Background: NDE1 and NDEL1 are neurodevelopmental and mitotic proteins with extended coiled-coil N termini, but unknown C-terminal structure.
Results: Recombinant NDE1/NDEL1 form dimers and tetramers in which their C termini interact with their N-terminal domains.
Conclusion: NDE1/NDEL1 each adopt a sharply bent back structure.
Significance: This explains the existence of two distinct dynein-binding domains on NDE1/NDEL1 and instability of disease-associated mutants lacking C termini.
Keywords: Electron Microscopy (EM), Homology Modeling, Mass Spectrometry (MS), Neurodevelopment, Protein Cross-linking, Protein Folding, Protein Structure, NDE1, NDEL1
Abstract
Paralogs NDE1 (nuclear distribution element 1) and NDEL1 (NDE-like 1) are essential for mitosis and neurodevelopment. Both proteins are predicted to have similar structures, based upon high sequence similarity, and they co-complex in mammalian cells. X-ray diffraction studies and homology modeling suggest that their N-terminal regions (residues 8–167) adopt continuous, extended α-helical coiled-coil structures, but no experimentally derived information on the structure of their C-terminal regions or the architecture of the full-length proteins is available. In the case of NDE1, no biophysical data exists. Here we characterize the structural architecture of both full-length proteins utilizing negative stain electron microscopy along with our established paradigm of chemical cross-linking followed by tryptic digestion, mass spectrometry, and database searching, which we enhance using isotope labeling for mixed NDE1-NDEL1. We determined that full-length NDE1 forms needle-like dimers and tetramers in solution, similar to crystal structures of NDEL1, as well as chain-like end-to-end polymers. The C-terminal domain of each protein, required for interaction with key protein partners dynein and DISC1 (disrupted-in-schizophrenia 1), includes a predicted disordered region that allows a bent back structure. This facilitates interaction of the C-terminal region with the N-terminal coiled-coil domain and is in agreement with previous results showing N- and C-terminal regions of NDEL1 and NDE1 cooperating in dynein interaction. It sheds light on recently identified mutations in the NDE1 gene that cause truncation of the encoded protein. Additionally, analysis of mixed NDE1-NDEL1 complexes demonstrates that NDE1 and NDEL1 can interact directly.
Introduction
NDE1 (nuclear distribution factor E-homolog 1, or nuclear distribution element 1) and NDEL1 (NDE1-like 1) are highly similar proteins, originating from a common ancestral gene (1, 2). Through their interactions with LIS1 (Lissencephaly 1, encoded by PAFAH1B1) and dynein they play crucial roles in microtubule organization and are required for cell cycle progression and neuronal development (reviewed in Ref. 3). They have recently been implicated in the pathology of major mental illness (reviewed in Ref. 4).
NDE1 (335 amino acid residues) and NDEL1 (345 amino acid residues) both consist of distinct N- and C-terminal regions (Fig. 1, A and B). X-ray crystallography, backed by other biophysical techniques, revealed how the N-terminal region of NDEL1 dimerizes by forming extended, continuous parallel α-helical coiled-coils that account for approximately half the length of the protein (residues 8–167) (Fig. 1C) (5, 6). Within this region lie sites that interact with LIS1 (7, 8). Two NDEL1 dimers associate in an anti-parallel fashion to form four-helix coiled-coil tetramers (Fig. 1D) (5). With ∼71% sequence identity, the NDE1 N-terminal region is likely to form similar structures (9). The C-terminal portions of both proteins are known to harbor numerous phosphorylation sites (7, 10–12) (Fig. 1A). These C-terminal regions are predicted to be largely unstructured, with the exception of a single strongly predicted α-helix (11) (Fig. 1A). This predicted α-helix in NDEL1 is responsible for interaction with its key binding partner DISC1 (disrupted-in-schizophrenia 1) (13), as well as, potentially, dynein and dynactin (14, 15). Frameshift mutations that lead to a loss of this C-terminal region in NDE1 are known to cause severe microcephaly (16, 17).
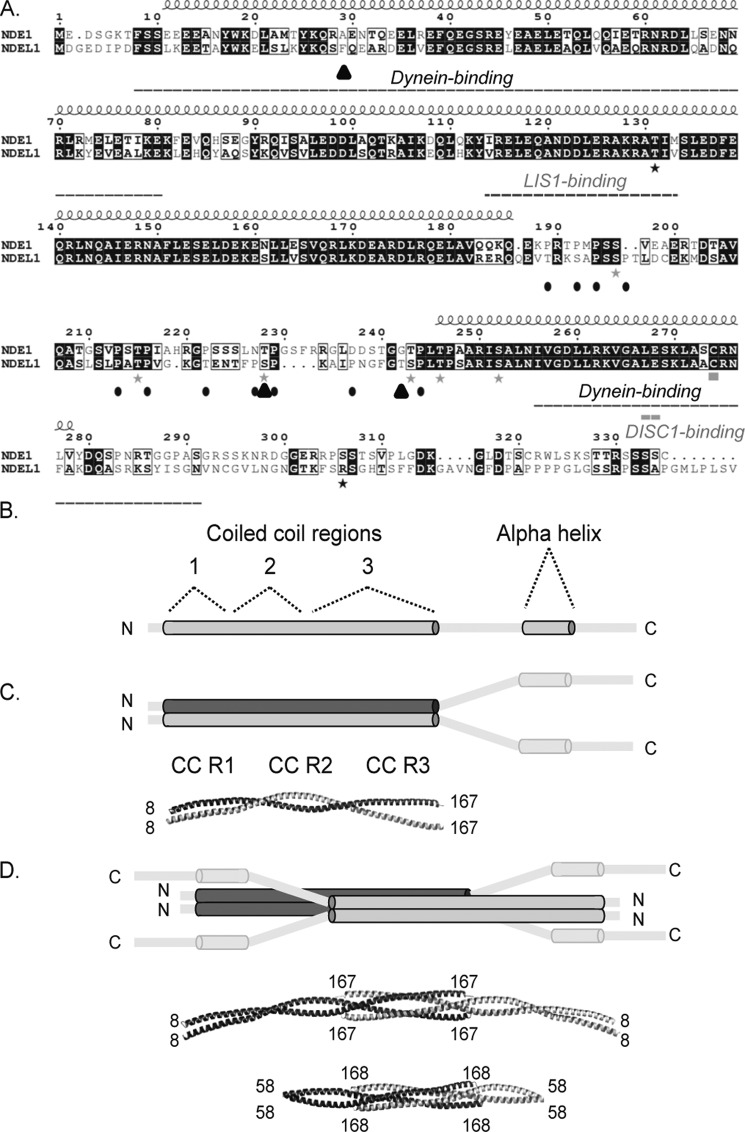
FIGURE 1.
Sequence and structure of NDE1 and NDEL1. A, sequence alignment between NDE1 and NDEL1. Indicated above the alignment is the consensus coiled-coil domain and predicted C-terminal α-helix for the two proteins (shown schematically in B). Phosphorylation sites for NDE1 and NDEL1 are shown as black and gray stars, respectively. NDE1 microcephaly frameshift mutations are shown as filled triangles. Binding sites described in text for dynein, DISC1, and LIS1 are indicated by dashed lines below alignment. Prolines lying between coiled-coil region and C-terminal α-helix are denoted by filled ovals. A palmytolation site is indicated by a filled rectangle. B, NDE1 and NDEL1 have an N-terminal coiled-coil domain (longer gray cylinder), which is made up of three coiled-coil regions (CC R): CC R1, CC R2, and CC R3. The two proteins are predicted to have a disordered C terminus (gray lines) containing a strongly predicted α-helix (shorter gray cylinder). The N and C termini are indicated. C, the NDEL1 N-terminal region was shown by crystallography (Protein Data Bank code 2V71) to dimerize via a parallel coiled-coil utilizing CC R1 (amino acids 9–44) and CC R2 (amino acids 44–99) (5). The structure of the C terminus remains unsolved. D, previously favored model of tetrameric assembly of full-length NDE1/NDEL1 (18) and, below, a structure determined by crystallography (the shorter tetramer fragment, consisting of amino acids 58–168 of Protein Data Bank code 2V66 and the longer tetramer fragment, consisting of amino acids 8–167 derived using symmetry operations from Protein Data Bank code 2V71). In the crystal structures, two NDEL1 dimers associate using CC R3 (amino acids 107–167) in an anti-parallel arrangement, to form tetramers (5).
Current understanding of the structures of NDE1 and NDEL1 is insufficient to explain important observations made in the recent literature. Notably, each of NDEL1 and NDE1 possess two distinct regions at their N and C termini that interact with intermediate chains of dynein (18, 19). Thus, despite being separated by ∼150 amino acids residues in the primary structure, these two regions could be juxtaposed because of the overall architecture of the folded protein. Given that the disordered C-terminal regions are unlikely to make good targets for crystallization, we have combined negative stain EM with a strategy of chemical cross-linking, MS, and database searching (6, 20, 21) to gain structural insight into full-length NDE1 and NDEL1 molecules in solution. This extends a previous application of the cross-linking/MS methodology to a truncated fragment of NDEL1 (6). We found that the C-terminal region of each protein bends back on to the main coiled-coil domain; this observation sheds light on the molecular architecture of the complex between NDE1, NDEL1, and dynein that is crucial for mitosis and neurodevelopment.
EXPERIMENTAL PROCEDURES
Primer Sequences and Cloning
Constructs encoding V5-tagged NDE1 and NDEL1 have been described previously (22). Open reading frames from these were subcloned in a two-step PCR process that introduced an N-terminal His6 tag. The primers used were: CCGAAAACCTGTATTTTCAGGGCGAGGACTCCGGAAAGACTTTCAG (NDE1, first step 5′), GGGGCCGAAAACCTGTATTTTCAGGGCGATGGTGAAGATATACCAG (NDEL1, first step 5′), GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGCAGGAGCTGGACGACCTGGTTG (NDE1, both steps, 3′), GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACACACTGAGAGGCAGCATACCCG (NDEL1, both steps 3′), and GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATATACATATGTCGTACTACCATCACCATCACCATCACGATTACGATATCCCAACGACCGAAAACCTGTATTTTCAGGGC (NDE1/NDEL1, second step 5′). The clones were sequentially transferred first into the pDONR-221 entry vector and then into pDEST-14 destination vector using BP clonase and LR clonase (Invitrogen). Correct cloning and insertion was confirmed by sequencing.
Protein Expression and Purification
To generate recombinant NDE1 or NDEL1 constructs were transformed into C41 (DE3) cells (Lucigen Corporation, Middleton, WI) and grown in either LB (NDE1) or 2×TY (NDEL1) medium containing 100 μg/ml carbenicillin to A600 of ∼0.6. Recombinant protein production was induced by adding 1 mm IPTG and incubating at 30 °C for 3 h with shaking at 250 rpm.
To label NDE1 by stable isotope labeling by amino acids in cell culture (SILAC)6 (23) for co-purification experiment with unlabeled NDEL1, constructs were transformed into BL21 (DE3) LysA ArgA cells (24) and grown in M9 minimal medium (6 mg/ml Na2HPO4, 3 mg/ml KH2PO4, 1 mg/ml NH4Cl, 500 μg/ml NaCl, 200 μg/ml glucose, 50 ng/ml vitamin B1, 6 ng/ml CaCl2, 1 mm MgSO4) containing 100 μg/ml carbenicillin and 10 μg/ml each of l-lysine and l-arginine that were 99% enriched for the stable isotopes 13C and 15N (CK Gas Products). The labeled NDE1 culture was induced as above and grown at 30 °C for 3 h.
Bacterial pellets were extracted, resuspended in 30 ml of immobilized metal affinity chromatography buffer (25 mm sodium phosphate, 500 mm NaCl, 20 mm imidazole, 1 mm DTT, pH 7.4) plus Complete protease inhibitor mixture (Roche Applied Science) per liter of original culture and lysed using a TS Cell Disruptor (Constant Systems) at 172 MPa. The lysates were centrifuged at 50,000 × g for 1 h, and the resultant supernatants were passed through 0.2-μm Whatman cellulose nitrate membrane filters. The filtered supernatant was then loaded onto a 1-ml HisTrap HP column (GE Healthcare), charged with 1 ml of 100 mm nickel chloride, using an ÄKTApurifier (GE Healthcare). Bound proteins were eluted using immobilized metal affinity chromatography buffer containing an imidazole gradient rising from 20 to 500 mm over 20 column volumes.
Fractions containing recombinant NDE1 or NDEL1-sized bands (confirmed by SDS-PAGE) were further purified by size exclusion chromatography using a 320-ml HiLoad 26/60 Superdex 200 preparative grade column (GE Healthcare) and PBS, pH 7.4 (for cross-linking analysis), or 25 mm HEPES, 200 mm NaCl, pH 7.4 (for electron microscopy analysis). Where appropriate, protein fractions were concentrated using Vivaspin Concentrators (Sartorius Stedim Biotech). For each recombinant protein, identity was confirmed by MALDI-TOF mass spectrometry (Applied Biosystems).
Biophysical Characterization
The mean hydrodynamic radii of the purified proteins were determined by dynamic light scattering using a Zetasizer auto plate sampler (Malvern Instruments Ltd.) at 10 °C with a 830-nm laser. Each dynamic light scattering measurement was performed on protein solution that was initially centrifuged for 15 min at 13,000 rpm.
For determination of secondary structure, protein was buffer-exchanged into 10 mm sodium phosphate buffer, pH 7.4, using pre-equilibrated PD-10 columns (GE Healthcare). Protein concentration was determined using a V-550 UV-visible spectrophotometer (JASCO) based on absorbance at 280 nm and the theoretical extinction coefficient of the respective protein sequence. Circular dichroism (CD) analysis was performed on a J-810 Spectropolarimeter (JASCO) at 25 °C using a path length of 1 mm, over the far UV range 185–280 nm. Five scans were acquired and averaged for each CD spectrum using the following parameters: data pitch, 0.1 nm; bandwidth, 1 nm; scan speed, 10 nm/min; and response time, 2 s). Solvent correction was achieved by subtraction of a spectrum of the buffer solution acquired with identical parameters.
Chemical Cross-linking and Mass Spectrometry
Cross-linking experiments were carried out using bis(sulfosuccinimidyl)suberate (BS3) (Thermo Fisher Scientific) as cross-linker in a 1:1000 protein:BS3 molar ratio by incubating for 2 h on ice. The reactions were quenched by the addition of 2.73 m ammonium bicarbonate in a 10:1 molar ratio with BS3 and incubation on ice for a further hour. The proteins were separated by SDS-PAGE, and bands corresponding to cross-linked complexes were excised, and the proteins therein were reduced using 100 mm DTT for 30 min at room temperature, alkylated with 55 mm iodoacetamide for 30 min in the dark at room temperature, and digested using 12.5 ng/μl trypsin (sequencing grade; Promega) overnight at 37 °C (6).
The supernatant containing the cross-linked peptides were fractionated on SCX-StageTips (25), desalted using C18-StageTips (26), and analyzed on either an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific) or an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) as described previously (20, 27). The peptides were analyzed using a high/high strategy; both MS spectra and MS-MS spectra were acquired in the Orbitrap (20). The data were processed, generating peak lists by MaxQuant (28) and matching cross-linked peptides to spectra using in-house developed Xi software.
Homology Modeling
Modeling of the NDE1-NDEL1 heterotetramer was undertaken using Modeller version 9.10 (29) based on the template fragments of NDEL1 solved by crystallography (5) deposited in the Protein Data Bank, i.e., the shorter fragment (regions 58–168) (Protein Data Bank code 2V66, chains B–E) and the longer (regions 8–167) fragment (Protein Data Bank code 2V71, chains A and B). Two heterotetramer NDE1-NDEL1 models were built based on the (shorter) tetramer and the (longer) tetramer sequence generated by symmetry operations, as described previously (9). Two sequences each for NDE1 and NDEL1 were aligned appropriately (Fig. 1A) with the template NDEL1 sequences (2V66 and 2V71) to form an anti-parallel arrangement. Thirty models were generated corresponding to each of the alignments, and the ones with the lowest objective function score (29) were selected as the representative NDE1-NDEL1 model in each case. The models were checked for valid stereochemistry using PROCHECK (30) and additionally assessed for valid packing quality using WHAT IF (31).
Negative Stain Electron Microscopy
NDE1 and NDEL1 protein samples were diluted to between 3 and 18 μg/ml before being placed on thin continuous carbon film electron microscopy grids and stained with 2% (w/v) uranyl acetate solution. To rule out DNA contamination for observation of high molecular weight species, NDE1 and NDEL1 protein (0.15 mg/ml) were independently incubated for 1 h at 37 ºC with 1.3 Kunitz units of DNAse1 (Sigma) in a buffer containing 20 mm MgCl2 and then diluted to 10 μg/ml protein immediately before being prepared for EM (as above). The grids were imaged in low dose mode using a F20 electron microscope (FEI, Eindhoven, The Netherlands) fitted with an 8192 × 8192 digital camera (F816; TVIPS GmbH, Germany). The images were collected at a nominal magnification of 29,000 at 200 kV (the calibrated scale is 0.269 nm/pixel). The data were collected only in thinly stained areas to maximize the contrast from the particles. The particles were manually selected (705 for NDE1 and 1299 for NDEL1), normalized, and band pass-filtered before being input into an iterative reference-free alignment and classification algorithm (EMAN (32) and IMAGIC (33)). The class averages include ∼40 images/class.
RESULTS
Biophysical Characterization and Oligomeric State Determination of NDE1 and NDEL1 in Solution
Recombinant full-length human NDE1 and NDEL1, incorporating an N-terminal His6 tag, were each purified from bacterial lysates using immobilized metal affinity chromatography followed by size exclusion chromatography. The two purifications yielded similar size exclusion chromatograms that each featured three peaks (Fig. 2A, referred to as P1, P2, and P3) all containing full-length NDE1 or NDEL1, as determined by mass spectrometry. The elution positions of these peaks were approximately similar, but the relative abundance of each peak varied between preparations. Dynamic light scattering indicated that both P2 and P3 contained discrete oligomers of defined size (Fig. 2B), whereas P1 consisted of highly multimeric protein. We therefore focused on P2 and P3. In some preparations an even higher molecular weight (MWt) peak of aggregated protein was observed. CD revealed that, for both NDE1 and NDEL1, the components of P2 and P3 were predominantly α-helical in composition: both exhibit near identical pronounced double minima at ∼208 and ∼222 nm and one positive peak at ∼192 nm in the far-UV range, characteristic of folded proteins with high α-helical content (Fig. 2C). This agrees with previous work showing that α-helical coiled-coils predominate in the NDEL1 N-terminal domain (5) and is consistent with CD spectra of recombinant full-length NDEL1 (34, 35). Thermal denaturation experiments on both purified protein samples recorded melting temperatures of ∼47 °C, in agreement with ∼50 °C reported for the N-terminal coiled-coil domain of NDEL1 (5).
FIGURE 2.
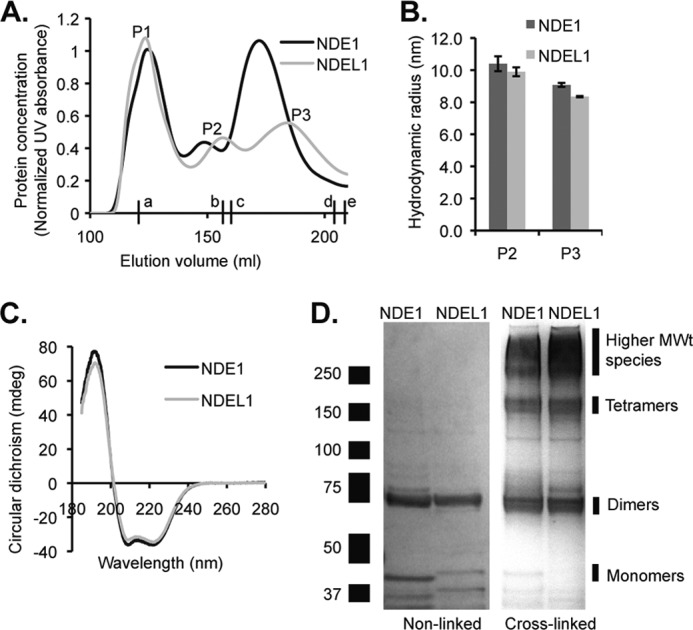
Oligomeric state of recombinant NDE1 and NDEL1. A, typical size exclusion chromatography profile of NDE1 and NDEL1 (column volume = 320 ml), immediately following affinity chromatography purification from bacterial lysate. Three peaks (P1–P3) have similar elution positions for both proteins, but relative abundance varies. Smaller proteins were also eluted after these peaks; however, these proteins were confirmed to be bacterial contaminants (and not monomeric NDE1 or NDEL1). Absorbance readings were normalized after 125 ml of elution. Lowercase letters indicate the elution volumes of protein standards: a, ferritin, 440 kDa; b, catalase, 232 kDa; c, aldolase, 158 kDa; d, bovine serum albumin, 67 kDa; and e, ovalbumin, 44 kDa. B, mean hydrodynamic radii of P2 and P3 for each of NDE1 and NDEL1, determined by dynamic light scattering analysis. C, circular dichroism spectra demonstrating that P2 of NDE1 (black) and NDEL1 (gray) share a similar, primarily α-helical secondary structure. D, SDS-PAGE of non-cross-linked recombinant NDE1 and NDEL1 (left panel) reveals monomeric species and incompletely reduced dimers (all bands confirmed to represent NDE1 or NDEL1 by mass spectrometry). In comparison, cross-linking of NDE1 and NDEL1 using BS3 (right panel) reveals mainly dimers, tetramers, and higher MWt species.
Samples of P2 and P3 from both the NDE1 and NDEL1 preparations (Fig. 2A) were subjected to limited chemical cross-linking using BS3, and the resultant products were resolved using SDS-PAGE. BS3 was used at the minimum molecular ratio with NDE1/NDEL1 (1000:1) required to cross-link all of the protein molecules under the conditions used. Application of higher concentrations of BS3 up to a 2500:1 ratio did not further alter the observed pattern of Coomassie-stained SDS-PAGE bands seen. The pattern of bands obtained on the gels for cross-linked NDE1 and for NDEL1 appeared very similar (Fig. 2D) with ∼75- and ∼150-kDa species detected, i.e., ∼2- and 4-fold larger than monomeric NDE1 or NDEL1 (Fig. 2D), along with higher MWt species (>250 kDa, corresponding to large multimers or aggregates). The size exclusion chromatograms collected in the absence of any cross-linker suggest that P1 is likely to correspond to the high MWt species, P2 is likely to correspond to the tetramers, and P3 is likely to correspond to the dimers (Fig. 2A); however, all fractions show a mixture of these species (Fig. 2D), implying that the proteins can dynamically alter their oligomeric state post elution.
Negative Stain Electron Microscopy Reveals That NDE1 and NDEL1 Exist as Needle-like Structures and Novel Polymers in Solution
Analysis of recombinant NDE1 and NDEL1 by negative stain EM revealed that both of these recombinant full-length proteins form extended “needle-like” structures in solution (Fig. 3, A and B). These observations are consistent with the tight, elongated coiled-coil crystal structure of the NDEL1 N-terminal domain (5), and they therefore extend this finding to NDE1, in keeping with a recent study based on comparative modeling (9). Class averages of the particle images revealed an average width of 2–3 nm and a mode length of ∼29 nm, with a relatively wide distribution (Fig. 3C). The mode length is close to what would be expected for parallel dimers (∼26 nm) if the extra residues not seen in the crystal structure but predicted to be helical (12, 36) up to amino acid position ∼190 are included (Figs. 1A and 3D). Some longer particles of NDE1 and NDEL1 seen by EM are consistent with the ∼36-nm-long anti-parallel tetramers of the N-terminal coiled-coil domains, based on previous studies (5) (see also Fig. 1). Dimers may predominate in the EM data because of the low protein concentration used, potentially causing disassociation of tetrameric species. Additionally, for each protein, occasional examples of longer end-to-end linear polymers were visible (Fig. 3E), not unlike other longer, but thicker filamentous coiled-coil proteins such as remorins (37). These structures were still seen following treatment with DNase1, implying that these are indeed NDE1/NDEL1 and not artifacts from DNA contamination. C-terminal regions either did not form compactly folded structures (in line with predictions of high content of random coil) or were folded but highly extended and intimately associated with the N-terminal coiled-coil domain and thus could not be resolved by EM.
FIGURE 3.

Electron microscopy of negatively stained NDE1 and NDEL1 proteins. A, class average images of NDE1 show a long thin structure with varying lengths that confirm the highly elongated structure predicted from sequence analysis and homology modeling (9). The C-terminal regions, predicted to be largely unstructured (11), are not discernible in the EM data, indicating that they do not form regular stable domains. Scale bar, 20 nm. B, class average images of NDEL1, which show very similar features. C, histogram of lengths measured from EM class average images for NDE1 (left panel) and NDEL1 (right panel). Following alignment and classification, particle images were measured manually in EMAN. In ∼10% of images the lengths were not discernible, and so these images were excluded from the analysis. Both proteins show a mode length of ∼29 nm, with NDEL1 showing more, longer species. Shorter species could be the result of partial protein degradation. D, comparison of two selected NDE1 (left panels) and NDEL1 (right panels) class average images with the crystal structures of dimeric (top panels) and tetrameric (bottom panels) NDEL1 (Protein Data Bank code 2V71) with additional predicted helical extensions (∼3 nm) shown as gray boxes and drawn to scale, in each case. E, sections of raw micrographs of NDE1 (top panels) and NDEL1 (bottom panels) show two examples each of long filamentous polymers. The start and end positions of the polymers are indicated by black asterisks. Scale bar, 50 nm.
The NDE1 and NDEL1 C-terminal Regions Are Intimately Associated with Their N-terminal Coiled-coil Domains
To infer the architecture of NDE1 and NDEL1 in solution, we deployed our previously established strategy of chemical cross-linking followed by mass spectrometry of proteolytic fragments. This methodology provides opportunities for mapping structural details of functional complexes in solution. The structure of proteins or protein complexes is ascertained by identifying amino acid peptide pairs that are positioned in close proximity to each other (21). We utilized an amine-to-amine BS3 cross-linker that is homobifunctional, water-soluble, and noncleavable. It contains amine-reactive N-hydroxysulfosuccinimide esters that are capable of reacting with primary amine side chain groups predominantly from Lys, but also with the hydroxyl end groups of Ser, Thr, and Tyr at suitable pH (38).
This technique was applied to samples that had been derived from size exclusion chromatography (Fig. 2D). Bands of cross-linked NDE1 and NDEL1, with inferred MWts that corresponded to dimeric, tetrameric, and higher MWt species, were excised individually from the SDS-PAGE gel and subjected to tryptic digestion. The resultant BS3-cross-linked peptides were enriched using strong cation exchange StageTips (25). A combination of mass spectrometry and database searching was then deployed to identify the cross-linked peptide pairs; these represent regions in NDE1 or in NDEL1 or between NDE1 and NDEL1 that are in sufficiently close spatial proximity (<11.4 Å between primary amines) to interact simultaneously with the same cross-linker. We identified a total of 300 (NDE1) and 75 (NDEL1) unambiguous linkage pairs from mass spectra matching to cross-linked peptides (supplemental Tables S1 and S2). Example fragmentation spectra are shown in Fig. 4, whereas cross-linked peptides detected in each oligomeric state are shown schematically in Fig. 5 (also listed in supplemental Table S2). A total of 64 and 23 were seen in the dimeric species, 99 and 34 in the tetramer, and 137 and 18 in the high MWt species for NDE1 and NDEL1, respectively. Some cross-linked pairs were observed more than once (supplemental Tables S1 and S2), adding credence to their physiological relevance in solution.
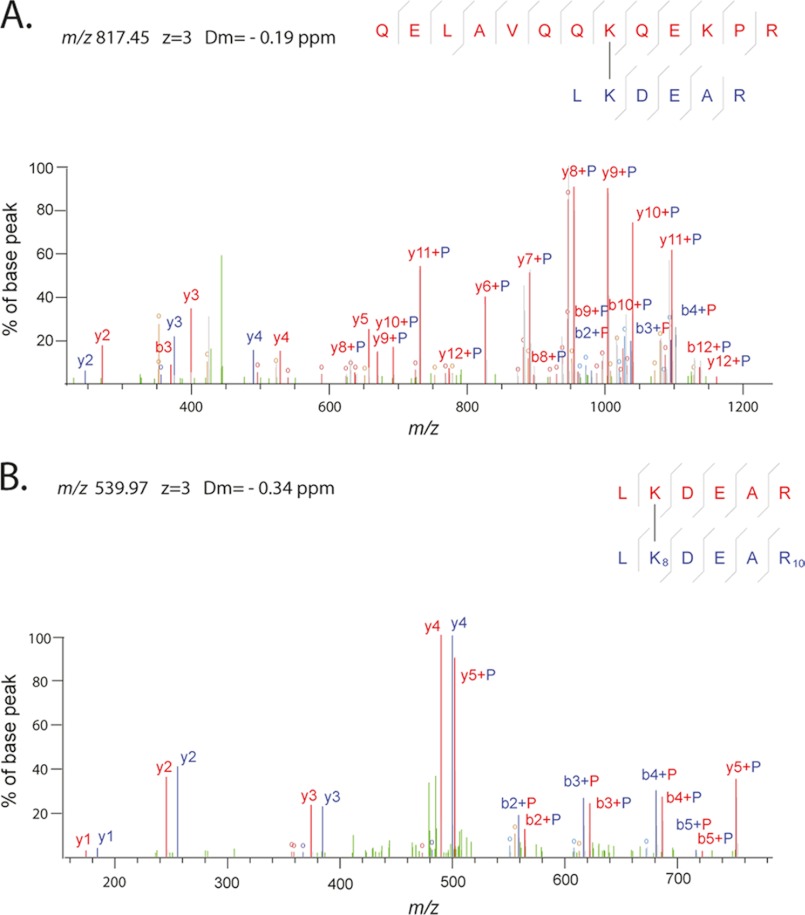
FIGURE 4.
Example fragmentation spectra. A, fragmentation spectrum of a cross-linked peptide that includes a link between Lys-185 and Lys-170 in NDE1. Peaks supporting QELAVQQKQEKPR are annotated in red, and those supporting LKDEAR are annotated in blue. Fragments that contain parts of both peptides lead to mixed color annotation, where a P denotes an intact peptide (red P represents red peptide and blue P represents blue peptide). Loss of water or ammonia from fragments is annotated with an O in the color of the respective peptide. Unexplained peaks are shown in green. B, fragmentation spectrum of a cross-linked peptide that includes a link between a SILAC-labeled NDE1 peptide lysine (K8) with an unlabeled but identical NDEL1 peptide lysine (Lys-170–Lys-170), is shown. The difference in mass (doublets) is apparent for the labeled versus unlabeled lysine, even though the peptide sequences are otherwise identical LKDEAR. Annotation and color scheme are otherwise, as shown in A.
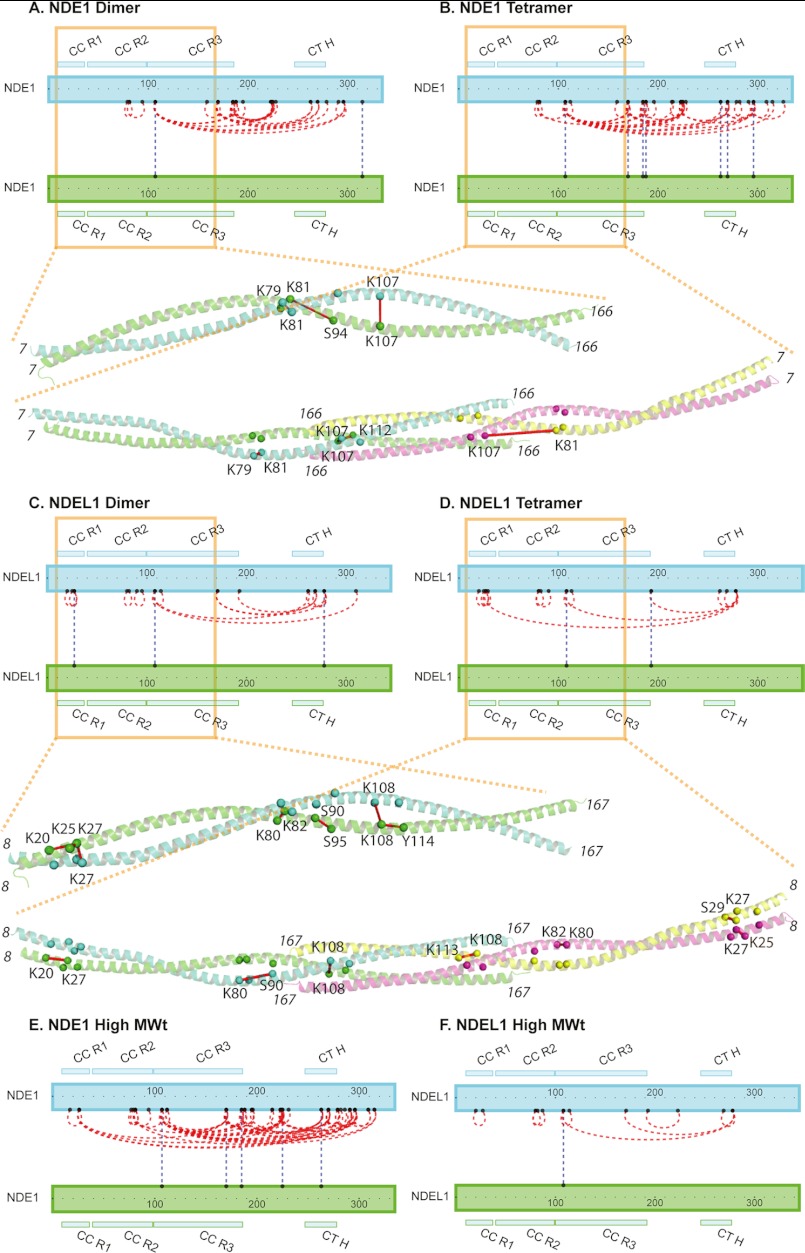
FIGURE 5.
Structural analysis of NDE1 and NDEL1 by chemical cross-linking and mass spectrometry. NDE1 and NDEL1 are represented by blue or green rectangular boxes, drawn to scale, running N terminus (left side) to C terminus (right side). The locations of CC R1, CC R2, and CC R3 as per the crystal structures (5) and the predicted C-terminal helix (CT H) (11) are shown. Pairs of residues shown to be cross-linked together are linked by lines. Blue dashed lines connecting different NDE1 or NDEL1 boxes indicate cross-links that are known to be intermolecular. Red dashed lines indicate cross-links that may be intra- or intermolecular. Only cross-links that passed the validation process are displayed. Some unambiguous, nonredundant cross-links were derived from experiments involving mixed NDE1-NDEL1 samples. Details of this, and the cross-linked peptide pairs, can be found in supplemental Table S2. Notably, the C-terminal regions of both NDE1 and NDEL1 are shown to interact with their N-terminal domains. Where cross-linking distances may be inferred from the solved crystal structure of NDEL1 (Protein Data Bank code 2V71) or homology model of NDE1, these cross-links are shown on the respective longer fragments using cartoon representation, with the α-carbon atoms of the residues involved shown as spheres and labeled, and the closest measured α-carbon distances (see supplemental Table S3) between them shown by a red line. The data are shown for samples of: NDE1 dimer (A), NDE1 tetramer (B), NDEL1 dimer (C), NDEL1 tetramer (D), NDE1 high MWt species (E), and NDEL1 high MWt species (F).
The data demonstrate that the N-terminal domain of NDE1 forms parallel dimers and that two such dimers can associate to form tetramers, consistent with our EM data showing that NDE1 closely resembles NDEL1 (Fig. 3). More importantly, the data demonstrated that in both oligomeric states, the C-terminal region of the protein interacts with the putative coiled-coil N-terminal domain (Fig. 5, A and B). We predict that this interaction would be mediated mainly by the single predicted α-helix (residues ∼246–278) near the C terminus, which would be expected to pack favorably to form an α-helical bundle with the coiled-coil domain. Note that the N- (acidic) and C-terminal (basic) regions of NDE1 and NDEL1 have complementary theoretical pI values (18) that could enable favorable electrostatic interaction. Several individual Lys residues in the C-terminal region of both proteins were observed to each form cross-links with multiple Lys residues in the N-terminal domain (e.g., Lys-270 of NDE1 cross-linked to both Lys-107 and Lys-170 of NDE1); such cross-links could not co-exist within the same conformation. Thus, multiple conformations of the inferred “bent back” architecture must co-exist (or undergo conformational exchange) in solution. The bend needed to explain the cross-linking results must occur within the region linking the N-terminal coiled-coil domain and the putative α-helix. This “linker” region contains eight proline residues in each of NDE1 and NDEL1 (Fig. 1A) and is predicted not to have a regular structure. These observations are consistent with the lack of visible density for any C-terminal domain by EM following particle averaging. The extreme C termini of subunits in dimeric NDE1 also appear to interact with each other, as demonstrated by cross-linking between the Lys-315 residues of two NDE1 molecules.
In the higher MWt species, similar results were obtained except that additional cross-linking occurred between the C-terminal region of NDE1 and another region proximal to its N terminus; for example, Lys-270 cross-links with Lys-26 (Fig. 5). This may reflect the existence of the extended polymers seen by EM, which could arise when the flexible C-terminal arms of one NDE1 dimer (or tetramer) interact with the extreme N terminus of another (Fig. 3E). Given their inconsistent appearance and their novelty, no high resolution structural interpretations can be provided at this time.
For NDEL1, the C-terminal region also bends back to interact with the N-terminal coiled-coil domain (Fig. 5, C and D, and supplemental Table S2). It is noteworthy that a previous study that probed a NDEL1 peptide array consisting of overlapping 25-mer peptides with full-length NDEL1 did not find any C-terminal fragment association with the N terminus (39). In contrast, more recently published data indicated that a longer 47-mer peptide derived from the NDEL1 C terminus was indeed capable of interacting with full-length NDEL1 (40). Our data, which utilize full-length constructs, is consistent with the latter study and provides definitive proof for this occurrence in solution.
Mapping Cross-linking Data on the Crystal Structure Fragment of NDEL1 and Homology Model of NDE1
The cross-linked peptide pairs that originate from within the N-terminal coiled-coil domain for the dimer and tetramer-sized bands were analyzed with respect to the existing crystallographic data for the N-terminal truncated fragments of NDEL1 (amino acids 8–167 in Protein Data Bank code 2V77 and amino acids 58–168 in Protein Data Bank code 2V61) (5) and the homology-derived models of the NDE1 N-terminal fragment (9).
For the NDE1 dimer, three of the observed cross-linked peptide pairs occur solely within the coiled-coil N-terminal domain (amino acids 7–166) including a self-residue cross-link at Lys-107; so these directly address the existence of this structural motif in solution. Additionally, the aforementioned self-residue cross-link at Lys-315 is in agreement with the inference from the homology model of NDE1; namely that, like NDEL1, two NDE1 chains associate unambiguously as a parallel dimer.
Within the context of the NDEL1 dimer, on the other hand, seven pairs of cross-linked peptides were obtained within the coiled-coil domain. Two of these, Lys-27–Lys-27 (7.8 Å, between α carbons in 2V71) and Lys-108–Lys-108 (13.9 Å between α carbons in 2V71), correspond to self-residue cross-linked pairs, in further agreement with a parallel arrangement for the coiled-coil helices in solution. Measurements in the crystal structure for the five remaining pairs (Lys-20–Lys-27, Lys-25–Lys-27, Lys-80–Lys-82, Ser-90–Ser-95, and Lys-108–Tyr-114) allow distinction between feasible interchain and intrachain cross-link formation within the NDEL1 dimer. Supplemental Table S3 contains a list of the lowest calculated distances measured in three-dimensional Cartesian space for all possible combinations of peptide residue linkages in the crystal structures (NDEL1) and homology models (for NDE1) with their respective oligomeric states.
With respect to the NDEL1 tetramer, seven of the cross-linked peptide pairs reside within the coiled-coil domain. As in the dimer, the presence of the self-residue Lys-108–Lys-108 cross-link (supplemental Tables S2 and S3) reinforces the evidence for a parallel dimer. The Lys-20–Lys-27, Lys-25–Lys-27, and Lys-80–Lys-82 cross-links that were observed in the dimer also appear in the tetrameric sample. In addition, novel cross-links were observed between Lys-27 and Ser-29 (5.5 Å between α carbons in 2V71), Lys-80 and Ser-90 (15.2 Å between α carbons in both 2V71 and 2V66), and between Lys-108 and Lys-113 (8.9 Å and 8.6 Å between α carbons in 2V71 and 2V66, respectively); these cross-links are again consistent with the crystal structure. We did not observe any cross-link that was in conflict with the published crystal structures, i.e., greater than the maximum theoretical distance from the cross-link of 27.4 Å (20).
In the NDE1 tetramer, four cross-linked peptide pairs could be identified and used for comparisons with the homology models. Three of these, Lys-79–Lys-81, Lys-107–Lys-107, and Lys-107–Lys-112, had previously been identified in the NDEL1 dimer and tetramer (i.e., Lys-80–Lys-82 and Lys-108–Lys-108) and the NDEL1 tetramer (i.e., Lys-108–Lys-113), respectively. The remaining cross-link, Lys-81–Lys-107, was novel to NDE1, but not in agreement with the homology model. Here the α carbon atoms are ∼36 Å apart from each other when mapped onto the homology models (Fig. 5B), and this is addressed under “Discussion.”
NDE1 and NDEL1 Can Co-complex in Both Parallel and Anti-parallel Arrangements in Solution
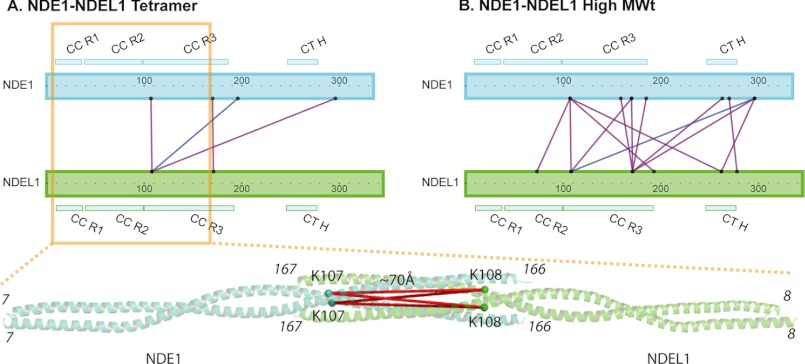
NDE1 and NDEL1 proteins have been shown to interact with each other in mammalian cells (22, 41) by an unknown mechanism that is modulated by a phosphorylation event that has been implicated in neurite outgrowth (9). The two proteins are highly similar in sequence within their respective N-terminal coiled-coils known to facilitate tetramerization (87% identity, 99% similarity, NDEL1 residues 107–192) (5); they are less similar in the dimer-forming regions (57% identity, 87% similarity, NDEL1 residues 9–99). Based on these similarities it was hypothesized that homodimers of NDE1 and NDEL1 might interact to form heterotetramers (9). To test this, recombinant NDE1 and NDEL1 in bacterial lysates were mixed together prior to co-purification (to best reflect the process by which the proteins were purified individually) and analysis using the cross-linking, tryptic digestion, and mass spectrometry strategy described above, yielding six unambiguous cross-link pairs. Stable isotope labeled-NDE1 was used to facilitate differentiation of NDE1 from NDEL1 peptides in some experiments, given the high sequence conservation between the two proteins (Figs. 1A and 4B). Interestingly, cross-linked peptides were detected that implied both parallel (Lys-107(NDE1)–Lys-108(NDEL1) and Lys-170(NDE1)–Lys-171(NDEL1)) and anti-parallel (Ser-196(NDE1)–Lys-108(NDEL1)) arrangements of NDE1 and NDEL1 in the heterotetrameric state (Fig. 6A and supplemental Table S2). Although the Ser-196–Lys-108 cross-link is likely, when an anti-parallel arrangement is considered for the heterotetramers (formed by two parallel homodimers that bend-back on themselves after region 3, similar to NDE1 and NDEL1 homotetramers), the parallel arrangement could derive from a putative heterodimeric arrangement between the two proteins that may form. This is because the distances between the two residues involved in the cross-link Lys-107–Lys-108 are ∼70 Å apart, when mapped on the modeled heterotetrameric structure that is formed by anti-parallel association between the two proteins (Fig. 6A). Only a parallel arrangement between the two proteins in their dimeric formation would allow for a cross-link between the two residues in question. We subsequently processed dimer-sized bands but did not obtain any unambiguous cross-links between the two proteins. We conclude that NDE1 and NDEL1 are capable of interacting to form both putative parallel heterodimers and anti-parallel heterotetramers, resembling their respective homomeric counterparts. Higher molecular weight species containing mixed NDE1/NDEL1 were also observed that yielded 35 unambiguous cross-link pairs (Fig. 6B).
FIGURE 6.
Analysis of mixed NDE1-NDEL1 by chemical cross-linking and mass spectrometry. Co-purified populations of SILAC-labeled NDE1 and unlabeled NDEL1, as well as co-purified unlabeled NDE1 and unlabeled NDEL1, were analyzed using our cross-linking/mass spectrometry paradigm as in Fig. 5. Only those cross-linked residues confirmed to be between NDE1 and NDEL1 molecules unambiguously in the heterotetramers (A) and high MWt species (B) are shown (purple lines in the case of SILAC-labeled NDE1-NDEL1 data and blue line for unlabeled NDE1-NDEL1 data). For the sole cross-link (Lys-107 in NDE1 with Lys-108 in NDEL1) that is present within the boundaries of the three-dimensional modeled structure of the NDE1-NDEL1 tetramer, this is shown on the respective longer homology model fragment using cartoon representation, with the α-carbon atoms of the residues involved shown as spheres and labeled, and the α-carbon distances (see supplemental Table S3) between them shown by a red line.
DISCUSSION
In this study, we demonstrated that recombinant NDE1 forms tetramers, as has previously been described for NDEL1 (5, 35). We confirmed the presence of dimeric forms of both proteins, despite such species not being detected by Narayanan et al. (35); this is the principal form that interacts with LIS1 (8). We also detected higher MWt forms of the proteins in solution, which may correspond to the long linear aggregates detected by negative stain EM, and/or with larger (∼400 kDa) forms of recombinant NDEL1 described previously (34). We did not, however, find any evidence of NDE1/NDEL1 monomers, in contrast to a previously reported 40-kDa form of NDEL1, described as having a cysteine peptidase activity (42) and possessing ∼40% β-sheet by CD analysis. The predominantly helical, self-associating species described here and elsewhere (5, 34, 35) seems to be the physiologically relevant form of the protein. Our results are consistent with previously published bioinformatic analysis (11) and support the notion that a putative monomeric coiled-coil helix would be unstable in isolation. That some protein partners may interact preferentially with distinct oligomeric states of NDE1 and NDEL1 (e.g., LIS1 to dimers (8) and potentially DISC1 to tetramers (35)) raises the intriguing possibility that the oligomeric state of NDE1/NDEL1 may be specifically regulated within different cellular environments. Investigating this is an important avenue for future research.
We previously undertook chemical cross-linking/MS analysis on a truncated fragment of NDEL1 17–174, in an early demonstration of the utility of our integrated structural analysis toolkit (6) to shed light on the directionality of the homodimeric coiled-coil of NDEL1, in agreement with the crystal structure of truncated NDEL1 (5). In this study, we extended this analysis to the full-length constructs of NDE1, NDEL1, and co-purified NDE1-NDEL1 to gain insight into their three-dimensional structural architecture. We also explored the use of SILAC labeling to differentiate between NDE1 from NDEL1 sequences, which is the first successful application of this labeling approach coupled to our cross-linking/MS methodology.
We obtained, in total, 416 cross-linked peptide pairs that could be assigned to oligomeric states of NDE1, NDEL1, and NDE1-NDEL1 with high confidence. We showed, with additional input from EM studies, that the disordered C-terminal regions of NDEL1 (11) have a locally flexible but overall “bent back” architecture, presumably facilitated by the presence of numerous proline residues in the “linker,” allowing interaction between a strongly predicted single α-helix that also resides therein and the known N-terminal coiled-coil domain (5). Furthermore, we have shown via a range of techniques that its paralog NDE1 shares both the α-helical N-terminal coiled-coil domain, as predicted previously (9), but also the bent back C-terminal structure; thus we provide the first experimental insights into the structure of this protein. This architecture sheds light on how the experimentally demonstrated presence of two dynein-interacting sites, ∼150 amino acid residues apart in the sequence of NDEL1, can be reconciled (18, 19). The two binding sites are in fact brought into spatial proximity in at least one conformation of NDEL1 (captured by cross-linking) and most likely can contribute jointly to a common site of interaction with dynein (Fig. 7). An alternative interpretation, that the C-terminal arms of two anti-parallel NDEL1 dimers (and NDE1 dimers) form a tetrameric arrangement via a continuous extension (18), cannot be formally discounted on the basis of the data pertaining to tetrameric species. However, only our new model was able to explain our cross-linking data for dimeric species of NDE1 and NDEL1.
FIGURE 7.
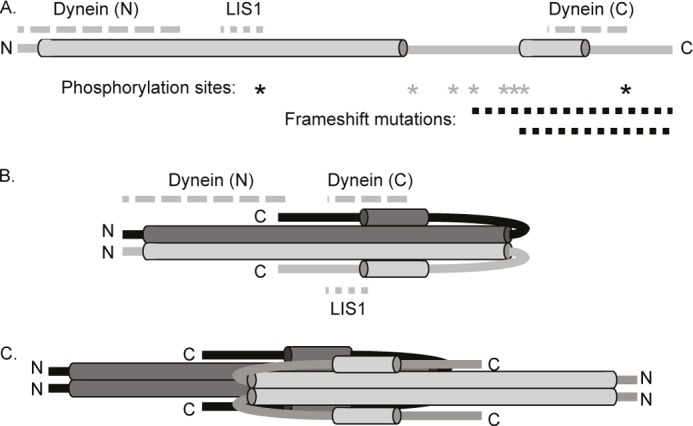
A bent back model for NDEL1 and NDE1 and its implications for dynein interaction. A, a linear cartoon of NDEL1 showing the locations of its two known dynein-binding domains (gray dashed lines), located more than 150 amino acids apart in the primary structure (18, 19, 45). The known LIS1 interaction domain is also shown (5, 7, 8). The N-terminal coiled-coil domain and predicted C-terminal helix are displayed, as in Fig. 1. Asterisks indicate known phosphorylation sites in NDE1 (black) (9, 11) and NDEL1 (gray) (7, 10, 12). Dashed black lines indicate C-terminal regions deleted by two frameshift mutations that cause severe microcephaly (16, 17). The N and C termini are indicated. B, our “bent back” model of the NDE1/NDEL1 dimer C terminus brings the two dynein-binding domains into close physical proximity. C, hypothetical model of the NDE1/NDEL1 tetramer; note that it is unclear whether all four C-terminal regions can “bend back” simultaneously or how they interact with each other. This has the potential, however, to bring many dynein-binding domains from the different chains into close proximity with each other.
Interactions of NDEL1 with LIS1, Cdc42GAP, and 14-3-3ϵ are all modulated by phosphorylation events (7, 43, 44), mainly within the regions of the largely disordered NDEL1 C terminus (11) (Figs. 1A and 7A). This is despite good experimental evidence that these proteins bind to NDEL1 at more “structured” sites (5, 7, 13, 18, 19, 45), distinct from the location of the phosphorylation sites (7, 43, 44, 46). Our observation of dynamic interactions between N- and C-terminal domains provides an explanation for this because it allows for the possibility that phosphorylation in the putatively unstructured, flexible, parts of the C-terminal region can influence the overall dynamics and architecture of the protein sufficiently to impact on multiple protein-protein interaction events.
The functional importance of this C-terminal region is further highlighted by the recent identification of stop mutations in the NDE1 gene, which result in deletion of this region (Figs. 1A and 7A). Phenotypic consequences in homozygous patients included severe microcephaly. Likely mechanisms include the abolition of dynein binding in these mutants, as well as the loss of their centrosomal functions as revealed by subsequent in vitro analysis (16, 17). These mutant proteins also displayed reduced stability (16). We suggest that instability in the disease-associated mutation is likely to arise, at least in part, from the loss of the C-terminal α-helix and the extra stability that it would confer on the main coiled-coil domains of NDE1.
Another notable observation from our cross-linking studies is that there appears to be a relative dearth of data within the N-terminal 100 amino acid residues of NDE1 and NDEL1, the region that facilitates dimerization (5). This is in keeping with our previous study on NDEL1 17–174, where we only obtained three cross-linked peptide pairs (6). This is despite subsequent improvement in MS techniques including high resolution fragmentation spectra and an additional fractionation step applied in this study. This is characteristic of the “undemocratic” nature of the cross-linking technique (21), which requires reactive sites of the residues in question to be available, accessible, and in linkable geometry. In the case of NDE1 and NDEL1, the majority of lysine residues occur from coiled-coil region 3 onwards. These regions encompass both the tetramerization domain of the coiled-coil (5) and the C-terminal regions of the proteins, for which no structural information was previously forthcoming.
Of the cross-linked pairs located within the N-terminal coiled-coil region, only one in NDE1 (Lys-81–Lys-107) of a total of 21, cannot be reconciled with either the crystal structures for N-terminal NDEL1 or the homology models of NDE1. There are several possible explanations for this: (a) we utilized full-length protein constructs, whereas the homology models (9) were based on constructs corresponding to only the coiled-coil domain (5); (b) we used physiological pH while undertaking our cross-linking experiments, whereas the crystal structures were solved at acidic pH (5); and (c) the coiled-coil domain and/or the lysine side chains might be sufficiently flexible to enable alteration in the overall structure or local environment.
Our study indicates that there are clear dynamic conformational changes for the proteins with respect to the levels of “reach,” flexibility, and/or rigidity of the C-terminal regions when they bend back to interact with the N-terminal coiled-coil domain; cross-linking can capture these dynamic structures as we recently demonstrated (20). There exists a precedent for dynamic change in the conformation of NDEL1 (39), the biophysical nature of which is unknown. Additionally, we recently reported a protein kinase A phosphorylation site at Thr-131 within the coiled-coil region 3 in NDE1 (9). The side chains of this residue face inwards within two opposing α-helices and are consequently not readily accessible (9). Conformations different from those seen in crystal structures to allow this phospho-site to be more accessible may for example ratify the single cross-linked pair that is in conflict in the homology models. Finally, much of the C terminus of NDE1 and NDEL1 is predicted to lack a fixed structure (11), consistent with the notion that the C terminus might bend back in multiple ways.
Our circular dichroism, electron microscopy, and cross-linking data demonstrate that NDE1 and NDEL1 share a highly similar overall structure, reinforcing the suggestion that they fulfill similar roles in the cell (47), known to include neurite outgrowth and mitotic functions (reviewed in Refs. 3 and 4), although the possibility that they are differentially regulated, in terms of expression, post-translational modification or protein-protein interaction partners, remains. The question of whether NDE1 and NDEL1 perform these roles in parallel or in partnership is less clear. Previously, it was determined that NDE1 and NDEL1 can be co-immunoprecipitated from both cell lines and brain lysates, although a direct interaction between them had not been shown (22, 41), leaving it unclear whether they form functional heteromultimers or simply interact simultaneously with common binding proteins such as DISC1 or LIS1. Here we demonstrate that such direct binding of NDE1 and NDEL1 can occur, at least at the level of heterotetramer formation and higher molecular weight species. Furthermore, it appears that the mechanisms by which NDE1 and NDEL1 interact with each other include those by which they each form homo-oligomers. Future work to dissect how differential phosphorylation of these two proteins functions synergistically to influence the overall architecture of the hetero-oligomers is now needed.
Finally, our results bear upon possible pathogenic mechanisms. Not only will mutations in NDE1 reported in microcephaly abrogate function, but they are also likely to influence NDEL1 interaction (we obtained a cross-link between NDEL1 108 and NDE1 296 in the heterotetramer), DISC1 binding, and other associated protein interaction pathways. Similarly, copy number variation for NDE1 has also been reported in association with both autism (48) and schizophrenia (49–54). That NDE1 and NDEL1 are shown by our analysis to form hetero-oligomers suggests that this propensity will be affected by the balance of NDE1 and NDEL1 expression that may be altered in disease-associated gene insertions or deletions.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Cronshaw for advice on MALDI-TOF, Dr. Lauri Peil for BL21 (DE3) LysA ArgA cell lines, and Sandra Bruce and Dr. Cornelia Ludwig for technical support on protein expression and purification.
This work was supported by Wellcome Trust Grant 088179/A/09/Z (to D. C. S., N. J. B., J. K. M., P. N. B., M. D. W., J. R., and D. J. P.) and funds from the Wellcome Trust (to M. A. W., E. A. B., and J. B.), the Scottish University Life Sciences Alliance, and the Biotechnology and Biological Sciences Research Council.

This article contains supplemental Tables S1–S3.
- SILAC
- stable isotope labeling by amino acids in cell culture
- BS3
- bis(sulfosuccinimidyl)suberate
- CC R
- Coiled-coil region
- CD
- circular dichroism
- MWt
- molecular weight.
REFERENCES
- 1. Shmueli A., Segal M., Sapir T., Tsutsumi R., Noritake J., Bar A., Sapoznik S., Fukata Y., Orr I., Fukata M., Reiner O. (2010) Ndel1 palmitoylation. A new mean to regulate cytoplasmic dynein activity. EMBO J. 29, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Efimov V. P., Morris N. R. (2000) The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 150, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chansard M., Hong J. H., Park Y. U., Park S. K., Nguyen M. D. (2011) Ndel1, Nudel (Noodle). Flexible in the cell? Cytoskeleton 68, 540–554 [DOI] [PubMed] [Google Scholar]
- 4. Bradshaw N. J., Porteous D. J. (2012) DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology 62, 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Derewenda U., Tarricone C., Choi W. C., Cooper D. R., Lukasik S., Perrina F., Tripathy A., Kim M. H., Cafiso D. S., Musacchio A., Derewenda Z. S. (2007) The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure 15, 1467–1481 [DOI] [PubMed] [Google Scholar]
- 6. Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 7. Yan X., Li F., Liang Y., Shen Y., Zhao X., Huang Q., Zhu X. (2003) Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 23, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tarricone C., Perrina F., Monzani S., Massimiliano L., Kim M. H., Derewenda Z. S., Knapp S., Tsai L. H., Musacchio A. (2004) Coupling PAF signaling to dynein regulation. Structure of LIS1 in complex with PAF-acetylhydrolase. Neuron 44, 809–821 [DOI] [PubMed] [Google Scholar]
- 9. Bradshaw N. J., Soares D. C., Carlyle B. C., Ogawa F., Davidson-Smith H., Christie S., Mackie S., Thomson P. A., Porteous D. J., Millar J. K. (2011) PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J. Neurosci. 31, 9043–9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mori D., Yano Y., Toyo-oka K., Yoshida N., Yamada M., Muramatsu M., Zhang D., Saya H., Toyoshima Y. Y., Kinoshita K., Wynshaw-Boris A., Hirotsune S. (2007) NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol. Cell. Biol. 27, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradshaw N. J., Ogawa F., Antolin-Fontes B., Chubb J. E., Carlyle B. C., Christie S., Claessens A., Porteous D. J., Millar J. K. (2008) DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophys. Res. Commun. 377, 1091–1096 [DOI] [PubMed] [Google Scholar]
- 12. Niethammer M., Smith D. S., Ayala R., Peng J., Ko J., Lee M. S., Morabito M., Tsai L. H. (2000) NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711 [DOI] [PubMed] [Google Scholar]
- 13. Brandon N. J., Handford E. J., Schurov I., Rain J. C., Pelling M., Duran-Jimeniz B., Camargo L. M., Oliver K. R., Beher D., Shearman M. S., Whiting P. J. (2004) Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex. Implications for schizophrenia and other major neurological disorders. Mol. Cell Neurosci. 25, 42–55 [DOI] [PubMed] [Google Scholar]
- 14. Liang Y., Yu W., Li Y., Yang Z., Yan X., Huang Q., Zhu X. (2004) Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J. Cell Biol. 164, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. (2006) Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell 17, 680–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alkuraya F. S., Cai X., Emery C., Mochida G. H., Al-Dosari M. S., Felie J. M., Hill R. S., Barry B. J., Partlow J. N., Gascon G. G., Kentab A., Jan M., Shaheen R., Feng Y., Walsh C. A. (2011) Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected]. Am. J. Hum. Genet. 88, 536–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bakircioglu M., Carvalho O. P., Khurshid M., Cox J. J., Tuysuz B., Barak T., Yilmaz S., Caglayan O., Dincer A., Nicholas A. K., Quarrell O., Springell K., Karbani G., Malik S., Gannon C., Sheridan E., Crosier M., Lisgo S. N., Lindsay S., Bilguvar K., Gergely F., Gunel M., Woods C. G. (2011) The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am. J. Hum. Genet. 88, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zylkiewicz E., Kijanska M., Choi W. C., Derewenda U., Derewenda Z. S., Stukenberg P. T. (2011) The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J. Cell Biol. 192, 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S., Zheng Y. (2011) Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract. J. Biol. Chem. 286, 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rappsilber J. (2011) The beginning of a beautiful friendship. Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradshaw N. J., Christie S., Soares D. C., Carlyle B. C., Porteous D. J., Millar J. K. (2009) NDE1 and NDEL1. Multimerisation, alternate splicing and DISC1 interaction. Neurosci. Lett. 449, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24. Matic I., Jaffray E. G., Oxenham S. K., Groves M. J., Barratt C. L., Tauro S., Stanley-Wall N. R., Hay R. T. (2011) Absolute SILAC-compatible expression strain allows Sumo-2 copy number determination in clinical samples. J. Proteome Res. 10, 4869–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 26. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 27. Braun N., Zacharias M., Peschek J., Kastenmüller A., Zou J., Hanzlik M., Haslbeck M., Rappsilber J., Buchner J., Weinkauf S. (2011) Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc. Natl. Acad. Sci. U.S.A. 108, 20491–20496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31. Vriend G. (1990) WHAT IF. A molecular modeling and drug design program. J. Mol. Graphics 8, 52–56, 29 [DOI] [PubMed] [Google Scholar]
- 32. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 33. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 34. Leliveld S. R., Bader V., Hendriks P., Prikulis I., Sajnani G., Requena J. R., Korth C. (2008) Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 28, 3839–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narayanan S., Arthanari H., Wolfe M. S., Wagner G. (2011) Molecular characterization of disrupted in schizophrenia-1 risk variant S704C reveals the formation of altered oligomeric assembly. J. Biol. Chem. 286, 44266–44276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng Y., Olson E. C., Stukenberg P. T., Flanagan L. A., Kirschner M. W., Walsh C. A. (2000) LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679 [DOI] [PubMed] [Google Scholar]
- 37. Bariola P. A., Retelska D., Stasiak A., Kammerer R. A., Fleming A., Hijri M., Frank S., Farmer E. E. (2004) Remorins form a novel family of coiled-coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol. Biol. 55, 579–594 [DOI] [PubMed] [Google Scholar]
- 38. Kalkhof S., Sinz A. (2008) Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 392, 305–312 [DOI] [PubMed] [Google Scholar]
- 39. Collins D. M., Murdoch H., Dunlop A. J., Charych E., Baillie G. S., Wang Q., Herberg F. W., Brandon N., Prinz A., Houslay M. D. (2008) Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through protein kinase A (PKA). Cell Signal. 20, 2356–2369 [DOI] [PubMed] [Google Scholar]
- 40. Segal M., Soifer I., Petzold H., Howard J., Elbaum M., Reiner O. (2012) Ndel1-derived peptides modulate bidirectional transport of injected beads in the squid giant axon. Biol. Open 1, 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burdick K. E., Kamiya A., Hodgkinson C. A., Lencz T., DeRosse P., Ishizuka K., Elashvili S., Arai H., Goldman D., Sawa A., Malhotra A. K. (2008) Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia. Evidence of epistasis and competitive binding. Hum. Mol. Genet. 17, 2462–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hayashi M. A., Portaro F. C., Bastos M. F., Guerreiro J. R., Oliveira V., Gorrão S. S., Tambourgi D. V., Sant'Anna O. A., Whiting P. J., Camargo L. M., Konno K., Brandon N. J., Camargo A. C. (2005) Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc. Natl. Acad. Sci. U.S.A. 102, 3828–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toyo-oka K., Shionoya A., Gambello M. J., Cardoso C., Leventer R., Ward H. L., Ayala R., Tsai L. H., Dobyns W., Ledbetter D., Hirotsune S., Wynshaw-Boris A. (2003) 14-3-3ϵ is important for neuronal migration by binding to NUDEL. A molecular explanation for Miller-Dieker syndrome. Nat. Genet. 34, 274–285 [DOI] [PubMed] [Google Scholar]
- 44. Shen Y., Li N., Wu S., Zhou Y., Shan Y., Zhang Q., Ding C., Yuan Q., Zhao F., Zeng R., Zhu X. (2008) Nudel binds Cdc42GAP to modulate Cdc42 activity at the leading edge of migrating cells. Dev. Cell 14, 342–353 [DOI] [PubMed] [Google Scholar]
- 45. Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., Hirotsune S. (2000) A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696 [DOI] [PubMed] [Google Scholar]
- 46. Johnson C., Crowther S., Stafford M. J., Campbell D. G., Toth R., MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lam C., Vergnolle M. A., Thorpe L., Woodman P. G., Allan V. J. (2010) Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J. Cell Sci. 123, 202–212 [DOI] [PubMed] [Google Scholar]
- 48. Ullmann R., Turner G., Kirchhoff M., Chen W., Tonge B., Rosenberg C., Field M., Vianna-Morgante A. M., Christie L., Krepischi-Santos A. C., Banna L., Brereton A. V., Hill A., Bisgaard A. M., Müller I., Hultschig C., Erdogan F., Wieczorek G., Ropers H. H. (2007) Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum. Mutat. 28, 674–682 [DOI] [PubMed] [Google Scholar]
- 49. Need A. C., Ge D., Weale M. E., Maia J., Feng S., Heinzen E. L., Shianna K. V., Yoon W., Kasperavici[overbar]ute D., Gennarelli M., Strittmatter W. J., Bonvicini C., Rossi G., Jayathilake K., Cola P. A., McEvoy J. P., Keefe R. S., Fisher E. M., St Jean P. L., Giegling I., Hartmann A. M., Möller H. J., Ruppert A., Fraser G., Crombie C., Middleton L. T., St Clair D., Roses A. D., Muglia P., Francks C., Rujescu D., Meltzer H. Y., Goldstein D. B. (2009) A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 5, e1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ikeda M., Aleksic B., Kirov G., Kinoshita Y., Yamanouchi Y., Kitajima T., Kawashima K., Okochi T., Kishi T., Zaharieva I., Owen M. J., O'Donovan M. C., Ozaki N., Iwata N. (2010) Copy number variation in schizophrenia in the Japanese population. Biol. Psychiatry 67, 283–286 [DOI] [PubMed] [Google Scholar]
- 51. Ingason A., Rujescu D., Cichon S., Sigurdsson E., Sigmundsson T., Pietiläinen O. P., Buizer-Voskamp J. E., Strengman E., Francks C., Muglia P., Gylfason A., Gustafsson O., Olason P. I., Steinberg S., Hansen T., Jakobsen K. D., Rasmussen H. B., Giegling I., Möller H. J., Hartmann A., Crombie C., Fraser G., Walker N., Lonnqvist J., Suvisaari J., Tuulio-Henriksson A., Bramon E., Kiemeney L. A., Franke B., Murray R., Vassos E., Toulopoulou T., Mühleisen T. W., Tosato S., Ruggeri M., Djurovic S., Andreassen O. A., Zhang Z., Werge T., Ophoff R. A., Rietschel M., Nöthen M. M., Petursson H., Stefansson H., Peltonen L., Collier D., Stefansson K., St Clair D. M. (2011) Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol. Psychiatry 16, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Magri C., Sacchetti E., Traversa M., Valsecchi P., Gardella R., Bonvicini C., Minelli A., Gennarelli M., Barlati S. (2010) New copy number variations in schizophrenia. PLoS One 5, e13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. International Schizophrenia Consortium (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kirov G., Grozeva D., Norton N., Ivanov D., Mantripragada K. K., Holmans P., International Schizophrenia Consortium, Wellcome Trust Case Control Consortium, Craddock N., Owen M. J., O'Donovan M. C. (2009) Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet. 18, 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.