Background: Poly(ADP-ribose) polymerase 1 (PARP-1) modulates chromatin structure and is activated upon DNA damage.
Results: PARP-1 differentiates between nucleosomes and DNA in its binding affinity and is activated to different degrees.
Conclusion: PARP-1 engages different DNA-binding modules with nucleosomes and DNA.
Significance: The role of PARP-1 as a chromatin architectural protein and responder in DNA repair is reflected in different binding modes.
Keywords: DNA Repair, DNA/Protein Interaction, Enzymes, Fluorescence Resonance Energy Transfer (FRET), Nucleosome, PARP-1
Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is an abundant nuclear protein that binds chromatin and catalyzes the transfer of ADP-ribose groups to itself and to numerous target proteins upon interacting with damaged DNA. The molecular basis for the dual role of PARP-1 as a chromatin architectural protein and a first responder in DNA repair pathways remains unclear. Here, we quantified the interactions of full-length PARP-1 and its N-terminal half with different types of DNA damage and with defined nucleosome substrates. We found that full-length PARP-1 prefers nucleosomes with two linker DNA extensions over any other substrate (including several free DNA models) and that the C-terminal half of PARP-1 is necessary for this selectivity. We also measured the ability of various substrates to activate PARP-1 activity and found that the most important feature for activation is one free DNA end rather than tight interaction with the activating nucleic acid. Our data provide insight into the different modes of interaction of this multidomain protein with nucleosomes and free DNA.
Introduction
Poly(ADP-ribose) polymerase (PARP-1)3is a conserved multidomain enzyme that is present in all eukaryotes except yeast. With an estimated abundance of ∼106 molecules/cell, there is approximately one PARP-1 molecule/20 nucleosomes (1). Historically, its role in DNA damage detection has received much attention. More recently, PARP-1 has been linked to the regulation of chromatin structure and transcription (reviewed in Refs. 2 and 3). In its enzymatically inactive form, PARP-1 binds chromatin and contributes to the formation of transcriptionally silent chromatin domains (4). Recent data indicate a role in promoting the formation of chromatin structures that are permissive to gene expression (5). Upon sensing DNA damage, PARP-1 catalyzes the cleavage of its substrate NAD+ into nicotinamide and ADP-ribose and polymerizes long ADP-ribose chains onto core histones, linker histone H1, and many other nuclear proteins (heteromodification), as well as onto itself (automodification), with itself as the vastly preferred substrate (6). Mutational studies have revealed several automodification sites in PARP-1 (see Fig. 1) (7, 8). Because of its well described role in DNA damage repair, PARP-1 is an attractive drug target to augment cancer therapy (9, 10). However, little quantitative information is available on the many interactions of unmodified and modified PARP-1. For example, it is not known how strongly PARP-1 interacts with nucleosomes compared with nucleosome-free DNA and whether PARP-1 can recognize DNA damage in the context of chromatin. This limits our understanding of PARP-1 function in chromatin structure maintenance and DNA repair.
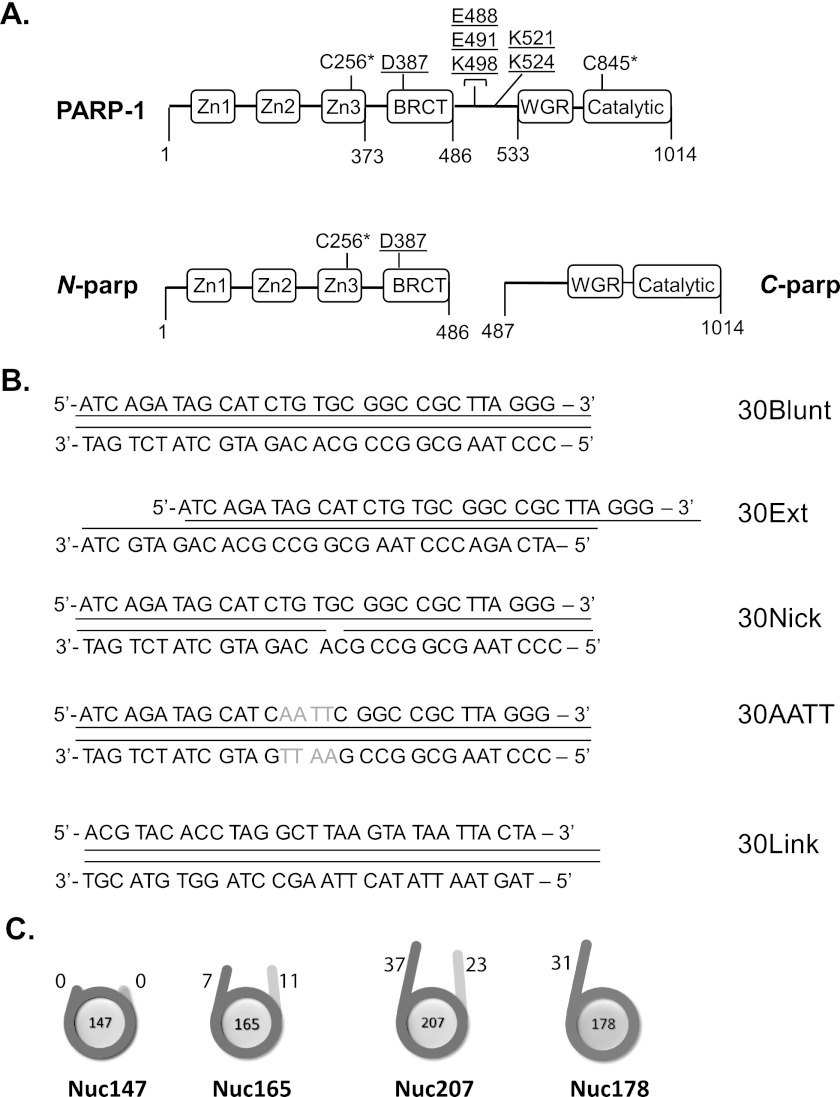
FIGURE 1.
PARP-1 constructs and substrates assayed in this study. A, full-length PARP-1 contains all six domains; N-parp encompasses zinc fingers Zn1–Zn3 and the BRCT domain (amino acids 1–486); and C-parp spans residues 487-1014 and includes the WGR and CAT domains. Surface-exposed native cysteine residues (positions 256 and 845; indicated by asterisks) were labeled with Alexa Fluor 488. Underlined residues denote auto-PARylation sites (8). B, DNA models used for PARP-1 binding and activity assays. 30Blunt, 30Ext, and 30Nick are identical in sequence. 30AATT replaces 4 central bp with AATT. 30Link is identical in sequence to the linker in Nuc178. All DNA models were labeled at the 5′-end with Cy5 or ATTO 647N. C, nucleosome substrates were labeled with ATTO 647N at histone H4 E63C on the histone octamer (21). All nucleosomal DNA is based on the 601 positioning sequence (34). The length of the linker DNA in each particle is indicated in base pairs.
PARP-1 contains three N-terminal zinc finger domains and a BRCA1 C-terminal (BRCT) domain that is linked to the tryptophan/glycine/arginine-rich (WGR) domain and catalytic (CAT) domain through a flexible linker (see Fig. 1A). Structural information on all individual domains is available. Zinc finger (Zn) 1 and Zn2 bind DNA with high affinity in a sequence-independent and structure-dependent manner (11, 12), with the strongest interaction observed for Zn2. Zn3 does not bind DNA on its own but is essential for DNA-dependent stimulation of PARP-1 activity (13). It has been proposed that DNA binding by the zinc fingers triggers a conformational change in the full-length protein, which then activates the CAT domain (12). The impressive structure of a nearly full-length PARP-1·DNA complex (14) provides a detailed view of the domain arrangements upon DNA damage and explains the propensity of PARP-1 for PARylating itself rather than target protein substrates. The crystallized PARP-1 construct, which lacks only Zn2 and the BRCT domain, binds DNA as a monomer, consistent with earlier studies (11, 12), and displays extensive contacts between the DNA damage interface and the CAT domain. Importantly, the interaction with a single DNA fragment is afforded by residues from Zn1, Zn3, and the WGR domain. This latter domain had previously not been implicated in DNA binding, and it was generally believed that amino acids 1–486 are solely responsible for the interaction with DNA (11, 15).
In addition to recognizing DNA damage, PARP-1 also binds chromatin and protects an additional ∼10–20 bp of nucleosomal DNA near the entry-exit sites, reminiscent of the pattern observed for H1/nucleosome interaction (16). Only a moderate contribution of the C-terminal domain of PARP-1 to the interaction with DNA or chromatin was reported (13, 17). However, this domain is essential for chromatin compaction, independent of its catalytic activity (17).
There are reports that PARP-1 activity is stimulated not only by free DNA but also by chromatin and isolated histones (16–18). Consistent with the qualitative observation that PARP-1 also binds mixtures of histones in vitro, even in the absence of DNA, PARP-1 is reportedly activated by the N-terminal tail of histone H4 (18). However, readout of the binding affinities and catalytic activity was indirect. Additionally, no systematic quantitative comparisons of the degree of PARP-1 activation by the various allosteric activators have been made.
To fill these significant gaps in our understanding of PARP-1 function, we measured the interactions of highly pure full-length PARP-1, its N-terminal half (amino acids 1–486, referred to as N-parp), and its CAT domain (amino acids 487–1014, referred to as C-parp) with defined DNA fragments, as well as with nucleosome substrates with various extensions of linker DNA (see Fig. 1). We also quantified the ability of the various binding substrates to stimulate the enzymatic activity of PARP-1. Our data suggest fundamental differences in the mode of interaction between chromatin and free DNA, consistent with the two roles of PARP-1 as a chromatin architectural protein and a sensor of DNA damage. Furthermore, our data demonstrate that PARP-1 is capable of recognizing DNA double-strand breaks in the context of a nucleosome.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Fluorescent Labeling of PARP-1 and N-parp
N-parp was expressed, purified, and labeled as described (19). Full-length human PARP-1 V762A was expressed in Sf9 insect cells (20). Cell pellets were thawed from −80 °C and sonicated (3 × 5 s, output 6.5, and duty cycle 65% on a Branson 450 Sonifier) on ice in lysis buffer (300 mm NaCl, 25 mm Tris-HCl (pH 8), 1 mm β-mercaptoethanol, and 1 mm PMSF). Cell lysates were then cleared by centrifugation at 14,000 rpm for 30 min at 4 °C, and the pellet was discarded. DNA was removed by the addition of 1.0 mg/ml salmon sperm protamine sulfate (Sigma-Aldrich), followed by centrifugation at 14,000 rpm for 30 min at 4 °C. The supernatant was precipitated by a two-step ammonium sulfate treatment at 4 °C while stirring overnight. In the first step, the supernatant was incubated with 30% ammonium sulfate (164 g/1000 ml) and centrifuged as described above. In the second step, the supernatant from 30% ammonium sulfate was brought up to 70% ammonium sulfate saturation (249 g/1000 ml). The precipitate was resuspended in heparin chromatography buffer A (100 mm NaCl, 25 mm Tris-HCl (pH 8), and 1.0 mm β-mercaptoethanol), loaded onto a HiTrap heparin column (GE Healthcare), and eluted with a linear gradient (0–100% buffer B) of heparin chromatography buffer B (1.5 m NaCl, 25 mm Tris-HCl (pH 8), and 1.0 mm β-mercaptoethanol). Further purification included size exclusion chromatography and cation exchange using a HiTrap SP column (GE Healthcare). This homogeneous preparation of PARP-1 tested negatively for automodification by Western blotting (data not shown).
Purified full-length PARP-1 was fluorescently labeled at its native surface-exposed cysteine residues (Cys-256 and Cys-842). 10 mm Alexa Fluor 488 fluorophore (Invitrogen) in Me2SO was added to PARP-1 in 300 mm NaCl and 25 mm Tris (pH 7.5) in equimolar amounts three times over 3 h and allowed to mix overnight at 4 °C. Excess fluorophore was removed using a HiTrap heparin HP column as described above. Labeled PARP-1 and N-parp run on a 4–12% gradient SDS-polyacrylamide gel (Criterion XT) appeared as homogeneous bands (see Fig. 2, A and B). A typical labeling efficiency of 10–25% was routinely obtained.
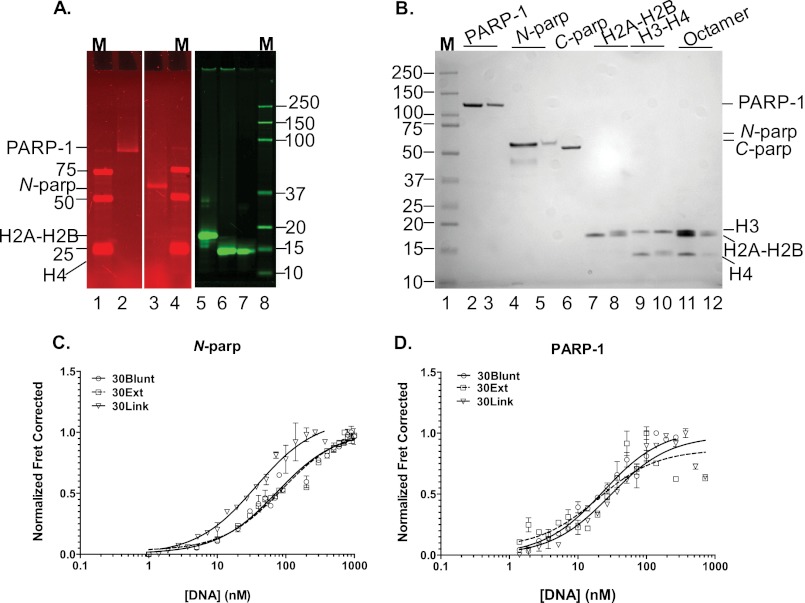
FIGURE 2.
The CAT domain of PARP-1 contributes moderately to the interaction with DNA. A, fluorescently labeled PARP-1 constructs and histones. All samples were run on a Criterion XT 4-12% gradient SDS-polyacrylamide gel that was scanned on a Typhoon Imager at wavelengths appropriate for measuring donor (488 nm excitation and 520 nm emission) for the left panels depicting PARP-1 constructs and for measuring acceptor (633 nm excitation and 670 nm emission) for the right panel with labeled histones. Lanes 1, 4, and 8, molecular weight markers (M); lane 2, Alexa Fluor 488-labeled PARP-1 (at Cys-256 and Cys-845); lane 3, Alexa Fluor 488-labeled N-parp (at Cys-256); lane 5, H2A-H2B dimer (with ATTO 647N-labeled at histone H2B T112C); lane 6, (H3-H4)2 tetramer (labeled with ATTO 647N at histone H4 E63C); lane 7, histone octamer (labeled with ATTO 647N at histone H4 E36C). B, the same gel was visualized with Imperial stain. Lane 1, protein size marker; lane 2, unlabeled PARP-1; lane 3, labeled PARP-1; lane 4, unlabeled N-parp; lane 5, labeled N-parp; lane 6, C-parp; lane 7, unlabeled H2A-H2B; lane 8, labeled H2A-H2B; lane 9, unlabeled H3-H4; lane 10, labeled H3-H4; lane 11, unlabeled histone octamer; lane 12, labeled histone octamer. C, N-parp (labeled with Alexa Fluor 488 at Cys-256) binding to selected free DNA models shown in B measured by HI-FI FRET. D, PARP-1 binding curves for the same DNA fragments. The concentrations of the titrated acceptor species are plotted on the x axis, and normalized FRET-corrected values are plotted on the y axis (19, 24). Affinities from this and similar experiments are listed in Table 1. Error bars shown here were obtained from duplicates from individual representative experiments.
Histone Labeling
Histone H4 E63C and H2B T112C mutants were labeled with ATTO 647N and refolded as described (see Fig. 2, A and B) (21). A typical labeling efficiency of 10–25% was routinely obtained.
DNA Oligomers
30-bp blunt-ended, nicked, and overhang DNAs, all containing the template sequence 5′-ATC AGA TAG CAT CTG TGC GGC CGC TTA GGG-3′ either with or without a 5′-Cy5 or 5′-ATTO 647N fluorophore, were ordered from Integrated DNA Technologies (see Fig. 1B). Annealing was carried out by mixing equimolar amounts of template and reverse strand and heating at 95 ºC for 2 min, followed by slow cooling to room temperature.
All DNAs used for nucleosome assembly contained the 601 positioning sequence with variable linker arms (see Fig. 1C) and were expressed and purified as described (22). The 147-bp DNA represents the minimal nucleosomal DNA; Nuc165 has linker arms of 7 and 11 bp, and Nuc207 exhibits linker lengths of 23 and 37 bp, respectively. We also generated an asymmetric linker arm by digesting the 207-bp DNA with BsiEI, followed by mung bean nuclease digestion, producing the 178-bp DNA (see Fig. 1C).
Chromatin Assembly and Characterization
Labeled nucleosomes were assembled on DNAs of varying lengths as described (22) using ATTO 647N-labeled histone octamer (see Fig. 3, A and B). The nucleosome preparations typically had <1% free DNA present.
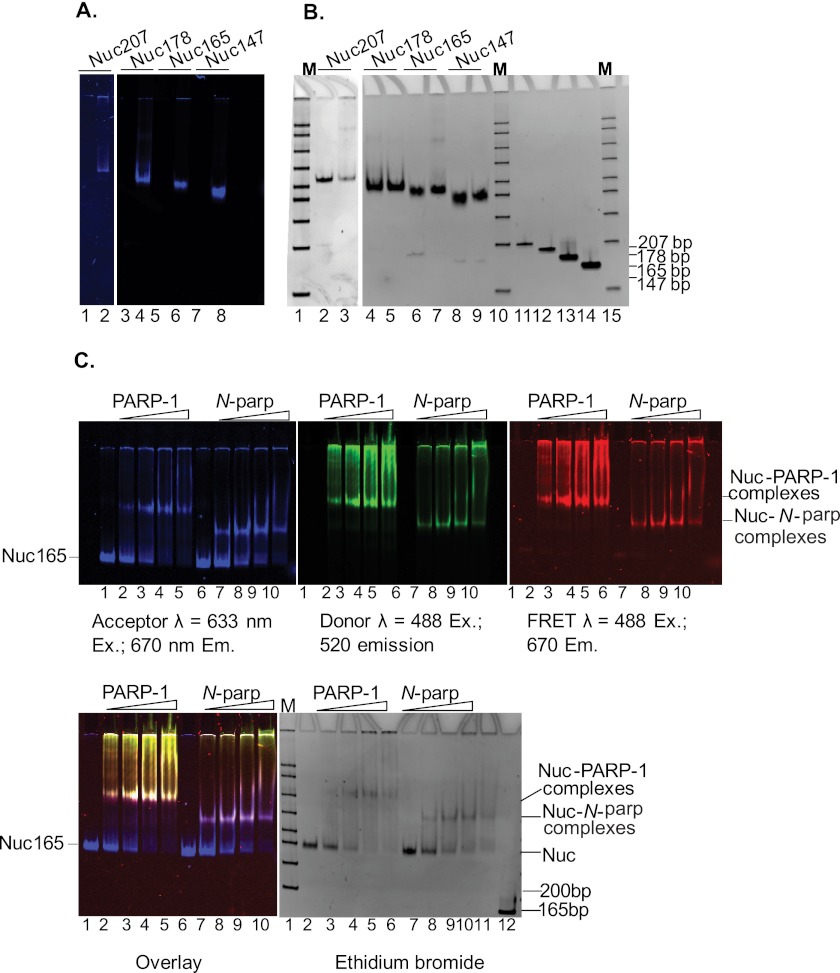
FIGURE 3.
PARP-1 interacts with nucleosomes. A, fluorescently labeled nucleosome substrates. DNA fragments 207, 178, 165, and 147 bp in length, all containing the 601 positioning sequence, were assembled into nucleosomes with histone octamers labeled at histone H4 E63C with ATTO 647N. Nucleosomes were run on 5% native polyacrylamide gel and scanned on a Typhoon Imager at an emission wavelength of 670 nm. Lanes 2, 4, 6, and 8 are nucleosomes assembled on 207-, 178-, 165-, and 147-bp DNAs, respectively. Lanes 1, 3, 5, and 7 are unlabeled nucleosomes assembled on 207, 178, 165, and 147 bp DNA respectively. B, the same gel stained with ethidium bromide. Lanes 1 and 2 are labeled and unlabeled Nuc207, respectively. Lanes 3 and 4 are labeled and unlabeled Nuc178, respectively. Lanes 5 and 6 are labeled and unlabeled Nuc165, respectively. Lanes 7 and 8 are labeled and unlabeled Nuc147, respectively. Lanes 9–12 are 207-, 178-, 165-, and 147-bp DNA fragments, respectively. Note the absence of free DNA (<1%) in the nucleosome samples. C, ATTO 647N (acceptor)-labeled nucleosomes (Nuc165) were incubated with increasing amounts of Alexa Fluor 488-labeled PARP-1 or N-parp and analyzed by native PAGE. Gels were scanned on a Typhoon Imager at the indicated wavelengths. Lower left panel, acceptor, donor, and FRET channels are overlaid. Lanes 1 and 6, Nuc165; lanes 2–5, nucleosomes incubated with increasing molar ratios of PARP-1 (0.5-, 1-, 1.5-, and 2-fold excess); lanes 7–10, nucleosomes incubated with increasing molar ratios of N-parp (0.5-, 1-, 1.5-, and 2-fold excess); lane 11 in the lower right panel, free 165-bp DNA.
HI-FI FRET Assay
We used the previously developed HI-FI FRET assay (19) for measuring the affinities and stoichiometries of PARP-1 and N-parp labeled with the donor dye Alexa Fluor 488 and titrated in substrates labeled with the acceptor dye ATTO 647N. The buffer used for setting up the binding reactions contained 25 mm Tris (pH 7.5), 200 mm NaCl, and 0.01% (v/v) each Nonidet P-40 and CHAPS. FRET calculations and corrections were performed as described (19). The data were plotted in GraphPad Prism and fitted using one-site binding + background or one site-specific binding with Hill slope. The data were represented by plotting titrated species labeled with the acceptor on the x axis and normalized FRET-corrected values on the y axis. The Hill coefficient was held constant at 1 unless mentioned otherwise.
EMSA
Labeled Nuc165 (1 μm) was titrated with increasing molar ratios of PARP-1 or N-parp labeled with Alexa Fluor 488 in the binding buffer described above and incubated for 30 min at room temperature. Samples were subsequently run on a 22 × 20-cm native Tris borate/EDTA (TBE) gel and run in 0.5× TBE at 4 °C for 120 min at 300 V and 10 watts. The gel was scanned on a Typhoon Imager at wavelengths appropriate for measuring acceptor (633 nm excitation and 670 nm emission), donor (488 nm excitation and 520 nm emission), and FRET (488 nm excitation and 670 nm emission). Gels were then stained with ethidium bromide to visualize the DNA.
Unlabeled nucleosomes (1 μm) were incubated with increasing amounts of labeled or unlabeled PARP-1 constructs (PARP-1, N-parp, and C-parp) in 25 or 50 mm Tris (pH 7.5), 150 mm NaCl, 2 mm arginine, 0.01% CHAPS, and Nonidet P-40. The DNA/chromatin/PARP-1 samples were incubated at room temperature for 30 min, loaded on a prerun 5% native TBE gel, and run at 150 V for 60 min at 4 °C for 8 × 8-cm gels in 0.2× TBE. Gels were stained with ethidium bromide, followed by Imperial protein stain.
Size Exclusion Chromatography-Multiangle Light Scattering (SEC-MALS)
Nucleosomes (Nuc147, Nuc165, and Nuc207) and their complexes with PARP-1 were assembled in 50 mm Tris (pH 7.5), 150 or 300 mm NaCl, and 2 mm arginine and analyzed by SEC-MALS as described (23).
PARP-1 Enzymatic Assay
PARP-1 (constant at 1 μm) and “activators” (DNA or nucleosomes; 1–2 μm) were mixed to a final volume of 30 μl in 50 mm Tris (pH 8), 50 mm NaCl (or 100 mm NaCl for chromatin activators), 10 mm MgCl2 (or 1 mm MgCl2 for chromatin activators), and 1 mm DTT and allowed to incubate for 1 h at 30 °C. 30 μl of the various NAD+ stocks (0–400 μm) were added to the above tubes. Reactions were quenched after 30 s with either Laemmli buffer or ice-cold 20% TCA. Reactions quenched with Laemmli buffer were analyzed by 8% SDS-PAGE and Western blotting. 1–5% of the reactions quenched with 20% TCA were loaded onto a Zeta-Probe membrane (Bio-Rad) using a Bio-Rad dot blot apparatus (20). A poly(ADP-ribose) (PAR) standard curve was also included in each blot to correlate the amount of PAR generated by automodification directly to a known amount of standard PAR. After loading the sample, the wells were washed once with 10% TCA, followed by washing with 70% ethanol. The membrane was then dried on a gel dryer at 80 °C for 1 h and blocked with 5% milk in 1× TBS overnight. The blot was incubated with anti-PAR primary antibody (Abcam) for 1 h, followed by five washes with 1× TBS and 0.01% (v/v) Tween 20. ATTO 647N-conjugated goat anti-mouse secondary antibodies (Sigma) were incubated for 1 h, followed by five washes with 1× TBS containing 0.01% Tween 20. The blots were scanned on a Typhoon Imager at wavelength appropriate for Atto647N, as described above, and quantified using ImageQuant (GE Healthcare). Michaelis-Menten parameters were derived using GraphPad Prism v5® nonlinear regression.
RESULTS
PARP-1 Exhibits a Slight Preference for Flexible DNA
We have previously shown by agarose gel mobility shift assays that a fragment of PARP-1 encompassing the three zinc fingers and the BRCT domain (N-parp) (Fig. 1A) binds tightly to various DNA damage models (11). We wanted to investigate how full-length PARP-1 compares with N-parp using a more rigorous solution-state assay that we recently developed in our laboratory (19, 24). PARP-1 and N-parp were purified to homogeneity and labeled with fluorophores (Fig. 2, A and B) as described (19). Electrophoretic mobility shifts were observed when a 30-bp DNA fragment (referred to as 30Blunt DNA) was titrated with either full-length PARP-1 or N-parp, qualitatively confirming that both fluorescently labeled proteins form defined complexes with DNA (data not shown). Quantitative information on the interactions was obtained by monitoring binding reactions through FRET in a plate assay (HI-FI FRET) (19); representative data are shown in Fig. 2 (C and D). Table 1 summarizes the affinities of the two PARP-1 constructs for the free DNA models listed in Fig. 1B. Both full-length PARP-1 and its N-terminal half (N-parp) exhibited a slight preference for DNA containing an internal nick or an AATT insert. These features are thought to induce a curved or bent conformation into double-stranded DNA (25).
TABLE 1.
Relative affinities of N-parp and PARP-1 for various free DNA models and nucleosomes
Data were obtained using the HI-FI FRET assay (19). Buffer for all binding reactions contained 25 mm Tris (pH 7.5), 200 mm NaCl, and 0.01% (v/v) each Nonidet P-40 and CHAPS (with the exception of the value indicated in Footnote a). S.D. values are reported for two to five independent experiments (with the exception of the value indicated in Footnote b).
| Binding substrate | N-parp |
PARP-1 |
||
|---|---|---|---|---|
| Kd(app) | R2 | Kd(app) | R2 | |
| nm | nm | |||
| 30Blunt | 62.2 ± 10.2 | 0.97 | 31.7 ± 6.9 | 0.95 |
| 30Ext | 111.5 ± 30.5 | 0.98 | 66.0 ± 11.0 | 0.89 |
| 30Nick | 27.8 ± 5.6 | 0.95 | 23.4 ± 4.8a | 0.98 |
| 30AATT | 25.7 ± 0.9 | 0.92 | 8.5 ± 2.1 | 0.87 |
| 30Link | 33.1 ± 1.5 | 0.98 | 24.0 ± 2.0 | 0.96 |
| Nuc147 | >500 | 0.99 | >500 | 0.98 |
| Nuc165 | 57.8 ± 6.1 | 0.88 | 2.2 ± 1.5 | 0.86 |
| Nuc207 | 48.8 ± 21.2 | 0.97 | 1.0 ± 0.2 | 0.90 |
| Nuc178 | 238.0 ± 26.5 | 0.92 | 84.6 ± 7.7b | 0.98 |
a 250 mm NaCl was used instead of 200 mm NaCl. At 200 mm NaCl, the affinity of PARP-1 for 30Nick was in the low nanomolar range (data not shown).
b Errors are derived from one data set only.
The dissociation constants of N-parp·DNA complexes, as determined by HI-FI FRET, compare well with the previously reported affinities for the various DNA models 30Blunt DNA, 30Ext DNA, and 30Nick DNA (11). The overall 3–5-fold tighter affinities of N-parp in this study are likely due to differences in binding conditions (200 mm NaCl here versus 300 mm NaCl in previous studies). This is in keeping with the previously observed strong dependence of PARP-1/DNA interactions on ionic strength (19). Compared with N-parp, full-length PARP-1 exhibited 1.4–3-fold tighter affinity for all free DNA models (Table 1). This indicates that the C-terminal half of PARP-1 contributes moderately to the binding event, consistent with structural data demonstrating interactions between the WGR domain (not contained in N-parp) and DNA (14). The C-terminal half of PARP-1 on its own is unable to interact measurably with DNA (data not shown).
A Single PARP-1 Molecule Interacts Strongly with a Nucleosome Containing Symmetric Linker DNA
We next wanted to test N-parp and PARP-1 affinities for defined mononucleosomes that vary in length and symmetry of their linker DNA (Fig. 1C). Nuc147 is a mononucleosome that completely lacks DNA linker arms, whereas Nuc165 and Nuc207 contain two linker arms each. Nuc178 was designed to have only one exposed linker arm. The sequence of this 30-bp extension is identical to that of 30Link (Fig. 1). According to our analysis by native PAGE, all nucleosomes are uniquely positioned, and the percentage of free DNA in each of these nucleosome preparations was <1% (Fig. 3, A and B). The addition of fluorophore to histones did not change the electrophoretic mobility of the reconstituted nucleosomes, indicating that they are structurally intact. The interaction of N-parp and PARP-1 with Nuc165 was first tested by EMSA. When fluorescently labeled Nuc165 was titrated with either labeled PARP-1 or N-parp, distinct bands exhibiting both acceptor and donor fluorescence were observed (Fig. 3C). These bands also displayed FRET (pink bands in lower left panel), providing further proof of defined complex formation.
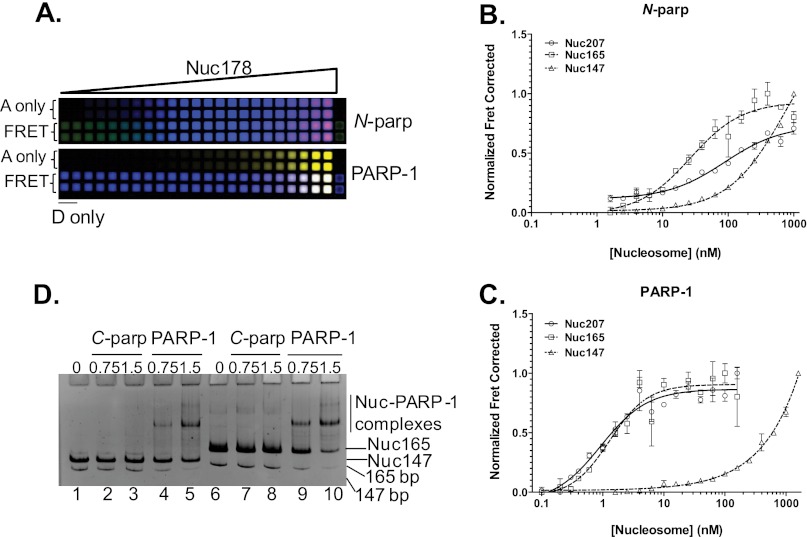
We next quantified the interaction of N-parp and PARP-1 with the various nucleosome substrates in solution using HI-FI FRET (Fig. 4A). Nuc165 and Nuc207 bound N-parp with 50–60 nm affinity, whereas no plateau was achieved with Nuc147 (Fig. 4B), characteristic of very weak interaction. Because regions outside of N-parp are known not to interact with DNA on their own, we were surprised to see that full-length PARP-1 bound nucleosomes with two DNA linker ends 25–50-fold tighter than N-parp (Fig. 4C). In light of the moderate difference in the binding affinity of the two PARP-1 constructs for free DNA, this suggests a substantial contribution of the CAT domain to the interaction with nucleosomes containing two DNA linker arms. This is despite the inability of the C-terminal domain of PARP-1 (C-parp) to bind mononucleosomes when tested by EMSA (Fig. 4D). Like N-parp, full-length PARP-1 bound Nuc147 only weakly (Fig. 4C). Importantly, the interaction of full-length PARP-1 with Nuc207 and Nuc165 was significantly tighter than that with any of the free DNA substrates.
FIGURE 4.
Quantification of interactions between PARP-1 and nucleosomes. A, HI-FI FRET plate assay. A portion of a typical 384-well plate is shown for Nuc178 and N-parp (upper panel) and full-length PARP-1 (lower panel). Increasing amounts of Nuc178 labeled with ATTO 647N at histone H4 E63C were titrated with a constant amount of either N-parp or PARP-1 labeled with Alexa Fluor 488. The upper two rows in each panel represent acceptor-only (A only) controls. The first two wells in the lower two rows in each panel are donor-only (D only) wells. FRET between the interacting partners is shown in the lower two rows in each panel (pink/purple). The plate was scanned using a Typhoon Imager as described for the gel in Fig. 2. Data from experiments were normalized, and the resulting curves were fit as described (19). Results from this plate are shown in Table 1. B and C, N-parp and PARP-1 interactions, respectively, with the various mononucleosome substrates. All values from this and similar experiments are summarized in Table 1. D, C-parp does not bind nucleosomes. Nucleosomes were incubated with C-parp or PARP-1 at increasing molar excess (as indicated) and loaded on a prerun 5% native TBE gel. Gels were stained with ethidium Bromide. C-parp did not interact with Nuc147 (lanes 2 and 3) or Nuc165 (lanes 7 and 8), whereas PARP-1 caused an upshift in both (lanes 4 and 5 for Nuc147 and lanes 9 and 10 for Nuc165).
To further test whether both DNA linker arms are required for a stable PARP-1 interaction, we generated a nucleosome with a single asymmetric ∼30-bp extension of DNA linker (Nuc178) (Fig. 1C). Both PARP-1 constructs bound this nucleosome substrate with significantly reduced affinity compared with Nuc165 or Nuc207 (Table 1). The data suggest that both linker arms are required for optimal PARP-1 binding. Binding of both PARP-1 constructs to Nuc178 was also 3–7-fold weaker than to the corresponding “free” 30-mer with identical sequence (30Link), indicating steric hindrance of PARP-1 binding to Nuc178. Finally, full-length PARP-1 bound these nucleosomes only 3-fold tighter than did N-parp, in contrast with the 25–50-fold increase in affinity for nucleosomes with two linker ends. This suggests that the CAT domain contributes to positioning PARP-1 in a way that allows engagement of both DNA linker arms and thus does not contribute as much to the interaction with a nucleosome with only one linker arm.
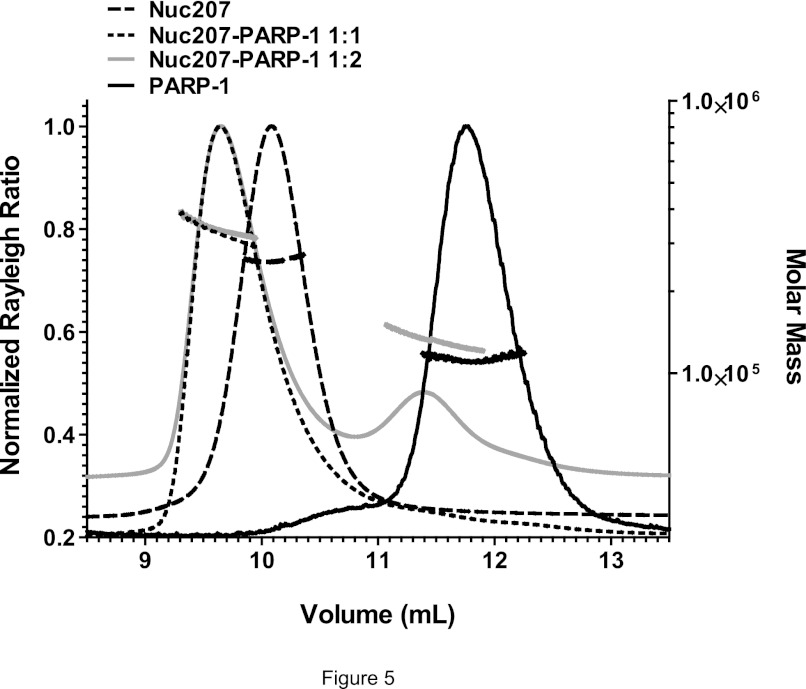
We next wanted to determine the stoichiometry of the various PARP-1·nucleosome complexes. Nuc207, Nuc165, or Nuc147 was mixed with varying amounts of PARP-1 and analyzed by SEC-MALS (Fig. 5 and Table 2). For the complexes between Nuc207 or Nuc165 and PARP-1, the observed molecular weights matched the calculated value for a 1:1 complex even when excess PARP-1 was added. In this case, a second peak for free PARP-1 was observed. A stoichiometry of 1:1 was also measured for N-parp/nucleosome complexes (data not shown). Consistent with the low binding affinity, Nuc147 and PARP-1 eluted as two separate peaks (Table 1), despite the residual interactions observed by native PAGE (Fig. 4D).
FIGURE 5.
One PARP-1 molecule binds per nucleosome. Shown are SEC-MALS profiles for Nuc207 and its complexes with PARP-1. Nuc207 formed a 1:1 complex with PARP-1 even when excess PARP-1 was added to the reaction mixture. The molecular weights for the various complexes derived from this and similar SEC-MALS experiments are listed in Table 2.
TABLE 2.
Molecular mass analysis of nucleosomes and their complexes with PARP-1 as determined by SEC-MALS (Fig. 5)
| Observed Mr | Calculated Mr | |
|---|---|---|
| Nuc207 | ||
| Nuc207 | 2.45 × 105 | 2.36 × 105 |
| PARP-1 | 1.1 × 105 | 1.13 × 105 |
| Nuc207 + PARP-1 (1:1) | 3.5 × 105 | 3.49 × 105 |
| Nuc207 + PARP-1 (1:2) | 3.5 × 105 | 4.62 × 105 |
| Nuc165 | ||
| Nuc165 | 2.17 × 105 | 2.10 × 105 |
| PARP-1 | 1.01 × 105 | 1.13 × 105 |
| Nuc165 + PARP-1 (1:1) | 2.6 × 105 | 3.23 × 105 |
| Nuc165 + PARP-1 (1:2) | 3.08 × 105 | 4.4 × 105 |
| Nuc147 | ||
| Nuc147 | 2.02 × 105 | 1.99 × 105 |
| PARP-1 | 1.01 × 105 | 1.13 × 105 |
| Nuc147 + PARP-1 (1:1) | 1.95 × 105 | 3.12 × 105 |
| Nuc147 + PARP-1 (1:2) | 1.82 × 105 | 4.25 × 105 |
Together, our quantitative analysis of PARP-1 nucleosome binding and stoichiometry reveals a strong contribution of the WGR-CAT domains to the interaction with nucleosomes with two linker arms (25–50-fold increased affinity), whereas the contribution of these domains to the interaction with free DNA is moderate at best (1.4–3-fold). Similarly, the affinity of full-length PARP-1 for Nuc178 is increased only 3-fold compared with N-parp. Because nucleosomes without linker DNA show no significant PARP-1 binding, we conclude that the contributions of the “nucleosome core” itself are minimal. Thus, high-affinity binding of full-length PARP-1 is provided by specific arrangement of two linker DNA arms that are provided only in the context of a nucleosome.
PARP-1 Is Activated by DNA and Nucleosomes
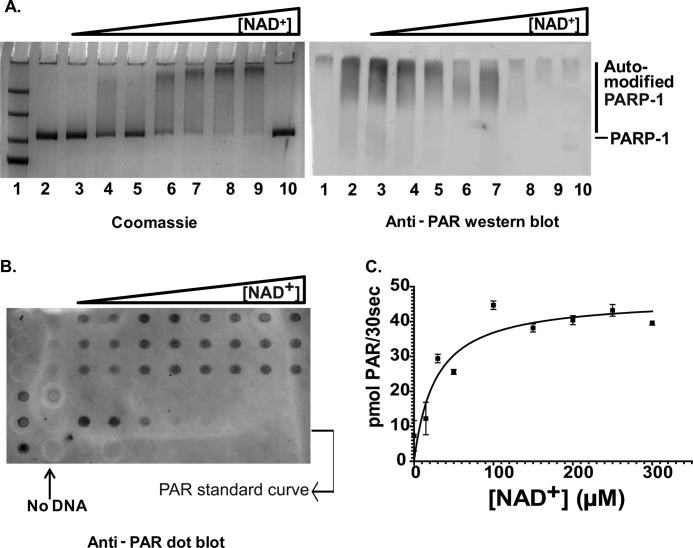
In light of the tight interaction of PARP-1 with a variety of DNA and chromatin substrates, we wanted to know what triggers the catalytic activity of PARP-1 and whether there is a quantitative difference in the degree of activation by the different DNA and chromatin substrates. To address these questions, we measured PARP-1 activity in the presence of various DNA and chromatin activators using the slot blot method (20). A representative case of PARP-1 activation by 30Blunt DNA is shown in Fig. 6. SDS-PAGE followed by Western blot analysis with anti-PAR antibody clearly demonstrated an upshift in the PARP-1 band with increasing NAD+ concentrations, indicative of the addition of PAR chains to PARP-1 (Fig. 6A). To quantify the amount of PAR generated in each reaction, samples were analyzed by slot blot and probed with the same anti-PAR antibody used above (Fig. 6B). The data were plotted in GraphPad Prism using Michaelis-Menten curve fitting (Fig. 6C). The enzymatic parameters for PARP-1 in the absence and presence of the various activators are summarized in Table 3. kcat values in the absence of DNA reflect the low basal background activity of PARP-1.
FIGURE 6.
PARP-1 is activated by DNA. A, SDS-PAGE showing a shift in PARP-1 mobility as it undergoes auto-PARylation in the presence of 1 μm 30Blunt DNA and increasing concentrations (0, 10, 20, 50, 100, 200, and 400 μm in lanes 3–9, respectively) of NAD+ (left panel). The right panel is a Western blot of an identical gel probed with anti-PAR antibodies. Lane 1, protein size marker; lane 2, no NAD+; lanes 3–9, increasing amounts of NAD+; lane 10, no DNA in the presence of 400 μm NAD+. B, a slot blot of the above reaction was probed with anti-PAR antibody and ATTO 647N-conjugated secondary antibodies and visualized on a Typhoon scanner at 633-nm excitation and 670-nm emission wavelengths. C, the data were quantified using ImageQuant TL and analyzed in a Michaelis-Menten plot. A complete list of all parameters for this and other activators is listed in Table 3.
TABLE 3.
Enzymatic parameters for PARP-1 upon activation by DNA or nucleosomes
Reaction conditions were as follows: DNA(50) = 50 mm Tris (pH 8.0), 50 mm NaCl, 10 mm MgCl2, and 1 mm DTT; Chromatin(50) = 50 mm Tris (pH 8.0), 50 mm NaCl, 1 mm MgCl2, and 1 mm DTT; Chromatin(100) = 50 mm Tris (pH 8.0), 100 mm NaCl, 1 mm MgCl2, and 1 mm DTT; and DNA(100) = 50 mm Tris (pH 8.0), 100 mm NaCl, 10 mm MgCl2, and 1 mm DTT.
| Allosteric activators | Kd(app)a | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| nm | pmol/min/μg | μm NAD+ | s−1 | s−1 m−1 | |
| No DNA | 9.40 | 19.4 ± 30.8 | 0.02 | 0.09 | |
| DNA(50) | |||||
| pGEM-3Z | 188.0 | 62.0 ± 17.7 | 0.35 | 0.56 × 104 | |
| 30Blunt | 32 | 541.1 | 27.1 ± 6.03 | 1.02 | 3.95 × 104 |
| 30Ext | 66 | 541.4 | 62.9 ± 9.9 | 1.01 | 4.31 × 104 |
| 30Nick | 23 | 311.0 | 33.0 ± 7.6 | 0.88 | 1.89 × 104 |
| 30AATT | 8.5 | 568.2 | 25.7 ± 6.4 | 1.06 | 6.55 × 104 |
| 30Link | 24 | 1070.6 | 33.9 ± 18.9 | 1.99 | 6.03 × 104 |
| Chromatin(50) | |||||
| Nuc147 | 642.4 | 23.1 ± 7.52 | 1.2 | 5.20 × 104 | |
| Nuc178 | 1408.2 | 16.4 ± 4 | 2.65 | 16.15 × 104 | |
| Chromatin(100) | |||||
| Nuc147 | >500 | 82.3 | 37.4 ± 21.0 | 0.15 | 0.42 × 104 |
| Nuc207 | 1.0 | 786.4 | 23.6 ± 8.0 | 1.47 | 6.22 × 104 |
| Nuc178 | 85.0 | 709.8 | 7.6 ± 10.5 | 1.33 | 1.76 × 104 |
| DNA(100) | |||||
| 30Blunt | 148.1 | 20.6 ± 9.3 | 0.28 | 1.40 × 104 | |
a Kd values are taken from Table 1.
PARP-1 was significantly activated over background levels by all linear DNA substrates, as evident by increases in Vmax; our values are in good agreement with those obtained using a similar approach (20). Closed circular plasmid DNA caused residual enzyme turnover, presumably due to the unavoidable contamination with nicked or linear DNA in most plasmid preparations. Although PARP-1 bound NAD+ even in the absence of DNA (no significant changes in Km), kcat values ranged between 0.9 and 2/s for all linear DNA fragments but were near 0 in the absence of DNA (Table 3). Nucleosomes with either one or two linker ends (Nuc178 and Nuc207) activated PARP-1 to a similar degree, despite the difference in binding affinity and presumably binding mode. Nuc147, which lacks free linker ends, had only reduced ability to stimulate PARP-1. Chromatin with at least one free DNA end activated PARP-1 to a higher degree than a linear DNA fragment with the same sequence (compare 30Link and Nuc178) (Table 3).
Although Nuc147 was rather inefficient at activating PARP-1 at 100 mm NaCl, it became a better activator at 50 mm NaCl, consistent with the idea that lower ionic strength promotes spontaneous “breathing” of the DNA ends (26, 27). The degree of activation resembled that achieved by short free DNA segments under the same conditions, where Nuc178 was still superior to either substrate. No PARP-1 activation was observed in the presence of any of the histone subcomplexes in the absence of DNA (data not shown). Thus, under our conditions, there is no direct correlation between activation and binding affinity; however, the presence of a nucleosome in addition to a DNA double-strand break appears to contribute to PARP-1 activation.
DISCUSSION
PARP-1 is a highly abundant nuclear protein with a multitude of biological functions (reviewed in Ref. 3). PARP-1 contributes to the compaction of chromatin through direct interactions with nucleosomes but also binds various forms of damaged DNA. Although its interaction with free DNA has been reasonably well studied (e.g. Refs. 11, 12, and 14), much less is known about the interaction of PARP-1 with chromatin. To fill this significant gap, we measured the affinity of PARP-1 for defined DNA and chromatin substrates and quantified the degree of stimulation of its enzymatic activity by the various ligands. Together, our data demonstrate (i) a significant contribution of the WGR-CAT domains to the interaction of PARP-1 with nucleosomes, but not with free DNA; (ii) a requirement for a pair of linker DNAs for high-affinity binding to nucleosomes; and (iii) a requirement for at least one free DNA end on the nucleosome for enzymatic activation.
Our analysis of the interaction of PARP-1 with short DNA fragments revealed that PARP-1 prefers DNA substrates with a propensity to bend. EM studies have shown that PARP-1 induces a bend into nicked or gapped DNA (28). The recognition of the weakened base stacking and the increased flexibility at DNA lesion sites has been proposed as a first step in DNA damage recognition by many repair proteins (29). Consistent with previous qualitative reports (16) and with the recent crystal structure of nearly full-length PARP-1 in complex with a short DNA fragment (14), we found a modest contribution of the C-terminal half of PARP-1 (presumably due to the interactions made by the WGR domain) to the interaction with each of the short DNA fragments tested.
Full-length PARP-1 binds very tightly to mononucleosomes that contain at least 10 bp of linker DNA extending on either side. This is consistent with the result that 160 bp of nucleosomal DNA are protected from micrococcal nuclease digestion in the presence of PARP-1 (16) but contradicts indirect evidence that linker DNA does not contribute to the interaction (18). The strong contribution of the C-terminal half of PARP-1 to the interaction with nucleosomes is striking because this region on its own does not measurably bind nucleosomes and because full-length PARP-1 interacts only very marginally with nucleosomes lacking DNA linkers (Nuc147). Because histones are also subject to a low degree of PARylation, this interaction might also entail substrate recognition by the CAT domain. However, the interaction of PARP-1 with nucleosomal histones or histone tails is not sufficient for robust binding in the absence of linker DNA.
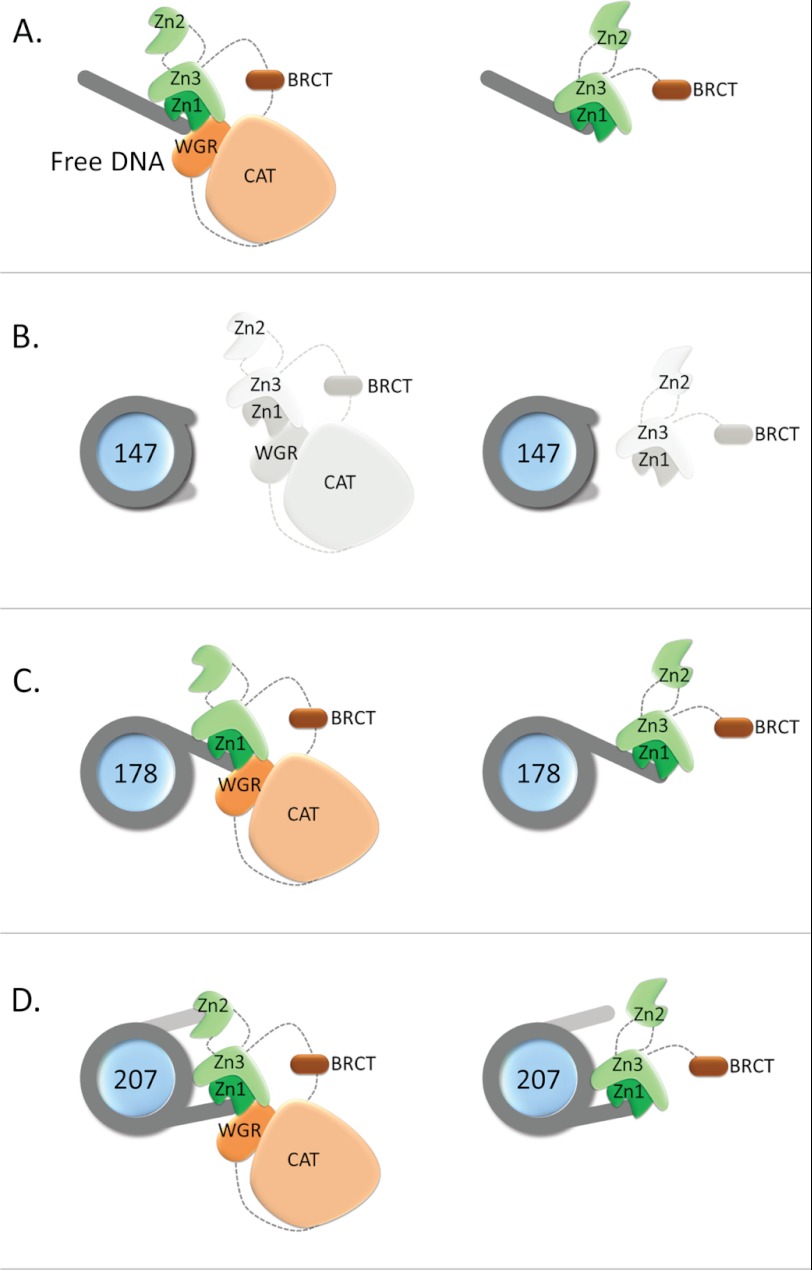
PARP-1 likely engages both DNA linker ends because the deletion of one of the two linker arms in a nucleosome (leaving one 30-bp DNA linker, Nuc178) resulted in a significant reduction in binding and in a much reduced contribution of the WGR-CAT domains to the interaction. The model for the structure of full-length PARP-1 in complex with a single DNA fragment proposed by Langelier et al. (14) suggests that Zn2 might be the domain responsible for interacting with the second DNA linker end (Fig. 7). This domain binds linear DNA on its own and with tighter affinity than Zn1, but it is not needed for the enzymatic activation of PARP-1 (13). The 25–50-fold increase in binding affinity for nucleosome substrates of full-length PARP-1 compared with N-parp suggests that the engagement of the WGR domain (and perhaps the CAT domain) is required for Zn2 to position itself optimally for interaction with the second linker arm (Fig. 7). This effect is not observed on nucleosomes with just one linker arm or on free DNA. Both N-parp and full-length PARP-1 bind less tightly to these nucleosomes than they bind to 30Link DNA, which has the same sequence as the linker extension in Nuc178, suggesting steric interference with binding by the nucleosome core.
FIGURE 7.
Model for N-parp and PARP-1 interactions with DNA and nucleosomes. A, interaction with free DNA as shown in Ref. 14. (The schematic is adapted from Ref. 35.) B, nucleosomes without linker do not interact with either PARP-1 construct. C, nucleosomes with one asymmetric linker arm likely interact similarly with PARP-1 as they do with free DNA, with some steric inhibition from the nucleosome, as indicated by the weaker binding affinities. D, interaction with nucleosomes with two linker arms. Both linker arms are engaged in PARP-1 binding, with Zn2 binding the second DNA linker. Regions from C-parp are required for correct orientation of Zn2.
Some controversy exists over the stoichiometry of PARP-1 in solution (8, 30) and on free DNA (11, 31, 32). Here, we have shown that a single PARP-1 molecule binds per nucleosome, consistent with the idea that PARP-1 and linker histone H1 interact similarly with PARP-1 (16). Using the same approach, we found that H1 bound Nuc207 with higher affinity than full-length PARP-1 (19).
PARP-1 activity is reportedly induced by DNA damage, chromatin, and even isolated histones (16, 18). Using highly pure recombinant PARP-1 and well defined DNA and nucleosome substrates, we found that the activity of PARP-1 was stimulated by free DNA and by nucleosomal linker DNA, irrespective of its affinity for the allosteric activator. For example, PARP-1 bound nucleosomes with one single linker arm with rather low affinity, yet its enzymatic activity was stimulated to a similar extent as by nucleosomes with two symmetric linker arms. This result is consistent with the observation that Zn2 is not required for PARP-1 activation (13). Thus, Zn2 appears to contribute mainly to PARP-1 in its role as a chromatin architectural protein. Our data demonstrate that PARP-1 is able to recognize DNA double-strand breaks in the context of chromatin and is potently activated, consistent with its role as a first responder to DNA damage in eukaryotic cells. The high affinity of PARP-1 to nucleosomes and its activation by DNA and nucleosomes explain how PARP-1 regulates chromatin structure, transcription, and DNA repair pathways. Additionally, the requirement of NAD+ for PARP-1 activation implies that other pathways utilizing NAD+ will further regulate PARP-1 activity in the various cellular processes (33). However, to understand if and how PARP-1 redistributes from undamaged chromatin to sites of DNA damage, we have to quantify the interactions of PARP-1 with complex chromatin structures and chromatin components in the absence of DNA damage.
Acknowledgments
We thank the W. M. Keck Protein Expression and Purification Facility at Colorado State University (directed by H. Scherman) for providing histones, A. White for various DNA constructs, and P. Dyer for labeled histone samples. We also thank M. Dechassa, S. Bergeron, and A. Hieb for comments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM088409. This work was also supported by the Howard Hughes Medical Institute.
- PARP-1
- poly(ADP-ribose) polymerase
- BRCT
- BRCA1 C-terminal
- WGR
- tryptophan/glycine/arginine-rich
- CAT
- catalytic
- Zn
- zinc finger
- TBE
- Tris borate/EDTA
- SEC-MALS
- size exclusion chromatography-multiangle light scattering
- PAR
- poly(ADP-ribose).
REFERENCES
- 1. Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lord C. J., Ashworth A. (2012) The DNA damage response and cancer therapy. Nature 481, 287–294 [DOI] [PubMed] [Google Scholar]
- 3. Krishnakumar R., Kraus W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ji Y., Tulin A. V. (2010) The roles of PARP-1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 20, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 6. Gibson B. A., Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 7. Altmeyer M., Messner S., Hassa P. O., Fey M., Hottiger M. O. (2009) Molecular mechanism of poly(ADP-ribosyl)ation by PARP-1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 37, 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tao Z., Gao P., Liu H. W. (2009) Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 [DOI] [PubMed] [Google Scholar]
- 9. Mangerich A., Bürkle A. (2011) How to kill tumor cells with inhibitors of poly(ADP-ribosyl)ation. Int. J. Cancer 128, 251–265 [DOI] [PubMed] [Google Scholar]
- 10. Javle M., Curtin N. J. (2011) The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer 105, 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lilyestrom W., van der Woerd M. J., Clark N., Luger K. (2010) Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J. Mol. Biol. 395, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eustermann S., Videler H., Yang J. C., Cole P. T., Gruszka D., Veprintsev D., Neuhaus D. (2011) The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 407, 149–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langelier M. F., Planck J. L., Roy S., Pascal J. M. (2011) Crystal structures of poly(ADP-ribose) polymerase 1 (PARP-1) zinc fingers bound to DNA. Structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 286, 10690–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langelier M. F., Planck J. L., Roy S., Pascal J. M. (2012) Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schreiber V., Molinete M., Boeuf H., de Murcia G., Ménissier-de Murcia J. (1992) The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 11, 3263–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 17. Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) The DNA-binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinnola A., Naumova N., Shah M., Tulin A. V. (2007) Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 282, 32511–32519 [DOI] [PubMed] [Google Scholar]
- 19. Hieb A. R., D'Arcy S., Kramer M. A., White A. E., Luger K. (2012) Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res. 40, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beneke S., Scherr A. L., Ponath V., Popp O., Bürkle A. (2010) Enzyme characteristics of recombinant poly(ADP-ribose) polymerases 1 of rat and human origin mirror the correlation between cellular poly(ADP-ribosyl)ation capacity and species-specific life span. Mech. Ageing Dev. 131, 366–369 [DOI] [PubMed] [Google Scholar]
- 21. Winkler D. D., Luger K. (2011) The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dyer P. N., Edayathumangalam R. S., White C. L., Bao Y., Chakravarthy S., Muthurajan U. M., Luger K. (2004) Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 [DOI] [PubMed] [Google Scholar]
- 23. Yang C., van der Woerd M. J., Muthurajan U. M., Hansen J. C., Luger K. (2011) Biophysical analysis and small-angle x-ray scattering-derived structures of MeCP2-nucleosome complexes. Nucleic Acids Res. 39, 4122–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winkler D. D., Luger K., Hieb A. R. (2012) Quantifying chromatin-associated interactions: The HI-FI system. Methods Enzymol. 512, 1–32 [DOI] [PubMed] [Google Scholar]
- 25. Koo H. S., Wu H. M., Crothers D. M. (1986) DNA bending at adenine-thymine tracts. Nature 320, 501–506 [DOI] [PubMed] [Google Scholar]
- 26. Li G., Widom J. (2004) Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11, 763–769 [DOI] [PubMed] [Google Scholar]
- 27. Gurunathan K., Levitus M. (2009) Single-molecule fluorescence studies of nucleosome dynamics. Curr. Pharm. Biotechnol. 10, 559–568 [DOI] [PubMed] [Google Scholar]
- 28. Le Cam E., Fack F., Ménissier-de Murcia J., Cognet J. A., Barbin A., Sarantoglou V., Révet B., Delain E., de Murcia G. (1994) Conformational analysis of a 139-base pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J. Mol. Biol. 235, 1062–1071 [DOI] [PubMed] [Google Scholar]
- 29. Yang W. (2006) Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair 5, 654–666 [DOI] [PubMed] [Google Scholar]
- 30. Langelier M. F., Servent K. M., Rogers E. E., Pascal J. M. (2008) A third zinc-binding domain of human poly(ADP-ribose) polymerase 1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 283, 4105–4114 [DOI] [PubMed] [Google Scholar]
- 31. Ali A. A., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P. O., Hassler M., Ladurner A. G., Pearl L. H., Oliver A. W. (2012) The zinc finger domains of PARP-1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 19, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langelier M. F., Ruhl D. D., Planck J. L., Kraus W. L., Pascal J. M. (2010) The Zn3 domain of human poly(ADP-ribose) polymerase 1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285, 18877–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim M. Y., Zhang T., Kraus W. L. (2005) Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 34. Lowary P. T., Widom J. (1998) New DNA sequence rules for high-affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 35. Gagné J. P., Rouleau M., Poirier G. G. (2012) Structural biology. PARP-1 activation–bringing the pieces together. Science 336, 678–679 [DOI] [PubMed] [Google Scholar]