Abstract
Miniature inverted-repeat transposable elements (MITEs) are ubiquitous in high eukaryotic genomes. More than 178,000 MITE sequences of 338 families are present in the genome of rice (Oryza sativa) cultivar Nipponbare. Interestingly, only two of the 338 MITE families have homologous sequences in the genome of Brachypodium distachyon, a relative in the grass family. Therefore, the vast majority of MITEs in the rice genome were originated and amplified after the divergence of Oryza and Brachypodium. Comparison between rice cultivar Nipponbare and another rice cultivar 93–11 showed 14.8% of MITEs exhibit presence/absence (P/A) polymorphism. The P/A polymorphism was mainly attributed to recent MITE transpositions, while less than 10% of the P/A polymorphism was caused by MITE excisions. Therefore, the high P/A polymorphisms of MITEs may generate considerable gene expression and phenotypic diversity for O. sativa.
Keywords: amplification, diversity, excision, origin, transposition
Introduction
MITEs are short, non-autonomous transposons that are widespread in plant and animal genomes.1 MITE sequences may vary dramatically such that it is difficult to identify new MITE families using homology-based approach (see below). Instead, structural features such as terminal inverted repeat (TIR) flanked by target site duplication (TSD) have been used to identify novel MITE families.2,3 There had been limited success in de novo identification of MITEs until the recent development of programs, MITE-hunter and RSPB.4,5 Compared with previous programs, these two have considerably improved the efficiency and reliability on de novo identification of MITEs. Both programs could detect most MITE families in a genome. MITE-hunter is better in identifying MITE families with few copies in a database, while RSPB is better in detecting MITE families with unusual TIR and TSD sequences (such as short TIR sequences). The combination of MITE-hunter and RSPB is predicted to identify a vast majority of, if not all, MITE families in a genome, with no prior information required. The two programs can be applied to the dozens of eukaryotic genomes that have been sequenced, and comparative analysis of MITEs in multiple species in future will further our understanding of MITEs’ origin, amplification and roles on gene and genome evolution.
The programs MITE-hunter and RSPB detected a total of 178,000 MITE sequences in the genome of rice cultivar Nipponbare (subspecies japonica), which occupy approximately 8.6% of the rice genome. The MITE sequences in the rice genome were classified into 338 families. The data from our recent study showed that the large number of MITE sequences in the rice genome is due to the presence of many large MITE families, which were generated by amplification bursts.5 However, little is known on how and when these MITE families were originated and amplified.
The MITEs in rice genome are preferentially distributed on chromosome arms. However, further analysis showed that the MITEs are randomly distributed on chromosome arms and not preferentially associated with genes. The above observation is inconsistent with conclusions drew from previous studies.6,7 The currently active MITE family mPing in rice has been experimentally shown to be preferentially transposed to upstream of genes.8 Such preferred transposition is more likely due to mechanisms such as chromosome structure rather than selection, since transposition was investigated in a generation in which selection had not taken effects. However, selections might have played important roles in the locations of other MITEs, since MITEs can regulate the expression of their nearby genes. It is worth noting that MITEs can be selected for if inserted near some genes, while they may be selected against if inserted near other genes. Therefore, the overall effects of selection on MITE insertions may be neutralized and consequently MITEs seem to be distributed randomly on chromosome arms, as shown in rice.5
MITEs play important roles in gene expression, either upregulating or downregulating gene expression. It was reported that some MITEs can upregulate gene expression. For example, the insertion of a Kiddo MITE element into the promoter of rice gene Ubiquitin2, increased its expression by 20%.9 The insertions of mPing MITE family into the upstream of rice genes had a modest effect on gene transcription.8 In our previous study, we compared the expression of all genes with MITE insertion in one rice cultivar but absent in the other, and found no significant difference between them. However, genome-wide comparison clearly showed that genes with MITE insertions in a genome have significantly lower expression than genes with no MITE insertions.
The downregulation of gene expression by MITEs was postulated to be through the small RNA pathway since MITE-derived small RNAs are frequently present in plant genomes.10 Approximately one quarter of the small RNAs obtained from rice are generated by MITE sequences.5 The MITE-derived small RNA may be generated through natural sense/antisense transcripts.5,10 Alternatively, they can be generated through single-stranded stem-loop precursors, which are transcribed from the TIR sequences of MITEs.11
In this study, we studied the origin of rice MITE families by BLAST search of the genome of B. distachyon, a relative of rice in the grass family. Additionally, the diversity derived from MITE insertions/excisions in rice were investigated.
Results and Discussion
Most MITE families in rice originated and amplified after the divergence of Oryza and Brachypodium
To investigate the origin and amplification of MITEs, the seed sequences of the 338 rice MITE families were used as query in BLASTN search of the genome of B. distachyon, a relative of rice in the grass family. Of the 338 rice MITE families, 20 families have hits with e-values of e−10-e−47. The high scoring pairs (HSPs) for all but two families are most likely false positives since the corresponding sequences in B. distachyon have low copy and are not from MITE sequences. Alternatively, these detected sequences in B. distachyon might be part of decayed MITEs. Nevertheless, most (318) of the 338 MITE families have no significant hits in the B. distachyon genome (e−10). We conclude that the vast majority of extant MITE families in the rice genome were originated and amplified after the divergence of Oryza and Brachypodium. This conclusion is consistent with the estimated amplification time of some MITE families (~8–20 mya) and the divergence time between Oryza and Brachypodium (~50 mya).5,12 MITEs are widely distributed in eukaryotic genomes, and it is reasonable to believe that MITEs were abundantly present in the common ancestor of Oryza and Brachypodium. The ancient MITEs might have diverged dramatically and become part of the genome sequences of modern species that have no obvious reminiscence of ancient MITEs.
MITE family OsP3 and OsP20 originated before the divergence of Oryza and Brachypodium
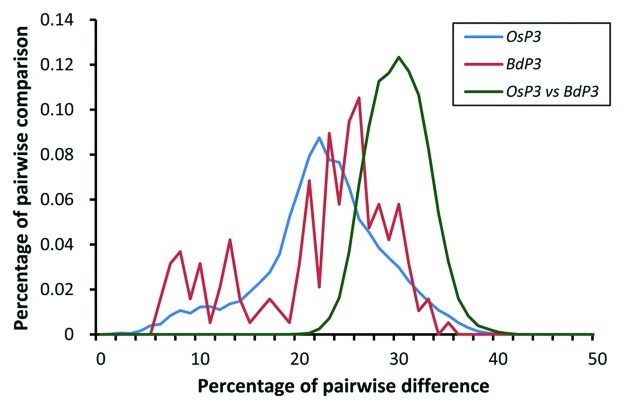
Two of the 338 rice MITE families (OsP3 and OsP20) exhibit significant homology with B. distachyon sequences that are also part of MITE. OsP20 has only a few diverse elements in the rice and B. distachyon genomes and were not analyzed further. The OsP3 family has 928 members in the rice genome and its corresponding MITE family in B. distachyon has 20 elements. Their pairwise nucleotide diversities were calculated and histograms were drawn (Fig. 1). The histogram for nucleotide diversity in the OsP3 family is a wave-like curve with unimodal distribution, suggesting that the OsP3 family has experienced one round of amplification burst.5 The amplification burst was estimated to be approximately 17–34 mya, after the divergence of the two species.13,14 The pairwise nucleotide diversities for the MITE family in B. distachyon vary considerably and their histogram does not show a wave-like curve, probably due to small sampling size. The average pairwise nucleotide diversities between OsP3 and the corresponding MITE family in B. distachyon is 0.31, suggesting a divergence time of 24–48 mya; and we cannot exclude the possibility of origin of OsP3 before the divergence of Oryza and Brachypodium (~50 mya).
Figure 1. Histograms of nucleotide diversities between MITEs. The wave-like curve for elements from the rice family OsP3 indicates that it has experienced one round of amplification burst, likely after the divergence of Oryza and Brachypodium. The pairwise nucleotide diversities between members from different species may reflect the divergence time between Oryza and Brachypodium.
To illustrate the phylogenetic relationship of the two MITE families, a phylogenetic tree was constructed for all full-length elements (Fig. 2). The MITE sequences from B. distachyon formed one clade and the rice MITE family OsP3 forms several clades. Interestingly, the Brachypodium clade is very close to one of the rice clade. The most similar sequences between the two species are from the two closely related clades. The most similar pair exhibits 70% nucleotide identity, which is higher than the nucleotide identities between some members within a species. Above data suggest that the two MITE families were originated from the same ancient family, though they have high sequence divergence and vary dramatically in copy number in the two species.
Figure 2. Phylogenetic tree for the MITE family OsP3 in rice and the corresponding MITE family in B. distachyon. All members from B. distachyon are grouped into one clade (BdP3). The rice OsP3 family are grouped into several tight clades (all except the BdP3 clade, and only OsP3-a is marked), indicating amplifications after the divergence of Oryza and Brachypodium. The close relationship of the B. distachyon clade (BdP3) and one rice clade (OsP3-a) suggests that they were originated before the divergence of the two species. The most similar pair between rice and B. distachyon is indicated by lines with arrowhead.
Many MITE loci are fixed in O. sativa
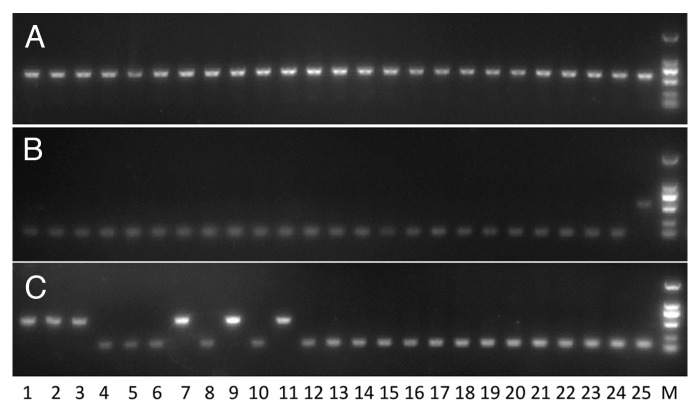
The large amount of MITE-derived small RNAs and the detectable effects of MITEs on gene expressions suggest that MITEs may play important roles in functional and phenotypic diversity of a species if different genotypes have different MITE loci. To investigate the potential roles of MITEs on species diversity, we studied the insertional polymorphism of MITEs between rice cultivars Nipponbare and 93–11 (subspecies indica). Comparison between the two cultivars showed that the vast majority (85.3%) of MITE loci are shared between the two rice cultivars. Eight MITE loci, which contain MITEs in both cultivars Nipponbare and 93–11, were randomly chosen to screen a panel of 25 rice cultivars (11 indica and 14 japonica) representing the entire species. PCR reactions showed that these eight loci have MITE insertions in all 25 rice cultivars (Fig. 3A). Therefore, most MITE insertions are predicted to be fixed in O. sativa.
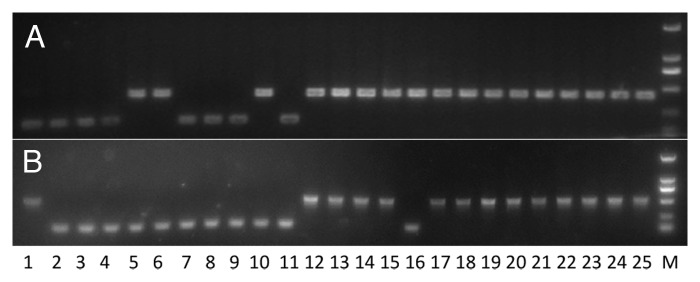
Figure 3. Polymorphisms generated by MITE insertions. PCR products were amplified using primers flanking a MITE locus. Larger fragments show presence of a MITE, while shorter fragments show absence of a MITE at the locus. The presence locus and the absence locus are identical except that the presence locus has a full-length MITE and a TSD sequence. (A) MITE locus fixed in O. sativa. (B) mPing locus present in japonica cultivars only. (C) mPing locus present in indica cultivars only. Lane 1–11 are indica cultivars (Wukezhan, Teqing, Zhongzao 18, Yanshuichi, H94, Heimichan, Nanjing 11, Minghui 63, Zhenshan 97, KDML 105, 93–11). Lane 12–25 are japonica cultivars (Laohuzhong, Shennong 606, Wuyujing-3, Qiuguang, Zidao, Muxiqiu, Zhonghua 11, Balilla, 58N, Mudanjiang 8, Lement, SLG, IRAT 109, Nipponbare). M, marker.
Most mPing loci were generated after the divergence of indica and japonica subspecies
To further study the origin time for polymorphic MITEs, ten polymorphic MITE loci were randomly chosen from different MITE families were randomly chosen from the mPing family to investigate their origin time. PCR screening of the 25 selected rice cultivars showed that the MITE insertion at nine of the ten loci occurred before the divergence of the two rice subspecies while the insertion at the 10th locus occurred after the divergence. Another set of 16 MITE loci were randomly chosen from the currently active mPing family to investigate their origin time. Nine of 16 mPing loci have MITE insertions in Nipponbare (japonica) but not in 93–11 (indica). PCR screening of the 25 rice cultivars showed that these nine loci have MITEs in 1–5 japonica cultivars but not in any indica cultivars (Fig. 3B). The other seven loci contain MITE insertions in 93–11 but not in Nipponbare. Similarly, these seven loci have MITE insertions in indica cultivars only (Fig. 3C). Therefore, most mPing loci were generated after the divergence of the two subspecies.
The presence/absence polymorphisms were mainly generated by MITE insertions rather than excisions
A total of 7,458 loci show MITE presence/absence polymorphism between rice cultivars Nipponbare and 93–11. The polymorphic sites might have been generated through either insertions or excisions of MITEs. To distinguish between two possibilities, 366 loci with presence/absence polymorphism between the two rice cultivars were randomly chosen for further analysis. Sequence comparison showed that 92.6% loci flanking the insertion site are identical in the two cultivars except that one cultivar has an extra MITE sequence and an additional TSD sequence. Assuming no precise excision,15 above structure indicated that the polymorphisms were generated through MITE insertions. The other 7.4% loci showed sequence variation resembling to the footprints of transposons, suggesting MITE excisions (Fig. 4).
Figure 4. Footprint derived from the same excision event. A MITE is present in cultivar Nipponbare but absent in the other ten cultivars. Its TSD sequences (TA) are underlined and part of the TIR sequences (framed) are shown. The central part of the MITE is omitted as show by “….” Dash in the alignment represents missing nucleotide. The footprint includes an inclusion of 10 extra nucleotides (shaded) and deletion of 7 nucleotides (shaded). The first column is cultivar name: 931 for 93–11, Nan for Nanjing 11, Min for Minghui 63, Yan for Yanshuichi, Teq for Teqing, Wuk for Wukezhan, Hei for Heimichan, Zho for Zhongzao 18, Zhe for Zhenshan 97 and Nip for Nipponbare.
Five loci that have full-length MITEs in cultivar Nipponbare but have patterns of footprints in cultivar 93–11, were chosen to investigate the contribution of excisions to rice diversity. PCR screening of the panel of 25 rice cultivars showed that the excisions at three of the five loci occurred after the divergence of the two subspecies (Fig. 5A), while the excisions at the other two loci happened before the divergence (Fig. 5B). Sequencing 27 PCR products showed that the footprints at each locus are generally identical in different cultivars with one exception only, indicating that they were originated from the same excision event (Fig. 4). Therefore, excisions of MITEs were infrequent and contributed little to the diversity of O. sativa.
Figure 5. Polymorphisms generated by MITE excision. See Figure 3 for description of PCR fragments and cultivar names. All absence locus showed footprint patterns, suggesting excisions (see Figure 4). (A) Excision occurred after the divergence of indica and japonica. (B) Excision occurred before the divergence of indica and japonica.
Materials and Methods
Materials
A total of 25 rice cultivars were included in this study. Besides rice cultivars Nipponbare and 93–11, which have been sequenced, another 10 rice indica cultivars (Wukezhan, Teqing, Zhongzao 18, Yanshuichi, H94, Heimichan, Nanjing 11, Minghui 63, Zhenshan 97 and KDML 105) and 13 japonica cultivars (Laohuzhong, Shennong 606, Wuyujing-3, Qiuguang, Zidao, Muxiqiu, Zhonghua 11, Balilla, 58N, Mudanjiang 8, Lement, SLG and IRAT 109) were randomly chosen from the core collections of rice germplasm to represent the species of Oryza sativa.
The genome sequences of Brachypodium distachyon were downloaded from (http://www.brachypodium.org/); the genome sequences of rice cultivar Nipponbare were downloaded from (http://rice.plantbiology.msu.edu/), and the genome sequences of rice cultivar 93–11 were downloaded from (http://rice.genomics.org.cn).
Sequence analysis
Sequence analysis follows the methods of Lu et al.5 MITE sequences of the same family were aligned using MUSCLE v3.8,16 and distance trees were constructed using MEGA 4.17 A Perl script was used to calculate pairwise nucleotide diversity among members of a MITE family and to construct its histogram. Substitution rate of 6.5 × 10−9-1.3 × 10−8 base substitutions/site/year was used to estimate the divergence time between two sequences.13,14
MITEs tansposition, excision of and presence/absence polymorphism
To understand the mechanism of the presence/absence polymorphism of a MITE sequence, 366 loci with MITE insertion in one of the two cultivars Nipponbare or 93–11 but not in both were randomly chosen for manual comparison. A locus may show a strict related empty site pattern (RESite)18: the presence allele has an additional TSD sequence besides a full-length MITE when compared with the absence allele. In this case, the polymorphism is considered to be generated by transposition of a MITE. On the other hand, if a locus does not show perfect RESite, the absence allele is considered as footprint generated by excision of MITEs.
To investigate the presence/absence polymorphism of a MITE, PCR primers flanking MITEs were designed (Table 1) and were used to screen the panel of 25 rice cultivars. The PCR was a 25 μl volume reaction, containing 2 × Taq PCR Master (Zomanbio), 30 ng genomic DNA as a template, and 0.12 μM of forward and reverse primers. The PCR cycles are 94°C for 5 min, followed by 32 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 30s, and a final extension of 72°C for 10 min. The PCR products were treated with Exo-SAP (Fermentas) and then sequenced directly using ABI 3730xl DNA Analyzer following manufacturer’s instruction.
Table 1. Oligonucleotide primers used in this study.
| Combination no. | Primer name | Sequence (5′–3′) | Purpose |
|---|---|---|---|
| 1 |
01g01360-F 01g01360-R |
CGAAAAGTTGTGCTTAAAGTACG TGGGAGATGCCGAGGAAGTTGTT |
Diversity |
| 2 |
mPingNIP-2F mPingNIP-2R |
CATGTCATCAGCAGTTCTTGTC CTGAGAATGGCTTGGGCTAACA |
Diversity |
| 3 |
mPing9311-4F mPing9311-4R |
AGTGGGAAGTGATGAGGAGGAGA CCACCACTCCTCCCTCACCTACA |
Diversity |
| 4 |
T3bp_79F T3bp_79R |
GATACCACTGCTTCCCGACGCT GTGTTGATCCAACTTGGAATGC |
Diversity and footprinting |
| 5 |
T8bp_46F T8bp_46R |
CTACAACCATCTGATTGCAAC AATTGGATGTAGTAGCACCT |
Diversity and footprinting |
| 6 |
T10bp_3F T10bp_3R |
TCACAAGTTGAACTCCGTGAC CTATTGGGCTGGTCCGTGTAT |
Footprinting |
| 7 | Tstowaway37F Tstowaway37R |
ATCGATCGGCAATTCGGCATC ACGTACGGCTGCATATGCGAC |
Footprinting |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by China Transgenic grant no.2009ZX08009-045B, the National Natural Science Foundation of China grant no. 30921002, and China Postdoctoral Science Foundation grant no. 20100471195
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/20773
References
- 1.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–41. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 2.Tu Z. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2001;98:1699–704. doi: 10.1073/pnas.041593198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang G, Hall TC. MDM-1 and MDM-2: two mutator-derived MITE families in rice. J Mol Evol. 2003;56:255–64. doi: 10.1007/s00239-002-2397-y. [DOI] [PubMed] [Google Scholar]
- 4.Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29:1005–17. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau TE, Ronald PC, Wessler SR. A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc Natl Acad Sci U S A. 1996;93:8524–9. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, et al. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci U S A. 2006;103:17620–5. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–4. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 9.Yang G, Lee YH, Jiang Y, Shi X, Kertbundit S, Hall TC. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–68. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Zhang Y, Yang K, Sun Z, Fu Y, Chen X, et al. Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J. 2011;65:820–8. doi: 10.1111/j.1365-313X.2010.04467.x. [DOI] [PubMed] [Google Scholar]
- 12.Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci U S A. 1996;93:10274–9. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 2004;101:12404–10. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104:10962–7. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 18.Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–49. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]