Background: The bacterial pathway of l-hydroxyproline metabolism has not been identified.
Results: Different types of d-hydroxyproline dehydrogenases and unique Δ1-pyrroline-4-hydroxy-2-carboxylate deaminase involved in the bacterial l-hydroxyproline pathway were identified and characterized for the first time.
Conclusion: l-Hydroxyproline degradation by bacteria was elucidated at the molecular level.
Significance: Our results suggest that d-hydroxyproline dehydrogenases evolved convergently, and we discovered a unique deaminase enzyme likely within the aldolase protein family.
Keywords: Dehydrogenase, Enzyme Catalysis, Flavoproteins, Hydroxyproline, Pseudomonas, Pseudomonas aeruginosa, d-Hydroxyproline Dehydrogenase
Abstract
l-Hydroxyproline (4-hydroxyproline) mainly exists in collagen, and most bacteria cannot metabolize this hydroxyamino acid. Pseudomonas putida and Pseudomonas aeruginosa convert l-hydroxyproline to α-ketoglutarate via four hypothetical enzymatic steps different from known mammalian pathways, but the molecular background is rather unclear. Here, we identified and characterized for the first time two novel enzymes, d-hydroxyproline dehydrogenase and Δ1-pyrroline-4-hydroxy-2-carboxylate (Pyr4H2C) deaminase, involved in this hypothetical pathway. These genes were clustered together with genes encoding other catalytic enzymes on the bacterial genomes. d-Hydroxyproline dehydrogenases from P. putida and P. aeruginosa were completely different from known bacterial proline dehydrogenases and showed similar high specificity for substrate (d-hydroxyproline) and some artificial electron acceptor(s). On the other hand, the former is a homomeric enzyme only containing FAD as a prosthetic group, whereas the latter is a novel heterododecameric structure consisting of three different subunits (α4β4γ4), and two FADs, FMN, and [2Fe-2S] iron-sulfur cluster were contained in αβγ of the heterotrimeric unit. These results suggested that the l-hydroxyproline pathway clearly evolved convergently in P. putida and P. aeruginosa. Pyr4H2C deaminase is a unique member of the dihydrodipicolinate synthase/N-acetylneuraminate lyase protein family, and its activity was competitively inhibited by pyruvate, a common substrate for other dihydrodipicolinate synthase/N-acetylneuraminate lyase proteins. Furthermore, disruption of Pyr4H2C deaminase genes led to loss of growth on l-hydroxyproline (as well as d-hydroxyproline) but not l- and d-proline, indicating that this pathway is related only to l-hydroxyproline degradation, which is not linked to proline metabolism.
Introduction
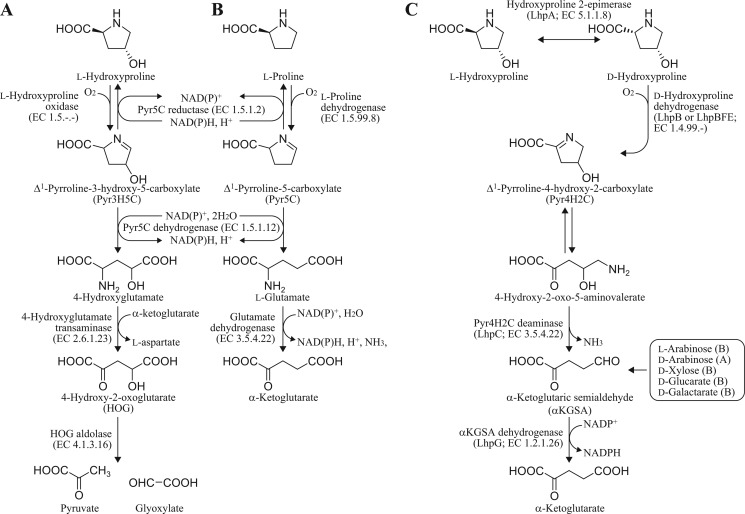
l-Hydroxyproline (4-hydroxyproline) exists in collagen and the cell wall of plants and algae. Although direct biosynthesis from l-proline has been known in a few bacteria (1), the free compound in nature is mainly produced from collagen by amino peptidase(s). Metabolism of l-hydroxyproline in mammals has been well studied and involves the stepwise action of four mitochondrial enzymes (see Fig. 1A) (2). l-Hydroxyproline oxidase (so-called PRODH2) first produces Δ1-pyrroline-3-hydroxy-5-carboxylate (Pyr3H5C).2 Pyr3H5C is a spontaneously opened ring and is converted to l-4-hydroxyglutamate by Δ1-pyrroline-5-carboxylate (Pyr5C) dehydrogenase (EC 1.5.1.12), which is also involved in l-proline metabolism (see next section). The final products, pyruvate and glyoxylate, are produced from l-4-hydroxyglutamate through d-4-hydroxy-2-oxoglutarate by the sequential action of aspartate aminotransferase (EC 2.6.1.1) and 4-hydroxy-2-oxoglutarate aldolase (EC 4.1.3.16). Recently, PRODH2 and 4-hydroxy-2-oxoglutarate aldolase were characterized genetically and enzymatically (3, 4).
FIGURE 1.
Known and proposed pathways of l-hydroxyproline and l-proline metabolism. A, known mammalian l-hydroxyproline pathway. B, known general l-proline pathway. C, novel l-hydroxyproline pathway of bacteria. Gray-colored reaction is hypothetical. αKGSA is produced from alternative metabolic pentose pathways and acid-sugar pathways from bacteria (B) and archaea (A).
The mammalian l-hydroxyproline pathway is partially linked to the general l-proline pathway in which l-proline is metabolized to l-glutamate by two enzymes (see Fig. 1B). The first enzyme, l-proline dehydrogenase (l-PDH; EC 1.5.99.8), catalyzes the flavin adenine dinucleotide (FAD)-dependent oxidation of l-proline to Pyr5C. It is also known that Pyr5C reductase (EC 1.5.1.2) can reduce either Pyr5C or Pyr3H5C to l-proline or l-hydroxyproline, respectively (5). The Pyr5C intermediate is spontaneously hydrolyzed to glutamate γ-semialdehyde, which is subsequently oxidized to l-glutamate by the second enzyme, Pyr5C dehydrogenase. Intriguingly, whereas l-PDH and Pyr5C dehydrogenase are separate enzymes in eukaryotes and some bacteria (6), they are fused into a single polypeptide chain in other bacteria that is known as PutA (proline utilization A) (7). On the other hand, three l-PDHs from hyperthermophilic archaea differing from the known l-PDHs have been recently reported, although the reaction product is the same, Pyr5C (summarized in supplemental Table S6). First, l-PDH of Thermococcus profundus (Tp/l-PDH) forms a complex consisting of four different subunits with molecular masses of 54 (α), 42 (β), 19 (γ), and 11 kDa (δ), respectively, with a heterotetrameric structure of αβγδ containing two FADs (in α- and β-subunits) (8). Second, l-PDH isozyme 1 of Pyrococcus horikoshii (Ph/l-PDH1) consists of two different subunits with molecular masses of 56 (α) and 43 kDa (β) with a heterooctameric structure of α4β4 containing one FAD (β), one FMN (between α- and β-subunits), and one ATP (α) (9, 10). P. horikoshii also possesses l-PDH isozyme 2 (Ph/l-PDH2), which is similar to Tp/l-PDH. Third, l-PDH of Pyrobaculum calidifontis is a homodimeric enzyme containing one FAD per subunit (11). These enzymes can utilize artificial electron acceptors such as 2,6-dichloroindophenol and ferricyanide instead of the natural acceptors.
Although l-hydroxyproline is a non-utilizable amino acid for most microorganisms, a few bacteria, including Pseudomonas putida (12), Pseudomonas aeruginosa (13, 14), and Pseudomonas striata (15, 16), can rapidly grow on l-hydroxyproline as a sole carbon source, and it is believed that the hypothetical pathway, different from that in mammals, is operational (Fig. 1C). The first two steps in this pathway involve epimerization to d-hydroxyproline catalyzing l-hydroxyproline 2-epimerase (EC 5.1.1.8) and oxidization to Δ1-pyrroline-4-hydroxy-2-carboxylate (Pyr4H2C) catalyzing d-hydroxyproline dehydrogenase (oxidase) (d-HypDH; EC 1.4.3.-). The cyclic imine Pyr4H2C is spontaneously hydrolyzed to 4-hydroxy-2-oxo-5-aminovalerate, which is subsequently deaminated to α-ketoglutaric semialdehyde (αKGSA) by Pyr4H2C deaminase (EC 3.5.4.22). The last step is the NAD(P)+-dependent dehydrogenation of the semialdehyde to α-ketoglutarate by αKGSA dehydrogenase (KGSADH) (2,5-dioxovalerate dehydrogenase; EC 1.2.1.26). The αKGSA intermediate is also produced from alternative pentose and/or acid-sugar pathways of bacteria and/or archaea (17–19), and the metabolic fate is the same as that of the bacterial l-hydroxyproline pathway. Previously, we first identified three KGSADH isozymes induced by l-arabinose (KGSADH-I), d-glucarate/d-galactarate (KGSADH-II), and l-hydroxyproline (KGSADH-III; LhpG in this study) in Azospirillum brasilense, a nitrogen-fixing bacteria (20, 21).
Although genetic and enzymatic characterizations of l-hydroxyproline 2-epimerases from several bacteria, including P. aeruginosa, were recently reported (4), only the genes encoding d-HypDH and Pyr4H2C deaminase have been identified so far. In particular, FAD-dependent amino-acid dehydrogenases, including d-HypDHs from P. aeruginosa (13) and P. striata (15), generally associate with cell and/or organelle membranes and are unstable in solution, which has made purification, preservation, and characterization very difficult. In this study, we identified different types of putative gene clusters related to l-hydroxyproline metabolism of P. putida and P. aeruginosa by bioinformatics analysis. Furthermore, characterization of the recombinant proteins expressed in P. putida or Escherichia coli cells revealed that d-HypDHs from P. putida and P. aeruginosa (referred to as Pp/d-HypDH and Pa/d-HypDH, respectively) had clearly evolved convergently and that Pyr4H2C deaminase belongs to a unique member of the protein family mainly consisting of aldolase proteins.
MATERIALS AND METHODS
General Procedures
Basic recombinant DNA techniques were performed as described by Sambrook et al. (23). Bacterial genomic DNA was prepared using a DNeasy Tissue kit (Qiagen). PCR was carried out using GeneAmp PCR System 2700 (Applied Biosystems) for 30 cycles in a 50-μl reaction mixture containing 1 unit of KOD FX DNA polymerase (Toyobo), 15 pmol of the appropriate primers, and template DNA under the following conditions: denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and extension at 68 °C for time periods calculated as an extension rate of 1 kbp/min. Protein concentrations were determined by the method of Lowry et al. (24) with bovine serum albumin as the standard. SDS-PAGE was performed as described by Laemmli (25). Non-denaturing PAGE was performed by omitting SDS and 2-mercaptoethanol from the solution used in SDS-PAGE. Proteins on the gel were stained with Coomassie Brilliant Blue R-250 and destained with 7.5% (v/v) acetic acid in 25% methanol. High pressure liquid chromatography (HPLC) was performed using an Agilent 1120 Compact LC system (Tosoh). NMR spectra were recorded on a Jeol JNM-EC400 NMR spectrometer (Jeol Ltd., Tokyo, Japan) operating at 400 MHz. 2,2-Dimethyl-2-silapentane-5-sulfonate was used as an internal standard.
Bacterial Strain, Culture Conditions, and Preparation of Cell-free Extracts
Pseudomonas strains were cultured aerobically with vigorous shaking at 30 °C in LB medium (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl/liter) or minimal medium (pH 6.8) containing 0.8 g of KH2PO4/K2HPO4, 2 g of (NH4)2SO4, 0.002 g of FeSO4, 0.080 g of MgSO4·7H2O, and 2 g of carbon source/liter. The grown cells were harvested by centrifugation at 30,000 × g for 20 min, suspended in 50 mm Tris-HCl (pH 8.0) containing 2 mm MgCl2, and disrupted by sonication for 20 min at appropriate intervals on ice using an Ultra Sonic Disruptor Model UR-200P (Tomy Seiko Co., Ltd, Tokyo, Japan) and then centrifuged at 108,000 × g for 20 min at 4 °C to obtain cell-free extracts. If necessary, 1% (v/v) Tween 20 was added to the buffer for solubilization of d-HypDH.
Plasmid Construction for Expression of Recombinant Proteins
Primer sequences used in this study are shown in supplemental Table S1. In this report, the prefixes P. putida (Pp), P. aeruginosa (Pa), and A. brasilense (Ab) have been added to gene symbols or protein designations when required for clarity. PpLhpB (PP_1255) PaLhpB-LhpF-LhpE (PA1266 and PA1267), PpLhpC (PP_1257), and PpLhpG (PP_1256) (GenBankTM accession numbers are given in parentheses) genes were amplified by PCR using primers containing appropriate restriction enzyme sites at the 5′- and 3′-ends and genomic DNA of P. putida KT2442 or P. aeruginosa PAO1 as a template. In the case of the PaLhpBFE gene, SacI and HindIII sites were attached to the 5′-end of the PaLhpB gene and the 3′-end of the PaLhpE gene, respectively. Each amplified DNA fragment was introduced into BamHI-HindIII sites (for all others except the PaLhpBFE gene) or SacI-HindIII (for the PaLhpBFE gene) in pQE-80L (Qiagen), a plasmid vector for conferring an N-terminal His6 tag on expressed proteins, to obtain pQE/PpLhpB, pQE/PaLhpBFE, pQE/PpLhpC, and pQE/PpLhpG, respectively.
pUCP26Km (26) was previously constructed by inserting the Tn5-derived 1.3-kbp BamHI kanamycin resistance (Kmr) cassette of pUC4K (Amersham Biosciences) into the BamHI site in pUCP26, an Escherichia-Pseudomonas shuttle plasmid (27). The promoter and partial open reading frame region (∼240 bp) of the ahpC gene from P. putida that encodes a small subunit of alkyl hydroperoxide reductase was amplified and introduced into the SalI site in pUCP26Km to yield pUCP26KmAhpCp. Each DNA fragment of His6-PpLhpB-t0 terminator and His6-PaLhpBFE-t0 terminator was amplified by PCR using pQE/PpLhpB and pQE/PaLhpBFE as a template, respectively, and introduced into SalI-EcoRI sites in pUCP26KmAhpCp to obtain pUCP/PpLhpB and pUCP/PaLhpBFE, respectively (see Fig. 2F).
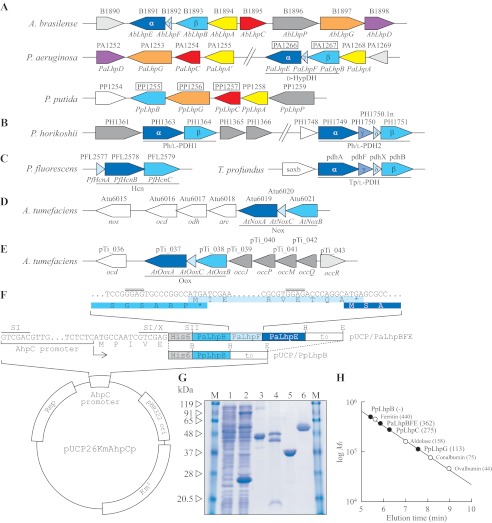
FIGURE 2.
Schematic gene clusters related to l-hydroxyproline metabolism of A. brasilense, P. aeruginosa, and P. putida (A); l-proline metabolism of hyperthermophilic archaea P. horikoshii and T. profundus (B); hydrogen cyanide biosynthesis of Pseudomonas fluorescens (C); and d-nopaline metabolism (D) and d-octopine metabolism (E) of Agrobacterium tumefaciens are shown. The gene cluster of A. brasilense was assumed from the genome sequence of Burkholderia sp. 383 because it is known that both nucleotide sequences around the AbLhpG gene, previously characterized as KGSADH (21), are very homologous. Putative genes in the boxes were purified and characterized in this study (see G). F, plasmid construction for expression of PpLhpB and PaLhpBFE genes in P. putida cells. The inset shows the nucleotide sequence of the PaLhpB, PaLhpF, and PaLhpE genes at the intergenic regions along with the corresponding deduced amino acid sequence (same colors as in A). Rep, replication protein. Putative ribosomal binding sites are indicated by double overlines. SI, X, SII, B, H, and E indicate SalI, XhoI, SacII, BamHI, HindIII, and EcoRI restriction enzyme sites, respectively. G, purification of recombinant PpLhpB (lane 3; 20 μg), PaLhpBFE (lane 4; 50 μg), PpLhpC (lane 5; 20 μg), and PpLhpG (lane 6; 20 μg). Lanes 1 and 2 are cell-free extracts (100 μg) of P. putida KT2442 (parent strain) and KT2442-oxyR1 mutant, respectively, and the major band with a molecular mass of ∼24 kDa is inherent AhpC protein in KT2442-oxyR1 mutant. M represents marker proteins. H, molecular mass determination for PpLhpBFE, PpLhpC, and PpLhpG (closed circles) using calibration obtained with a molecular mass standard (open circles). Values in parentheses are native molecular masses estimated using calibration. The elution profile is shown in supplemental Fig. S1.
Expression and Purification of His6-tagged Recombinant Proteins
PpLhpC and PpLhpG genes were expressed in E. coli cells, whereas PpLhpB and PaLhpBFE genes were expressed in P. putida cells. E. coli DH5α harboring pQE/PpLhpC or pQE/PpLhpG was grown at 37 °C to a turbidity of 0.6 at 600 nm in Super broth medium (pH 7.0; 12 g of tryptone, 24 g of yeast extract, 5 ml of glycerol, 3.81 g of KH2PO4, and 12.5 g of K2HPO4/liter) containing 50 mg/liter ampicillin. After the addition of 1 mm isopropyl β-d-thiogalactopyranoside, the culture was further grown for 6 h to induce the expression of His6-tagged protein. On the other hand, P. putida KT2442-oxyR1 (28) harboring pUCP/PpLhpB or pUCP/PaLhpBFE obtained using the the heat-shock transformation method was grown at 30 °C overnight in LB medium containing 50 mg/liter rifampicin and 50 mg/liter kanamycin. Cells were harvested and resuspended in Buffer A (50 mm sodium phosphate buffer (pH 8.0) containing 2 mm MgCl2, 300 mm NaCl, 1% (v/v) Tween 20, and 10 mm imidazole). The cells were then disrupted by sonication, and the solution was centrifuged. The supernatant was loaded onto a nickel-nitrilotriacetic acid Superflow column (Qiagen), which was equilibrated with Buffer A, linked to the BioAssist eZ system (Tosoh). The column was washed with Buffer B (50 mm sodium phosphate buffer (pH 8.0) containing 2 mm MgCl2, 300 mm NaCl, 0.1% (v/v) Tween 20, 10% (v/v) glycerol, and 50 mm imidazole). The enzymes were then eluted with Buffer C (pH 8.0; Buffer B containing 250 mm imidazole instead of 50 mm imidazole). For purification of PpLhpC and PpLhpG, a buffer system without Tween 20 was used. PpLhpB and PpLhpC were stored at 4 °C and used within 1 day in further experiments, whereas PaLhpBFE and PpLhpG were concentrated by ultrafiltration with Centriplus YM-30 (Millipore), dialyzed against 50 mm Tris-HCl buffer (pH 9.0) containing 2 mm MgCl2 and 50% (v/v) glycerol, and stored at −35 °C until use.
The native molecular mass of recombinant proteins was estimated by gel filtration, which was carried out using an HPLC system at a flow rate of 1 ml/min. The purified enzyme was loaded onto a TSKgel G3000SWXL column (Tosoh) equilibrated with 50 mm Tris-HCl buffer (pH 8.0). A high molecular weight gel filtration calibration kit (GE Healthcare) was used as molecular markers.
Enzyme Assays
All enzyme assays were performed at 30 °C.
d-HypDH Assay
d-HypDH activity was assayed by monitoring the reduction rate of p-iodonitrotetrazolium violet (INT) (artificial electron acceptor) and phenazine methosulfate (PMS) (electron transfer intermediate) using a Shimadzu UV-1800 spectrophotometer (Shimadzu GLC Ltd., Tokyo, Japan). The standard reaction mixture contained 0.25 mm INT and 0.06 mm PMS in 50 mm Tris-HCl buffer (pH 9.0). The reaction was started by the addition of 100 mm d-hydroxyproline (100 μl) with a final reaction volume of 1 ml. The millimolar absorption coefficient (ϵ) for INT was 15.0 mm−1·cm−1 at 490 nm. To estimate the specificity of electron acceptors, nitroblue tetrazolium (NBT) together with PMS (ϵ = 36.0 mm−1·cm−1 at 530 nm), 2,6-dichloroindophenol (ϵ = 21.5 mm−1·cm−1 at 600 nm), ferricyanide (ϵ = 1.04 mm−1·cm−1 at 405 nm), horse liver cytochrome c (ϵ = 15.3 mm−1·cm−1 at 553 nm), and NAD(P)+ (ϵ = 6.2 mm−1·cm−1 at 340 nm) at a final concentration of 0.25 mm were used. The kinetic parameters, Km and kcat values, were calculated by a Lineweaver-Burk plot.
Pyr4H2C Deaminase Assay
Pyr4H2C deaminase was assayed spectrophotometrically in the coupling system with KGSADH. The recombinant AbLhpG protein (KGSADH-III) was prepared by the method described previously (21). The reaction mixture (1 ml) consisted of 50 mm Tris-HCl buffer (pH 8.0), 10 mm Pyr4H2C, 1.5 mm NADP+, and 1 unit of purified AbLhpG. Pyr4H2C was enzymatically synthesized from d-hydroxyproline by Pa/d-HypDH as described below. No change in absorbance at 340 nm was found at this stage. The reaction was started by the addition of a small amount of protein, and the increasing absorption caused by NADPH produced in the reaction was measured at 340 nm.
KGSADH Assay
KGSADH activity was assayed by the method described previously (20, 21).
l-Hydroxyproline 2-Epimerase Assay
The reaction mixture consisted of 50 mm Tris-HCl (pH 8.0), 0.25 mm INT, 0.06 mm PMS, and 10 mm l-hydroxyproline. After the addition of a small amount of cell-free extract from P. putida or P. aeruginosa, the mixture was incubated for 5 min at 30 °C. No change in absorbance at 490 nm was found at this stage. The reaction was started by the addition of 1 unit of Pa/d-HypDH, and the increasing absorption was measured at 490 nm. l-Hydroxyproline 2-epimerase in cell-free extract converts l-hydroxyproline to d-hydroxyproline, which is detectable by the d-HypDH assay system.
Zymogram Staining Analysis
Cell-free extract or purified d-HypDH was separated by non-denaturing PAGE with a 10% (w/v) gel at 4 °C. The gels were then soaked in 10 ml of staining solution consisting of 50 mm Tris-HCl (pH 9.0), 2 mm MgCl2, 0.25 mm INT, 0.06 mm PMS, and 10 mm d-hydroxyproline at room temperature for 15 min in the dark. Dehydrogenase activity appeared as a red band.
Internal Peptide Analysis of Recombinant Pa/d-HypDH
As described under “Results,” when the PaLhpBFE gene cluster was expressed in P. putida cells, the purified enzyme consisted of three bands with different molecular masses in SDS-PAGE analysis (see Fig. 2G, lane 4). To identify these proteins with molecular masses of 39, 8, and 44 kDa as PaLhpB, PaLhpF and PaLhpE, respectively, each protein band on the 12% (w/v) SDS-PAGE gel was excised, and the gel pieces were incubated with 10 mm dithiothreitol at 56 °C for 45 min. Then protein in the gel was incubated with 55 mm iodoacetamide at room temperature for 45 min and subjected to in-gel digestion with trypsin (Promega). The digests were applied to a liquid chromatography (LC)-electrospray ionization-tandem mass spectrometer (LCQ FleetTM LC-ESI-MSn system, ThermoFisher Scientific) in the positive ion mode. The data were collected in the data-dependent tandem mass spectrometry (MS/MS) scan mode and analyzed using SEQUEST software (ThermoFisher Scientific).
Determination of Flavin Cofactor in d-HypDHs
Purified Pp/d-HypDH and Pa/d-HypDH were dialyzed against H2O, and cofactors were released by heat denaturation. After removal of the precipitate formed by centrifugation, the supernatant was used to identify the flavin cofactor(s) by HPLC. HPLC analysis was carried out using a TSKgel ODS-80Tm column (4.6 × 150 mm; Tosoh). An isocratic elution (10 min) with 10 mm potassium phosphate (pH 6.0) followed by a linear gradient (30 min) between 0 and 70% methanol in the same solution was used for the elution. The flow rate was 1.0 ml/min, and elution was monitored by the absorbance at 260 nm. Non-heme iron contents in Pa/d-HypDH were estimated by the method of Fish (29).
Identification of Reaction Product of d-HypDHs
The reaction mixture (200 ml) consisted of 50 mm Tris-HCl buffer (pH 9.0), 2 mm MgCl2, 50 mm d-hydroxyproline, and 0.05 mm PMS. After the addition of 50 mg of purified Pp/d-HypDH or Pa/d-HypDH, the mixture was left at 30 °C in the dark overnight. Remaining PMS was removed by adding charcoal and filtered (referred to as Solution X). Solution X was applied to a Dowex 1X2 Cl− form (100–200 mesh) resin column followed by a 0–1 m gradient of HCl. To identify a putative Δ1-pyrroline product, 125 μl of each fraction was added to 500 μl of 5% (w/v) p-dimethylaminobenzaldehyde in n-propanol followed by 1.25 ml of 3 n H2SO4. The mixtures were shaken thoroughly and incubated at 70 °C for 5 min, and the absorbance was measured at 550 nm. Active fractions were combined, adjusted to ∼pH 11 with NaOH, and lyophilized. The 1H NMR spectrum of d-hydroxyproline (400 MHz, D2O) was as follows: 4.41 (1H, m), 4.04 (1H, dd, J1 = 10 Hz, J2 = 4 Hz), 3.30 (1H, dt, J1 = 12 Hz, J2 = 2 Hz), 3.20 (1H, dd, J1 = 12 Hz, J2 = 4 Hz), 2.34 (1H, m), and 2.09 (1H, m). As described under “Results,” Pyr4H2C was not purified by this procedure. However, when Solution X was used as the substrate of d-HypDH, no significant activity was found, indicating the complete conversion of d-hydroxyproline to the reaction product. Therefore, this sample was used as 50 mm Pyr4H2C solution for measuring the activity of PpLhpC.
Target Disruption of l-Hydroxyproline Gene Cluster
The Kmr cassette of pUC4K was inserted into the single SalI site in the coding sequence of the PpLhpC gene of pQE/PpLhpC to yield pQE/PpLhpC::Km. To introduce the restriction site for BamHI at the 5′- and 3′-ends of the DNA fragment containing the Kmr gene in the PpLhpC gene, PCR was carried out using pQE/PpLhpC::Km as a template and two primers, P7 and P8 (supplemental Table S1). The 2.3-kbp BamHI DNA fragment was subcloned into the BamHI site of the suicide vector pKNG101 (sacB Smr) (30) to yield pKNG/PpLhpC::Km (see Fig. 6C). This plasmid was introduced into parent strain P. putida KT2442, and the resulting transformants were selected for integration of the Kmr marker in the chromosome on an LB agar plate containing 50 mg/liter kanamycin. The mutant was analyzed by colony PCR with P15 and P16 primers designed to target the flanking region of the PpLhpC gene to confirm the presence of its disrupted copy of the PpLhpC gene.
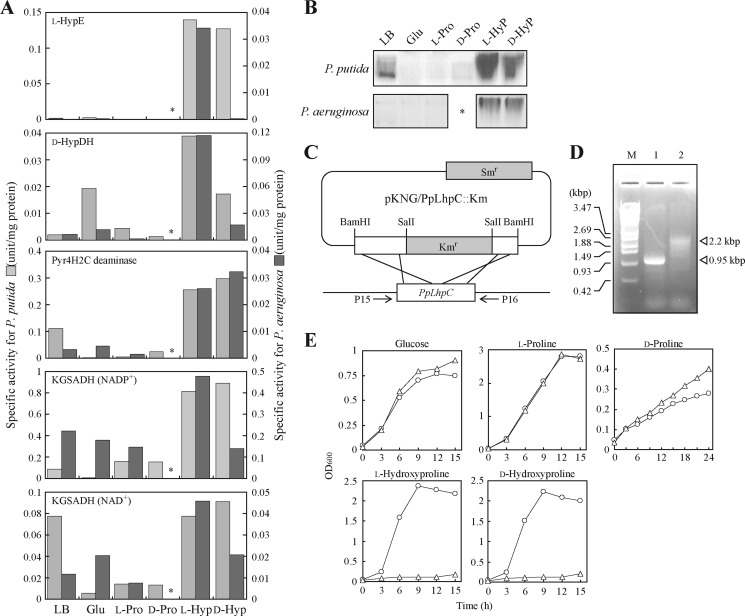
FIGURE 6.
Physiological analysis of Lhp gene cluster. A, enzyme activities of cell-free extracts prepared from cells grown on LB medium and minimal medium supplemented with 2% (w/v) glucose (Glu), l-proline (l-Pro), d-proline (d-Pro), l-hydroxyproline (l-Hyp), or d-hydroxyproline (d-Hyp). Light gray bar, P. putida for left axis; dark gray bar, P. aeruginosa for right axis. P. aeruginosa could not grow on d-proline (asterisks). KGSADH activity was measured using NAD+ or NADP+ as coenzymes. B, zymogram staining analysis of d-HypDH using the same cell-free extracts as in A. Fifty micrograms of cell-free extract was analyzed. Upper panel, P. putida; lower panel, P. aeruginosa. C, construction of an LhpC disruptant of P. putida. The introduction of pKNG/PpLhpC::Km into P. putida parent strain KT2442 resulted in the disruption of chromosomal PpLhpC by homologous recombination. D, genomic PCR analysis of KT2442 (lane 1) and LhpC− mutant strains (lane 2). M represents DNA size markers. E, growth curves of P. putida KT2442 (circles) and LhpC− mutant strains (triangles) on the indicated carbon sources.
Amino Acid Sequence Alignment and Phylogenetic Analysis
Protein sequences were analyzed using Protein BLAST and the ClustalW program distributed by DDBJ (DNA Data Bank of Japan). The phylogenetic tree was produced using the TreeView 1.6.1. program.
RESULTS
Gene Cluster Related to Bacterial l-Hydroxyproline Metabolism
As described in the Introduction, we previously identified l-hydroxyproline-inducible KGSADH-III (referred to as LhpG in this report) from A. brasilense (18) (Fig. 2A). Protein BLAST analysis revealed that the homologous gene exists as a gene cluster on the genomes of many bacteria, including P. putida and P. aeruginosa, with the ability to grow on l-hydroxyproline. The gene cluster of P. putida consisted of five hypothetical genes, LhpB (FAD-dependent oxidoreductase), LhpG (KGSADH), LhpC (dihydrodipicolinate synthase), LhpA (epimerase), and LhpP (amino-acid permease) among which LhpB was a likely candidate for d-HypDH (Fig. 2A). Surprisingly, the gene cluster of P. aeruginosa contained several additional genes, LhpB (glycine/d-amino-acid oxidase), LhpE (heterotetrameric sarcosine oxidase α-subunit), LhpF (ferredoxin), and LhpD (malate/lactate dehydrogenase), in the order of LhpB-F-E-D (PaLhpF was not identified in the database, but we found an AbLhpF gene homolog between PaLhpB and PaLhpE genes). The 3′-part of PaLhpB and PaLhpF was slightly overlapped by the 5′-part of PaLhpF and PaLhpE, respectively (Fig. 2F, inset). A similar tendency was also found in heteromeric Tp/l-PDH (8) (Fig. 2B). This suggested the possibility that d-HypDHs of P. putida and P. aeruginosa are largely different and that PaLhpB-F-E encode each subunit in the heteromeric structure.
Expression of Recombinant Proteins in P. putida Cells
Although we first attempted to express PpLhpB, PaLhpB, PaLhpF, and PaLhpE proteins in E. coli cells, all mainly appeared in the fraction of the insoluble form despite efforts involving a survey of E. coli strains and vectors (data not shown). No improvement was found in the co-expression of PaLhpB-F-E. We previously revealed that P. putida KT2442-oxyR1 strain constitutively produces a soluble AhpC protein, a small subunit of alkyl hydroperoxide reductase, with a molecular mass of 24 kDa (Fig. 2G, lanes 1 and 2) (28). Therefore, we expressed His6-tagged PpLhpB protein under regulation of the ahpC promoter inserted into the Escherichia-Pseudomonas shuttle plasmid in P. putida KT2442-oxyR1 cells (Fig. 2F) and purified it to homogeneity in a single step using immobilized metal (Ni2+) affinity chromatography. SDS-PAGE gave a single band with a molecular mass of ∼47 kDa, which corresponded to the theoretical value of PpLhpB (46,856.49 Da) (Fig. 2G, lane 3). On the other hand, it was impossible to determine the native molecular mass because of the elution in the void volume of gel filtration column (Fig. 2H and supplemental Fig. S1). The purified enzyme was yellow, which is typical of flavoproteins, and showed 6.41 units/mg of protein specific activity toward d-hydroxyproline using standard assay conditions in the artificial electron acceptor system (PMS-INT; Table 1), indicating the function of d-hydroxyproline dehydrogenase (Pp/d-HypDH).
TABLE 1.
Kinetic parameters of d-hydroxyproline dehydrogenases from P. putida and P. aeruginosa
| Substrates | Electron acceptors | Specific activitya | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| units/mg protein | mm | min−1 | min−·mm−1 | ||
| PpLhpB | |||||
| d-Hydroxyproline | PMS-INT | 6.41 ± 0.30 | 0.276 ± 0.012b | 280 ± 11b | 1,010 ± 4 |
| PMS-NBT | 0.463 ± 0.003 | 0.0802 ± 0.008c | 19.7 ± 1.3c | 246 ± 9 | |
| d-Proline | PMS-INT | 1.28 ± 0.20 | 6.58 ± 1.15d | 59.9 ± 7.5d | 9.15 ± 0.45 |
| PMS-NBT | 0.185 ± 0.002 | 3.49 ± 0.34d | 9.95 ± 0.66d | 2.86 ± 0.09 | |
| PaLhpBFE | |||||
| d-Hydroxyproline | PMS-INT | 21.1 ± 0.70 | 0.109 ± 0.009e | 2230 ± 122e | 20,500 ± 530 |
| PMS-NBT | 5.66 ± 0.40 | 0.121 ± 0.002e | 871 ± 10e | 7,200 ± 50 | |
| d-Proline | PMS-INT | 7.05 ± 0.02 | 8.55 ± 0.35f | 924 ± 28f | 108 ± 1 |
| PMS-NBT | 2.80 ± 0.05 | 11.0 ± 0.3f | 507 ± 18f | 46.0 ± 0.3 | |
a Under standard assay conditions as described under “Materials and Methods.”
b Seven different concentrations of d-hydroxyproline between 0.01 and 0.1 mm were used.
c Seven different concentrations of d-hydroxyproline between 0.02 and 1 mm were used.
d Six or seven different concentrations of d-proline between 1 and 6 mm were used.
e Nine different concentrations of d-hydroxyproline between 0.01 and 10 mm were used.
f Eight or 10 different concentrations of d-proline between 0.5 and 10 mm were used.
PaLhpB-F-E operon in which the His6 tag sequence was attached at the N terminus of the LhpB gene was also expressed in ahpC promoter in P. putida KT2442-oxyR1 cells (Fig. 2F), and the recombinant protein was successfully purified to homogeneity. Native PAGE gave a single band (see Fig. 3B, inset), and a symmetric peak appeared in gel filtration analysis (supplemental Fig. S1). On SDS-PAGE analysis, two major distinct bands (referred to as α- and β-subunits, respectively) along with one minor band (referred to as γ-subunit) were observed (Fig. 2G, lane 4). The estimated molecular masses of α-, β-, and γ-subunits (48, 42, and 10 kDa, respectively) were similar to the theoretical values of PaLhpE, PaLhpB, and PaLhpF (44,055.06, 41,133.39, and 8,184.55 Da, respectively). Furthermore, LC-electrospray ionization-MS/MS profiles using tryptic peptides of each protein band revealed that α-, β-, and γ-subunits corresponded to PaLhpE, PaLhpB, and PaLhpF, respectively (supplemental Fig. S2 and supplemental Tables S2–S4). Because the His6 tag sequence was attached only to the N terminus of PaLhpB, co-purification using Ni2+ affinity chromatography suggested that this protein consists of three different subunits. The molar ratio of α:β:γ was ∼1:1:1 as judged by image scanning of the SDS-PAGE peak areas using NIH ImageJ Version 1.45s software. The native molecular mass estimated by gel filtration using HPLC was ∼360 kDa (Fig. 2H), indicating that this protein molecule may be composed of a heterododecameric structure of subunits, i.e. α4β4γ4. Under the same assay conditions as PpLhpB, specific activity toward d-hydroxyproline was 21.1 units/mg of protein (Table 1). When compared with PpLhpB, the purified enzyme from P. aeruginosa was orange-brown rather than light yellow, indicating that some chromophores must be bound to protein (see below). These results suggested that PaLhpBFE is a different type of d-HypDH (Pa/d-HypDH).
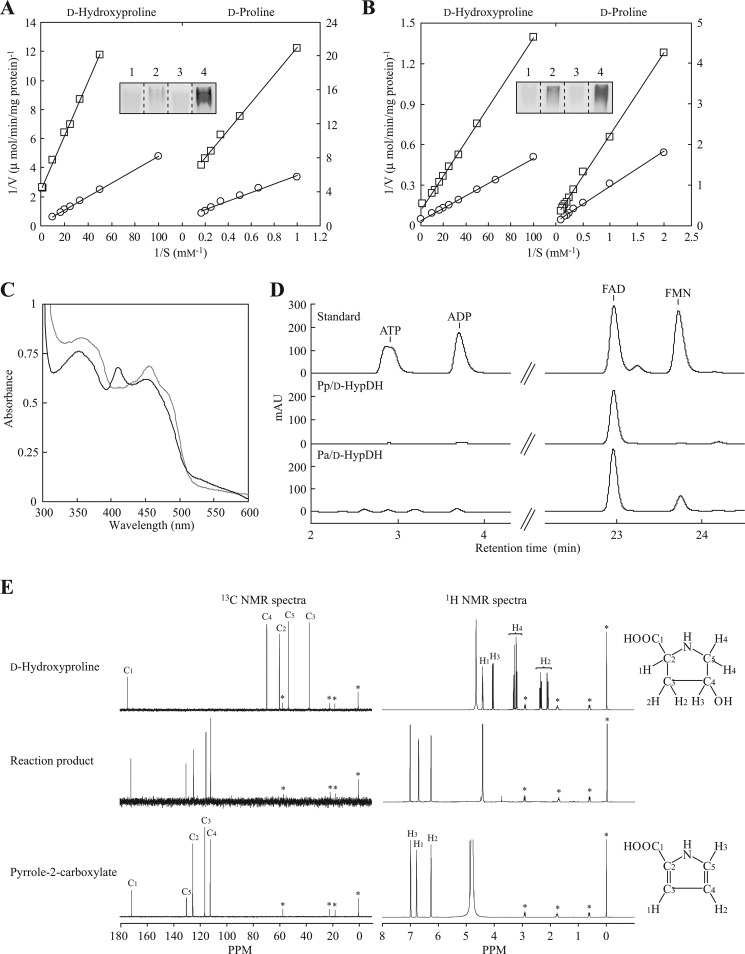
FIGURE 3.
Characterization of Pp/d-HypDH and Pa/d-HypDH. Lineweaver-Burk plots of Pp/d-HypDH (A) and Pa/d-HypDH (B) toward d-hydroxyproline and d-proline using PMS-INT (circle) and PMS-NBT (square) assay systems, respectively, are shown. The inset shows zymogram staining analysis using l-proline (lane 1), d-proline (lane 2), l-hydroxyproline (lane 3), and d-hydroxyproline (lane 4) as substrates together with the PMS-INT assay system. Purified enzymes (10 μg) were applied. C, absorption spectra of Pp/d-HypDH (gray line) and Pa/d-HypDH (black line). Each enzyme solution used was 5 mg/ml. D, HPLC analysis of prosthetic groups. Elution profiles of the standard mixture (top) and extracts of Pp/d-HypDH (middle) and Pa/d-HypDH (bottom) are shown. E, identification of reaction product of Pa/d-HypDH by NMR. Upper panels, authentic d-hydroxyproline; middle panels, reaction product; right panels, authentic pyrrole-2-carboxylate. Left panels, 1H NMR spectra showing the assignments of protons in D2O; right panels, 13C NMR spectra showing the assignments of carbons in D2O. Asterisks indicate peaks derived from an internal standard. In this analysis, pyrrole-2-carboxylate was identified as the reaction product (see in text). Similar results were obtained when Pp/d-HypDH was used as an alternative (data not shown). mAU, milliabsorbance units.
Successful purification of both Pp/d-HypDH and Pa/d-HypDH from P. putida cells was achieved using a buffer system containing Tween 20 (see “Materials and Methods”). Furthermore, when P. putida and P. aeruginosa cells grown on l-hydroxyproline were disrupted using buffer in the absence of Tween 20, no significant d-HypDH activity was found in cell-free extracts (data not shown). These results indicated that both native and recombinant proteins of Pp/d-HypDH and Pa/d-HypDH might be tightly bound to the cytoplasmic membrane as described in preliminary reports (13, 15).
Substrate and Electron Acceptor Specificity of d-HypDHs
In addition to d-hydroxyproline, l-hydroxyproline, l-proline, and d-proline were tested as substrates for Pp/d-HypDH and Pa/d-HypDH. Among them, significant activity was observed only with d-proline (see Fig. 3, A and B, insets), and the kinetic parameters of d-hydroxyproline and d-proline determined from the Lineweaver-Burk plot (Fig. 3, A and B) are shown in Table 1. The catalytic efficiency (kcat/Km) values of Pp/d-HypDH and Pa/d-HypDH (20,500 and 1010 min−1·mm−1) with d-hydroxyproline were 190- and 110-fold higher than those with d-proline (108 and 9.15 min−1·mm−1), mainly caused by 78- and 24-fold lower Km values, respectively. Both Pp/d-HypDH and Pa/d-HypDH showed electron acceptor activity only in PMS-INT and PMS-NBT but not 2,6-dichloroindophenol, ferricyanide, cytochrome c, and NAD(P)+. kcat/Km values with d-hydroxyproline of Pp/d-HypDH and Pa/d-HypDH using PMS-INT were 2.8- and 4.1-fold, respectively, of those using PMS-NBT, mainly due to lower kcat values. The preference of PMS-INT over PMS-NBT was also observed when d-proline was used as a substrate instead of d-hydroxyproline.
Among the characterized amino-acid dehydrogenases, only d-PDH from a hyperthermophilic archaeon, Pyrobaculum islandicum (31), showed significant activity with d-hydroxyproline and close Km values between d-proline and d-hydroxyproline (4.2 and 9.5 mm, respectively) (supplemental Table S6). This enzyme has broad substrate specificity with many d-amino acids in addition to d-hydroxyproline. Furthermore, 2,6-dichloroindophenol and/or ferricyanide is the most preferred electron acceptor(s) for known dl-PDHs, including P. islandicum d-PDH. To our knowledge, this is the first report of amino-acid dehydrogenase(s) with high specificity for d-hydroxyproline.
Effect of pH on Activity of d-HypDHs
pH activity profiles of Pp/d-HypDH and Pa/d-HypDH were similar: their maximal activities were observed at the range of pH 9–9.75 (supplemental Fig. S2). Furthermore, the enzymes maintained over 70% of their maximal activity after 2 h of incubation at pH 8–10.5 (data not shown). Apparent pKa values determined from the acidic limb of the profiles were 8.5–8.9. It is likely that these properties are similar to eukaryotic d-amino-acid oxidases (32) rather than archaeal dl-PDHs (supplemental Fig. S3).
Prosthetic Groups in d-HypDHs
As described above, both Pp/d-HypDH and Pa/d-HypDH were yellow (or orange-brown), and the addition of FAD or FMN in the standard assay mixture had no effect on the activity (data not shown). The UV-visible absorption spectrum of Pp/d-HypDH exhibited two pronounced absorption peaks at 354–360 and 454–456 nm, indicating typical flavoproteins (Fig. 3C). On the other hand, that of Pa/d-HypDH revealed absorption maxima at 353, 409, and 449–453 nm. Because the presence of [2Fe-2S] iron-sulfur cluster motifs was predictable on α- and γ-subunits on the primary structure (see below), this spectrum is likely consistent with the sum of the spectra of typical flavoproteins (maxima at about 370 and 450 nm) and an oxidized [2Fe-2S] center (maxima at about 320, 415, and 455 nm): the latter was particularly similar to that of [2Fe-2S] ferredoxin from E. coli (cf. Fig. 5 in Ref. 33).
FIGURE 5.
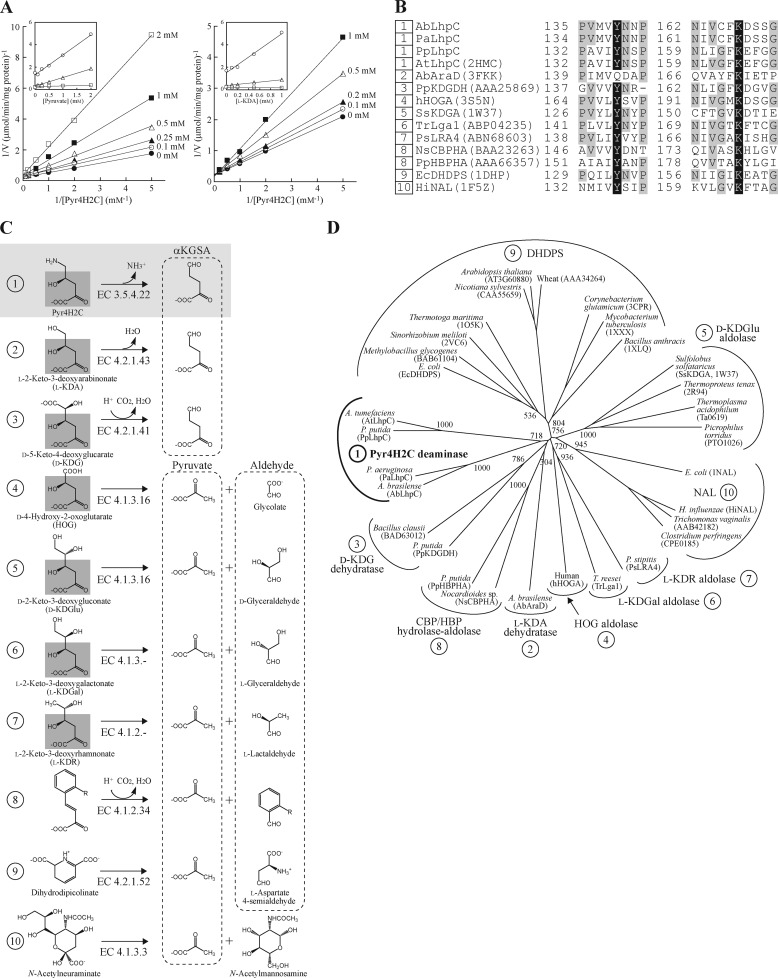
Characterization of Pyr4H2C deaminase within dihydrodipicolinate synthase (DHDPS)/N-acetylneuraminate lyase (NAL) protein family. A, inhibition of Pyr4H2C deaminase by pyruvate (right) and l-KDA (left). The Pyr4H2C concentrations were changed with the fixed concentrations of pyruvate (0, 0.1, 0.25, 0.5, 1, and 2 mm) or l-KDA (0, 0.1, 0.2, 0.5, and 1 mm). The inset shows the slopes (triangles), 1/v axis intercepts (squares), and Km values (circles) versus [pyruvate] or [l-KDA]. B, partial multiple sequence alignment of deduced amino acid sequences of Pyr4H2C deaminase. Catalytic lysine and tyrosine residues are shown as white letters in black boxes. Other highly conserved amino acid residues are shown as gray-shaded letters. Sequence names correspond to those in C. Numbers with the sequences indicate subfamily in B and C. C, schematic reactions catalyzed by dihydrodipicolinate synthase/N-acetylneuraminate lyase proteins. The common moiety with Pyr4H2C is shaded in gray. D, phylogenetic tree of the dihydrodipicolinate synthase/N-acetylneuraminate lyase protein family consisting of nine subfamilies. The number on each branch indicates the bootstrap value. HOG, 4-hydroxy-2-oxoglutarate.
We attempted to identify the prosthetic cofactors in Pp/d-HypDH and Pa/d-HypDH using HPLC (Fig. 3D). Authentic FAD, FMN, ATP, and ADP were used as standards. As a result, Pp/d-HypDH as well as most of the other amino-acid dehydrogenases contained only FAD. Although the molar value per mole of enzyme was unclear because the native molecular mass was not determined by gel filtration (supplemental Fig. S1), ∼0.02 μmol of FAD was released from 1 mg of the purified enzyme. Because one FAD-binding motif was predictable on the primary structure (see below), the quaternary structure may be a homodimer. Interestingly, both FAD and FMN were detected within Pa/d-HypDH. Moreover, we determined that there were ∼8 mol of FAD and ∼4 mol of FMN/mol of enzyme. On the other hand, iron contents per mole of enzyme were ∼7.8. Because the quaternary structure is α4β4γ4, two FADs, one FMN, and one [2Fe-2S]-iron sulfur cluster might be contained within the αβγ heterotrimer.
Identification of Reaction Product by d-HypDH
The reaction products of d-hydroxyproline dehydrogenation catalyzed by Pp/d-HypDH and Pa/d-HypDH were purified using anion exchange chromatography and identified by NMR. Although the reaction product was postulated to be Pyr4H2C or Pyr3H5C, the NMR spectra were very similar to those of pyrrole-2-carboxylate (Fig. 3E). Furthermore, when the reaction product and authentic pyrrole-2-carboxylate were co-solved in D2O, the NMR spectra overlapped completely (data not shown). It is known that in rats l-hydroxyproline is converted to Pyr4H2C by l-amino-acid oxidase (dehydrogenase) followed by pyrrole-2-carboxylate upon acidification of the reaction mixture via the unstable oxidation intermediate, probably Pyr4H2C (34). In this study, we observed that a pyrroline-related compound detected using p-dimethylaminobenzaldehyde is eluted from a column resin in the range of 0.7–1 m HCl (see “Materials and Methods”). Furthermore, when the reaction product (under alkaline conditions) was used directly for the PpLhpC-PpLhpG coupling assay, the final reaction product was identified as α-ketoglutarate (see below). These results suggested that the reaction product from d-hydroxyproline by Pp/d-HypDH and Pa/d-HypDH is Pyr4H2C, not Pyr3H5C. Although only P. islandicum d-PDH shows a similar conversion from d-proline to Δ1-pyrroline-2-carboxylate (but not Δ1-pyrroline-5-carboxylate) (31), its metabolic fate is unclear.
Amino Acid Sequence Analysis of d-HypDHs
Pp/d-HypDH showed only ∼20% sequence similarity to β-subunit of Pa/d-HypDH (Pa/d-HypDHβ) and other homomeric d-amino-acid dehydrogenases (31, 35) (Fig. 4A). On the other hand, the N-terminal 30 amino acid residues among these proteins had somewhat higher homology (∼30%) than other parts of the proteins. This region contains a highly conserved typical ADP-binding motif with a βαβ-fold (Gly-X-Gly-X2-Gly) (shaded in red), described by Wierenga et al. (36), confirming that Pp/d-HypDH contains only FAD as a prosthetic group (probably one FAD per subunit).
FIGURE 4.
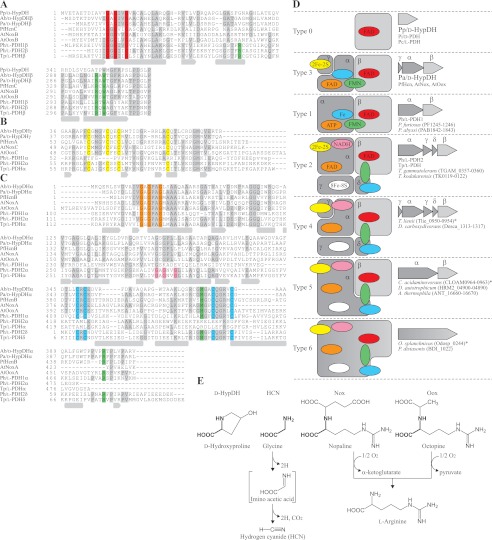
Partial multiple sequence alignment of deduced amino acid sequences of d-HypDHs. GenBank accession numbers are shown in Fig. 2, A–E. Gray-shaded letters indicate highly conserved amino acid residues. Secondary structures of Ph/l-PDH1 (Protein Data Bank code 1Y56) are shown under the sequence: α-helices, gray cylinders; β-strands, gray arrows. A, alignment of Pp/d-HypDH, β-subunits of Pa/d-HypDH, Hcn (PfHcnC), Nox (AtNoxB), Oox (AtOoxB), Ph/l-PDH1, Ph/l-PDH2, and Tp/l-PDH. The ADP-binding motif and FMN-binding sites (also in C) are shaded in red and green, respectively. B, alignment of β-subunits of Pa/d-HypDH, Hcn (PfHcnA), Nox (AtNoxC), Oox (AtOoxC), and N-terminal regions of α-subunits of Ph/l-PDH1, Ph/l-PDH2, and Tp/l-PDH. The [2Fe-2S]-binding motif is shaded in yellow. C, alignment of α-subunits of Pa/d-HypDH, Hcn (PfHcnB), Nox (AtNoxA), Oox (AtOoxA), Ph/l-PDH1, Ph/l-PDH2, and Tp/l-PDH and δ-subunits of Ph/l-PDH2 and Tp/l-PDH. Two ADP-binding motifs and the [2Fe-2S]-binding motif are shaded in orange, pink, and cyan, respectively. D, schematic subunit assembly of dehydrogenases. The colors of putative binding sites for prosthetic groups correspond to those in A–C. Figures on the right indicate the gene cluster encoding these proteins on the genomes of microorganisms. The deduced amino acid sequences with asterisks are shown in supplemental Table S3. E, structural formulas for d-hydroxyproline, glycine, d-nopaline, and d-octopine and schematic reactions by d-HypDH, Hcn, Nox, and Oox. The common moiety with d-hydroxyproline is shown in bold.
Relatively high homology with Pa/d-HypDH (30–40%) was found in the HcnABC complex of hydrogen cyanide synthase, NoxABC complex of d-nopaline oxidase, and OoxABC complex of d-octopine oxidase from bacteria (Fig. 4, A, B, and C, PfHcn, AtNox, and AtOox, respectively; Refs. 37–39). “HcnB, NoxA, and OoxA,” “HcnC, NoxB, and OoxB,” and “HcnA, NoxC, and OoxC” correspond to α-, β-, and γ-subunits, respectively, of Pa/d-HypDH. Furthermore, α- and β-subunits of Ph/l-PDH1 and Tp/l-PDH were also homologous with those of Pa/d-HypDH. Two ADP-binding motifs as described above were found at the N terminus of α- and β-subunits of Pa/d-HypDH (shaded in orange and red, respectively), indicating that the binding sites of two FADs in the Pa/d-HypDHαβγ complex may be here. In the case of Tp/l-PDHα, another ADP-binding motif is located in the middle region (shaded in pink) that may be related to the unique NADH dehydrogenation activity (8). In fact, Pa/d-HypDH showed no NADH dehydrogenation activity (data not shown). Interestingly, the homologous motifs of Ph/l-PDH1α and Tp/l-PDHα are located at over 120 amino acid residues in the internal region from the N terminus (see next section). On the other hand, two different types of [2Fe-2S]-binding motifs were found in Pa/d-HypDHα and Pa/d-HypDHγ (shaded in cyan and yellow, respectively). The former consisted of Cys-X-Cys-X15–20-Cys-X4-Cys near the C terminus, whereas the latter consisted of Cys-X4-Cys-X2-Cys-X11–12-Cys and matched the iron-sulfur binding signature of [2Fe-2S] ferredoxins. Therefore, the either of the two may be the [2Fe-2S]-binding site in the Pa/d-HypDHαβγ complex.
Functional Characterization of PpLhpC as Pyr4H2C Deaminase
The LhpC gene is commonly contained within l-hydroxyproline operons of P. putida and P. aeruginosa (sequence identity of ∼30%) (Fig. 2A). To estimate enzyme function, PpLhpC was overexpressed in E. coli cells by induction with isopropyl β-d-thiogalactopyranoside as a His6-tagged protein and purified to homogeneity with a nickel-chelating affinity column (Fig. 2G, lane 5). The apparent molecular mass estimated by SDS-PAGE was 35 kDa, coinciding with the theoretical value of 35,341.67 Da. The native molecular mass estimated by gel filtration was ∼275 kDa (Fig. 2H and supplemental Fig. S1), indicating a homooctameric structure.
As described above, LhpC belongs to the dihydrodipicolinate synthase/N-acetylneuraminate lyase family (22). Because most members of this protein family produce pyruvate and several aldehydes in the aldol cleavage reaction (see Fig. 5C), potential aldolation activity with Pyr4H2C in PpLhpC was first measured using lactic dehydrogenase as a coupling enzyme but was not detected (data not shown). Therefore, we alternatively used the purified recombinant AbLhpG (KGSADH), which shows high specificity with αKGSA, as a coupling enzyme (21) (also see next section). Under this assay system, PpLhpC showed specific activity of 2.41 units/mg of protein in the presence of 10 mm Pyr4H2C (and 1.5 mm NADP+). Furthermore, when NADP+-dependent l-glutamate dehydrogenase co-existed in the reaction mixture, the increase in absorbance at 340 nm was completely eliminated (data not shown) as a result of cooperative reactions by PpLhpC (Pyr4H2C → αKGSA + NH3), KGSADH (αKGSA + NADP+ → α-ketoglutarate + NADPH) and l-glutamate dehydrogenase (α-ketoglutarate + NH3 + NADPH → l-glutamate + NADP+). These results suggested that PpLhpC plays a role as a Pyr4H2C deaminase. The Km and kcat values with Pyr4H2C were estimated to be 0.693 ± 0.031 mm and 71.5 ± 2.1 min−1, respectively. Pyruvate is a common substrate for dihydrodipicolinate synthase/N-acetylneuraminate lyase proteins as described above. On the other hand, l-2-keto-3-deoxyarabinonate (l-KDA) has a hydroxyl group at the C5 position of Pyr4H2C (4-hydroxy-2-oxo-5-aminovalerate) instead of an amino group. Because l-KDA dehydratase also belongs to the dihydrodipicolinate synthase/N-acetylneuraminate lyase family (see “Discussion”), one possibility was that l-KDA is an alternative substrate for potential dehydration of Pyr4H2C deaminase. However, both pyruvate and l-KDA inhibited the enzyme competitively, and the Ki values were 0.492 and 0.592 mm, respectively (Fig. 5A).
Functional Characterization of PpLhpG as KGSADH
In addition to PpLhpC, His6-tagged PpLhpG was overexpressed in E. coli cells and purified to homogeneity with a nickel-chelating affinity column (Fig. 2G, lane 6). The apparent molecular mass of His6-tagged PpLhpC was estimated by SDS-PAGE analysis as 55 kDa, coinciding with the theoretical value of 56,244.13 Da. The native molecular mass estimated by gel filtration was ∼113 kDa (Fig. 2H and supplemental Fig. S1), indicating a homodimeric structure. As expected from the significant sequence similarity to AbLhpG (66% identity), the recombinant PpLhpG protein showed specific activities for αKGSA of 55.5 and 25.2 units/mg of protein in the presence of NADP+ and NAD+, respectively (Table 2). The kcat/Km with NADP+ (83,500 min−1·mm−1) was 134-fold higher than that with NAD+, mainly caused by the lower Km values (0.0230 and 6.15 mm−1, respectively; Table 3). Under the same assay conditions, AbLhpG showed similar kcat/Km values to αKGSA in the presence of NAD+ or NADP+, whereas kcat/Km with NADP+ was ∼22-fold higher than with NAD+ (21). Various aldehydes, including αKGSA, were tested as substrates for dehydrogenation by PpLhpG in the presence of NADP+. αKGSA was clearly the best substrate, whereas activities with a number of aliphatic aldehydes from C3 to C8 were only 10% less than those with αKGSA (Table 2). These results indicated that PpLhpG possesses NADP+ preference and high specificity toward αKGSA similar to AbLhpG.
TABLE 2.
Substrate specificity of KGSADH from P. putida
| Substrate | Coenzyme | Specific activitya |
|
|---|---|---|---|
| PpLhpG | AbLhpGb | ||
| units/mg protein | |||
| αKGSA | NADP+ | 55.5 (100)c | 16.4 (53.4)d |
| NAD+ | 25.2 (45.4) | 30.7 (100) | |
| Propionaldehyde (C3) | NADP+ | 0.045 (0.08) | 0.7 (2.3) |
| Butylaldehyde (C4) | NADP+ | 2.5 (4.5) | 2.8 (9.0) |
| Valeraldehyde (C5) | NADP+ | 2.9 (5.2) | 2.6 (8.3) |
| Hexylaldehyde (C6) | NADP+ | 4.5 (8.1) | 6.8 (22.2) |
| Heptylaldehyde (C7) | NADP+ | 1.1 (2.0) | 6.4 (20.8) |
| Octylaldehyde (C8) | NADP+ | 0.98 (1.8) | 3.8 (12.3) |
a Under standard assay conditions as described under “Materials and Methods.” All aldehyde concentrations were 1 mm.
b KGSADH-III from A. brasilense (21).
c Values relative to αKGSA in the presence of NADP+ (100%).
d Values relative to αKGSA in the presence of NAD+ (100%).
TABLE 3.
Kinetic parameters for coenzymes of KGSADH from P. putida
| Coenzyme | Km | kcat | kcat/Km | |
|---|---|---|---|---|
| mm | min−1 | min−1·mm−1 | ||
| NADP+ | 0.0230 ± 0.001a | 1,990 ± 34a | 83,500 ± 4160 (100)b | |
| NAD+ | 6.15 ± 0.57c | 3,850 ± 360c | 625 ± 12 (7.0) | |
a Six different concentrations of NADP+ between 0.01 and 0.1 mm were used. αKGSA was 1 mm.
b Values relative to NADP+ (100%).
c Six different concentrations of NAD+ between 1 and 5 mm were used. αKGSA was 1 mm.
Gene Regulation and Disruptant Analysis
When P. putida and P. aeruginosa were grown on l-hydroxyproline or d-hydroxyproline, almost all four metabolic enzymes were induced in cell-free extracts (Fig. 6A). The induction of d-HypDH was also detected in zymogram staining analysis (Fig. 6B). Although (partially) incomplete induction of KGSADH activity was found in P. aeruginosa, it has been reported that this bacterium possesses a constitutively expressed KGSADH isozyme(s) and/or other aldehyde dehydrogenase(s) with activity for αKGSA (14). To estimate the physiological role(s) of the Lhp gene cluster in l-hydroxyproline metabolism, we carried out gene disruption experiments by introducing a Kmr gene at the center of the PpLhpC gene (encoding Pyr4H2C deaminase) (Fig. 6, C and D). The obtained LhpC− mutant strain was distinct from the wild-type strain (KT2442) in that neither l-hydroxyproline nor d-hydroxyproline as a sole carbon source supported growth (Fig. 6E). On the other hand, there was no difference in growth on d-proline (an alternative substrate for d-HypDH), glucose, and l-proline between the two strains. Indeed, only l-hydroxyproline 2-epimerase activity was significantly found in the cell-free extract prepared from LhpC− mutant cells grown on LB medium supplemented with l-hydroxyproline; d-HypDH, Pyr4H2C deaminase, and KGSADH activities were not found (data not shown). These results indicated that LhpC disruption also led to the inactivation of LhpB and LhpG genes within the Lhp operon (Fig. 2A). Overall, these results clearly suggested that the Lhp gene cluster is involved in l-hydroxyproline (as well as d-hydroxyproline) but not d-proline metabolism.
DISCUSSION
Comparisons between Pa/d-HypDH and Pp/d-HypDH
Although Pa/d-HypDH and Pp/d-HypDH show very high specificity for d-hydroxyproline (as a substrate) and PMS-INT or PMS-NBT (as an electron acceptor), the former heteromeric enzyme contains FAD, FMN, and a [2Fe-2S] iron-sulfur cluster as prosthetic groups, which is different from the latter homomeric enzyme (only FAD), strongly indicating that d-HypDHs appeared convergently in P. putida and P. aeruginosa during an evolutionary stage. Only P. islandicum d-PDH shows significant activity for d-hydroxyproline in addition to d-proline, and the manner of dehydrogenation reaction is homologous to that of d-HypDH (31). However, it is likely that their substrate specificities are also acquired independently of each other because their sequence similarities are not high.
In this study, we first revealed that Pa/d-HypDH is phylogenetically related to hydrogen cyanide synthase (Hcn), d-nopaline oxidase (Nox), and d-octopine oxidase (Oox) of bacteria (37–39). Hcn, Nox, and Oox complexes are encoded by hcnA-B-C, noxB-C-A, and ooxB-C-A gene clusters, respectively, on the bacterial genome (Fig. 2, C, D, and E). Furthermore, each substrate of Pa/d-HypDH, Hcn, Nox, and Oox possesses structural formulas similar to the secondary amine (Fig. 4E), clearly indicating that they evolved from a common ancestor. Although it is likely that Hcn, Nox, and Oox are membrane-associated flavoproteins as are most amino-acid dehydrogenases (31, 35), the detailed features have not been reported. Pa/d-HypDH is the first of this type of protein to be experimentally characterized at the molecular level.
Comparisons with dl-PDHs from Archaea
d-HypDHs, in particular Pa/d-HypDH, show high sequence similarities to several dehydrogenases for l- and d-proline from archaea (8–11, 31). Their schematic structures are illustrated in Fig. 4D. The β-subunits of the heteromeric enzymes correspond to homomeric enzymes and commonly function as catalytic subunits, which contain one ADP-binding motif in the N-terminal region (for FAD binding) (shaded in red). In fact, Tp/l-PDHβ shows dehydrogenation activity for l-proline by itself (8). On the other hand, the α-subunits may contribute to the regulation of catalysis because of the binding of several prosthetic group(s), including FAD (Pa/d-HypDHα), ATP (Ph/l-PDH1α), and FAD and NADH (Tp/l-PDHα) at the ADP-binding motif(s) (shaded in orange and pink). The FMN molecule is located at the interface of α- and β-subunits in Ph/l-PDH1 (11), and the isoalloxazine ring is sandwiched by two hydrophobic residues, βTrp304 and αMet444, whereas the phosphate group is fixed by three hydrophilic residues, βArg46, βArg302, and αArg477. Pa/d-HypDH (as well as Hcn, Nox, and Oox) possesses the corresponding residues except for βArg46 (shaded in green), suggesting a similar manner of FMN binding to Ph/l-PDH1 (10). When compared with FAD and FMN, the functional roles of the iron-sulfur cluster in catalysis are unclear. Cysteine-cluster ([2Fe-2S]-binding motif) in Ph/l-PDH1α (shaded in cyan) may be structurally conserved in Pa/d-HypDHα and Tp/l-PDHδ, whereas the homologous [2Fe-2S]-binding motifs in Pa/d-HypDHγ and Tp/l-PDHα (shaded in yellow) are not found in Ph/l-PDH1α. Furthermore, only Tp/l-PDHγ contains the [8Fe-8S] cluster. This suggests that the components and structural location of iron-sulfur clusters in Pa/d-HypDH may be different from Ph/l-PDH1 and Tp/l-PDH.
We found that putative l-PDH or d-HypDH of microorganisms is classified into various types by bioinformatics analysis (Fig. 4D and supplemental Table S5). Interestingly, Pa/d-HypDHγ and Tp/l-PDHγ, which belong to “Types 3 and 2,” respectively, correspond to the first ∼100 and last ∼70 amino acid residues, respectively, of Ph/l-PDH1α (Fig. 4A). Therefore, it is likely that that these heteromeric proteins evolved from a common ancestral flavoenzyme (Type 1), such as homomeric Pp/d-HypDH, by genetic duplication, division, and/or fusion and that they eventually acquired a new function(s).
Pyr4H2C Deaminase within Aldolase Protein Family
It is likely that Pyr4H2C deaminase belongs to the dihydrodipicolinate synthase/N-acetylneuraminate lyase family based on the phylogenetic analysis. Generally, members of this protein family possess a common (β/α)8-barrel fold and share a common reaction step in their reactions, namely the formation of a “Schiff base” intermediate between a strictly conserved lysine residue and C2 carbon of the common α-keto acid moiety of the substrate (22) (Fig. 5B). The phenolic hydroxyl group of the tightly conserved tyrosine residue forms double hydrogen bonds to carboxyl and hydroxyl groups at position 4 of the substrate. Interestingly, 4-hydroxy-2-oxoglutarate aldolase, which is involved in mammalian l-hydroxyproline metabolism (Fig. 1A), is also a member of this protein family (4). Although lysine (Lys164) and tyrosine (Tyr136) align well with these residues in the sequence of PpLhpC (Fig. 5B), there is a poor phylogenetic relationship to any subfamilies of the other members (Fig. 5D), which is probably the reason for the unique enzyme catalysis (Fig. 5C). Site-directed mutagenesis and/or crystallographic analysis of Pyr4H2C deaminase would provide clearer evidence for assignment to the dihydrodipicolinate synthase/N-acetylneuraminate lyase protein family and help to elucidate novel catalytic mechanism in this protein family.
Acknowledgment
We thank Dr. Seiji Yamauchi (Ehime University, Japan) for technical advice.
This work was supported in part by a Grant-in-aid for Young Scientists (B) 18760592 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S. W.), a JGC-S Scholarship Foundation (to S. W.), the Nippon Life Insurance Foundation (to S. W.), the Asahi Glass Foundation (to S. W.), and the Takahashi Industrial and Economic Research Foundation (to S. W.).

This article contains supplemental Figs. S1–S3 and Tables S1–S6.
- Pyr3H5C
- Δ1-pyrroline-3-hydroxy-5-carboxylate
- l-PDH
- l-proline dehydrogenase
- Pyr5C
- Δ1-pyrroline-5-carboxylate
- Tp/l-PDH
- l-PDH of T. profundus
- Ph/l-PDH1
- l-PDH isozyme 1 of P. horikoshii
- Ph/l-PDH2
- l-PDH isozyme 2 of P. horikoshii
- Pyr4H2C
- Δ1-pyrroline-4-hydroxy-2-carboxylate
- d-HypDH
- d-hydroxyproline dehydrogenase
- αKGSA
- α-ketoglutaric semialdehyde
- KGSADH
- αKGSA dehydrogenase
- Pp/d-HypDH
- d-HypDH from P. putida
- Pa/d-HypDH
- d-HypDH from P. aeruginosa
- Hcn
- hydrogen cyanide synthase
- Nox
- d-nopaline oxidase
- Oox
- d-octopine oxidase
- Pp
- P. putida
- Pa
- P. aeruginosa
- Ab
- A. brasilense
- Kmr
- kanamycin resistance
- INT
- p-iodonitrotetrazolium violet
- PMS
- phenazine methosulfate
- NBT
- nitroblue tetrazolium
- l-KDA
- l-2-keto-3-deoxyarabinonate.
REFERENCES
- 1. Shibasaki T., Mori H., Chiba S., Ozaki A. (1999) Microbial proline 4-hydroxylase screening and gene cloning. Appl. Environ. Microbiol. 65, 4028–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu G., Bazer F. W., Burghardt R. C., Johnson G. A., Kim S. W., Knabe D. A., Li P., Li X., McKnight J. R., Satterfield M. C., Spencer T. E. (2011) Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids 40, 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooper S. K., Pandhare J., Donald S. P., Phang J. M. (2008) A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J. Biol. Chem. 283, 10485–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riedel T. J., Johnson L. C., Knight J., Hantgan R. R., Holmes R. P., Lowther W. T. (2011) Structural and biochemical studies of human 4-hydroxy-2-oxoglutarate aldolase: implications for hydroxyproline metabolism in primary hyperoxaluria. PLoS One 6, e26021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adams E., Goldstone A. (1960) Hydroxyproline metabolism. III. Enzymatic synthesis of hydroxyproline from Δ1-pyrroline-3-hydroxy-5-carboxylate. J. Biol. Chem. 235, 3499–3503 [PubMed] [Google Scholar]
- 6. White T. A., Krishnan N., Becker D. F., Tanner J. (2007) Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J. Biol. Chem. 282, 14316–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava D., Schuermann J. P., White T. A., Krishnan N., Sanyal N., Hura G. L., Tan A., Henzl M. T., Becker D. F., Tanner J. J. (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 107, 2878–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawakami R., Sakuraba H., Ohshima T. (2004) Gene and primary structures of dye-linked l-proline dehydrogenase from the hyperthermophilic archaeon Thermococcus profundus show the presence of a novel heterotetrameric amino acid dehydrogenase complex. Extremophiles 8, 99–108 [DOI] [PubMed] [Google Scholar]
- 9. Kawakami R., Sakuraba H., Tsuge H., Goda S., Katunuma N., Ohshima T. (2005) A second novel dye-linked l-proline dehydrogenase complex is present in the hyperthermophilic archaeon Pyrococcus horikoshii OT-3. FEBS J. 272, 4044–4054 [DOI] [PubMed] [Google Scholar]
- 10. Tsuge H., Kawakami R., Sakuraba H., Ago H., Miyano M., Aki K., Katunuma N., Ohshima T. (2005) Crystal structure of a novel FAD-, FMN-, and ATP-containing l-proline dehydrogenase complex from Pyrococcus horikoshii. J. Biol. Chem. 280, 31045–31049 [DOI] [PubMed] [Google Scholar]
- 11. Satomura T., Zhang X. D., Hara Y., Doi K., Sakuraba H., Ohshima T. (2011) Characterization of a novel dye-linked L-proline dehydrogenase from an aerobic hyperthermophilic archaeon, Pyrobaculum calidifontis. Appl. Microbiol. Biotechnol. 89, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 12. Gryder R. M., Adams E. (1969) Inducible degradation of hydroxyproline in Pseudomonas putida: pathway regulation and hydroxyproline uptake. J. Bacteriol. 97, 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bater A. J., Venables W. A., Thomas S. (1977) Allohydroxy-d-proline dehydrogenase. An inducible membrane-bound enzyme in Pseudomonas aeruginosa PA01. Arch. Microbiol. 112, 287–289 [DOI] [PubMed] [Google Scholar]
- 14. Manoharan H. T. (1980) J. Biosci. 2, 107–120 [Google Scholar]
- 15. Yoneya T., Adams E. (1961) Hydroxyproline metabolism. V. Inducible allohydroxy-d-proline oxidase of Pseudomonas. J. Biol. Chem. 236, 3272–3279 [PubMed] [Google Scholar]
- 16. Singh R. M., Adams E. (1965) Enzymatic deamination of Δ1-pyrroline-4-hydroxy-2-carboxylate to 2,5-dioxovalerate (α-ketoglutaric semialdehyde). J. Biol. Chem. 240, 4344–4351 [PubMed] [Google Scholar]
- 17. Watanabe S., Shimada N., Tajima K., Kodaki T., Makino K. (2006) Identification and characterization of l-arabonate dehydratase, l-2-keto-3-deoxyarabonate dehydratase, and l-arabinolactonase involved in an alternative pathway of l-arabinose metabolism. Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 281, 33521–33536 [DOI] [PubMed] [Google Scholar]
- 18. Brouns S. J., Barends T. R., Worm P., Akerboom J., Turnbull A. P., Salmon L., van der Oost J. (2008) Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-d-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily. J. Mol. Biol. 379, 357–371 [DOI] [PubMed] [Google Scholar]
- 19. Aghaie A., Lechaplais C., Sirven P., Tricot S., Besnard-Gonnet M., Muselet D., de Berardinis V., Kreimeyer A., Gyapay G., Salanoubat M., Perret A. (2008) New insights into the alternative d-glucarate degradation pathway. J. Biol. Chem. 283, 15638–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe S., Kodaki T., Makino K. (2006) A novel α-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial l-arabinose metabolism. J. Biol. Chem. 281, 28876–28888 [DOI] [PubMed] [Google Scholar]
- 21. Watanabe S., Yamada M., Ohtsu I., Makino K. (2007) α-Ketoglutaric semialdehyde dehydrogenase isozymes involved in metabolic pathways of d-glucarate, d-galactarate, and hydroxy-l-proline. Molecular and metabolic convergent evolution. J. Biol. Chem. 282, 6685–6695 [DOI] [PubMed] [Google Scholar]
- 22. Dobson R. C., Valegård K., Gerrard J. A. (2004) The crystal structure of three site-directed mutants of Escherichia coli dihydrodipicolinate synthase: further evidence for a catalytic triad. J. Mol. Biol. 338, 329–339 [DOI] [PubMed] [Google Scholar]
- 23. Sambrook J., Fritsch E. F., Maniatis T. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 25. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi Y., Ohtsu I., Fujimura M., Fukumori F. (2011) A mutation in dnaK causes stabilization of the heat shock sigma factor σ32, accumulation of heat shock proteins and increase in toluene-resistance in Pseudomonas putida. Environ. Microbiol. 13, 2007–2017 [DOI] [PubMed] [Google Scholar]
- 27. West S. E., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J. (1994) Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86 [DOI] [PubMed] [Google Scholar]
- 28. Hishinuma S., Yuki M., Fujimura M., Fukumori F. (2006) OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 8, 2115–2124 [DOI] [PubMed] [Google Scholar]
- 29. Fish W. W. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158, 357–364 [DOI] [PubMed] [Google Scholar]
- 30. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 31. Satomura T., Kawakami R., Sakuraba H., Ohshima T. (2002) Dye-linked d-proline dehydrogenase from hyperthermophilic archaeon Pyrobaculum islandicum is a novel FAD-dependent amino acid dehydrogenase. J. Biol. Chem. 277, 12861–12867 [DOI] [PubMed] [Google Scholar]
- 32. Pollegioni L., Caldinelli L., Molla G., Sacchi S., Pilone M.S. (2004) Catalytic properties of d-amino acid oxidase in cephalosporin C bioconversion: a comparison between proteins from different sources. Biotechnol. Prog. 20, 467–473 [DOI] [PubMed] [Google Scholar]
- 33. Ta D. T., Vickery L. E. (1992) Cloning, sequencing, and overexpression of a [2Fe-2S] ferredoxin gene from Escherichia coli. J. Biol. Chem. 267, 11120–11125 [PubMed] [Google Scholar]
- 34. Heacock A. M., Adams E. (1975) Formation and excretion of pyrrole-2-carboxylic acid. Whole animal and enzyme studies in the rat. J. Biol. Chem. 250, 2599–2608 [PubMed] [Google Scholar]
- 35. Tanigawa M., Shinohara T., Saito M., Nishimura K., Hasegawa Y., Wakabayashi S., Ishizuka M., Nagata Y. (2010) d-Amino acid dehydrogenase from Helicobacter pylori NCTC 11637. Amin. Acids 38, 247–255 [DOI] [PubMed] [Google Scholar]
- 36. Wierenga R. K., Terpstra P., Hol W. G. (1986) Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187, 101–107 [DOI] [PubMed] [Google Scholar]
- 37. Laville J., Blumer C., Von Schroetter C., Gaia V., Défago G., Keel C., Haas D. (1998) Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180, 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zanker H., Lurz G., Langridge U., Langridge P., Kreusch D., Schröder J. (1994) Octopine and nopaline oxidases from Ti plasmids of Agrobacterium tumefaciens: molecular analysis, relationship, and functional characterization. J. Bacteriol. 176, 4511–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blumer C., Haas D. (2000) Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173, 170–177 [DOI] [PubMed] [Google Scholar]