for 48 h in IMDM supplemented with 20% (v/v) horse serum, murine GM-CSF (2.5 ng/ml; Immunex, Thousand Oaks, CA), human IL-6 (20 ng/ml; Peprotech, Rocky Hill, NJ), and murine IL-1b (10 ng/ml Peprotech). The cells were transduced with a truncated retinoic acid receptor α (RARα403) by spinfection using supernatants derived from a stable GP+E86 producer line. Spinfections were performed over 2 h with the addition of polybrene (4 μg/ml). Following spinfection, the cells were transferred to IMDM supplemented with 20% horse serum, rat stem cell factor (200 ng/ml; Peprotech), either Wehi-3B conditioned medium (0.25%) as a source of IL-3 or recombinant mouse IL-3 (2.5–5 ng/ml; Peprotech) and human erythropoietin (8 units/ml; Amgen, Thousand Oaks, CA). Transduced cells were passaged every 2–3 days in IMDM supplemented with 20% (v/v) horse serum and 15% BHK-MKL conditioned medium as a source of stem cell factor and 0.3 ng/ml of IL-3. The cells were exposed to G418 for an initial period of 10 days. After a month in this medium, the cells were weaned off stem cell factor but could not be weaned off IL-3. Over time we have been able to grow the cells in medium supplemented with low concentrations of IL-3 (0.3 ng/ml; R & D Systems). However, complete removal from IL-3 results in apoptosis. Thus, within 2 months rapidly proliferating IL-3-dependent cell lines were generated. Although we did not isolate single cell clones from these cell lines for further analysis, it has been our experience that over time, two or three clones expressing high levels of the transduced truncated RARα gene predominate. All the experiments described below have been performed on these pooled clones. These MNPM1+/− and MNPM1+/+ cells (Myeloid) were maintained at a density of 5 × 105/ml in IMDM containing 20% horse serum and 10% Wehi-3B conditioned medium. Induction to mature neutrophils was conducted by the addition of either 10 μm all-trans-retinoic acid (ATRA; Sigma) for 48–72 h or by the addition of 10% HM-5 conditioned medium (as a source of GM-CSF) with or without 10 μm ATRA, in the absence of IL-3. Terminal neutrophil differentiation was monitored by Wright-Giemsa staining. Cultures were harvested at the indicated times for total RNA and whole cell protein extract preparation.
Real Time PCR Analysis
0.25–0.5 μg of total RNA from MNPM1+/− and MNPM1+/+ cells were used to generate first strand cDNA using oligo(dT) primers with the Superscript II reverse transcription kit (Invitrogen). The resulting cDNA was then diluted at 1:20 and analyzed by real time PCR in triplicate for each primer pair, and the corresponding threshold cycle (Ct) values were determined. All of the reactions were performed in a 20-μl volume using 100 ng of each primer and the iQ TM SYBR green Supermix from Bio-Rad as per the manufacturer's instructions using default cycling parameters in an iQ5 Cycler (Bio-Rad). Transcript levels of each mRNA were normalized to that of β-actin and then expressed as a fold change over the signal observed in the control sample. The primer pairs used are presented in supplemental Table S1.
Overexpression of Expression Plasmids for eIF4E, C/EBPαEx, and mut C/EBPαEx in HEK293 Cells
HEK293 cells were co-transfected with an expression plasmid for C/EBPα (C/EBPαEx, harboring the 5′-UTR region of the C/EBPα gene, a gift from Dr Calkhoven (21)), or mutant C/EBPαEx in which the AUG for C/EBPαp30 (supplemental Fig. S1) was mutated (Met to Ala) using the Stratagene QuikChange kit as per manufacturer's instructions, and an expression plasmid for eIF4E (pMSCV pgk GFP-eIF4E, a gift from Kathrine Borden, University of Montreal, Montreal, Canada) or vector (pMSCV pgk GFP) using FuGENE (Roche Applied Science) as per the manufacturer's instructions. 48 h post-transfection, whole cell protein extracts were prepared and subjected to Western blot analysis as described below.
Western Blot Analysis
Western blotting and detection was performed as previously described (22) in TBS-T at 4 °C overnight. The antibodies used were NPM1 (Santa Cruz, catalogue number sc-2004, 1:2000); C/EBPα (C terminus, Abcam catalogue number ab15048); eIF4E (C46H8) and eIF4E-P (Ser-209) (Cell Signaling); c-Myc (N-262, sc-746) and β-actin (C4; sc-47778) (Santa Cruz Biotechnology); GAPDH (catalogue number 2118; Cell Signaling, 1:5000); C/EBPα (N terminus, catalogue number ab15047; Abcam; 1:500 dilution); and C/EBPα (C terminus, catalogue number sc-61, 14AA; Santa Cruz; 1:2000). Comparative expression levels were measured as a ratio of expression of the two isoforms of C/EBPα, eIF4E, or eIF4E-P to that of β-actin using the image analysis software Image J (23).
Transient Transfection Analysis in HEK293 Cells
HEK293T cells were transiently co-transfected with an eIF4E promoter plasmid (harboring ∼1 kb of the human eIF4E promoter upstream of the firefly luciferase reporter gene; a gift from Kathrine Borden, University of Montreal) alone or with expression plasmids for c-Myc (2 μg), C/EBPαp30 (1 μg), or C/EBPαp42 (1 μg) as previously described (24). The cells were harvested 48 h post-transfection, and luciferase reporter gene activity was measured using a dual luciferase reporter gene assay kit (Promega Biotech). Normalized luciferase (to that of a co-transfected Renilla luciferase plasmid) values were used to calculate the fold change over the enzyme activity of the eIF4E promoter reporter plasmid alone (equal to 1). The statistical significance of fold differences was measured using a two-tailed Student's t test.
Add Back of C/EBPαp30 and C/EBPαp42 in NPM1+/− Cells
2 × 106 exponentially growing MNPM1+/− cells were transfected with 2 μg of empty vector or with an expression plasmid for mouse C/EBPα p42 and p30 lacking the C/EBPα 5′-UTR (25) by Amaxa-based nucleofection (Lonza) using the following program: T-020/Solution V. Following incubation, with or without GM-CSF for 48 h post-nucleofection, the cells were harvested, washed once with PBS, and transferred to TRIzol reagent (Invitrogen). Total RNA was isolated and subjected to quantitative RT-PCR as described above using gene specific oligomers (supplemental Table S1).
RESULTS
Generation of a Factor-dependent Myeloid Cell Line (MNPM1+/−) from the Bone Marrow of Npm1 Heterozygous Mice
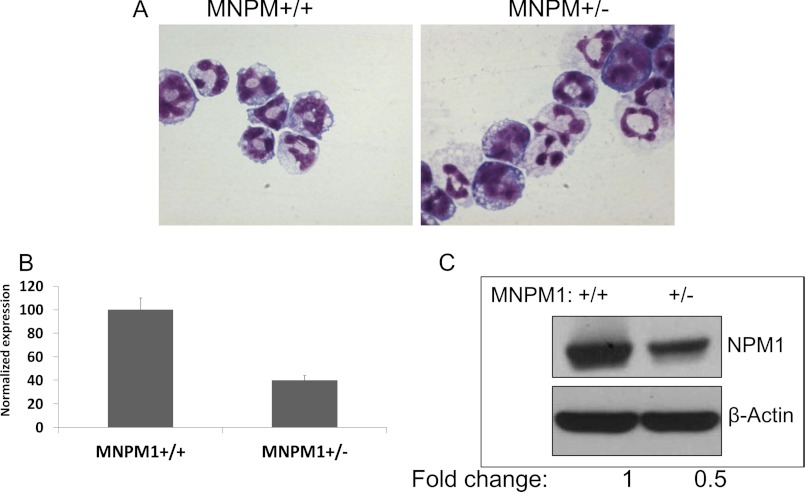
It has been previously demonstrated that Npm1 heterozygous mice develop a human MDS-like disease with dysplastic megakaryocytes and erythroid defects, including morphologic dysplasia and defective erythroid maturation (5). However, no myeloid defects were described in these mice. The mice subsequently develop myeloid and lymphoid malignancies, with a preponderance of the former (9). Because one copy of the NPM1 gene is often deleted in MDS (del)5q, haploinsufficiency of Npm1 may contribute to this form of MDS. In light of this, we generated IL-3-dependent myeloid cell lines (MNPM1+/− and MNPM1+/+) from cells harvested from the bone marrow of Npm1+/− and Npm1+/+ mice by methods outlined previously (24). The cells proliferate in IL-3-containing media and undergo neutrophil maturation upon removal of IL-3 and the addition of GM-CSF or ATRA separately or in combination. Both the MNPM1+/+ and MNPM1+/− cells undergo morphologic neutrophil maturation upon the addition of GM-CSF and ATRA as judged by Wright-Giemsa staining (Fig. 1A). Additionally, FACS analysis of both cell lines demonstrated an equivalent up-regulation of the myeloid markers Gr-1 and Mac-1 following 48-h GM-CSF induction of these cells (data not shown). As shown in Fig. 1 (B and C), the expression of NPM1 mRNA and protein in the MNPM1+/− cells is approximately half that of the MNPM1+/+ cells, consistent with their derivation from Npm1 heterozygous mice.
FIGURE 1.
A, Wright-Giemsa staining of MNPM+/+ and MNPM1+/− cells. The cells were grown as described under “Experimental Procedures” and treated with 10% HM-5 conditioned medium (as a source of GM-CSF) and 10 μm ATRA for 72 h. The cells were cytospun and stained with Wright-Giemsa stain. B, real time PCR analysis of MNPM1+/+ and MNPM1+/− cells. Total RNA isolated from the MNPM1 cells was converted to cDNA and subjected to quantitative PCR analysis using primers specific for the NPM1 gene. Expression data have been normalized to that of β-actin and expressed as a percentage assuming 100% expression in MNPM+/+ cells. C, Western blot analysis of NPM1 expression in MNPM1+/+ and MNPM1+/− cells. Total protein isolated from MNPM1+/+ and MNPM1+/− cells was subjected to Western blot analysis using anti-NPM1 and anti-β-actin antibodies sequentially. NPM1 protein levels in MNPM1+/− cells (lane 2) were ∼50% lower than that in MNPM1+/+ cells (lane 1).
Neutrophil-specific Gene Expression Is Impaired in MNPM1+/− Myeloid Cells
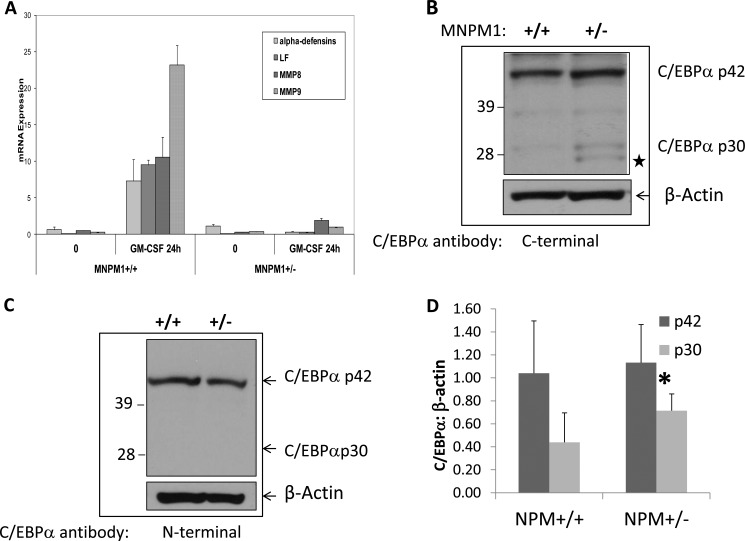
We next investigated neutrophil-specific gene expression in MNPM1+/− cells induced toward mature neutrophils. We performed real time PCR on reverse transcribed RNA derived from uninduced (0 h) and GM-CSF-induced (24 h) MNPM1 cells. As shown in Fig. 2A, the expression levels of both a neutrophil primary granule protein gene (α-defensins) and neutrophil secondary granule protein genes (LF, MMP8, and MMP9) were reduced to negligible levels in neutrophils derived from GM-CSF-induced MNPM1+/− cells compared with the MNPM1+/+ cells. This suggests that although neutrophils derived from NPM1-deficient cells appear morphologically normal (Fig. 1A), they are biochemically defective. We noted that most of the genes that are deregulated in the NPM1+/− cells are regulated by the myeloid master transcription factor C/EBPα. We therefore examined the levels of C/EBPα protein in the MNPM1+/− cells compared with the MNPM1+/+ cells by Western blot analysis. As shown in Fig. 2B, the levels of the full-length C/EBPαp42 isoform remained unchanged in the two cell lines, but protein levels of a 30-kDa protein were increased on average 1.63-fold in the MNPM1+/− cells (Fig. 2D, p = 0.05). To demonstrate that this protein was indeed the dominant negative C/EBPαp30 isoform, we probed the Western blot with an N-terminal specific C/EBPα antibody that recognizes only the p42 isoform of C/EBPα as opposed to the C-terminal specific antibody that recognizes both isoforms (Fig. 2B). As shown in Fig. 2C, the 30-kDa band clearly visible in Fig. 2B was absent in Fig. 2C in the NPM1+/− lane, suggesting that the shorter band is C/EBPαp30. We further confirmed the 30-kDa protein to be C/EBPαp30 by (a) comparing its migration in the SDS gel with a positive C/EBPαp30 control (not shown), (b) using the different C/EBPα antibodies to identify p30 in HEK293 cells overexpressing C/EBPα p30 and p42 (supplemental Fig. S2), and (c) mutating the p30 translational start site in a full-length C/EBPα expression plasmid followed by expression in HEK293 cells (see Fig. 4C). Expression levels of p30 remained high following GM-CSF induction of MNPM1+/− cells (data not shown). Interestingly, in addition to the C/EBPα p30 isoform, a shorter band was also consistently observed in extracts of MNPM1+/− cells (Fig. 2B, ★). We speculate that this shorter band likely arises from translation of an in-frame AUG downstream of the p30 translational start site in the C/EBPα mRNA transcript (supplemental Fig. S1). Thus, expression levels of the shorter C/EBPαp30 isoform appear to be up-regulated in cells expressing haploinsufficient levels of NPM1. We and others have shown that increased concentrations of C/EBPαp30 negatively alters the transactivation ability of the wild type C/EBPαp42 protein (supplemental Fig. S3) and hence the expression of its downstream targets.
FIGURE 2.
A, neutrophil-specific gene expression is impaired in NPM1+/− myeloid cells. Expression levels of four neutrophil-specific genes, α-defensins, LF, neutrophil collagenase (MMP8), and neutrophil gelatinase (MMP9) were measured using real time PCR analysis using cDNA derived from total RNA from uninduced (0 h) and 48-h GM-CSF-induced MNPM1+/+ and MNPM1+/− cell lines. Expression levels were normalized to that of β-actin. The expression of each gene was measured in triplicate, and the S.E. values are indicated. These data are representative of a total of three independent experiments. B, NPM1 haploinsufficient cells express increased levels of the C/EBPα p30 isoform. Expression levels of C/EBPα were measured by Western blot analysis in uninduced MNPM1+/+ and MNPM1+/− cell lines and probed sequentially with a C-terminal specific C/EBPα antibody and β-actin antibody. This experiment has been repeated on three independent occasions. ★ represents a shorter C/EBPα “isoform” (see supplemental Fig. S1 for details) C, Western blot analysis of cell lysates prepared from MNPM1+/+ and MNPM1+/− cells was performed. The blot was probed sequentially using an antibody specific for C/EBPαp42 recognizing the N terminus of the C/EBPα protein and β-actin antibody as a loading control. Note that no C/EBPαp30 was observed in this blot. D, quantitative analysis of the relative expression levels of C/EBPαp30 and C/EBPαp42 from three independent experiments compared with β-actin expression using Image J software in MNPM1+/+ versus MNPM1+/− cells. No change in expression of p42 was observed. p30 levels increased 1.63-fold in MNPM1+/− cells (*, p = 0.05).
FIGURE 4.
Overexpression of the translation initiation factor eIF4E contributes to elevated levels of C/EBPαp30. A, HEK293 cells were transfected with empty expression vector or an eIF4E expressing plasmid. 24 h post-transfection, cell lysates were prepared and subjected to Western blot analysis. The blot was probed sequentially for phospho-eIF4E (Ser-209), eIF4E, and β-actin. B, HEK293 cells were co-transfected with the C/EBPαEx expression plasmid, empty vector, and an eIF4E expression plasmid. Total protein was collected 48 h post-transfection and subjected to Western blot analysis as in A. The blot was probed with anti-C/EBPα and β-actin antibodies. C, HEK293 cells were co-transfected as above with wild type C/EBPαEx (WT) or p30 mutant C/EBPαEx (Mut) plasmids with or without an expression plasmid for eIF4E (see “Experimental Procedures”). The Western blot was processed as above and probed sequentially with anti-C/EBPα and GAPDH antibodies. D, expression plasmids for C/EBPα p30 (1 and 2 μg) and p42 (2 μg) were transfected into HEK293 cells. 24 h post-transfection, the cell lysates were prepared and subjected to Western blot analysis. The blot was probed C/EBPα and β-actin. E, cell lysates overexpressing C/EBPαp30 and p42 (D) were analyzed by Western blot analysis and probed sequentially with anti phospho-eIF4E, eIF4E, and β-actin antibodies. The numbers indicate normalized (to β-actin) fold change in expression of phospho-eIF4E and eIF4E in extracts overexpressing C/EBPαp30 compared with those expressing C/EBPαp42 (equal to 1). Un, untransfected lysates; Vec, vector.
To rule out the possibility that the observed changes in gene expression were not due to decreased expression of the GM-CSF receptor, we performed quantitative RT-PCR analysis using RNA prepared from uninduced and 48-h GM-CSF induced MNPM1+/+ and MNPM1+/− cells. A 5-fold increase in the mRNA expression levels of the GM-CSF receptor was observed in both the GM-CSF treated MNPM1 wild type and haploinsufficient cells, suggesting that the observed changes do not reflect GM-CSF receptor-mediated effects (supplemental Fig. S4). These observations suggest that NPM-1 does not control neutrophil specific gene expression and function via aberrant GM-CSF receptor expression in the MNPM+/− cells. On the other hand, expression of the GCSF-R, a canonical C/EBPα target, was sensitive to NPM-1 haploinsufficiency (supplemental Fig. S4).
Increased Levels of the Active Form of the Translation Initiation Factor eIF4E in MNPM1+/− Cells Contribute to Elevated Levels of C/EBPα p30
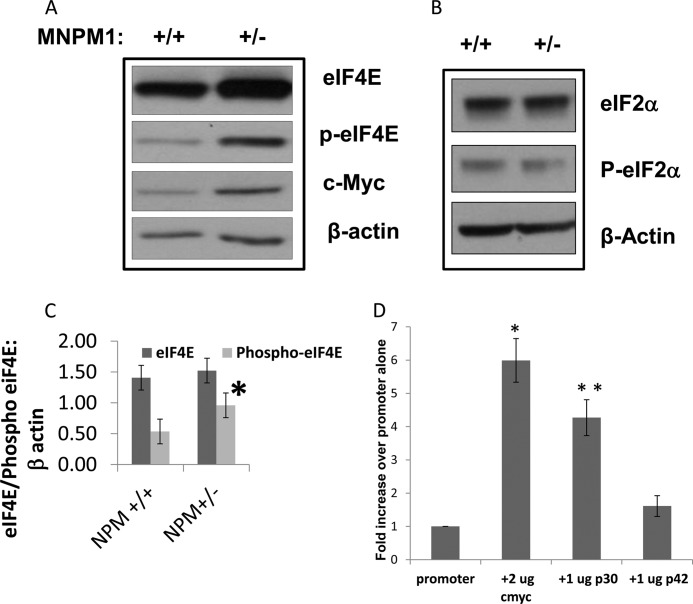
Elevated levels of the mRNA translation cap-binding factor eIF4E and the initiation factor eIF2 have been shown to be associated with increased expression of truncated isoforms of members of the C/EBP family, including C/EBPα (12) and C/EBPβ (26). To determine whether this pathway might contribute to the increased levels of C/EBPαp30 in MNPM1+/− cells, we performed Western blot analysis for eIF4E and phosphorylated eIF4E on cellular extracts prepared from MNPM1+/+ and MNPM1+/− cells. As is evident in Fig. 3A, although endogenous levels of eIF4E remained unchanged, its activated phosphorylated form was up-regulated on average 1.8-fold in MNPM1+/− cells compared with wild type cells (p = 0.044). We attribute the translation of the shorter C/EBPα “isoform” observed in Fig. 2B (★) to the activity of increased expression of eIF4E in NPM1-deficient cells (12) (see supplemental Fig. S1 for explanation).
FIGURE 3.
EIF4E-P but not eIF2α-P protein levels are elevated in NPM1+/− cells. A, MNPM1+/− cells express elevated levels of eIF4E-P and c-Myc compared with MNPM1+/+ cells, as measured by Western blot analysis. Equal protein loading was confirmed by probing the blot with β-actin. This experiment has been repeated four times. B, no change in protein levels of eIF2α or P-eIF2α were observed in MNPM+/− cells compared with wild type MNPM1+/+ cells as measured by Western blot analysis. Equal protein loading was confirmed by probing the blot with β-actin. C, quantitative analysis of the relative expression levels of eIF4E and phopsho-eIF4E in four separate experiments compared with β-actin expression using Image J software comparing MNPM1+/+ and MNPM1+/− cells. Expression levels of eIF4E remained unchanged, but those of eIF4E-P increased 1.8-fold in MNPM1+/− cells (p = 0.044). D, C/EBPαp30 up-regulates the eIF4E promoter. Transient co-transfection analysis of eIF4E promoter-reporter and expression plasmids for c-Myc (2 μg) as a positive control, C/EBPαp30 (1 μg), and C/EBPαp42 (1 μg) in HEK293 cells. 48 h post-transfection, reporter gene activity was measured by the dual luciferase assay kit (see “Experimental Procedures”). Normalized firefly luciferase (to that of a co-transfected Renilla luciferase plasmid) values have been represented as a fold change over the enzyme activity of eIF4E promoter reporter plasmid alone (equal to 1). The figure represents the normalized means ± S.E. obtained from an experiment performed in triplicate. This experiment was repeated three times. *, p = 0.009; **, p = 0.016.
Increased eIF4E activity is not associated with a global increase in mRNA translation but affects the translation of a specific subset of mRNAs with complex 5′-UTRs (27). Among these mRNAs is c-Myc, a multifunctional oncogene that plays a role in controlling cell growth and proliferation. We show that levels of c-Myc, a downstream translational target of eIF4E, are also elevated in MNPM1+/− cells (Fig. 3A). Interestingly, it has been shown that eIF4E is a transcriptional target of c-Myc (18, 28), suggesting here that a feed-forward loop may keep levels of eIF4E elevated in MNPM1+/− cells. Western blot analysis of eIF2α, a second rate-limiting factor in mRNA translation and the catalytic component of the eIF2 complex, and phospho-eIF2α did not show any increase in MNPM1+/− cells as compared with MNPM1+/+ cells (Fig. 3B). This suggests that NPM1 deficiency specifically up-regulates eIF4E.
We next asked whether C/EBPα activity contributed in any way to the observed increased levels of eIF4E. To this end we utilized the HEK293 cell line, which does not express C/EBPα endogenously, thus providing an unbiased system for analysis. HEK293 cells were co-transfected with an eIF4E promoter-reporter plasmid (harboring both c-Myc and C/EBP-binding sites; data not shown) and either c-Myc, C/EBPαp30, or C/EBPαp42 expression plasmids. Normalized luciferase activity was measured 48 h post-transfection (Fig. 3D). As expected, c-Myc transactivated the eIF4E promoter 6.2-fold over promoter alone (p = 0.009). Surprisingly, C/EBPαp30 also transactivated this promoter ∼4.8-fold over promoter alone (p = 0.016), whereas C/EBPαp42 was capable of only a 1.6-fold increase in reporter gene activity (p = 0.2). This suggests that eIF4E is transactivated more efficiently by C/EBPαp30 compared with C/EBPαp42 and is likely a C/EBPαp30 target gene.
Overexpression of eIF4E in HEK293 Cells Increases Expression Levels of C/EBPαp30 at the Protein Level
To demonstrate that increased protein expression of eIF4E could lead to increased C/EBPαp30 levels, we co-transfected HEK293 cells with an expression plasmid for eIF4E along with a C/EBPα expression plasmid harboring the C/EBPα 5′-UTR, termed C/EBPαEx, capable of expressing all of the C/EBPα isoforms (21). Western blot analysis demonstrated that eIF4E was indeed overexpressed and correlated with increased levels of phospho-eIF4E in HEK293 cells (Fig. 4A). Increased protein expression of C/EBPαp30 was observed only in cells expressing phospho-eIF4E (Fig. 4B). However, low levels of C/EBPαp30 were also observed in the vector transfected lane (Fig. 4B). This may be due to the relatively high levels of phosphorylated eIF4E in these transfected HEK293 cells (Fig. 4A). To further demonstrate the specificity of eIF4E on the expression of C/EBPαp30, we mutated the p30 AUG site (Met to Ala) in the full-length C/EBPαEx plasmid (supplemental Fig. S1). As is evident in Fig. 4C, mutating the p30 AUG site abrogates expression of C/EBPαp30 but not C/EBPαp42 in transfected HEK293 cells (Fig. 4C, Mut lanes). Co-expression of eIF4E increases levels of p30 in the presence of wild type C/EBPαEx (Fig. 4, B and C, WT lanes), thereby confirming previous studies that correlate up-regulation of C/EBPαp30 with elevated levels of phospho-eIF4E (reviewed in Ref. 13). Interestingly, the lower band of the “p30 doublet” in Fig. 4B appears to be expressed in the p30 mutant lanes (Fig. 4C), presumably because of translation initiation at a conserved in-frame AUG downstream of the p30 AUG in the full-length C/EBPα gene (supplemental Fig. S1). Expression of eIF4E also increases protein levels of this shorter band upon co-expression with mutant-C/EBPαEx (Fig. 4C), thus arguing against this band being a nonspecific or degradation-associated protein product.
Having demonstrated a specific increase in C/EBPαp30 protein levels upon up-regulation of eIF4E (Fig. 4, A–C), as well as increased eIF4E promoter activity upon C/EBPαp30 expression (Fig. 3D), we further confirmed the C/EBPαp30-eIF4E connection at the protein level (Fig. 4, D and E). Transfection of increasing concentrations of C/EBPαp30 (1 and 2 μg; Fig. 4D) resulted in a graded increase in not only eIF4E expression (1.9- and 3.5-fold normalized increase in expression compared with C/EBPαp42, respectively) but also in eIF4E-P protein levels (4.0- and 9.1-fold normalized increase in expression compared with C/EBPαp42) in HEK293 cells (Fig. 4E). These observations point to a feed-forward regulatory loop involving the C/EBPαp30-eIF4E axis.
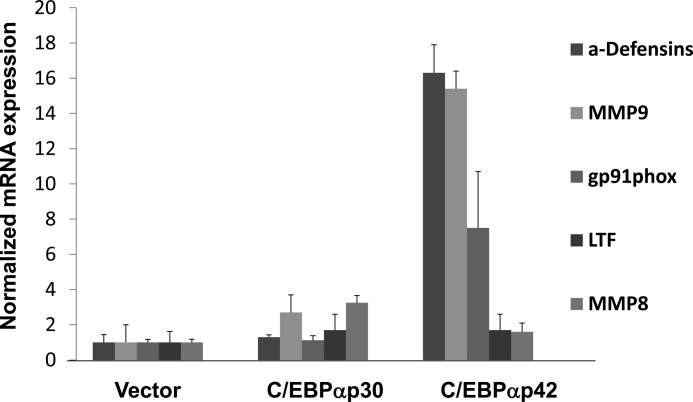
To determine whether reduced activity of C/EBPα caused by increased expression of C/EBPαp30 was responsible for the observed defects in neutrophil-specific gene expression, we nucleofected MNPM1+/− cells with C/EBPα expression plasmids lacking the C/EBPα 5′-UTR, that express either the full-length p42 or the shorter p30 isoform, in the MNPM1+/− cells and performed quantitative RT-PCR expression analysis using total RNA isolated 24 h post-transfection. Overexpression of C/EBPαp42 in MNPM1+/− cells was associated with increased expression of a panel of neutrophil-specific mRNAs, including α-defensins, cybb (gp91phox), and MMP9 but not MMP8, and LF (Fig. 5). Interestingly, we found that the loss of expression of gp91phox, a critical component of the neutrophil function-associated NADPH oxidase complex, in MNPM1+/− cells corresponds to functional defects in neutrophils derived from these cells as measured by the respiratory burst and chemotaxis assays (data not shown). Expression of C/EBPαp30 in MNPM1+/− cells, on the other hand, did not result in the same level of up-regulation of these myeloid genes (Fig. 5), confirming our observations that elevated expression levels of C/EBPαp30 do not support normal myeloid gene expression.
FIGURE 5.
Adding p42 but not p30 C/EBPα expression plasmid back in NPM1 haploinsufficient cells restores neutrophil specific gene expression. MNPM1+/− cells were nucleofected with empty vector or expression plasmids for C/EBPαp30 or C/EBPαp42. 48 h post-transfection, total RNA was isolated, converted to cDNA, and subjected to quantitative RT-PCR analysis using gene specific oligomers. Expression levels have been normalized to that of β-actin and calculated using the ΔΔCT method. Fold change in expression has been calculated for each gene by assuming its expression in untransfected cells is equal to 1.
We also overexpressed NPM-1 in the MNPM1+/− line (supplemental Fig. S5). Expression levels of α-defensins, MMP9, and cybb (gp91phox) were significantly elevated upon expression of NPM1. Interestingly, the expression of LF and MMP8 remained unaffected in both the NPM1 and C/EBPαp42 overexpressing MNPM1+/− cells. However, induction of MNPM1+/− cells overexpressing NPM1 or p42 with GM-CSF restored expression of these two genes (data not shown). These observations suggest that the expression of LF and MMP8 requires transcription factors induced by GM-CSF, such as C/EBPϵ, for high level expression during neutrophil maturation. The panel of genes up-regulated by C/EBPα (Fig. 5) closely parallels that observed for NPM1 up-regulation in the MNPM1+/− cells (supplemental Fig. S5), suggesting that NPM1 influences neutrophil-specific gene expression via modulation of C/EBPα expression.
DISCUSSION
In this report we demonstrate that the myeloid defects associated with NPM1 deficiency are likely due to changes in expression of the myeloid master regulator C/EBPα. We have demonstrated elevated protein levels of the dominant negative C/EBPαp30 isoform in an inducible IL-3-dependent myeloid cell line generated from the bone marrow of NPM1+/− mice. We further show in our MNPM1+/− factor-dependent cell line that neutrophil-specific gene expression is severely impaired. Re-expression of either C/EBPαp42 or NPM1 but not C/EBPαp30 in the MNPM1+/− cell line partially restores normal neutrophil gene expression, suggesting that NPM1 plays a role in mediating myeloid homeostasis and likely contributes to myeloid maturation by modulating C/EBPα activity.
NPM1 is a ubiquitously expressed and abundant nucleolar phosphoprotein with a diverse portfolio of cellular functions. These include regulation of chromatin structure including centrosome stability, the synthesis and processing of ribosomal RNA, and nucleocytoplasmic transport of ribosomal RNA and ribosomal proteins (reviewed in Refs. 1 and 2). NPM1 has recently been described as a histone chaperone, which upon acetylation is capable of acting as a co-activator of chromatin-associated transcription (29). Although NPM1 is not itself a transcription factor, it has been shown to act as a co-activator of transcriptional regulation by transcription factors such as NF-κB (30), c-Myc and some of its downstream target genes (31), and p53 (2). The propensity of NPM1 haploinsufficient mice to develop myeloid leukemias (5) has a counterpart in human myeloid malignancy. NPM1 is involved in chromosomal translocations associated with myeloid leukemias (such as t(5;17) NPM1-RARα in APL and t(3;5) NPM-MLF1 in AML/MDS), and the region on chromosome 5q harboring NPM1 is a frequent target of deletions in de novo and therapy-related MDS. Furthermore, mutations in the NPM1 gene account for nearly 50% of AML cases with normal karyotype (reviewed in Ref. 1). These data together indicate that alterations in NPM-1 expression lead to dysmyelopoiesis. Our data suggest that the myeloid-specific effects associated with NPM1 haploinsufficiency are induced in part by changes in expression of the two C/EBPα isoforms.
Changes in the ratio of isoforms of C/EBPα (C/EBPα p42:p30) contribute to changes associated with normal and abnormal myelopoiesis (15). We and others have shown that increasing p30 expression exerts a dominant negative effect on its full-length counterpart (p42) by an unclear mechanism. Recent studies, however, suggest two possibilities. First, p30 may displace p42 at a canonical C/EBP-binding site in target gene promoters or may sequester p42 by the formation of p30-p42 dimers. Alternatively, gene regulation may occur through a unique repertoire of downstream targets of C/EBPαp30. For example, it has recently been shown that p30 up-regulates the expression of Ubc9 (32), an E2-Sumo-conjugating protein, and PIN1, peptidyl-prolyl cis/trans-isomerase (33). The up-regulation of Ubc9 does not require binding of p30 to the promoter of this gene. In comparison, it has been demonstrated that p30 regulates the PIN1 promoter by augmenting the transactivating capacity of the transcription factor E2F1, which binds to and regulates the PIN1 promoter. Our data indicate that p30 preferentially transactivates the eIF4E promoter. This is unlikely to involve a canonical C/EBPα-binding site, although a putative site has been identified in the eIF4E promoter (not shown), because p30 has been shown to bind very inefficiently to such elements (34). Although we have identified two putative E2F-binding sites in the eIF4E promoter, their role, if any, in the p30-mediated up-regulation of eIF4E remains to be elucidated. Additionally, a recent study has shown that mice homozygously expressing p30 C/EBPα from the C/EBPΑ locus develop AML with complete penetrance (35). Thus, alteration in p30:p42 ratios can alter cell fate.
Translational control of C/EBPα-isoform expression is orchestrated by a conserved cis-regulatory upstream ORF in the 5′-UTR that is out of frame with the coding region of C/EBPα and is thought to be responsive to the activities of the translation initiation factors eIF4E and eIF2. Two rate-limiting events take place during the initiation of cap-dependent mRNA translation: (a) the recruitment of mRNA to the ribosomal complex followed by (b) the selection of the AUG translation initiation codon. Both processes are mediated by multiprotein complexes. The first involves the assembly of the eukaryotic translation-initiation-factor complex, eIF4F, at the mRNA 5′ cap. Availability of the cap-binding protein, eIF4E, is the first rate-limiting step in the assembly of the eIF4F complex. Under basal conditions, eIF4E is sequestered by the 4E-binding proteins, and release of eIF4E only occurs after phosphorylation of the 4E-binding proteins in response to growth factor signaling. The selection of the translation initiation site requires the formation of a ternary complex between the G-protein, eIF2, the initiator Met-tRNAiMet, and GTP, thereby allowing AUG codon recognition and protein synthesis to proceed. Phosphorylation of the α subunit of eIF2 by stress-induced kinases leads to the potent inhibition of translation. Control of C/EBP-isoform expression is mediated via a conserved cis-regulatory upstream ORF in the 5′-UTR that is sensitive to the activities of both eIF4E and eIF2. Although it has been shown that enhanced eIF2 or eIF4E activity up-regulates the C/EBPα p30 isoform in mammary-epithelial and intestinal-epithelial cancer cells, attenuated translation of C/EBPα in bronchial smooth muscle cells from asthmatic patients was found to be associated specifically with diminished levels of eIF4E (36). We observed elevated levels of C/EBPαp30 in MNPM1+/− cells. However, no changes in the protein levels of eIF2α or its phosphorylated form were observed in MNPM1+/− cells. Protein levels of the active phosphorylated form of eIF4E, on the other hand, were increased in MNPM1+/− cells, suggesting that increased eIF4E activity may contribute to higher p30 expression. Co-expression of eIF4E and a C/EBPα expression plasmid harboring the 5′-UTR in HEK293 cells (which normally do not express C/EBPα) resulted in the specific up-regulation of the C/EBPαp30 but not the C/EBPαp42 isoform, thus confirming the role of eIF4E in contributing to the up-regulation of C/EBPαp30 protein expression. Conversely, graded overexpression of C/EBPαp30 in HEK293 cells up-regulated endogenous protein levels of both eIF4E and phosphorylated-eIF4E, an effect not observed with the C/EBPαp42 isoform. It appears that overexpression of C/EBPαp30 in HEK293 cells leads to up-regulation of both eIF4E and eIF4E-P, whereas the subtle but significant endogenous increase of C/EBPαp30 in MNPM1+/− cells results in a significant increase in levels of eIF4E-P. This apparent discrepancy is likely due to a C/EBPαp30 gene dosage effect. Based on these observations, it is tempting to speculate that C/EBPαp30 may also regulate the expression of the MAP kinase-interacting kinases (Mnk1 and Mnk2), which are known to regulate phosphorylation of eIF4E (reviewed in Ref. 37).
Our observations led us to postulate that NPM1 deficiency leads to a subtle but significant increase in eIF4E/P-eIF4E levels, which in turn result in an increase in expression of p30. Because C/EBPαp30 in turn up-regulates eIF4E expression, we propose a feed-forward loop, where increased p30 protein levels further increase expression of eIF4E. Thus, NPM1 haploinsufficiency triggers a cascade of events that lead to an increase in expression levels of the mRNA translation initiation control factor eIF4E and its phosphorylated form, as well as the leukemogenic C/EBPαp30 isoform.
NPM1 deficiency is also associated with increased expression of the oncoprotein c-Myc in MNPM1+/− cells. c-Myc has previously been shown to up-regulate levels of eIF4E at the transcriptional level (18, 38), which may account for the increased expression of eIF4E in MNPM1+/− cells. eIF4E in turn, up-regulates c-Myc transcriptionally (18, 38), suggesting the presence of another aberrant feed-forward loop involving c-Myc and eIF4E. In addition, c-Myc is a negatively regulated target of C/EBPα (39), and its overexpression down-regulates C/EBPα-regulated gene expression (40). Thus, elevated levels of c-Myc in MNPM1+/− cells likely also contribute to impaired neutrophil-specific gene expression in these cells.
It has long been known that eIF4E activity is dysregulated in cancer. Although the regulation of eIF4E is modulated, in part, by phosphorylation, the physiologic significance of phosphorylated eIF4E in mammalian cells remained unclear until recently. Furic et al. (41) showed that mice expressing a “knock-in” of a nonphosphorylatable form of eIF4E were resistant to tumorigenesis in a prostate cancer model. They further demonstrated that phosphorylation of eIF4E was necessary for the increased translation of a number of mRNAs, including MMP3, CCL2, VEGFC, and BIRC2, all involved in tumorigenesis. These findings establish eIF4E-P as a critical player in tumorigenesis (41). Additionally, it has been suggested that phosphorylation of eIF4E may increase its affinity for the 5′-mRNA cap, thereby increasing translation of the mRNA in question (42). This has been specifically shown for c-Myc, where increasing phosphorylation of eIF4E increases c-Myc protein levels (43). We have now shown that increased expression of eIF4E-P in MNPM1+/− cells correlates with higher protein levels of both c-Myc and C/EBPα p30.
In conclusion, we have demonstrated that haploinsufficiency of NPM1 triggers the up-regulation of the translation initiation factor eIF4E, likely as a result of transcriptional up-regulation by c-Myc, which is also elevated at the protein level in MNPM1+/− cells (see model in Fig. 6). We propose that increased levels of eIF4E/eIF4E-P then up-regulates C/EBPαp30 expression. High C/EBPαp30 expression acts as a dominant negative inhibitor of full-length C/EBPαp42 activity and disrupts the normal neutrophil development program in MNPM1+/− cells. We postulate that the operation of two aberrant feed-forward loops, one involving c-Myc and eIF4E and the other eIF4E and C/EBPαp30 in MNPM1+/− cells, contributes to the MDS phenotype.
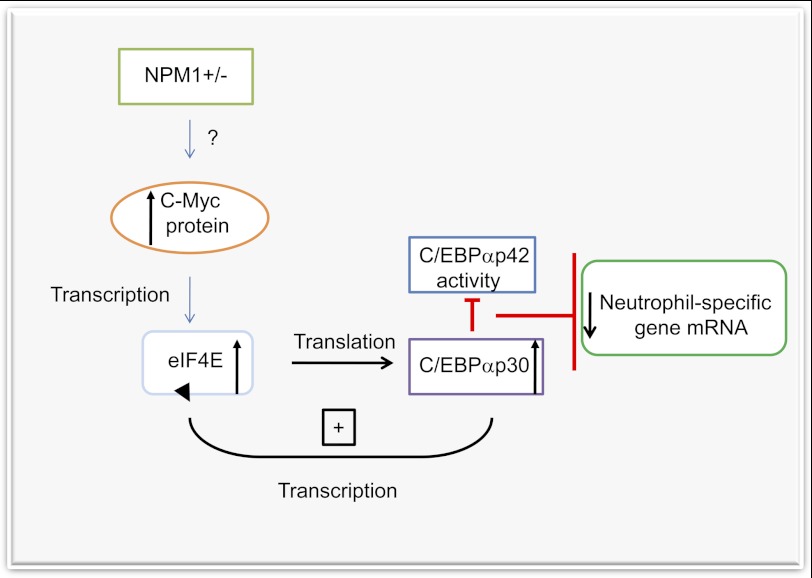
FIGURE 6.
Model for mechanism of disruption of neutrophil-gene regulation in NPM1 haploinsufficient cells. NPM1 haploinsufficient cells express higher levels of the proto-oncogene c-Myc via an unknown mechanism (question mark). Increased c-Myc protein in turn transcriptionally activates the mRNA rate-limiting translation initiation factor eIF4E. Elevated levels of eIF4E then increase translation of the C/EBPαp30 isoform, which in turn has a dominant negative effect on the activity of C/EBPαp42, resulting in reduced neutrophil-specific mRNA gene expression. Increased C/EBPαp30 protein further increases levels of eIF4E through a feed-forward loop (shown with black arrows) that works to sustain levels of the oncoproteins eIF4E and C/EBPαp30 in NPM1+/− cells. A similar feed-forward loop involving eIF4E and c-Myc has been previously observed. These aberrant pathways likely contribute to the MDS-phenotype associated with NPM1 deficiency.
Disruption of the expression and/or function of the master myeloid regulator C/EBPα is an emerging paradigm in the pathobiology of AML (reviewed in Ref. 16). Our data extend this model by demonstrating that NPM1 deficiency contributes to disruption of myeloid homeostasis via the C/EBPα axis, thereby contributing to the abnormal myeloid biology associated with MDS/AML.
Supplementary Material
Acknowledgments
We thank Dr. Cornelius Calkhoven (Leibniz Institute for Age Research, Fritz, Germany) and Dr. Kathrine Borden (University of Montreal) for valuable reagents and members of the Berliner and Pandolfi labs for encouragement and advice.
This work was supported, in whole or in part, by National Institutes of Health Grants CA-71692 and CA-74031 (to P. P. P.) and P01-HL63357 (to N. B.).

This article contains supplemental Table S1 and Figs. S1–S5.
- AML
- acute myeloid leukemia
- NPM1
- nucleophosmin1
- C/EBPα
- CCAAT enhancer-binding protein α
- eIF4E
- eukaryotic translation initiation factor 4E
- MDS
- myelodysplastic syndrome
- IMDM
- Iscove's modified Dulbecco's medium
- ATRA
- all-trans-retinoic acid
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- LF
- lactoferrin.
REFERENCES
- 1. Naoe T., Suzuki T., Kiyoi H., Urano T. (2006) Nucleophosmin. A versatile molecule associated with hematological malignancies. Cancer Sci. 97, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grisendi S., Mecucci C., Falini B., Pandolfi P. P. (2006) Nucleophosmin and cancer. Nat. Rev. Cancer 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 3. Okuda M. (2002) The role of nucleophosmin in centrosome duplication. Oncogene 21, 6170–6174 [DOI] [PubMed] [Google Scholar]
- 4. Colombo E., Marine J. C., Danovi D., Falini B., Pelicci P. G. (2002) Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 4, 529–533 [DOI] [PubMed] [Google Scholar]
- 5. Grisendi S., Bernardi R., Rossi M., Cheng K., Khandker L., Manova K., Pandolfi P. (2005) Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437, 147–153 [DOI] [PubMed] [Google Scholar]
- 6. Olney H. J., Le Beau M. M. (2007) Evaluation of recurring cytogenetic abnormalities in the treatment of myelodysplastic syndromes. Leuk. Res. 31, 427–434 [DOI] [PubMed] [Google Scholar]
- 7. Falini B., Martelli M., Mecucci C., Liso A., Bolli N., Bigerna B., Pucciarini A., Pileri S., Meloni G., Martelli M., Haferlach T., Schnittger S. (2008) Cytoplasmic mutated nucleophosmin is stable in primary leukemic cells and in a Xenotransplant model of NPMc+ acute myeloid leukemia in SCID mice. Hematologica 93, 775–779 [DOI] [PubMed] [Google Scholar]
- 8. Falini B., Gionfriddo I., Cecchetti F., Ballanti S., Pettirossi V., Martelli M. P. (2011) Acute myeloid leukemia with mutated nucleophosmin (NPM1). Any hope for a targeted therapy? Blood Rev. 25, 247–254 [DOI] [PubMed] [Google Scholar]
- 9. Sportoletti P., Grisendi S., Majid S. M., Cheng K., Clohessy J. G., Viale A., Teruya-Feldstein J., Pandolfi P. P. (2008) Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood 111, 3859–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tenen D. (2003) Disruption of differentiation in human cancer. AML shows the way. Nat. Rev. Cancer 3, 89–101 [DOI] [PubMed] [Google Scholar]
- 11. Khanna-Gupta A. (2008) Sumoylation and the function of CCAAT enhancer binding protein alpha (C/EBPα). Blood Cells Mol. Dis. 41, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calkhoven C. F., Müller C., Leutz A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Gene Dev. 14, 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 13. Khanna-Gupta A. (2011) Regulation and deregulation of mRNA translation during myeloid maturation. Exp. Hematol. 39, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mueller B. U., Pabst T. (2006) C/EBPα and the pathophysiology of acute myeloid leukemia. Curr. Opin. Hematol. 13, 7–14 [DOI] [PubMed] [Google Scholar]
- 15. Pabst T., Mueller B. U., Zhang P., Radomska H. S., Narravula S., Schnittger S., Behre G., Hiddemann W., Tenen D. G. (2001) Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27, 263–270 [DOI] [PubMed] [Google Scholar]
- 16. Koschmieder S., Halmos B., Levantini E., Tenen D. (2009) Dysregulation of the C/EBPα differentiation pathway in human cancer. J. Clin. Oncol. 27, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruggero D., Pandolfi P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 18. Jones R. M., Branda J., Johnston K. A., Polymenis M., Gadd M., Rustgi A., Callanan L., Schmidt E. V. (1996) An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-Myc. Mol. Cell. Biol. 16, 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calkhoven C. F., Müller C., Leutz A. (2002) Translational control of gene expression and disease. Trends Mol. Med. 8, 577–583 [DOI] [PubMed] [Google Scholar]
- 20. Tsai S., Bartelmez S., Sitnicka E., Collins S. (1994) Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 8, 2831–2841 [DOI] [PubMed] [Google Scholar]
- 21. Müller C., Bremer A., Schreiber S, Eichwald S., Calkhoven C. (2010) Nucleolar retention of a translational C/EBPα isoform stimulates rDNA transcription and cell size. EMBO J. 29, 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khanna-Gupta A., Zibello T., Idone V., Sun H., Lekstrom-Himes J., Berliner N. (2005) Human neutrophil collagenase expression is C/EBP-dependent during myeloid development. Exp. Hematol. 33, 42–52 [DOI] [PubMed] [Google Scholar]
- 23. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image Processing with Image J. Biophotonics Int. 11, 36–42 [Google Scholar]
- 24. Khanna-Gupta A., Sun H., Zibello T., Lee H. M., Dahl R., Boxer L. A., Berliner N. (2007) Growth factor independence-1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene-inactivating mutation in the C/EBPepsilon gene. Blood 109, 4181–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller M., Shuman J. D., Sebastian T., Dauter Z., Johnson P. F. (2003) Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein α. J. Biol. Chem. 278, 15178–15184 [DOI] [PubMed] [Google Scholar]
- 26. Li S., Pal R., Monaghan S. A., Schafer P., Ouyang H., Mapara M., Galson D. L., Lentzsch S. (2011) IMiD immunomodulatory compounds block C/EBPβ translation through eIF4E down-regulation resulting in inhibition of MM. Blood 117, 5157–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richter J. D., Sonenberg N. (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 [DOI] [PubMed] [Google Scholar]
- 28. Lin C. J., Cencic R., Mills J. R., Robert F., Pelletier J. (2008) c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 68, 5326–5334 [DOI] [PubMed] [Google Scholar]
- 29. Gadad S. S., Shandilya J., Swaminathan V., Kundu T. K. (2009) Histone chaperone as coactivator of chromatin transcription. Role of acetylation. Methods Mol Biol. 523, 263–278 [DOI] [PubMed] [Google Scholar]
- 30. Dhar S. K., Lynn B. C., Daosukho C., St Clair D. K. (2004) Identification of nucleophosmin as an NF-κB co-activator for the induction of the human SOD2 gene. J. Biol. Chem. 279, 28209–28219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z., Boone D., Hann S. (2008) Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci. U.S.A. 105, 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geletu M., Balkhi M. Y., Peer Zada A. A., Christopeit M., Pulikkan J. A., Trivedi A. K., Tenen D. G., Behre G. (2007) Target proteins of C/EBPαp30 in AML. C/EBPαp30 enhances sumoylation of C/EBPαp42 via up-regulation of Ubc9. Blood 110, 3301–3309 [DOI] [PubMed] [Google Scholar]
- 33. Pulikkan J. A., Dengler V., Peer Zada A. A., Kawasaki A., Geletu M., Pasalic Z., Bohlander S. K., Ryo A., Tenen D. G., Behre G. (2010) Elevated PIN1 expression by C/EBPα-p30 blocks C/EBPα-induced granulocytic differentiation through c-Jun in AML. Leukemia 24, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hankey W., Silver M., Sun H., Zibello T., Berliner N., Khanna-Gupta A. (2011) Differential effects of Sumoylation on the activity of CCAAT enhancer binding protein alpha (C/EBPα) p42 versus p30 may contribute to aberrant C/EBPα activity in acute myeloid leukemias. Hematol. Rep. 5, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirstetter P., Schuster M. B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T. A., Pabst T., Schrock E., Porse B. T., Jacobsen S. E., Bertone P., Tenen D. G. (2008) Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 13, 299–310 [DOI] [PubMed] [Google Scholar]
- 36. Borger P., Miglino N., Baraket M., Black J. L., Tamm M., Roth M. (2009) Impaired translation of CCAAT/enhancer binding protein α mRNA in bronchial smooth muscle cells of asthmatic patients. J Allergy Clin Immunol 123, 639–645 [DOI] [PubMed] [Google Scholar]
- 37. Hou J., Lam F., Proud C., Wang S. (2012) Targeting Mnks for cancer therapy. Oncotarget 3, 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Makhlouf A. A., Namboodiri A. M., McDermott P. J. (2001) Transcriptional regulation of the rat eIF4E gene in cardiac muscle cells. The role of specific elements in the promoter region. Gene 267, 1–12 [DOI] [PubMed] [Google Scholar]
- 39. Johansen L. M., Iwama A., Lodie T. A., Sasaki K., Felsher D. W., Golub T. R., Tenen D. G. (2001) c-Myc is a critical target for c/EBPα in granulopoiesis. Mol. Cell. Biol. 21, 3789–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mink S., Mutschler B., Weiskirchen R., Bister K., Klempnauer K. (1996) A novel function for Myc. Inhibition of C/EBP-dependent gene activation. Proc. Natl. Acad. Sci. U.S.A. 93, 6635–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Furic L., Rong L., Larsson O., Koumakpayi I. H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L. A., Pandolfi P. P., Saad F., Sonenberg N. (2010) eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahalingam M., Cooper J. (2001) Phosphorylation of mammalian eIF4E by Mnk1 and Mnk2. Tantalizing prospects for a role in translation. Prog. Mol. Subcell. Biol. 27, 132–142 [PubMed] [Google Scholar]
- 43. Li Y., Yue P., Deng X., Ueda T., Fukunaga R., Khuri F. R., Sun S. Y. (2010) Protein phosphatase 2A negatively regulates eukaryotic initiation factor 4E phosphorylation and eIF4F assembly through direct dephosphorylation of Mnk and eIF4E. Neoplasia 12, 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.