Abstract
Dengue is emerging as the most important mosquito borne viral disease in the world. In mainland China, sporadic and large outbreaks of dengue illness caused by the four serotypes of dengue virus (DENV-1 to DENV-4) have been well documented. Guangdong province is the major affected area in China, and DENV-1 has dominantly circulated in Guangdong for a long time. In this study, a family cluster of DENV-3 infection in Guangzhou was described. Three cases were diagnosed as dengue fever based on clinical manifestation, serological and RT-PCR assays. Two DENV-3 strains were isolated in C6/36 cells and the complete genome sequences were determined. Phylogenetic analysis revealed that the new DENV-3 isolates from the family cluster were grouped within genotype III. Considering the fact that several DENV-3 strains within genotype V were also identified in Guangzhou in 2009, at least two genotypes of DENV-3 co-circulated in Guangzhou. Careful investigation and virological analysis should be warranted in the future.
Keywords: Dengue virus type 3, Family cluster, Genotype, Co-circulation
Background
Dengue is increasing in both frequency and magnitude worldwide, posing a heavy public health and economic burden especially in tropical and subtropical countries. Today, dengue ranks as the most important mosquito-borne viral disease in the world. Annually, up to 50 million human infections occur with 22, 000 deaths mainly in children [1]. Even, population growth, urbanization, international travel, and global warming continuously enhance vector transmission and disease outbreaks [2]. Dengue virus (DENV) contains four serotypes, and each of them can cause a wide spectrum of clinical manifestations, including mild dengue fever (DF), severe dengue haemorrhagic fever (DHF) and deadly dengue shock syndrome (DSS). Although intensive efforts have been made for decades, no preventive vaccines or antiviral drugs is currently available. The pathogenesis of DHF and DSS remains poorly understood. However, secondary infection with another DENV serotypes clearly increased the risk of severe diseases via the mechanism of antibody dependent enhancement (ADE) [3-5]. Epidemiological and in vivo data also indicated that anti-DENV antibodies mediated pathogenesis of a second heterotypic DENV infection [6-8].
Mainland China has experienced large outbreaks of DF during World War II, after that dengue disappeared for about 30 years. Since 1978, mainland China has seen a resurgence of dengue, epidemics involving hundreds of thousands of people have occurred in many provinces of Southern China, including Hainan, Guangdong, Guangxi, Fujian, Yunnan and Zhejiang provinces [9-14]. Currently, DF is listed as the notifiable infectious disease by the Ministry of Health, China. The recent epidemiology of dengue in China is characterized by a 3–5 year cycle. Most cases are DF, and only a few DHF or DSS cases have been reported over the last decade in mainland China [9,10,13].
In dengue endemic country, the presence of four serotypes of DENV is common, and co-circulation of multiple dengue serotypes in the same area has been well documented [15-17]. Guangdong province has been recognized as the major affected area of China. Although all four serotypes of DENV have been isolated in China, the dominant serotype circulating in Guangdong is DENV-1, no other serotypes has been recorded since 2001 [9,10,13,18]. Large DF outbreaks involving more than 1000 cases caused by DENV-1 have been described in Guangdong, China in 2002 and 2006, respectively [13,19].
In this study, we sought to determine the cause of a family cluster of DF in Guangzhou, Guangdong province, China in 2009, and analyze the possible origin of these emerging isolates responsible for the epidemic.
Materials and methods
Case description
On Aug 6, 2009, three adult family members admitted to Guangzhou No.8 People’s Hospital as suspected DF cases. The 30-year-old son firstly had a sudden fever with headache, then his father (56-year-old) and mother (50-year-old) fell ill subsequently in the following two days. All the three cases developed typical DF symptoms, including fever, headache, chills, rash, muscle and joint pain, and anorexia. The couples developed diarrhoea, and none of them showed vomiting. The tourniquet tests were all positive. All patients recovered uneventfully and discharged on Aug 11, 2009.
Ethics statement
The research was approved by the Review Board of Guangzhou No.8 People’s Hospital and the Ethical Committee of State Key Laboratory of Pathogen and Biosecurity. Informed consent was obtained from patients.
Serological assay and RT-PCR
Acute phrase sera were subjected to serological assays using IgM and IgG capture ELISA kit (PanBio, Queensland, Australia) according to the manufacturer’s instruction. RT-PCR assays were performed to detect and typing of DENVs as previously described [20].
Virus isolation and identification
Acute phase sera from the three patients were inoculated in C6/36 mosquito cells (Aedesalbopitus clone) and maintained in 1640 medium (Life Technologies, CA, USA) supplement with 2% fetal bovine serum (Life Technologies) at 28 °C in 5% CO2. When complete cytopathic effects (CPE) were observed, culture supernatants from positive samples were collected and stored at −70 °C until use. Indirect immunofluscence assay (IFA) was performed as previously described [21].
Sequencing of complete genome of DENV-3 isolates
The viral RNA was extracted from 200 μl of DENV-3 infected C6/36 culture supernatant using Purelink RNA mini kit (Life Technologies) in accordance with the manufacturer’s instructions. A total of 11 overlapping amplicons spanning the complete genomic region were amplified using 11 pairs of primers. The PCR products were sequenced and assembled. The 5’ and 3’ untranslated regions (UTRs) of viral genome of each isolate were determined using a rapid amplification of either 5’ or 3’ cDNA ends (RACE) kit (Roche, Mannheim, Germany) followed the manufacturer’s recommendation. All primers can be found in Table 1.
Table 1.
Primers used for sequencing reactions
| Primer Name | Locationa | Sequence (5'-3') | Product (bp) |
|---|---|---|---|
| F1 |
1-22 |
AGTTGTTAGTCTACGTGGACCG |
1054 |
| R1 |
1036-1054 |
CGTGGGCTTGTTCTTAGCC |
|
| F2 |
857-877 |
GCCCATTACATAGGCACTTCC |
1199 |
| R2 |
2036-2055 |
CTTTCCCCAAAAGGAGGTTC |
|
| F3 |
1946-1965 |
GAGGATGGACAAGGGAAAGC |
1403 |
| R3 |
3326-3348 |
CACCATTCGTGTATCAACTTCCC |
|
| F4 |
3224-3245 |
GGAAAATTGGAGCTGGACTTCA |
1001 |
| R4 |
4206-4224 |
GCCACTAATGGTCCAGCCA |
|
| F5 |
4112-4131 |
CTCAAAAGGAGAAGCTGGCC |
1208 |
| R5 |
5299-5319 |
CGCATTGTGAACGTTGCGTG |
|
| F6 |
5252-5274 |
GCAACAAAATCTGAACACACAGG |
1071 |
| R6 |
6301-6321 |
GAATAAGTGCGGGCATCAAGC |
|
| F7 |
6059-6077 |
AAGTCAGCCGCCATAGACG |
1389 |
| R7 |
7428-7447 |
AGGTGATCCTTCCCAGAGTG |
|
| F8 |
7032-7049 |
AGGCAGTGGTCCTGATGG |
1143 |
| R8 |
8153-8174 |
GCATTCCTCCATGTTTCCTTTG |
|
| F9 |
8015-8032 |
TCACCAAGCCCAACAGTG |
1528 |
| R9 |
9520-9542 |
TGGCCATTCTTTTTAACCTCTCC |
|
| F10 |
9270-9293 |
AGCTCACATACCAAAACAAAGTGG |
1060 |
| R10 |
10299-10318 |
AGCTTCTTCCGTACTGTGGC |
|
| F11b |
10118-10137 |
GGTCTCACTTCCAGAGCAAC |
590 |
| R11 |
10674-10696 |
AGAACCTGTTGATTCAACAGCAC |
|
| 5'RACE outer primer |
661-678 |
TTGCACGTTCCATAGGTC |
|
| 5'RACE inner primer | 267-284 | CTGCTGTTGGTGGAATGG |
a Primer location refers to DENV-3 strain 80–2 [GenBank: AF317645].
b Primer F11 was also used as 3’RACE primer.
Sequence alignment and phylogenetic analysis
The complete nucleotide sequences of the complete sequences of coding region or envelope (E) gene of global DENV-3 strains were retrieved from GenBank (Table 2). Multiple sequence alignment was carried out employing the CLUSTAL W program [22]. Phylogenetic analyses based on the nucleotide sequence of complete coding region of 44 DENV-3 or complete envelope gene of 58 DENV-3 were carried out by Neighbor-Joining method using MEGA version 5.05 or by Bayesian method using BEAST version 1.7.1 [23,24]. The Neighbor-Joining trees were constructed by Tamura-Nei model with gamma-distribution of among-site [25]. The Bayesian trees were inferred by Markov Chain Monte Carlo (MCMC) for 5,000,000 generations, sampling at every 100 the generations. Sequences of the DENV-1 strain WestPac [GenBank: U88535], DENV-2 strain NGC [GenBank: AF038403] and DENV-4 strain B5 [GenBank: AF289029] were used as outgroups.
Table 2.
DENV-3 isolates investigated in this study
| Isolate | Year of isolation | Geographical origin | GenBank Accession No. | Genotype |
|---|---|---|---|---|
| GZ1D3 |
2009 |
China: Guangdong |
In this study |
III |
| GZ2D3 |
2009 |
China: Guangdong |
In this study |
III |
| 09/GZ/11144 |
2009 |
China: Guangdong |
HM466966 (E) |
III |
| 09/GZ/11194 |
2009 |
China: Guangdong |
HM466967 (E) |
III |
| 09/GZ/13105 |
2009 |
China: Guangdong |
HM466968 (E) |
III |
| 09/GZ/10616 |
2009 |
China: Guangdong |
HM466964 (E) |
III |
| Zhejiang/08/09 |
2009 |
China: Zhejiang |
GU721065 |
III |
| Zhejiang/15/09 |
2009 |
China: Zhejiang |
GU721066 |
III |
| Zhejiang/17/09 |
2009 |
China: Zhejiang |
GU721067 |
III |
| Zhejiang/27/09 |
2009 |
China: Zhejiang |
GU721068 |
III |
| Zhejiang/31/09 |
2009 |
China: Zhejiang |
GU721069 |
III |
| ZJYW2009 |
2009 |
China: Zhejiang |
JF504679 |
III |
| 07CHLS001 |
2007 |
China: Zhejiang |
EU367962 |
II |
| DTID-ZJU04 |
2009 |
China: Zhejiang |
GU189648 |
II |
| 09/GZ/1081 |
2009 |
China: Guangdong |
HM466962 (E) |
V |
| 09/GZ/1483 |
2009 |
China: Guangdong |
HM466963 (E) |
V |
| 09/GZ/10806 |
2009 |
China: Guangdong |
HM466965 (E) |
V |
| 80-2 |
1980 |
China: Guangxi |
AF317645 |
V |
| ND143 |
2007 |
India |
FJ644564 |
III |
| DEL-72 |
2008 |
India |
GQ466079 |
III |
| DENV-3/LK/BID-V2405 |
1983 |
Sri Lanka |
GQ199887 |
III |
| DENV-3/LK/BID-V2409 |
1997 |
Sri Lanka |
GQ252674 |
III |
| DENV-3/MX/BID-V2989 |
2007 |
Mexico |
FJ898442 |
III |
| DENV-3/US/BID-V2103 |
2000 |
USA |
FJ547071 |
III |
| DENV-3/US/BID-V1080 |
2006 |
USA |
EU529699 |
III |
| DENV-3/US/BID-V1620 |
2005 |
USA |
FJ182010 |
III |
| DENV-3/US/BID-V1090 |
1998 |
USA |
EU529703 |
III |
| DENV-3/US/BID-V1043 |
2006 |
USA |
EU482555 |
III |
| DENV-3/VE/BID-V2179 |
2000 |
Venezuela |
FJ639750 |
III |
| DENV-3/VE/BID-V2481 |
2007 |
Venezuela |
GQ868586 |
III |
| DENV-3/NI/BID-V2419 |
1998 |
Nicaragua |
GQ199886 |
III |
| DENV-3/NI/BID-V2420 |
1994 |
Nicaragua |
FJ882576 |
III |
| DENV-3/NI/BID-V2649 |
2008 |
Nicaragua |
FJ873813 |
III |
| DENV-3/NI/BID-V4761 |
2009 |
Nicaragua |
HM181972 |
III |
| DENV-3/NI/BID-V3055 |
2008 |
Nicaragua |
GQ199860 |
III |
| DENV-3/NI/BID-V5498 |
2010 |
Nicaragua |
JF937633 |
III |
| DENV-3/LC/BID-V3929 |
2001 |
Saint Lucia |
GQ868616 |
III |
| DENV-3/BR/BID-V2400 |
2007 |
Brazil |
FJ850092 |
III |
| DENV-3/BR/BID-V2977 |
2001 |
Brazil |
FJ898446 |
III |
| DENV-3/BR/BID-V3606 |
2007 |
Brazil |
GU131876 |
III |
| DENV-3/IPC/BID-V3832 |
2007 |
Cambodia |
GU131917 |
II |
| DENV-3/KH/BID-V2082 |
2003 |
Cambodia |
FJ639725 |
II |
| DENV-3/IPC/BID-V3820 |
2006 |
Cambodia |
GU131908 |
II |
| DENV-3/IPC/BID-V4286 |
2007 |
Cambodia |
GU131937 |
II |
| DENV-3/KH/BID-V2080 |
2003 |
Cambodia |
FJ639723 |
II |
| DENV-3/VN/BID-V1013 |
2006 |
Viet Nam |
EU482457 |
II |
| DENV-3/VN/BID-V1911 |
2008 |
Viet Nam |
FJ547066 |
II |
| C0360/94 |
1994 |
Thailand |
AY923865 |
II |
| C0331/94 |
1994 |
Thailand |
AY876494 |
II |
| DENV-3/TH/BID-V2315 |
2001 |
Thailand |
FJ744729 |
II |
| DENV-3/TH/BID-V3360 |
1973 |
Thailand |
GQ868593 |
II |
| PF90/3050 |
1990 |
French Polynesia |
AY744679 |
I |
| PF92/4190 |
1992 |
French Polynesia |
AY744684 |
I |
| den3-88 |
1988 |
Indonesia |
AY858038 |
I |
| FW01 |
2004 |
Indonesia |
AY858040 |
I |
| H87 |
1956 |
Philippines |
M93130 |
V |
| BS-PRico63 |
1963 |
Puerto Rico |
AY146762 |
IV |
| 1339 | 1977 | Puerto Rico | AY146761 | IV |
Results
All three family members were diagnosed as DF according to the new guideline of World Health Organization [26]. Laboratory tests disclosed low WBC and lymphocytes counts for all the three cases. Normal platelet counts were recorded for two cases, while that of the mother was low. None of the patients presented plasma leakage, severe bleeding, or severe organ involvement. All cases recovered in a week post admission.
The acute phase sera from all the three family members were positive for dengue IgM antibody, but negative for IgG antibody. Two of the three cases were positive for DENV-specific RT-PCR. DNA sequencing of the PCR products and blast analysis revealed closely homologous with DENV-3. Considering the fact that DENV-3 has not been described in Guangdong for many years, all the three samples were inoculated into C6/36 cells to isolate the viruses. Typical CPE were observed six or seven days post inoculation for two of three samples. After another passage in C6/36 cells, two strains were isolated and named with GZ1D3 and GZ2D3, respectively. Both strains were further confirmed by IFA using dengue specific monoclonal antibody.
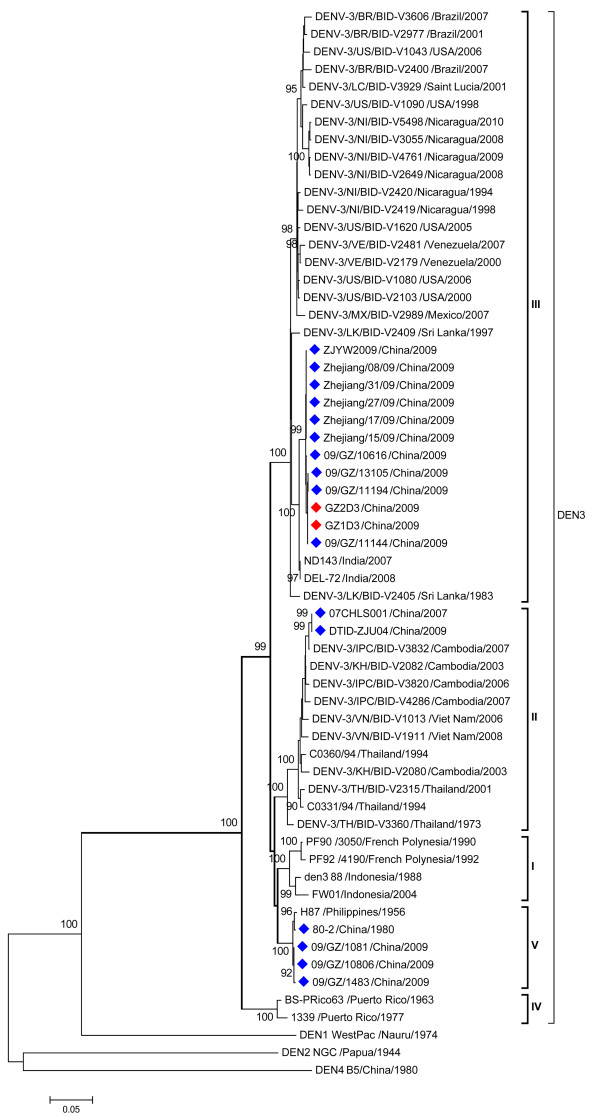
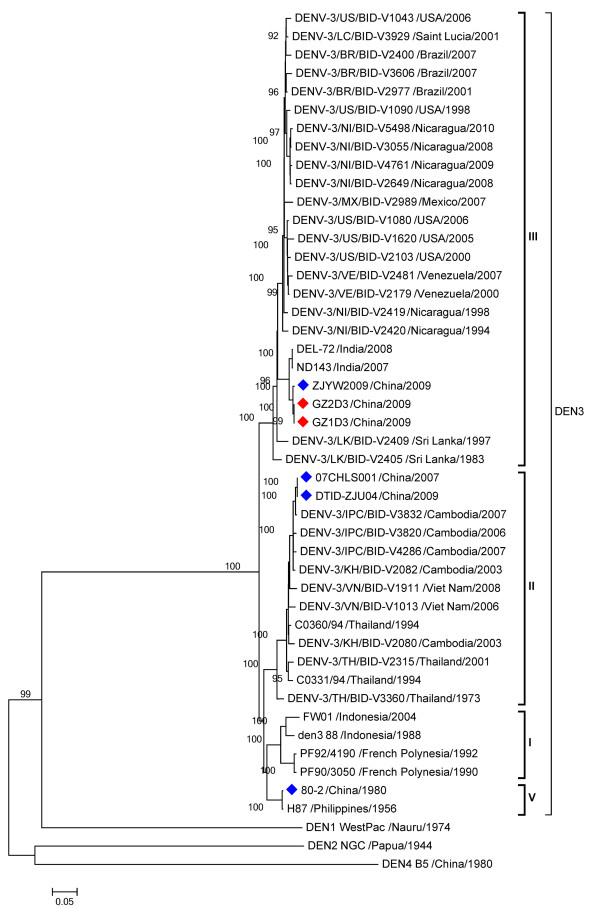
Finally, the complete genome sequences of the isolates were determined, assembled and submitted to GenBank [GenBank: GU363549; JN662391]. Both strains were highly homologous (99.9%) with only three nucleotide differences. Phylogenetic analysis based on the complete envelope gene classified DENV-3 isolates into five genotypes (Figure 1), which was confirmed by the Bayesian method (Additional file 1: Figure S1). Phylogenetic tree based on the complete sequence of coding region of DENV-3 genome showed the same genotype classification (Figure 2). The newly isolated DENV-3 strains belong to the genotype III, clustering with other DENV-3 isolates circulating in China in 2009 and in India in 2007 and 2008 (Figure 1). Interestingly, three additional DENV-3 strains isolated in Guangzhou in 2009 (09/GZ/1081, 09GZ/1483 and 09/GZ/10806) [27] belong to genotype V (Figure 1 & Additional file 1: Figure S1), which indicated that two genotypes of DENV-3 were co-circulating in Guangdong, 2009.
Figure 1.
Phylogenetic tree based on the complete envelope gene from 58 DENV-3 strains sampled globally. The evolutionary history was inferred using the neighbor-joining method with MEGA 5 software [24]. Each strain is abbreviated with strain name and country of origin followed by the year of isolation. Bootstrap values greater than 0.9 based on 1000 replicates are shown for key nodes. The tree was rooted using DENV-1 strain Nauru, DENV-2 strain New Guinea C and DENV-4 strain B5 as outgroups. The newly DENV-3 isolates in the study are marked with red squares and other Chinese DENV-3 isolates taken for comparison are marked with blue squares.
Figure 2.
Phylogenetic tree based on complete sequences of coding region of viral genome from 44 DENV-3 strains sampled globally. The evolutionary history was inferred using the neighbor-joining method with MEGA 5 software [24]. Bootstrap values greater than 0.9 based on 1000 replicates are shown for key nodes. The tree was rooted using DENV-1 strain Nauru, DENV-2 strain New Guinea C and DENV-4 strain B5 as outgroups. The newly DENV-3 isolates in the study are marked with red squares and other Chinese DENV-3 isolates taken for comparison are marked with blue squares.
Discussion
In the present study, a family cluster of DENV-3 infections in Guangzhou, China was described. Three family members were diagnosed as DF, and all recovered finally. All the three family members recalled mosquito biting before illness, and none of them went aboard or on trip recently. Although family cluster of vector-borne diseases have been intensively described [28,29], information regarding DF family cluster is limited. There is no doubt that any cluster of cases is of great concern and should be thoroughly investigated. Dengue can cause both the large epidemics and sporadic infections. The recognition of clustering of disease is important for medical providers and public health personnel in treating and controlling the disease, because multiple infections can occur simultaneously or following an index case. In this study, strict mosquito control measures were initiated immediately after confirmation of the DF cases, and no further cases were reported nearby thereafter.
Whether dengue is endemic in Guangdong remains disputable. Most dengue epidemics in Guangdong were initiated by the introduction of virus from imported cases [13,18,19,27,30], however, in this study none of the family member travelled aboard. Further epidemiology investigation also did not identify imported case nearby either. The origin of these DENV-3 isolate is really interesting. Since DENV-1 has circulated in Guangzhou for about ten years, the new DENV-3 has potential to increase the rate of secondary heterotypic infection. Furthermore, the previous studies showed that epidemic DHF has appeared in association with DENV-3 [31-33]. In the Americas, DENV-3 presented greater epidemic potential and virulence [20]. Whatever, the emerging DENV-3 in Guangzhou might represent a risk factor for severe dengue illness, careful investigation and surveillance should be warranted in the future.
Most importantly, phylogenetic analysis demonstrated that at least two different genotypes of DENV-3 were co-circulated in Guangdong, China in 2009, which partly agree with the findings of the previous study [27]. Five genotypes of DENV-3 have been reported [20,34]. The newly isolates in Guangdong form a distinct cluster with other Chinese isolates sampled from Zhejiang province in 2009 [35]. All these genotype III DENV-3 strains were closely related to those sampled in India in 2007 and 2008, suggesting these Chinese isolates might be imported from India. Previously, the introduction of new genotype III of DENV-3 has been recognized as one of the factors leading to the emergence of DHF in Pakistan and India [33,36]. These strains are therefore interesting and their virological characterization and virulence analyses are currently underway.
However, three additional DENV-3 strains (09/GZ/1081, 09GZ/1483, and 09/GZ/10806), belonging to genotype V, were also identified in Guangzhou, 2009 in a separate study [27]. In addition, two strains (07CSHL001 and DTID-ZJU04) sampled from China, were grouped within genotype II. The origin of these isolates is difficult to determine without further information. The situation that multiple genotypes of DENV-3 co-circulated in Guangzhou, China deserves close concern and careful investigation.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TJ, XDY, CFQ and FCZ: designed the study, did laboratory testing, analyzed the test results. TJ, EDQ and CFQ co-wrote and edited the manuscript. WXH and WZZ participated in the gene sequencing and phylogenetic analysis. MY, YQD, SYZ, EDQ and JW took samples and did laboratory testing and virus isolation. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Phylogenetic tree based on the complete envelope gene from 58 DENV-3 strains by Bayesian method. The evolutionary history was inferred using BEAST 1.7.1 software. The tree was rooted using DENV-1 strain Nauru, DENV-2 strain New Guinea C and DENV-4 strain B5 as outgroups. The newly described DENV-3 isolates in the study are marked with red squares and other Chinese DENV-3 isolates taken for comparison are marked with blue squares. (TIFF 1759 kb)
Contributor Information
Tao Jiang, Email: jiang_tao@126.com.
Xue-Dong Yu, Email: xuedongyu@163.com.
Wen-Xin Hong, Email: winsonhong@sina.com.
Wei-Ze Zhou, Email: weizezhou@163.com.
Man Yu, Email: yu_man@sohu.com.
Yong-Qiang Deng, Email: dengyq1977@yahoo.com.cn.
Shun-Ya Zhu, Email: zhu_duck@163.com.
E-De Qin, Email: qinede@sohu.com.
Jian Wang, Email: wj87905@163.com.
Cheng-Feng Qin, Email: qincf@bmi.ac.cn.
Fu-Chun Zhang, Email: zfc8y@yahoo.com.cn.
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (No.30972613 and No.81101243), the National 973 Project of China (No.2012CB518904) and the 39th Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry. CFQ was supported by Beijing Nova Program of Science and Technology (No.2010B041).
References
- Impact of Dengue. http://www.who.int/csr/disease/dengue/impact/en/
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried JR, Gibbons RV, Kalayanarooj S, Thomas SJ, Srikiatkhachorn A, Yoon IK, Jarman RG, Green S, Rothman AL, Cummings DA. Serotype-specific differences in the risk of dengue hemorrhagic fever: an analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoSNegl Trop Dis. 2010;4:e617. doi: 10.1371/journal.pntd.0000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Saito M, Oishi K, Inoue S, Dimaano EM, Alera MT, Robles AM, Estrella BD, Kumatori A, Moji K, Alonzo MT. et al. Association of increased platelet-associated immunoglobulins with thrombocytopenia and the severity of disease in secondary dengue virus infections. ClinExpImmunol. 2004;138:299–303. doi: 10.1111/j.1365-2249.2004.02626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S. et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy TP, Nisalak A, Chunsuttitwat S, Vaughn DW, Green S, Ennis FA, Rothman AL, Libraty DH. Relationship of preexisting dengue virus (DV) neutralizing antibody levels to viremia and severity of disease in a prospective cohort study of DV infection in Thailand. J Infect Dis. 2004;189:990–1000. doi: 10.1086/382280. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Engle RE, St Claire M, Purcell RH, Lai CJ. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. ProcNatlAcadSci U S A. 2007;104:9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Lun ZR, James AA, Chen XG. Dengue Fever in mainland China. AmJTrop Med Hyg. 2010;83:664–671. doi: 10.4269/ajtmh.2010.09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Bai Z, Zhou H, Tu Z, Fang M, Tang B, Liu J, Liu L, Chen W. Molecular epidemiology of dengue viruses in southern China from 1978 to 2006. Virol J. 2011;8:322. doi: 10.1186/1743-422X-8-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu FX, Gubler DJ, Liu JC, Chen QQ. Dengue in China: a clinical review. Bull World Health Organ. 1993;71:349–359. [PMC free article] [PubMed] [Google Scholar]
- Li FS, Yang FR, Song JC, Gao H, Tang JQ, Zou CH, Hu BN, Wen SR, Qiu FX. Etiologic and serologic investigations of the 1980 epidemic of dengue fever on Hainan Island, China. AmJTrop Med Hyg. 1986;35:1051–1054. doi: 10.4269/ajtmh.1986.35.1051. [DOI] [PubMed] [Google Scholar]
- He JF, Luo HM, Liang WJ, Zheng K, Min K, Liu LP. Epidemic situation of dengue fever outbreak in Guangdong province, China, 1990–2005. Dengue Bull. 2007;31:1–9. [Google Scholar]
- Wang Q, Xu Z, Dou FM, Zhou H, Wang XF, Yin WW, Li Q. Current situation and surveillance on dengue fever in China, 2005–2007. Zhonghua Liu Xing Bing XueZaZhi. 2009;30:802–806. [PubMed] [Google Scholar]
- Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, Kabra SK, Broor S. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi. India. Virol J. 2008;5:1. doi: 10.1186/1743-422X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Tio PH, Perera D, Muhi J, Cardosa J. Importation and co-circulation of multiple serotypes of dengue virus in Sarawak, Malaysia. Virus Res. 2009;143:1–5. doi: 10.1016/j.virusres.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Lorono-Pino MA, Cropp CB, Farfan JA, Vorndam AV, Rodriguez-Angulo EM, Rosado-Paredes EP, Flores-Flores LF, Beaty BJ, Gubler DJ. Common occurrence of concurrent infections by multiple dengue virus serotypes. AmJTrop Med Hyg. 1999;61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- Chen S. The origin of dengue viruses caused the DF outbreak in Guangdong province, China, in 2006. Infect Genet Evol. 2011;11:1183–1187. doi: 10.1016/j.meegid.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Zheng K, Zhou HQ, Yan J, Ke CW, Maeda A, Maeda J, Takashima I, Kurane I, Ma H, Xie XM. Molecular characterization of the E gene of dengue virus type 1 isolated in Guangdong province, China, in 2006. Epidemiol Infect. 2009;137:73–78. doi: 10.1017/S0950268808000617. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Lewis JG, Gubler DJ, Trent DW. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75(Pt 1):65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Deng YQ, Dai JX, Ji GH, Jiang T, Wang HJ, Yang HO, Tan WL, Liu R, Yu M, Ge BX. et al. A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS One. 2011;6:e16059. doi: 10.1371/journal.pone.0016059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC EvolBiol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. MolBiolEvol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. MolBiolEvol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- WHO. Dengue - Guidelines for diagnosis, treatment, prevention and control. 2009. http://www.who.int/entity/tdr/publications/documents/dengue-diagnosis.pdf. [PubMed]
- Di B, Bai ZJ, Wang YL, Luo L, Chen Y, Jiang LY, Yang ZC, Wang M. Molecular epidemiologic analysis on new emerged type 3 dengue virus in Guangzhou in 2009. Zhonghua Liu Xing Bing XueZaZhi. 2010;31:804–807. [PubMed] [Google Scholar]
- Papa A, Bino S, Llagami A, Brahimaj B, Papadimitriou E, Pavlidou V, Velo E, Cahani G, Hajdini M, Pilaca A. et al. Crimean-Congo hemorrhagic fever in Albania, 2001. Eur J ClinMicrobiol Infect Dis. 2002;21:603–606. doi: 10.1007/s10096-002-0770-9. [DOI] [PubMed] [Google Scholar]
- Jones TF, Craig AS, Paddock CD, McKechnie DB, Childs JE, Zaki SR, Schaffner W. Family cluster of Rocky Mountain spotted fever. Clin Infect Dis. 1999;28:853–859. doi: 10.1086/515213. [DOI] [PubMed] [Google Scholar]
- Yang F, Ma SQ, He JF, Mai ZJ, Liang WJ, Cai MX, Luo HM. Epidemiological analysis of imported cases of dengue fever in Guangdong province and Hong Kong during 2004–2006 in China. Zhonghua Liu Xing Bing XueZaZhi. 2009;30:42–44. [PubMed] [Google Scholar]
- Briseno-Garcia B, Gomez-Dantes H, Argott-Ramirez E, Montesano R, Vazquez-Martinez AL, Ibanez-Bernal S, Madrigal-Ayala G, Ruiz-Matus C, Flisser A, Tapia-Conyer R. Potential risk for dengue hemorrhagic fever: the isolation of serotype dengue-3 in Mexico. Emerg Infect Dis. 1996;2:133–135. doi: 10.3201/eid0202.960210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–809. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Dash PK, Agarwal S, Shukla J, Parida MM, Rao PV. Comparative complete genome analysis of dengue virus type 3 circulating in India between 2003 and 2008. J Gen Virol. 2011;92:1595–1600. doi: 10.1099/vir.0.030437-0. [DOI] [PubMed] [Google Scholar]
- Wittke V, Robb TE, Thu HM, Nisalak A, Nimmannitya S, Kalayanrooj S, Vaughn DW, Endy TP, Holmes EC, Aaskov JG. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301:148–156. doi: 10.1006/viro.2002.1549. [DOI] [PubMed] [Google Scholar]
- Sun J, Lin J, Yan J, Fan W, Lu L, Lv H, Hou J, Ling F, Fu T, Chen Z. et al. Dengue virus serotype 3 subtype III, Zhejiang Province, China. Emerg Infect Dis. 2011;17:321–323. doi: 10.3201/eid1702.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E, Hasan R, Mehraj V, Nasir A, Siddiqui J, Hewson R. Co-circulations of two genotypes of dengue virus in 2006 out-break of dengue hemorrhagic fever in Karachi, Pakistan. J ClinVirol. 2008;43:176–179. doi: 10.1016/j.jcv.2008.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree based on the complete envelope gene from 58 DENV-3 strains by Bayesian method. The evolutionary history was inferred using BEAST 1.7.1 software. The tree was rooted using DENV-1 strain Nauru, DENV-2 strain New Guinea C and DENV-4 strain B5 as outgroups. The newly described DENV-3 isolates in the study are marked with red squares and other Chinese DENV-3 isolates taken for comparison are marked with blue squares. (TIFF 1759 kb)