Background: Integrin α1 I domain undergoes conformational changes upon collagen binding.
Results: Deuterium exchange was used to measure the effects of cations, collagen, or an antibody on the α1I solution structure.
Conclusion: Full-length collagen and metal ions induce changes that differ in key aspects from previously proposed models for α1I activation.
Significance: These studies support a new model for integrin I domain activation.
Keywords: Adhesion, Cell Surface Receptor, Integrins, Mass Spectrometry (MS), Receptor Structure-Function, Activation, Hydrogen-Deuterium Exchange
Abstract
We have applied hydrogen-deuterium exchange mass spectrometry, in conjunction with differential scanning calorimetry and protein stability analysis, to examine solution dynamics of the integrin α1 I domain induced by the binding of divalent cations, full-length type IV collagen, or a function-blocking monoclonal antibody. These studies revealed features of integrin activation and α1I-ligand complexes that were not detected by static crystallographic data. Mg2+ and Mn2+ stabilized α1I but differed in their effects on exchange rates in the αC helix. Ca2+ impacted α1I conformational dynamics without altering its gross thermal stability. Interaction with collagen affected the exchange rates in just one of three metal ion-dependent adhesion site (MIDAS) loops, suggesting that MIDAS loop 2 plays a primary role in mediating ligand binding. Collagen also induced changes consistent with increased unfolding in both the αC and allosteric C-terminal helices of α1I. The antibody AQC2, which binds to α1I in a ligand-mimetic manner, also reduced exchange in MIDAS loop 2 and increased exchange in αC, but it did not impact the C-terminal region. This is the first study to directly demonstrate the conformational changes induced upon binding of an integrin I domain to a full-length collagen ligand, and it demonstrates the utility of the deuterium exchange mass spectrometry method to study the solution dynamics of integrin/ligand and integrin/metal ion interactions. Based on the ligand and metal ion binding data, we propose a model for collagen-binding integrin activation that explains the differing abilities of Mg2+, Mn2+, and Ca2+ to activate I domain-containing integrins.
Introduction
Because of their critical role in mediating cell/cell and cell/matrix interactions, integrins are among the most studied families of transmembrane receptors. In particular, great progress has been made in understanding the general structural features of these heterodimeric cell-surface receptors and the conformational changes that regulate affinity modulation, ligand binding, and signaling (1, 2). Crystal structures of both intact integrins and functional domains have been critical to this process. Of particular interest is the α subunit I domain that is primarily responsible for mediating ligand binding by integrins containing this domain. Among others, the co-crystal structures of disulfide-“locked” αL I domains with ICAM-1 (3), ICAM-3 (4), and ICAM-5 (5) and the α2 I domain (α2I) with a collagen triple-helical peptide (THP)4 (6) have provided snapshots of the closed (nonligand bound), open (ligand-bound), and intermediate conformations of the ligand-binding regions of these integrins. In many cases, direct structural comparisons between free and ligand-bound integrins (or integrin I domains) cannot be made on the basis of existing data. Even less data are available on the nature of the conformational changes that occur upon integrin-ligand binding in solution. Thus, it is important to develop solution-based methods that allow the direct assessment of the conformational dynamics of integrin/ligand interactions.
In humans, the 18 α and 8 β integrin subunits pair to form at least 24 known heterodimers (7). Nine of the α subunits contain I domains, which play a central role in ligand binding and selectivity. The four collagen-binding receptors (α1β1, α2β1, α10β1, and α11β1) fall into this subset of I domain-containing integrins. Of these, α1β1 (VLA1) and α2β1 (VLA2) have been the most thoroughly studied. The crystal structure of α2I has been determined in the presence of a heterotrimeric THP corresponding to its primary ligand, type I collagen (6). These studies, along with structural studies of the α I domains of the β2 integrins LFA-1 (4, 5, 8–11), Mac-1 (12, 13), and αXβ2 (14), have helped to establish and refine a model for the ligand-induced conformational changes that lead to “outside-in” signaling in I domain-containing integrins (1). In this model, ligand binding to the divalent cation within the I domain metal ion-dependent adhesion site (MIDAS) induces a shift from the so-called “closed” (unliganded) to the “open” (liganded) conformation and induces allosteric changes in the C-terminal α7 helix of the I domain, triggering a large scale rearrangement of the global topology of the receptor. This model was refined to include the concept of an “intrinsic ligand,” in which a glutamate from the C terminus of the I domain binds to the MIDAS site within a similar domain within the β subunit (the β I domain or “I-like” domain) (15, 16).
VLA1 is a major receptor for basement membrane-type IV collagen and is expressed on hematopoietic, neuronal, and mesenchymal cell types (17). Inhibition of VLA1 function, through the use of genetic knock-outs or pharmacological agents, blocks cell retention and activation in pathogenic sites and leads to reduced inflammation and amelioration of disease in a variety of animal models (for examples, see Refs. 18–25). Despite the wealth of biological and pharmacological data on VLA1, limited structural information is available for this integrin. Crystal structures of isolated α1 I domains (α1I) have been reported (26–28), as well as the structure of α1I in complex with the function-blocking antibody AQC2 (29, 30). Although computational models have been proposed for the α1I open conformation (27) and for the α1I/collagen interaction site (31), no structures of α1I in complex with any type of ligand or ligand-mimetic peptide have been reported (2). Recently, the structure of α1I containing an activating mutation (E317A) was determined (32). This protein adopted a novel conformation that differed from both the open and closed structures previously published for other integrins, indicating that the activation mechanism of collagen-binding integrins may differ from that of other integrins.
Peptide amide hydrogen-deuterium exchange mass spectrometry (DXMS) has been used to analyze protein/protein, protein/substrate, protein/inhibitor, protein/solvent, and protein/DNA interactions, as well as protein dynamics and protein conformational changes (33–40). Here, we utilized DXMS to directly study the solution binding and conformational dynamics of the interaction between α1I and full-length type IV collagen. DXMS allowed us to investigate the conformational changes that occur when α1I binds to collagen under conditions closely resembling the native physiological environment and to test the current structural models for integrin I domain/ligand interactions that are largely based on static crystallographic data. We also examined conformational changes induced upon binding of the function-blocking α1 monoclonal antibody AQC2 and compared them with those mediated by the native ligand. Our results reveal important differences from existing models for integrin I domain/ligand interactions and ligand-induced allosteric changes, and they suggest novel dynamic features of the α1I/collagen and α1I/metal ion interactions. The methods we developed to characterize the α1I domain should be easily adapted to the study of other integrins.
EXPERIMENTAL PROCEDURES
Protein Preparation
These studies utilized a chimeric α1I comprising elements of both the rat and human sequences (Fig. 1). The sequence numbering is based on the full-length mature human α1 integrin sequence (NCBI Reference Sequence NP_852478.1). Four human residues (Val-213, Gln-214, Arg-215, and Arg-218) were substituted into the MIDAS region of the corresponding rat α1I sequence. This form of α1I was used to establish the binding sites for anti-human α1 integrin antibodies using biochemical methods (41) and was crystallized in complex with a Fab fragment of the function-blocking anti-human α1 integrin antibody AQC2 (29, 30). The bacterially expressed chimeric α1I is significantly more soluble than its fully human GST-α1I fusion counterpart, making it more suitable for structural studies. Recombinant α1I was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein and was purified as described previously (41). Samples were analyzed using size exclusion chromatography on a Superdex 200 10/300 GL column using phosphate-buffered saline (PBS) as the mobile phase and compared with a reference standard mixture (Bio-Rad). For studies requiring removal of the GST tag, the protein was cleaved using thrombin and purified as described previously (41). The anti-α1β1 monoclonal antibody AQC2 was prepared as described previously (41).
FIGURE 1.
Amino acid sequence of the chimeric α1I protein used in this study. Boxed regions represent N- and C-terminal non-integrin sequences derived from the thrombin cleavage site and the multiple cloning site of the bacterial expression vector, respectively. Numbering is based on the full-length α1 integrin sequence and does not include the vector-derived residues. Underlined residues (Val-213, Gln-214, Arg-215, and Arg-218) are amino acids from the human α1I sequence that were inserted in place of the corresponding rat α1I residues (Gly-213, Arg-214, Gln-215, and Leu-218) to generate the chimeric molecule.
Ligand Binding Studies
The binding of GST-α1I to collagen was measured using an electrochemiluminescence assay (42). Type IV collagen from human placenta (Sigma) was coated onto Tosyl M-280 Dynabeads (Dynal, A.S., Oslo, Norway) at a ratio of 20 μg of collagen/mg of beads, as described previously (42). Beads were blocked by incubation with 8% rat plasma in assay buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 0.1% Triton X-100) for 15 min at 25 °C prior to use. A goat polyclonal antibody against GST (GE Healthcare) was labeled with ruthenium(II) tris(bipyridine)-NHS ester (TAG-NHS Ester; IGEN International, Inc., Gaithersburg, MD) as per the manufacturer's instructions. Serial dilutions of GST-α1I in assay buffer were incubated with 30 μg/ml labeled anti-GST antibody, 0.2 mg/ml collagen-coated beads, and 0.5 mm MnCl2, in a final volume of 120 μl. After gentle agitation at 25 °C for 60 min, 200 μl of assay buffer was added, and samples were read in an ORIGEN 5.1 detector according to the manufacturer's instructions (IGEN). To assess the blocking ability of AQC2, serial dilutions of AQC2 were incubated with 5 ng/ml GST-α1I, 2 μg/ml labeled anti-GST antibody, 0.2 mg/ml collagen-coated beads, and 1 mm MnCl2 in a final volume of 100 μl and processed as described above.
Differential Scanning Calorimetry
DSC was performed using an automated capillary DSC (capDSC, GE Healthcare). Protein and reference solutions were sampled automatically from 96-well plates using the robotic attachment. Prior to each sample measurement, two buffer scans were performed to define the base line for subtraction. All 96-well plates containing protein were stored within the instrument at 5 °C. Each I domain sample was diluted to 1 mg/ml in relevant buffer. Scans were performed from 10 to 100 °C at 2 °C/min using the high feedback mode for enhanced peak resolution. Scans were analyzed using the Origin software package supplied by the manufacturer. Subsequent to the subtraction of reference base-line scans, nonzero sample base lines were corrected using a third order polynomial. The unfolding parameters for the multidomain unfolding profiles of I domain were deconvoluted using the multipeak fitting routine within the software assuming non-two-state unfolding behavior. The measured melting temperature (Tm) for α1I in the presence of the chelating agent EDTA ranged from 54.5 to 57.9 °C, depending on the protein concentration, buffer composition, and scan rate, but it was reproducible ±1 °C within a given set of experimental parameters.
Denaturation Studies
The denaturation of α1I as a function of urea was measured by fluorescence spectroscopy (41). Briefly, samples containing 300 μg/ml α1I in 50 mm Tris-HCl, pH 7.5, 0.1 mm DTT with 10 mm EDTA, 10 mm CaCl2, or 1 mm MnCl2 and with varying amounts of urea were analyzed at 25 °C using an excitation wavelength of 280 nm. Emission spectra from 300 to 400 nm were collected using a Spectramax M5 fluorescence spectrometer (Molecular Devices Inc., Sunnyvale, CA). Fluorescence data at 350 nm were plotted as a function of urea and standardized using the change in fluorescence from 0 to 9 m urea for each of the test conditions as a measure of the total fraction folded.
Optimization of Fragmentation Conditions
Prior to conducting the hydrogen-deuterium exchange experiments, test digests prepared with undeuterated buffer in varying concentrations of guanidine hydrochloride (GdnHCl) were made to optimize proteolysis conditions for the best peptide coverage (43). For each sample, 6 μl of stock solution of α1 I domain (8 mg/ml) was first diluted with 18 μl of H2O buffer (150 mm NaCl, 8.3 mm Tris-HCl, pH 7.2). The sample was then quenched with 36 μl of a quench solution (0 °C) of 0.8% formic acid, 17% glycerol, and GdnHCl at final concentrations of 0.05, 0.5, 1.0, 2.0, or 4.0 m. This quenching step lowered the pH to 2.2–2.5 to minimize further hydrogen-deuterium exchange in subsequent deuterated samples. The quench-denaturation process was allowed to proceed on ice for 30 s, after which the sample was stored frozen at −80 °C. Procedures for pepsin digestion for DXMS have been described previously (44–48). Briefly, a sample was thawed at 5 °C and then immediately digested over a protease column (66-μl bed volume) filled with porcine pepsin (Sigma; immobilized on Poros 20 AL medium at 30 mg/ml following the manufacturer's instructions) at a flow rate of 100 μl/min with 0.05% trifluoroacetic acid. Peptic fragments were collected on a C18 trap column and separated on a C18 reversed phase column (Vydac, catalog no. 218MS5105) with a linear acetonitrile gradient from 6 to 38%. The column effluent was electrosprayed directly into an LCQ Classic (Thermo Finnigan, Inc.) or Q-TOF mass spectrometer (Micromass). Determination of pepsin-generated peptides from MS/MS data sets was facilitated through the use of SEQUEST (Thermo Finnigan, Inc.). This set of peptides was then further verified by DXMS Explorer (Sierra Analytics Inc., Modesto, CA). The peptide coverage maps for the different concentrations of GdnHCl were compared, and the condition with the best coverage map for each individual protein or protein complex was used for subsequent deuterium exchange experiments.
Hydrogen-Deuterium Exchange Experiments
Samples were prepared with three states of hydrogen-deuterium exchange as follows: nondeuterated (ND), partially deuterated, and equilibrium deuterated. All samples were processed as described above. Equilibrium deuterated samples were prepared by incubating protein at room temperature in D2O buffer (1% (v/v) formic acid) overnight. Procedures for individual experiments are described below. The hydrogen-deuterium exchange experiments performed with our automated apparatus produces measurements of deuteron incorporation where the standard deviation is less than 2% of the mean of triplicate determinations (49, 50).
α1 I Domain
α1 I domain solutions (8 mg/ml) were mixed with solutions of CaCl2, MnCl2, or EDTA to final concentrations of 1, 0.5, or 5 mm, respectively, or with MgCl2 at either 1 or 10 mm. The solutions were preincubated at room temperature for 10 min and then chilled to 0 °C. The ND sample was processed exactly as described under “Optimization of Fragmentation Conditions.” Hydrogen-deuterium exchange experiments were initiated by mixing 6 μl of preincubated samples with 18 μl of D2O buffer (8.3 mm Tris, 150 mm NaCl, pH 7.2, in D2O). The samples with 10 mm Mg2+ were incubated at 0 °C for 10, 30, or 100 s or at 25 °C for 30, 100, or 300 s. All other samples were incubated at 0 °C for 10, 30, or 300 s or at 25 °C for 30, 100, 300, 1000, 3000, 10,000, 30,000, 100,000, or 300,000 s. At the indicated times, the H/D exchange was quenched by adding 36 μl of ice-cold quench solution (0.8% formic acid, 16.6% glycerol, and 0.05 m GdnHCl). The samples were then transferred to ice-cooled autosampler vials, frozen on dry ice, and stored at −80 °C. As shown in previous studies (51–54), incubation at 0 °C for 10, 30, or 100 s is equivalent to incubation at 25 °C for 1, 3, or 10 s, respectively. For all ribbon diagrams, the data generated at 0 °C were converted to the data equivalent at 25 °C.
AQC2-α1 I Domain Complex
The complex between antibody AQC2 and α1I was prepared by mixing solutions of α1I (7 mg/ml) and antibody (6.4 mg/ml) at a 2:1 molar ratio at room temperature for 30 min and then chilling to 0 °C. The ND sample was processed by diluting 25 μl of the complex with 75 μl of H2O Buffer (8.3 mm Tris, 150 mm NaCl, pH 7.2) and then mixed with 100 μl of quench solution (0.8% formic acid, 16.6% glycerol, 0.5 m GdnHCl). For deuterated samples, 25 μl of preincubated sample was mixed with 75 μl of D2O buffer and incubated at various time intervals (10, 30, and 100 s at 0 °C and 30, 100, 300, 1000, 3000, 10,000, 30,000, 100,000, and 300,000 s at 25 °C) before being added to 100 μl of quench solution. All samples were frozen on dry ice. Incubation at 0 °C for 10, 30, or 100 s is equivalent to incubation at 25 °C for 1, 3, or 10 s, respectively.
GST-α1 I Domain
For the ND sample, 5 μl of GST-α1I (12.7 mg/ml with 10 mm MgCl2) was diluted with 64 μl of H2O buffer and quenched with 96 μl of quench solution (0.8% formic acid, 16.6% glycerol, 0.5 m GdnHCl). Deuterated samples were prepared by diluting 5 μl of GST-α1I with 11 μl of H2O buffer, followed by mixing with 48 μl of D2O buffer. Samples were incubated for 10, 100, and 1000 s at 0 °C and 300, 1000, 3000, 10,000, 30,000, and 100,000 s at 25 °C. At the indicated times, the samples were added to vials containing the 96 μl of quench solution. The samples were transferred to autosampler vials and frozen on dry ice. Incubation at 0 °C for 10, 100, or 300 s is equivalent to incubation at 25 °C for 1, 10, or 100 s, respectively.
GST-α1 I Domain-Collagen Complex
An on-column exchange process was used to eliminate the interference of collagen peptides during data acquisition. The collagen column was created by the following procedure: 5 mg of collagen (type IV collagen from human placenta, Sigma) was dissolved in 2 ml of 50 mm sodium citrate, pH 4.4. 140 mg of POROS AL 20 media (Applied Biosystems), 330 μl of 1 m sodium cyanoborohydride, and 3.45 ml of 1.5 m sodium sulfate were added, and the reaction was mixed continuously using a tube shaker at 25 °C for 16 h. The beads were washed with 50 mm sodium citrate, pH 4.4, and incubated with 6 ml of 0.1 m ethanolamine, 50 mm sodium citrate, and 330 μl of 1 m sodium cyanoborohydride, pH 4.4, at 25 °C for 2 h. The beads were further washed using H2O Buffer and packed into a 150-μl bed volume column at 10 ml/min with H2O Buffer. For ND samples, 20 μl of 12.7 mg/ml GST-α1I was loaded onto the collagen column following pre-equilibration with H2O Buffer and incubated at 0 °C for 5 min. To initiate the H/D exchange reaction, 500 μl of D2O buffer was loaded onto the column and incubated for various times (10, 100, and 1000 s at 0 °C and 300, 1000, 3000, 10,000, 30,000, and 100,000 s at 25 °C). 500 μl of 1% formic acid was applied to the column to quench the reaction and elute the GSTα1I. 150 μl of eluent was added to 12.7 μl of quench solution. Samples were stored at −80 °C prior to analysis. Incubation at 0 °C for 10, 100, or 1000 s is equivalent to incubation at 25 °C for 1, 10, or 100 s, respectively. The collagen column was regenerated between runs by washing it with 1% formic acid until the acid-eluted material measured by absorbance at 214 nm (A214) reached base line, followed by equilibration of the column with neutral pH buffer (8.3 mm Tris, 150 mm NaCl, pH 7.2, with appropriate divalent cations). The efficiency of regeneration was confirmed by observation of complete restoration of ligand binding capacity assessed by both saturable binding capacity and the magnitude of material eluted after binding (measured by A214). In control studies, when the acid-washed, re-equilibrated column was again eluted without protein re-loading, less than 2% of the maximum protein binding capacity of the column was eluted, demonstrating that the column had been cleared of previously bound GST-α1I by the regeneration procedure.
Calculations
DXMS Explorer was used for the data processing and reduction of hydrogen-deuterium exchange experiments. Back-exchange occurs immediately after addition of the quench solution, and corrections for it were determined by previously described methods (48) and as shown in Equation 1,
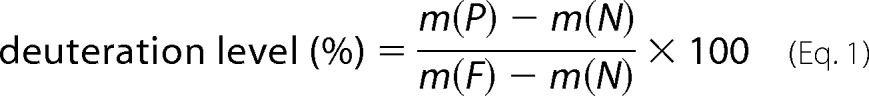 |
where m(P), m(N), and m(F) are the centroid value of the partially deuterated, ND, and equilibrium deuterated peptide, respectively.
Generation of Ribbon Diagrams
The deuteration levels of peptides were further sublocalized using overlapping peptides (44). First, fragments (fi) within each peptide were delineated in a set of overlapping peptides according to overlapping regions. At least five residues were included within each fragment to avoid overinterpreting the experimental data. Second, a variable si,t was defined to represent the number of deuterium of fragment fi values at the on-exchange time point t. Thus, a set of linear equations was established such that the sum of si,t in a specific peptide was equal to the total number of deuterons observed in DXMS experiments. The best solutions were determined by linear least square fitting, with the following stipulations: (a) that si,t is ≥0; and (b) for a given fragment fi, mass shift at the longer on-exchange time point cannot be smaller than the mass shift at the shorter on-exchange time point. These fitted values were colored according to the degree of deuterium exchange (ribbon diagrams).
Generation of α1I-Collagen Model
A predicted structure of the α1I-collagen complex was created by homology modeling with the program MODELLER (55, 56). The crystal structure of the α2 I-domain in complex with collagen (Protein Data Bank code 1DZI) (6) and a structure of the α1 I domain (Protein Data Bank code 1MHP) (30) were used as templates. Type IV collagen was modeled as a trimeric THP (31). Restraints for the MIDAS site and the αC helix were taken exclusively from the α2-collagen template structure so that the modeled I domain would remain in the open conformation. The model was refined by simulated annealing under MODELLER spatial restraints from the template structures.
RESULTS
Purity and Bioactivity of α1 Integrin I Domain Proteins
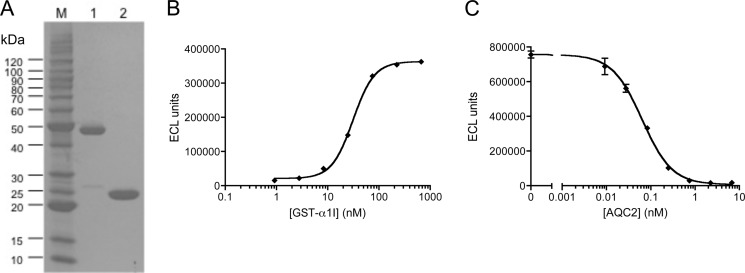
The α1 I domain was expressed as a GST fusion protein comprising elements of both the rat and human α1I sequences (Fig. 1) (41). To fully characterize the properties of this chimeric form of α1I, both the GST fusion (GST-α1I) and α1I generated by cleavage of the GST tag were purified (Fig. 2A). Under reducing conditions, GST-α1I migrated at ∼45 kDa (Fig. 2A, lane 1) and α1I migrated at ∼24 kDa (lane 2), consistent with their predicted molecular masses (51.1 and 24.9 kDa, respectively). Using size-exclusion chromatography, α1I migrated as a monomer (apparent mass ∼20 kDa), whereas GST-α1I eluted with an apparent molecular mass corresponding to a trimer (∼135 kDa), consistent with the native trimeric structure of GST (57). The purified GST-α1I protein bound to type IV collagen with an EC50 of 31 nm (Fig. 2B), and this interaction could be completely blocked by incubation with AQC2 (IC50 = 64 pm, Fig. 2C). The apparent affinity of chimeric GST-α1I for collagen was identical to what we have observed using the corresponding rat GST-α1I (data not shown), and is consistent with previously reported affinities for the human GST-α1I/collagen IV interaction (58, 59). The chimeric GST-α1I was utilized for these studies because the corresponding human protein was poorly soluble under the physiological conditions used to assess its biochemical and functional attributes.
FIGURE 2.
Biochemical characterization of α1I. A, purified GST-α1I (lane 1) and α1I (lane 2) were separated by 4–20% SDS-PAGE under reducing conditions and stained using Coomassie Brilliant Blue. Molecular weight markers (M) are shown on the left. B, binding of GST-α1I to collagen IV was measured using an electrochemiluminescence assay (EC50 = 31 nm). C, inhibition of GST-α1I/collagen IV binding by antibody AQC2 was measured using the electrochemiluminescence assay (IC50 = 64 pm).
Effect of Divalent Cations on the Stability of α1I
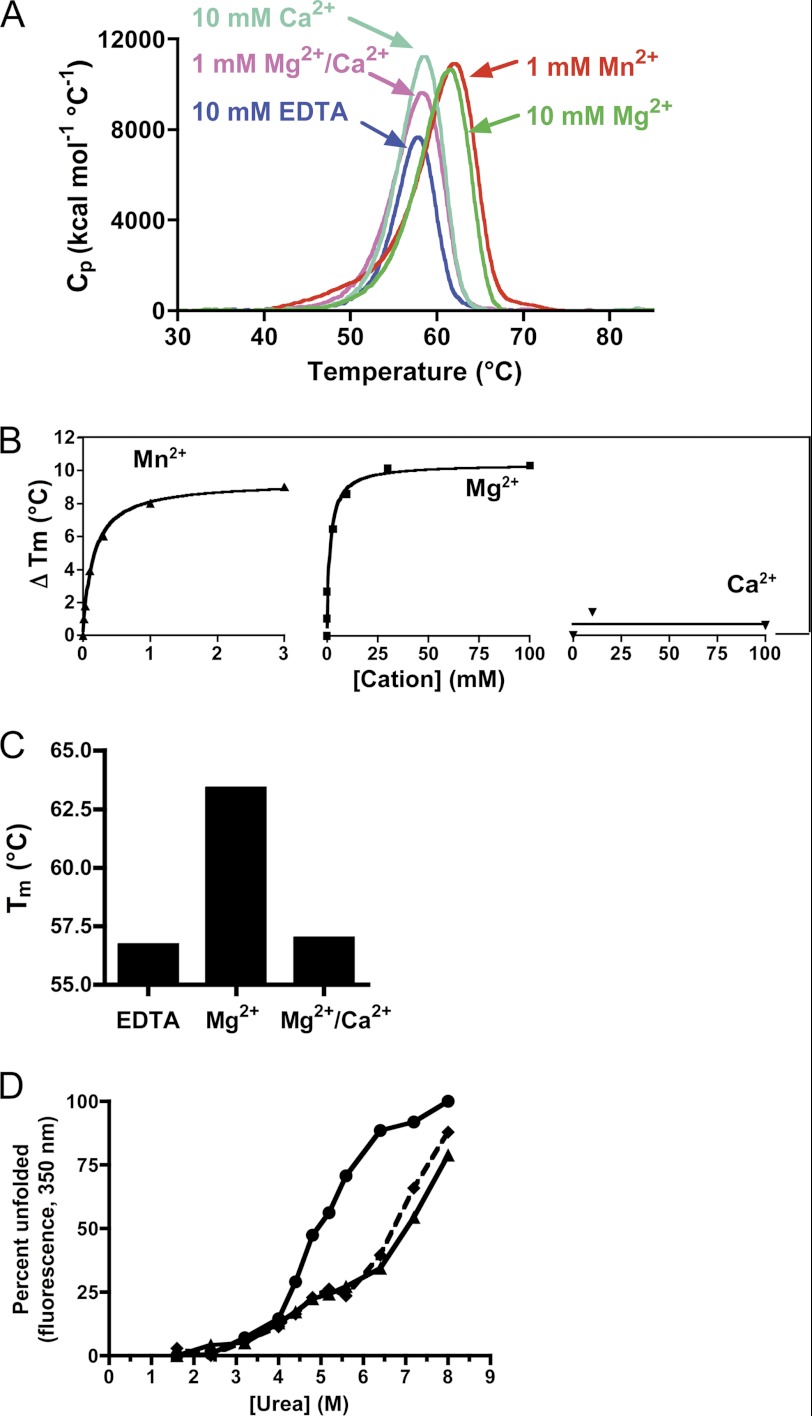
All integrins require divalent cations for ligand binding. A previous study, using circular dichroism, demonstrated the stabilizing effect of divalent cations (Mg2+ and Mn2+) on α1I (41). We have extended these observations using differential scanning calorimetry (DSC) (Fig. 3). The measured melting temperature (Tm) for α1I in the presence of the chelating agent EDTA was 57.9 °C. A single cooperative unfolding transition was observed (Fig. 3A). No significant change occurred in the presence of 10 mm CaCl2 alone (Tm = 58.5 °C) or in 1 mm CaCl2, 1 mm MgCl2, which approximates physiological cation concentrations (Tm = 58.2 °C, Fig. 3A). Addition of 10 mm Mg2+ (Tm = 61.4 °C) or 10 mm Mn2+ (Tm = 62.0 °C) alone increased the thermal stability of α1I significantly. The effects of Mn2+ and Mg2+ were concentration-dependent, as determined in a second set of experiments showing stabilizations of as much as 8–10 °C (Fig. 3B). The EC50 values for Mn2+ and Mg2+ were 0.16 and 1.8 mm, respectively. These values agree with Mn2+ and Mg2+ concentrations previously shown to support collagen binding to α1β1 (42, 60). Similar results were obtained using the GST fusion protein (GST-α1I), although under some conditions the I domain transition was masked by the GST unfolding transition at ∼57–59 °C (data not shown). To complement the thermal stability measurements, the impact of Mn2+ on susceptibility of α1I to the denaturant urea was measured by monitoring intrinsic tryptophan fluorescence (Fig. 3D). α1I has a single tryptophan residue (Trp-158), located near the MIDAS site. In the absence of divalent cation, ∼5 m urea was required for 50% denaturation, and Mn2+ significantly increased the stability of α1I domain, to an EC50 of ∼7 m urea.
FIGURE 3.
Metal ion-mediated stabilization of α1 I domain. A, thermal denaturation curves. Purified α1I was incubated for 1 h with either 10 mm EDTA (blue), 10 mm CaCl2 (light green), 1 mm MgCl2 + 1 mm CaCl2 (pink), 10 mm MgCl2 (dark green), or 1 mm MnCl2 (red) and analyzed using DSC. B, effect of varying concentrations of MnCl2, MgCl2, or CaCl2 on the melting temperature (Tm, in °C) of α1I measured by DSC. The results were plotted as change in Tm (ΔTm) relative to a reference sample containing 10 mm EDTA. C, reversal of Mg2+-induced stabilization of α1I by Ca2+, as measured by DSC. The Tm values for samples of α1I in the presence of 10 mm EDTA, 10 mm MgCl2, or 10 mm MgCl2 + 10 mm CaCl2 were determined using DSC. D, effect of metal ions on the chemical denaturation of α1I. Purified α1I in buffer containing 10 mm EDTA (circles), 1 mm MnCl2 (triangles), or 10 mm CaCl2 (diamonds) was incubated with urea at the indicated concentrations, and the fraction of unfolded protein was determined by monitoring tryptophan fluorescence (excitation 280 nm and emission 350 nm). Representative data confirmed by three independent experiments are shown.
Ca2+ does not support α1β1 collagen binding, and it has been shown to bind with poor (mm) affinity to integrin I domains (61–63). Unlike Mn2+ and Mg2+, Ca2+ did not stabilize α1I toward thermal denaturation at concentrations up to 100 mm (Fig. 3B). However, at equimolar concentrations (10 mm), Ca2+ completely reversed the stabilizing effect of Mg2+, indicating that Ca2+ is capable of binding to α1I and displacing Mg2+ from the MIDAS site. Interestingly, in urea denaturation studies, addition of 10 mm Ca2+ increased the urea concentration required for half-maximal denaturation to ∼7 m, similar to the amount of stabilization observed with Mn2+ (Fig. 3D). Similar results were observed with urea denaturation studies using the rat α1 I domain (Ref. 41 and data not shown). These results clearly indicate that Ca2+ is capable of binding to the α1I MIDAS, even though it does not effectively support collagen ligand binding. The difference between the thermal and chemical denaturation results suggests that Ca2+ binding affects the local environment around the MIDAS such that it can be detected by chemically induced unfolding but not by calorimetry, which is a more global measure of stability.
DXMS of α1I
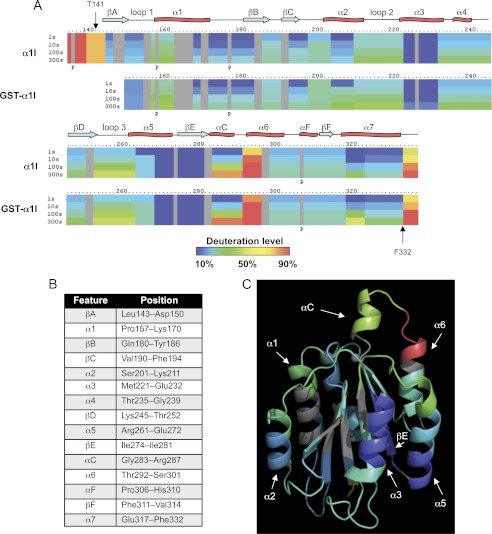
DXMS was utilized to evaluate the structural properties of unliganded α1I in solution and to provide a reference for subsequent experiments incorporating bound ligands. The DXMS method measures the solvent accessibility of main-chain amides in defined segments of a protein through a combination of time-dependent deuterium exchange, limited proteolysis, and mass spectrometry (33–40). Percent deuteration is operationally defined as the number of deuterium ions incorporated into a given peptide at a fixed time, divided by the maximum level incorporated at equilibrium. The DXMS profile of α1I in the presence of 10 mm MgCl2 is shown in Fig. 4. Briefly, a sample of α1I was diluted 1:3 into a deuterated buffer and incubated at 25 °C for defined time intervals from 1 to 1000 s. The exchange reaction was quenched by lowering the temperature and pH, and samples were digested with pepsin and analyzed using LC-MS. The sequence coverage of α1I was 93%. The data generated from 77 overlapping peptides was used to generate a one-dimensional ribbon map representing the average deuterium incorporation throughout the protein sequence (Fig. 4A). This linear map was annotated with the secondary structural elements identified from the α1I crystal structure (Fig. 4B). This experiment revealed several regions of α1I with very high incorporation of deuterium even at short incubation times, indicative of a highly flexible and dynamic structure. For example, N-terminal residues 134–143 appeared largely unfolded, exchanging >80% of the accessible amide protons within 1 s in D2O. This is consistent with the lack of any observed density N-terminal to residues 141–143 in α1 I domain crystal structures 1QC5 (28), 1PT6 (27), and 1CK4 (26). Similarly, residues at the C terminus (residues 333–336) were also exchanged very rapidly, consistent with the crystallographically determined terminus of the C-terminal α7 helix at position 332 (26). A third region comprising residues 291–295, a loop that connects the αC and α6 helices, also exchanged rapidly. As discussed below, the αC helix and adjacent residues play a key role in collagen binding and selectivity by this integrin. The αC helix itself (residues 283–287) exchanged more slowly than these other regions, but it still showed >75% deuterium incorporation within 300 s. The remainder of the α1I sequence, including the α-helical and β-strand regions forming the classic dinucleotide-binding fold, exchanged much more slowly, with most showing <50% deuterium incorporation even with longer D2O incubation times. The exchange data at 100 s were overlaid on the three-dimensional structure of α1I (Fig. 4C). From this overlay it is clear that the slowest exchanging regions are within well defined secondary structural elements (e.g. the α3 and α5 helices, and the βE strand), although the faster exchanging regions are contained within or at the interfaces with less structured loops. The DXMS profile of the GSTα1I fusion protein was also determined (Fig. 4A) and was very similar to the untagged α1I.
FIGURE 4.
Deuterium ion incorporation into α1I in the presence of 10 mm MgCl2. A, one-dimensional ribbon map. The H/D exchange profiles of α1I and GST-α1I are shown. The numbers above the map represent the primary sequence (residues 134–336 of α1I or 149–336 of GSTα1I, numbered as in Fig. 1), and the positions of I domain structural elements are annotated above (red twist, α-helix; blue arrow, β-strand). Each bar below the sequence is divided into rows corresponding to each time point from 1 to 300 s (top to bottom). Color coding indicates the percentage of deuterium incorporation into exchangeable amide protons after a given incubation time. Vertical arrows indicate residues Thr-141 and Phe-332, which are the N- and C-terminal residues, respectively, within which the α1 I domain crystal structure (Protein Data Bank code 1CK4) showed measurable electron density. P indicates the positions of proline residues, which have no exchangeable protons. Other areas in gray did not have sufficient coverage in the map to allow accurate quantitation of deuteration levels. B, annotated list of the primary structural features of α1I and the corresponding positions within the sequence (26). C, exchange rates at 100 s were mapped onto α1I crystal structure 1MHP. Color coding indicates the percentage of deuterium incorporation into exchangeable amide protons (identical scale as in A). The positions of prominent structural elements are indicated, and the metal ion in the MIDAS is represented by a gray sphere.
Effect of Divalent Cations on α1I Domain Solution Structure
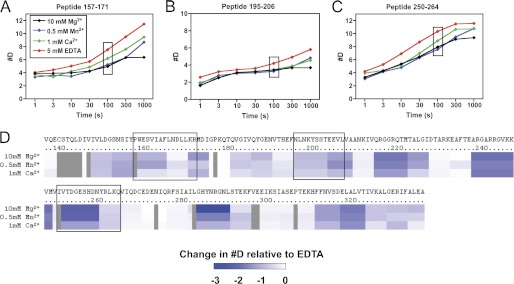
We next evaluated the impact of MIDAS site occupancy with different divalent cations on the DXMS exchange properties of α1I. The incorporation of deuterium into α1I was assessed in the presence of 5 mm EDTA (apo-α1I) or in 10 mm Mg2+, 0.5 mm Mn2+, or 1 mm Ca2+ (Fig. 5). Relative to apo-α1I, all three cations significantly decreased the exchange rates in multiple peptides, including residues 157–171, 195–206, and 250–264, as shown in Fig. 5A. No regions showed increased exchange rates in the presence of cation, suggesting an overall decrease in flexibility throughout the α1I structure. Difference maps comparing each of the cation-bound states versus apo-α1I (Fig. 5B) show that, at 100 s, slower exchange rates were observed over the majority of the α1I sequence. Larger decreases in exchange rates were observed with Mg2+ and Mn2+, consistent with the stabilizing effects measured using DSC and tryptophan fluorescence. Ca2+ led to smaller but still significant effects on exchange rates, further demonstrating that Ca2+ is competent to bind the MIDAS site and partially stabilize α1I.
FIGURE 5.
Divalent cations decrease the rate of H/D exchange in α1I. A–C, time courses of deuterium incorporation into peptides 157–171, 195–206, and 250–264, in the presence of 10 mm MgCl2, 0.5 mm MnCl2, 1 mm CaCl2, or 5 mm EDTA. Data are plotted as the number of deuterium ions (#D) incorporated into each peptide at a given time. D, difference map, indicating the change in #D in the presence of each metal ion relative to EDTA, after 100 s of incubation. The peptides described in A–C are indicated with boxes. The threshold for this plot was defined to resolve changes of up to ±3 deuterium ions/peptide.
Generally, the exchange rates of α1I in Mg2+ and Mn2+ were very similar (Fig. 5). This was confirmed by generating difference maps comparing these two conditions over a wide range of exchange times (1–300 s) (Fig. 6A). One set of peptides covering the αC helix region, however, exchanged much faster in Mn2+ than in Mg2+. This was exemplified by the 283–290-residue peptide, which incorporated notably higher levels of deuterium at exchange times <100 s in the presence of MnCl2 than it did in MgCl2 (Fig. 6B). This result was observed even when the protein was first treated with EDTA and reconstituted with excess MgCl2 or MnCl2, indicating that the result was not due to the effect of metals introduced during protein production or purification (data not shown). The observed differences in exchange rates in this region were surprising, because Mg2+ and Mn2+ were shown to bind integrin I domains with similar geometries (11, 62, 64) and showed essentially identical abilities, at saturating concentrations, to stabilize integrin structure (Fig. 3) and promote ligand binding (42, 60). The faster exchange rate in the presence of Mn2+ has led us to generate a new model to explain the differences in the relative abilities of various metal ions to activate integrin I domains, in which Mn2+ preferentially unfolds the αC helix by assuming a penta-coordinated geometry (discussed below). In any case, the ability of Mn2+ and Mg2+ to induce detectably different conformational changes in α1I indicates that, unlike previous suggestions (65), the choice of divalent cation impacts the local structure around the ligand-binding site.
FIGURE 6.
Mg2+ and Mn2+ induce different structures in the αC helix. A, difference map depicting deuterium ion incorporation into α1I in the presence of 0.5 mm MnCl2 or 10 mm MgCl2. The number of deuterium ions (#D) incorporated into each peptide at a given time was determined, and the value of #D in Mg2+ was subtracted from that in Mn2+. The threshold for this plot was defined to resolve changes of up to ±4 deuterium ions/peptide. Residues 283–290, corresponding to one of the peptides that exchanged notably faster in Mn2+ than in Mg2+, are indicated by a box. B, representative deuterium buildup curve for peptide 283–290 in the αC helix.
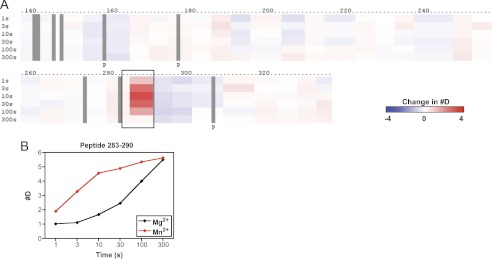
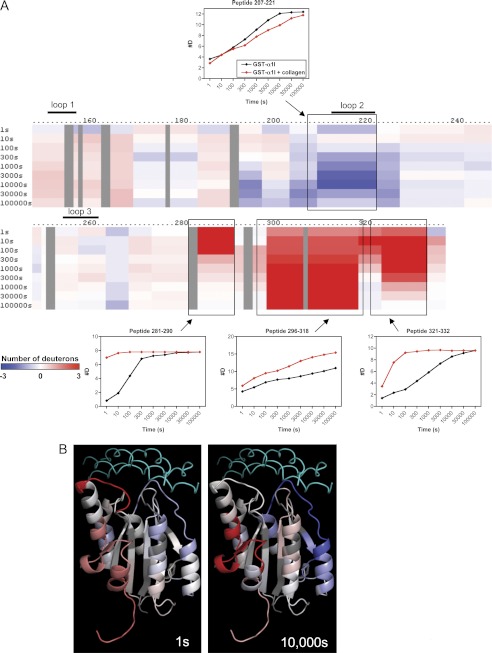
DXMS Profile of the Complex between α1I and Full-length Collagen
A crystal structure of α2I in complex with a triple-helical collagen-based peptide, but not a corresponding α1I-collagen peptide structure, has been reported (6). Because intact fibrillar collagen cannot be crystallized, no structure has been reported for full-length collagen bound to an integrin. To explore the nature of these interactions under native conditions, we determined the DXMS exchange pattern of the α1I protein bound to full-length type IV collagen. For these studies, a complex was formed by incubating the GST-α1I fusion protein with collagen IV that was immobilized on a bead-based support in the presence of 10 mm MgCl2. Deuterium exchange was then performed on the immobilized GST-α1I. The DXMS exchange rates in the presence of collagen were subtracted from those determined for GST-α1I alone to generate a series of difference maps (Fig. 7A). The exchange data at early (1 s) and late (10,000 s) time points were superimposed on a model of α1I bound to collagen IV (Fig. 7B), generated by docking α1I, in an open conformation, with a collagen THP as described previously (31).
FIGURE 7.
Effect of collagen binding on GST-α1I domain hydrogen-deuterium exchange rates. A, difference map indicating exchange rates in free versus collagen-bound GST-α1I in the presence of 10 mm MgCl2. Blue regions indicate decreased exchange (#D), and red regions indicate increased exchange, in the presence of collagen. The threshold for this plot was defined to resolve changes of up to ±3 deuterium ions/peptide. Exchange curves for representative peptides 207–221, 281–290, 296–318, and 321–332 are shown and indicated with boxes on the map. B, exchange rate differences at 1 and 10,000 s were mapped onto a model of the α1I-collagen complex. The THP used to generate the model is shown in cyan (at top of figure). Color coding on α1I is identical to that shown in A.
Regions at the interface of protein-protein binding surfaces are typically protected from amide exchange with water (66). Using DXMS, only residues in MIDAS loop 2 of α1I, exemplified by peptide 207–221, showed significant protection against deuterium exchange when bound to collagen (Fig. 7A). The structure of the α2I-collagen peptide complex suggested interactions between all three MIDAS loops (151–156, 212–220, and 253–260) and the collagen peptide (6). Similar interactions were proposed for the α1I-collagen IV structure, including stabilizing interactions with collagen involving Arg-218 in loop 2 and Glu-255 in loop 3 (31). The DXMS results indicate that little change in solvent accessibility occurs in either loop 1 or loop 3 upon binding to collagen. A closer examination of the model (Fig. 7B) suggested an explanation for this result. Loops 1 and 2 both make significant contacts with the collagen. Loop 2 has exposed backbone amides, particularly in residues 215–219, which are apparently protected upon collagen binding. Contacts of loop 1 with collagen are mediated mainly by the side chains of Asn-153 and Tyr-156, but the backbone amides of loop 1 are largely buried by folding within α1I itself. As a result, exchange is substantially slower in loop 1 even in the absence of collagen (see Fig. 4). Consequently, it is not surprising that collagen binding has a negligible effect on the loop 1 amides. In contrast, loop 3 has exposed backbone amides, but the model suggests it has very limited contact with collagen, which explains the limited protection observed in loop 3. This result does not support the proposal that Glu-255 forms a salt bridge with collagen (31) but is instead consistent with data showing that the corresponding glutamic acid in α2 (Glu-256) is not critical for ligand binding (67).
Several regions of α1I displayed significantly increased rates of exchange when complexed with collagen (Fig. 7). These increased rates of exchange reflect greater solvent exposure of the affected amide protons and suggest an increase in flexibility in these regions. The changes were clustered in the C-terminal region of the molecule. Residues 283–290, which include the αC helix, displayed very rapid exchange, with essentially complete deuterium incorporation within 1 s of solvent exposure, suggestive of extensive structural changes in this region. Residues 318–332, in the C-terminal α7 helix, and other intervening structural elements between αC and α7 (residues 298–317) also showed notable increases in the rates of deuterium incorporation, although exchange was not as fast as in the αC helix. These results provide direct evidence that α1I undergoes an allosteric conformational change in the C-terminal helix upon ligand binding, as has been postulated for other I domain-containing integrins on the basis of static crystal structures (3, 4, 6, 12), In addition, the striking increase in conformational flexibility of the α7 helix in the presence of ligand provides direct support for the proposal that the I domain C-terminal helix is unusually mobile (5), although this mobility appears to be induced by ligand binding rather than an intrinsic property of the unliganded I domain structure (discussed below).
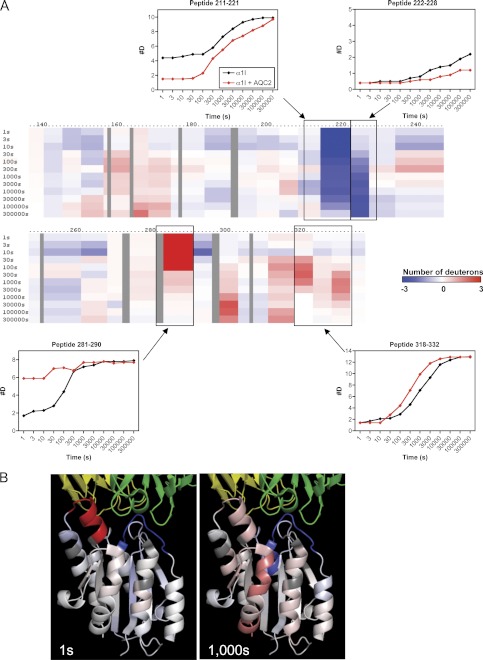
Effect of the “Pseudo-ligand Mimetic” Antibody AQC2 on α1I Exchange Rates
The α1 I domain was previously crystallized with a Fab fragment of the anti-α1β1 function-blocking antibody AQC2 (30). AQC2 can be classified as a pseudo-ligand mimetic antibody, because it binds to α1I in a ligand-mimetic fashion via direct interaction of an antibody aspartate residue with the MIDAS metal ion, but it does not induce a change to the open conformation (30, 68). The AQC2-α1I complex was generated in the presence of Mg2+, and DXMS studies were carried out in a similar manner as described above for the collagen-α1I complex (Fig. 8). A difference map is shown in Fig. 8A, with blue and red regions indicating decreased and increased exchange, respectively, upon antibody binding. The data at 1 and 1000 s were mapped onto the co-crystal structure of the AQC2-α1I complex (Fig. 8B) (30).
FIGURE 8.
DXMS exchange rates for α1I bound to antibody AQC2 in the presence of 1 mm MgCl2. A, difference map indicating exchange rates in free versus AQC2-bound α1I. Blue regions indicate decreased exchange (#D), and red regions indicate increased exchange, in the presence of antibody. The threshold for this plot was defined to resolve changes of up to ±3 deuterium ions/peptide. Exchange curves for representative peptides 211–221, 222–228, 281–290, and 318–332 are shown and indicated with boxes on the map. B, exchange rate differences at 1 and 1000 s were mapped onto the crystal structure of the AQC2-α1I complex. Antibody heavy and light chains are shown in green and yellow, respectively. Color coding on α1I is identical to that shown in A.
The most dramatic decreases in exchange rates were in residues 214–226, corresponding to MIDAS loop 2. This is consistent with the inability of AQC2 to bind to wild-type rat α1I (41), which has four amino acid differences in the 213–218-residue region, and with direct evidence from the crystal structure showing Arg-218 located inside of a cleft within the antibody (30). As in the collagen-α1I complex, no significant effects on MIDAS loops 1 or 3 were observed. The correlation between the collagen-α1I and AQC2-α1I exchange patterns within the MIDAS region indicates that the local interactions of AQC2 with α1I at the MIDAS site are very similar to those of the native ligand. This result is in striking contrast to the crystal structure of AQC2-α1I, which showed interactions of the antibody with all three loops, and suggests that the loop 2 region of α1I is predominantly responsible for the interaction of α1I with the antibody.
Similar to the collagen complex, the exchange rate in the αC helix (residues 283–290) increased upon AQC2 binding, indicating that the antibody induces a significant loss of secondary structure in this region. However, unlike collagen binding, the structural changes in the α7 C-terminal helix (residues 318–332) were modest. The crystal structure of α1I bound to AQC2 (30) adopted a closed conformation very similar to that of the unbound α1I (26, 28), with no evidence for a shift in the C-terminal helix. Thus, even though AQC2 binding to α1I leads to similar effects as the native ligand within the local region surrounding the MIDAS, it does not induce allosteric changes that would indicate a shift from the closed to the open conformation, characteristic of ligand binding.
DISCUSSION
We have successfully used hydrogen-deuterium exchange mass spectrometry (DXMS) to explore the conformational dynamics of cation, ligand, and antibody binding to the I domain of integrin α1 in aqueous solution. Most detailed structural information on integrin I domains is based on crystallographic data, so DXMS provided a unique opportunity to study the solution dynamics of the α1 I domain. The DXMS mapping method provided coverage of 93% of the α1I sequence with over 200 exchangeable hydrogens. The average peptide length was 15 amino acids. The observed H/D exchange rates for unliganded α1I were remarkably consistent with the structural elements observed in crystallographic studies of this domain. Regions at the N and C termini of the protein that did not show measurable density in the crystal structures incorporated deuterium rapidly, establishing the limits of the globular domain as comprising residues Asp-144 to Phe-332. Slow exchanging regions throughout the sequence largely correlated with defined secondary structural elements, although the unstructured loop regions, including the MIDAS loops involved in ligand binding, exchanged more rapidly.
In I domain-containing integrins, the I domain MIDAS coordinates a divalent cation such as Mg2+, which interacts directly with an acidic residue (Asp or Glu) in the ligand. Some nonphysiological cations, such as Mn2+, are capable of activating integrins much more strongly than Mg2+, whereas Ca2+ is incapable of mediating ligand binding, but the structural basis for this phenomenon is not well understood. Previous studies established that Mg2+ and Mn2+ at millimolar concentrations stabilize the I domain from α1 (41). Using DSC, we observed up to 8–10 °C increases in the Tm values for α1I at saturating concentrations of either Mn2+ or Mg2+. This overall stabilization by Mn2+ or Mg2+ was further established using DXMS, which revealed decreased H/D exchange rates throughout the α1I sequence in the presence of these cations.
Interpretation of the role of Ca2+ in integrin/ligand interactions is complicated (69). In α I domains, Ca2+ affinity for MIDAS, and its ability to regulate ligand binding, is both concentration- and activation-state dependent (61, 70, 71). The affinity of the αM I domain for Ca2+ was shown to be low (millimolar) (61–63), and quantum calculations on the αL I domain predicted that the Ca2+-bound state would have lower affinity than either the Mg2+- or Mn2+-bound state for ligand (71). However, some integrin I domains have shown differences in their relative abilities to bind calcium (70, 72), suggesting that the affinity of each I domain MIDAS site for Ca2+ may be different. Interestingly, even though we observed no effect of millimolar levels of Ca2+ by DSC, several other measures provided clear evidence for a stabilizing effect of Ca2+ on α1I. DXMS showed decreases in exchange rates throughout the α1I sequence. The concentration of urea required to denature α1I was increased by 2 m in the presence of 10 mm Ca2+, which was comparable in magnitude to the effect of Mn2+. Finally, Ca2+ at an equimolar concentration reversed the stabilizing effect of 10 mm Mg2+ ions. Thus, our results demonstrate that Ca2+ is capable of binding to the α1I MIDAS and that even though Ca2+ binding does not stabilize the I domain to thermal denaturation or support ligand binding, it significantly impacts the solution dynamics of the protein.
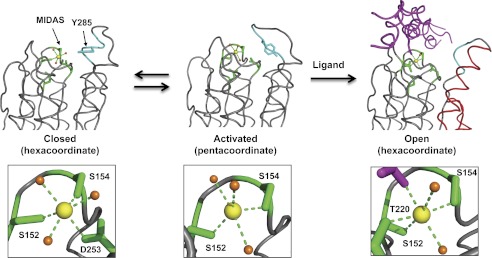
The DXMS patterns of α1I in Mg2+, Ca2+, and Mn2+ were very similar, except for a striking difference in the αC helix, a structural element unique to collagen-binding integrins that is believed to be critical for binding and collagen subtype selectivity (6, 59, 73). Relative to apo-α1I, saturating concentrations of Mg2+ stabilized the αC helix toward deuterium exchange to a much greater extent than either Mn2+ or Ca2+. This result was surprising, because both Mg2+ and Mn2+ support binding of collagen IV to α1β1 at these concentrations (42, 58), and crystallographic data on integrin I domains support the concept that Mg2+- and Mn2+-bound I domains form essentially identical structures (65). These data, in conjunction with the recently reported crystal structure of α1I with the activating mutation E317A (32), led us to propose a model for collagen-binding integrin I domain activation that provides a structural basis for the enhanced activating ability of Mn2+ and the inability of Ca2+ to mediate ligand binding (Fig. 9). The α1I(E317A) structure, which was the first to display an unwound αC helix in an I domain in the absence of a bound collagen peptide, contained a unique penta-coordinated Mg2+ in the MIDAS, suggesting that a change from penta- to hexa-coordinated metal ion upon ligand binding drives allosteric conformational changes within α1I. In our model, the closed conformation, with a hexa-coordinate metal ion and tightly wound αC helix, is in equilibrium with the activated conformation, containing a penta-coordinate metal and unwound αC helix. Collagen ligand binding leads to conversion back to the more stable hexa-coordinate geometry and induces allosteric changes in the C-terminal helices. Importantly, Mg2+ ions pay a significant energetic penalty for deviating from a hexacoordinate (octahedral) geometry, whereas Mn2+ is capable of adopting lower coordination states (32). Thus, Mn2+, with a lower energetic barrier to adopting the activated conformation, is more effective at driving the opening of the αC helix and enabling ligand binding. Ca2+, which is unable to adopt a pentavalent conformation, is incapable of mediating ligand binding.
FIGURE 9.
Model for collagen-binding integrin I domain activation. Structures of closed, activated, and open I domains were modeled based on Protein Data Bank entries 1QC5 (α1I), 4A0Q (α1I (E317A)), and 1DZI (α2I-THP complex). The MIDAS site (green) and αC helix (cyan) are highlighted, and the C-terminal α6 and α7 helices (red) and bound collagen (magenta) are indicated in the open structure. The side chain of Tyr-285 is shown for orientation in the closed and activated α1 conformations. Insets show the coordination states of the metal ion (yellow). Bound water molecules are shown in orange.
The observation of such striking metal ion-mediated differences in the αC helix conformation demonstrates the importance of studying the dynamics of integrins in solution as a complement to crystallographic studies. Because there are no crystal structures of free (not bound to ligand or antibody) α1 or α2 I domains in Mn2+, we cannot rule out the possibility that Mg2+ and Mn2+ would produce different conformations of αC even in a crystalline state. More likely, however, this difference is only detectable in solution, where crystal packing effects are eliminated. The concept that Mg2+ and Mn2+ induce different conformations of αC in solution is supported by tryptophan fluorescence studies on α1I, which demonstrated that binding of Mg2+ led to conformational changes in the vicinity of Trp-158, which is located at the end of MIDAS loop 1, very near the αC helix in the three-dimensional structure of α1I, and that these changes differed from the effects of added Mn2+ (72).
DXMS provided an ideal tool to characterize the conformational changes in α1I induced upon collagen binding and to test the existing models for integrin I domain/ligand interactions. The DXMS pattern of α1I bound to collagen showed significantly reduced exchange only in MIDAS loop 2, suggesting that this region of α1 integrin is a critical element mediating the interaction with collagen. Collagen also induced faster exchange in the αC, α6, and α7 helices, consistent with models in which ligand binding causes a downward shift of the C-terminal α7 helix of the I domain, leading to engagement of Glu-335 (in the case of α1 integrin) with the MIDAS site of the β subunit I-like domain (1). It has been proposed that the α7 helix is intrinsically mobile, allowing it to trigger this process with minimal energetic penalty (5, 10). Our DXMS data on α1I indicate that the flexibility of α7 is greatly increased upon ligand binding. We therefore propose a modification to this model in which ligand binding is responsible for relaxing the α7 helical structure and facilitating the conformational changes necessary for effective intersubunit communication, e.g. in outside-in signaling.
One of the key differences between the studies reported here and those previously described is the use of full-length collagen instead of a THP fragment. Solution binding studies on α2 I domain binding to a collagen THP have been reported using NMR (74). A number of residues located near the α7 helix showed changes in chemical shift pattern, although only one residue (Gly-329, equivalent to Gly-328 in α1I) within α7 itself was noted. The relatively small number of chemical shift changes observed in this region compared with our observations using DXMS on α1I-collagen could be due to limitations of the NMR method, such as the inherently reduced peak intensity observed upon peptide binding. Alternatively, the use of a THP instead of a full-length collagen ligand may have led to less robust conformational changes in the integrin. It is important to note that a single molecule of collagen contains multiple integrin-binding sites with differing affinities (75). This has the potential to lead to dispersion in the DXMS signal. It is unclear whether different residues on α1I are involved in binding to different collagen GXXGER motifs. If so, one could imagine that DXMS studies using α1I bound to individual THP units might be informative.
In contrast to collagen, binding of the function-blocking α1 antibody AQC2 led to much more limited changes in the DXMS pattern of α1I. In the crystal structure of the AQC2-α1I complex, Asp-101 from the antibody heavy chain bound directly to the metal ion in the α1I MIDAS, but the remainder of the I domain, including the C terminus, retained the closed conformation (30). We found that AQC2 binding selectively protected MIDAS loop 2, the same region protected from exchange by collagen. The similarity of AQC2 and collagen in their interactions with the metal ion and MIDAS loop 2, coupled with the stark differences in their abilities to transmit allosteric changes to the C terminus of α1I, is remarkable.
Two other examples of ligand-mimetic I domain antibodies have been reported, both of which also present Asp residues that interact with the MIDAS metal ion. One of these is the αL antibody AL-57 (76). Unlike AQC2, AL-57 binds to the open form of αL (68). It was suggested that a key salt bridge formed with Glu-241 of αL by either the native ligand ICAM-1 or AL-57 is necessary for the allosteric rearrangement to occur and that the reason AQC2 binds to the open form of α1I is that it does not form a similar bridge with the corresponding residue in α1I, Glu-255 (68). Unlike in the αL-ICAM interaction, this conserved glutamic acid (Glu-255 in α1 or Glu-256 in α2) is not critical for collagen binding by integrins (6, 67). Nonetheless, the finding that collagen but not AQC2 can induce long range conformational changes in α1I is consistent with the conclusion that AL-57 is a true ligand mimetic, whereas AQC2 co-opts a key element of the ligand-binding site (i.e. the MIDAS cation) without engaging other regions necessary for induction of signaling. One would predict that solution DXMS studies using AL-57 and the αL I domain should demonstrate pronounced antibody-induced conformational changes in the α7 helix. A true ligand-mimetic antibody would be less desirable as a therapeutic, however, because it could lead to agonistic effects through outside-in integrin signaling. Another related antibody, mAb107, preferentially binds to the αM I domain in Ca2+ via a bidentate interaction with Asp-107 (61, 77). Similarly to AQC2, ligation of mAb107 did not induce conversion to the open conformation. Interestingly, the bidentate mode of binding led to a heptavalent Ca2+ ion in the mAb107-I domain complex, highlighting the range of different geometries accessible by the MIDAS.
In summary, we have explored the dynamic conformational changes in the α1 integrin I domain induced by divalent cations and by ligand binding, and we tested several elements of the existing models for collagen-induced conformational changes and allosteric regulation, using DXMS. The data validated some elements of the models derived from crystallographic studies but importantly identified novel features of ligand and metal ion binding that were not detected using static methods. The application of DXMS to other integrin/ligand interactions should be very powerful, as it provides a method to explore the interaction of isolated I domains or even complete integrin heterodimers with native ligands, and it does not require the use of either conformationally restricted integrins (e.g. locked-open I domains) or engineered fragments of ligands (e.g. collagen THPs or RGD peptides).
Acknowledgments
We thank Mia Rushe for purification of the α1 I domain and Paul Rayhorn for experimental assistance and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA099835, CA118595, AI076961, AI072106, AI068730, AI081982, AI2008031, GM037684, GM020501, GM066170, GM093325, NS070899, and RR029388. This work was also supported by Discovery Grant UC10591 from the University of California IUCRP Program, Biogen Idec corporate sponsor (to V. L. W.).
- THP
- collagen triple-helical peptide
- VLA1
- very late antigen-1 (α1β1 integrin)
- VLA2
- very late antigen-2 (α2β1 integrin)
- MIDAS
- metal ion-dependent adhesion site
- DXMS
- hydrogen-deuterium exchange mass spectrometry
- DSC
- differential scanning calorimetry
- GdnHCl
- guanidine hydrochloride
- ND
- nondeuterated
- D/D
- hydrogen-deuterium.
REFERENCES
- 1. Luo B. H., Carman C. V., Springer T. A. (2007) Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takagi J. (2007) Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 19, 557–564 [DOI] [PubMed] [Google Scholar]
- 3. Shimaoka M., Xiao T., Liu J. H., Yang Y., Dong Y., Jun C. D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J. H., Springer T. A. (2003) Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell 112, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song G., Yang Y., Liu J. H., Casasnovas J. M., Shimaoka M., Springer T. A., Wang J. H. (2005) An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin αLβ2. Proc. Natl. Acad. Sci. U.S.A. 102, 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang H., Casasnovas J. M., Jin M., Liu J. H., Gahmberg C. G., Springer T. A., Wang J. H. (2008) An unusual allosteric mobility of the C-terminal helix of a high affinity αL integrin I domain variant bound to ICAM-5. Mol. Cell 31, 432–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Structural basis of collagen recognition by integrin α2β1. Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 7. Hynes R. O. (2002) Integrins. Bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 8. Huth J. R., Olejniczak E. T., Mendoza R., Liang H., Harris E. A., Lupher M. L., Jr., Wilson A. E., Fesik S. W., Staunton D. E. (2000) NMR and mutagenesis evidence for an I domain allosteric site that regulates lymphocyte function-associated antigen 1 ligand binding. Proc. Natl. Acad. Sci. U.S.A. 97, 5231–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimaoka M., Salas A., Yang W., Weitz-Schmidt G., Springer T. A. (2003) Small molecule integrin antagonists that bind to the β2 subunit I-like domain and activate signals in one direction and block them in the other. Immunity 19, 391–402 [DOI] [PubMed] [Google Scholar]
- 10. Legge G. B., Kriwacki R. W., Chung J., Hommel U., Ramage P., Case D. A., Dyson H. J., Wright P. E. (2000) NMR solution structure of the inserted domain of human leukocyte function associated antigen-1. J. Mol. Biol. 295, 1251–1264 [DOI] [PubMed] [Google Scholar]
- 11. Qu A., Leahy D. J. (1996) The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin. Structure 4, 931–942 [DOI] [PubMed] [Google Scholar]
- 12. Lee J. O., Rieu P., Arnaout M. A., Liddington R. (1995) Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18). Cell 80, 631–638 [DOI] [PubMed] [Google Scholar]
- 13. Li R., Rieu P., Griffith D. L., Scott D., Arnaout M. A. (1998) Two functional states of the CD11b A-domain. Correlations with key features of two Mn2+-complexed crystal structures. J. Cell Biol. 143, 1523–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie C., Zhu J., Chen X., Mi L., Nishida N., Springer T. A. (2010) Structure of an integrin with an αI domain, complement receptor type 4. EMBO J. 29, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alonso J. L., Essafi M., Xiong J. P., Stehle T., Arnaout M. A. (2002) Does the integrin αA domain act as a ligand for its βA domain? Curr. Biol. 12, R340–R342 [DOI] [PubMed] [Google Scholar]
- 16. Yang W., Shimaoka M., Salas A., Takagi J., Springer T. A. (2004) Intersubunit signal transmission in integrins by a receptor-like interaction with a pull spring. Proc. Natl. Acad. Sci. U.S.A. 101, 2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duband J. L., Belkin A. M., Syfrig J., Thiery J. P., Koteliansky V. E. (1992) Expression of α1 integrin, a laminin-collagen receptor, during myogenesis and neurogenesis in the avian embryo. Development 116, 585–600 [DOI] [PubMed] [Google Scholar]
- 18. Abraham W. M., Ahmed A., Serebriakov I., Carmillo A. N., Ferrant J., de Fougerolles A. R., Garber E. A., Gotwals P. J., Koteliansky V. E., Taylor F., Lobb R. R. (2004) A monoclonal antibody to α1β1 blocks antigen-induced airway responses in sheep. Am. J. Respir. Crit. Care Med. 169, 97–104 [DOI] [PubMed] [Google Scholar]
- 19. Chen L., Huq S., Gardner H., de Fougerolles A. R., Barabino S., Dana M. R. (2007) Very late antigen 1 blockade markedly promotes survival of corneal allografts. Arch. Ophthalmol. 125, 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cosgrove D., Rodgers K., Meehan D., Miller C., Bovard K., Gilroy A., Gardner H., Kotelianski V., Gotwals P., Amatucci A., Kalluri R. (2000) Integrin α1β1 and transforming growth factor-β1 play distinct roles in alport glomerular pathogenesis and serve as dual targets for metabolic therapy. Am. J. Pathol. 157, 1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Fougerolles A. R., Chi-Rosso G., Bajardi A., Gotwals P., Green C. D., Koteliansky V. E. (2000) Global expression analysis of extracellular matrix/integrin interactions in monocytes. Immunity 13, 749–758 [DOI] [PubMed] [Google Scholar]
- 22. Ianaro A., Cicala C., Calignano A., Koteliansky V., Gotwals P., Bucci M., Gerli R., Santucci L., Fiorucci S., Cirino G. (2000) Anti-very late antigen-1 monoclonal antibody modulates the development of secondary lesion and T-cell response in experimental arthritis. Lab. Invest. 80, 73–80 [DOI] [PubMed] [Google Scholar]
- 23. Krieglstein C. F., Cerwinka W. H., Sprague A. G., Laroux F. S., Grisham M. B., Koteliansky V. E., Senninger N., Granger D. N., de Fougerolles A. R. (2002) Collagen-binding integrin α1β1 regulates intestinal inflammation in experimental colitis. J. Clin. Invest. 110, 1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meharra E. J., Schön M., Hassett D., Parker C., Havran W., Gardner H. (2000) Reduced gut intraepithelial lymphocytes in VLA1 null mice. Cell. Immunol. 201, 1–5 [DOI] [PubMed] [Google Scholar]
- 25. Ray S. J., Franki S. N., Pierce R. H., Dimitrova S., Koteliansky V., Sprague A. G., Doherty P. C., de Fougerolles A. R., Topham D. J. (2004) The collagen binding α1β1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity 20, 167–179 [DOI] [PubMed] [Google Scholar]
- 26. Nolte M., Pepinsky R. B., Venyaminov S. Yu., Koteliansky V., Gotwals P. J., Karpusas M. (1999) Crystal structure of the αβ1 integrin I-domain. Insights into integrin I-domain function. FEBS Lett. 452, 379–385 [DOI] [PubMed] [Google Scholar]
- 27. Nymalm Y., Puranen J. S., Nyholm T. K., Käpylä J., Kidron H., Pentikäinen O. T., Airenne T. T., Heino J., Slotte J. P., Johnson M. S., Salminen T. A. (2004) Jararhagin-derived RKKH peptides induce structural changes in α1I domain of human integrin α1β1. J. Biol. Chem. 279, 7962–7970 [DOI] [PubMed] [Google Scholar]
- 28. Rich R. L., Deivanayagam C. C., Owens R. T., Carson M., Höök A., Moore D., Symersky J., Yang V. W., Narayana S. V., Höök M. (1999) Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, α1β1 integrin and Staphylococcus aureus cna MSCRAMM. J. Biol. Chem. 274, 24906–24913 [DOI] [PubMed] [Google Scholar]
- 29. Clark L. A., Boriack-Sjodin P. A., Eldredge J., Fitch C., Friedman B., Hanf K. J., Jarpe M., Liparoto S. F., Li Y., Lugovskoy A., Miller S., Rushe M., Sherman W., Simon K., Van Vlijmen H. (2006) Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci. 15, 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karpusas M., Ferrant J., Weinreb P. H., Carmillo A., Taylor F. R., Garber E. A. (2003) Crystal structure of the α1β1 integrin I domain in complex with an antibody Fab fragment. J. Mol. Biol. 327, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 31. Saccà B., Sinner E. K., Kaiser J., Lübken C., Eble J. A., Moroder L. (2002) Binding and docking of synthetic heterotrimeric collagen type IV peptides with α1β1 integrin. ChemBioChem 3, 904–907 [DOI] [PubMed] [Google Scholar]
- 32. Lahti M., Bligt E., Niskanen H., Parkash V., Brandt A. M., Jokinen J., Patrikainen P., Käpylä J., Heino J., Salminen T. A. (2011) Structure of collagen receptor integrin α1I domain carrying the activating mutation E317A. J. Biol. Chem. 286, 43343–43351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burke J. E., Hsu Y. H., Deems R. A., Li S., Woods V. L., Jr., Dennis E. A. (2008) A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2. J. Biol. Chem. 283, 31227–31236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coales S. J., Tuske S. J., Tomasso J. C., Hamuro Y. (2009) Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography, and mass spectrometry. Rapid Commun. Mass Spectrom. 23, 639–647 [DOI] [PubMed] [Google Scholar]
- 35. Engen J. R. (2009) Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 81, 7870–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamuro Y., Coales S. J., Hamuro L. L., Woods V. L., Jr. (2008) in Mass Spectrometry Analysis for Protein-Protein Interactions and Dynamics (Chance M., ed) pp. 123–155, Wiley-VCH, Hoboken, NJ [Google Scholar]
- 37. Hsu Y. H., Burke J. E., Li S., Woods V. L., Jr., Dennis E. A. (2009) Localizing the membrane binding region of Group VIA Ca2+-independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 284, 23652–23661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konermann L., Tong X., Pan Y. (2008) Protein structure and dynamics studied by mass spectrometry. H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass. Spectrom. 43, 1021–1036 [DOI] [PubMed] [Google Scholar]
- 39. Mendillo M. L., Hargreaves V. V., Jamison J. W., Mo A. O., Li S., Putnam C. D., Woods V. L., Jr., Kolodner R. D. (2009) A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 106, 22223–22228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Truhlar S. M., Torpey J. W., Komives E. A. (2006) Regions of IκBα that are critical for its inhibition of NF-κB. DNA interaction fold upon binding to NF-κB. Proc. Natl. Acad. Sci. U.S.A. 103, 18951–18956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gotwals P. J., Chi-Rosso G., Ryan S. T., Sizing I., Zafari M., Benjamin C., Singh J., Venyaminov S. Y., Pepinsky R. B., Koteliansky V. (1999) Divalent cations stabilize the α1β1 integrin I domain. Biochemistry 38, 8280–8288 [DOI] [PubMed] [Google Scholar]
- 42. Weinreb P. H., Yang W. J., Violette S. M., Couture M., Kimball K., Pepinsky R. B., Lobb R. R., Josiah S. (2002) A cell-free electrochemiluminescence assay for measuring β1-integrin-ligand interactions. Anal. Biochem. 306, 305–313 [DOI] [PubMed] [Google Scholar]
- 43. Hamuro Y., Zawadzki K. M., Kim J. S., Stranz D. D., Taylor S. S., Woods V. L., Jr. (2003) Dynamics of cAPK type IIβ activation revealed by enhanced amide H/2H exchange mass spectrometry (DXMS). J. Mol. Biol. 327, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 44. Burns-Hamuro L. L., Hamuro Y., Kim J. S., Sigala P., Fayos R., Stranz D. D., Jennings P. A., Taylor S. S., Woods V. L., Jr. (2005) Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 14, 2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hamuro Y., Anand G. S., Kim J. S., Juliano C., Stranz D. D., Taylor S. S., Woods V. L., Jr. (2004) Mapping intersubunit interactions of the regulatory subunit (RIα) in the type I holoenzyme of protein kinase A by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J. Mol. Biol. 340, 1185–1196 [DOI] [PubMed] [Google Scholar]
- 46. Pantazatos D., Kim J. S., Klock H. E., Stevens R. C., Wilson I. A., Lesley S. A., Woods V. L., Jr. (2004) Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc. Natl. Acad. Sci. U.S.A. 101, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spraggon G., Pantazatos D., Klock H. E., Wilson I. A., Woods V. L., Jr., Lesley S. A. (2004) On the use of DXMS to produce more crystallizable proteins. Structures of the T. maritima proteins TM0160 and TM1171. Protein Sci. 13, 3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Z., Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry. A new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hsu S., Kim Y., Li S., Durrant E. S., Pace R. M., Woods V. L., Jr., Gentry M. S. (2009) Structural insights into glucan phosphatase dynamics using amide hydrogen-deuterium exchange mass spectrometry. Biochemistry 48, 9891–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hsu Y. H., Burke J. E., Stephens D. L., Deems R. A., Li S., Asmus K. M., Woods V. L., Jr., Dennis E. A. (2008) Calcium binding rigidifies the C2 domain and the intradomain interaction of GIVA phospholipase A2 as revealed by hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 283, 9820–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Molday R. S., Englander S. W., Kallen R. G. (1972) Primary structure effects on peptide group hydrogen exchange. Biochemistry 11, 150–158 [DOI] [PubMed] [Google Scholar]
- 53. Oyeyemi O. A., Sours K. M., Lee T., Resing K. A., Ahn N. G., Klinman J. P. (2010) Temperature dependence of protein motions in a thermophilic dihydrofolate reductase and its relationship to catalytic efficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 10074–10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilderman P. R., Shah M. B., Liu T., Li S., Hsu S., Roberts A. G., Goodlett D. R., Zhang Q., Woods V. L., Jr., Stout C. D., Halpert J. R. (2010) Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and x-ray crystallography. J. Biol. Chem. 285, 38602–38611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 56. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2007) in Current Protocols in Protein Science (Colligan J. E., Dunn B. M., Speicher D.W., Wingfield P.T., eds) pp. 2.9.1–2.9.31, Wiley Interscience, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 57. Chasseaud L. F. (1979) The role of glutathione and glutathione S-transferases in the metabolism of chemical carcinogens and other electrophilic agents. Adv. Cancer Res. 29, 175–274 [DOI] [PubMed] [Google Scholar]
- 58. Calderwood D. A., Tuckwell D. S., Eble J., Kühn K., Humphries M. J. (1997) The integrin α1A-domain is a ligand-binding site for collagens and laminin. J. Biol. Chem. 272, 12311–12317 [DOI] [PubMed] [Google Scholar]
- 59. Dickeson S. K., Mathis N. L., Rahman M., Bergelson J. M., Santoro S. A. (1999) Determinants of ligand binding specificity of the α1β1 and α2β1 integrins. J. Biol. Chem. 274, 32182–32191 [DOI] [PubMed] [Google Scholar]
- 60. Luque A., Sánchez-Madrid F., Cabañas C. (1994) Functional regulation of the human integrin VLA-1 (CD49a/CD29) by divalent cations and stimulatory β1 antibodies. FEBS Lett. 346, 278–284 [DOI] [PubMed] [Google Scholar]
- 61. Ajroud K., Sugimori T., Goldmann W. H., Fathallah D. M., Xiong J. P., Arnaout M. A. (2004) Binding affinity of metal ions to the CD11b A-domain is regulated by integrin activation and ligands. J. Biol. Chem. 279, 25483–25488 [DOI] [PubMed] [Google Scholar]
- 62. Baldwin E. T., Sarver R. W., Bryant G. L., Jr., Curry K. A., Fairbanks M. B., Finzel B. C., Garlick R. L., Heinrikson R. L., Horton N. C., Kelley L. L., Mildner A. M., Moon J. B., Mott J. E., Mutchler V. T., Tomich C. S., Watenpaugh K. D., Wiley V. H. (1998) Cation binding to the integrin CD11b I domain and activation model assessment. Structure 6, 923–935 [DOI] [PubMed] [Google Scholar]
- 63. Michishita M., Videm V., Arnaout M. A. (1993) A novel divalent cation-binding site in the A domain of the β2 integrin CR3 (CD11b/CD18) is essential for ligand binding. Cell 72, 857–867 [DOI] [PubMed] [Google Scholar]
- 64. Liddington R., Bankston L. (1998) The integrin I domain. Crystals, metals, and related artifacts. Structure 6, 937–938 [DOI] [PubMed] [Google Scholar]
- 65. Bella J., Berman H. M. (2000) Integrin-collagen complex. A metal-glutamate handshake. Structure 8, R121–126 [DOI] [PubMed] [Google Scholar]
- 66. Tu T., Drăguşanu M., Petre B. A., Rempel D. L., Przybylski M., Gross M. L. (2010) Protein-peptide affinity determination using an h/d exchange dilution strategy. Application to antigen-antibody interactions. J. Am. Soc. Mass Spectrom. 21, 1660–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kamata T., Liddington R. C., Takada Y. (1999) Interaction between collagen and the α2 I-domain of integrin α2β1. Critical role of conserved residues in the metal ion-dependent adhesion site (MIDAS) region. J. Biol. Chem. 274, 32108–32111 [DOI] [PubMed] [Google Scholar]
- 68. Zhang H., Liu J. H., Yang W., Springer T., Shimaoka M., Wang J. H. (2009) Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1. Proc. Natl. Acad. Sci. U.S.A. 106, 18345–18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen J., Takagi J., Xie C., Xiao T., Luo B. H., Springer T. A. (2004) The relative influence of metal ion-binding sites in the I-like domain and the interface with the hybrid domain on rolling and firm adhesion by integrin α4β7. J. Biol. Chem. 279, 55556–55561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Griggs D. W., Schmidt C. M., Carron C. P. (1998) Characteristics of cation binding to the I domains of LFA-1 and MAC-1. The LFA-1 I domain contains a Ca2+-binding site. J. Biol. Chem. 273, 22113–22119 [DOI] [PubMed] [Google Scholar]
- 71. San Sebastian E., Mercero J. M., Stote R. H., Dejaegere A., Cossío F. P., Lopez X. (2006) On the affinity regulation of the metal-ion-dependent adhesion sites in integrins. J. Am. Chem. Soc. 128, 3554–3563 [DOI] [PubMed] [Google Scholar]
- 72. Obsil T., Hofbauerová K., Amler E., Teisinger J. (1999) Different cation binding to the I domains of α1 and α2 integrins. Implication of the binding site structure. FEBS Lett. 457, 311–315 [DOI] [PubMed] [Google Scholar]
- 73. Käpylä J., Ivaska J., Riikonen R., Nykvist P., Pentikäinen O., Johnson M., Heino J. (2000) Integrin α2I domain recognizes type I and type IV collagens by different mechanisms. J. Biol. Chem. 275, 3348–3354 [DOI] [PubMed] [Google Scholar]
- 74. Lambert L. J., Bobkov A. A., Smith J. W., Marassi F. M. (2008) Competitive interactions of collagen and a jararhagin-derived disintegrin peptide with the integrin α2-I domain. J. Biol. Chem. 283, 16665–16672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Siljander P. R., Hamaia S., Peachey A. R., Slatter D. A., Smethurst P. A., Ouwehand W. H., Knight C. G., Farndale R. W. (2004) Integrin activation state determines selectivity for novel recognition sites in fibrillar collagens. J. Biol. Chem. 279, 47763–47772 [DOI] [PubMed] [Google Scholar]
- 76. Shimaoka M., Kim M., Cohen E. H., Yang W., Astrof N., Peer D., Salas A., Ferrand A., Springer T. A. (2006) AL-57, a ligand-mimetic antibody to integrin LFA-1, reveals chemokine-induced affinity up-regulation in lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 13991–13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mahalingam B., Ajroud K., Alonso J. L., Anand S., Adair B. D., Horenstein A. L., Malavasi F., Xiong J. P., Arnaout M. A. (2011) Stable coordination of the inhibitory Ca2+ ion at the metal ion-dependent adhesion site in integrin CD11b/CD18 by an antibody-derived ligand aspartate. Implications for integrin regulation and structure-based drug design. J. Immunol. 187, 6393–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]