Abstract
Gastrointestinal microbiomes play important roles in the health and nutrition of animals and humans. The medicinal leech, Hirudo verbana, serves as a powerful model for the study of microbial symbioses of the gut, due to its naturally limited microbiome compared with other popular models, the ability to cultivate the most abundant microbes, and genetically manipulate one of them, Aeromonas veronii. This review covers the relevance and application of leeches in modern medicine as well as recent discoveries detailing the nature of the gut microbiome. Additionally, the dual life-style of A. veronii allows one to do direct comparisons between colonization factors for beneficial and pathogenic associations, and relevant findings are detailed with respect to their role within the host and pathogenicity to other animals.
Keywords: Hirudo verbana, medicinal leech, Aeromonas, Rikenella, symbiosis, microbiome
Introduction
Members of the kingdom Animalia live in a beneficial symbiotic relationship with microorganisms, most of which are members of the other two domains of life, Bacteria and Archaea. In many instances, such as symbioses associated with structures of the alimentary canal, these relationships are generally mutualistic, whereby both the host and symbiont(s) derive benefits that neither could achieve individually. Increasing recognition of the role that the gastrointestinal microbiome plays in human health has spurred widespread research into the relationships between a host and its microbial constituents, as well as between different members of the microbiome itself.1-10 Estimates of the number of different microorganisms found in mammalian gastrointestinal microbiomes range from 500 to over 10,000 distinct species or strains.11-13 Due to the immense complexity that these systems pose in detailing the nature of host/microbe and microbe/microbe interactions, animals that possess a limited microbiome are currently better suited to serve as model organisms for more complex interactions.
Gnotobiotic mice that possess a defined, usually synthetically derived gastrointestinal microbiome, are a popular model for analyzing gut symbioses. These mice are colonized by either a single species or a small defined microbial community and have been used by a number of researchers to clarify the roles of certain bacterial species with regards to both host nutrition and immune response.14-17 A notable example is colonization with the common mammalian gut bacterium Bacteroides thetaiotaomicrom, which is now known to metabolize complex carbohydrates and starches that the host cannot digest into short-chain fatty acids, thus providing an additional source of nutrients.18,19 However, experiments with gnotobiotic mice and defined microbial populations do not fully replicate the complexity and co-evolutionary intimacy of microbial symbioses in nature. Another popular model for the study of host/microbe symbioses is the light organ of the Hawaiian bobtail squid, Euprymna scolopes, which exists in a natural, mono-specific symbiosis with the bioluminescent bacterium Vibrio fischeri. This model system has yielded many important insights into how host/microbe symbioses are both established and maintained, as well as identifying potential natural selective pressures on the host and bacterium that drive the co-evolution of the symbiosis.20-22
The Hungarian medicinal leech, Hirudo verbana, is an intriguing model for host/microbe symbioses as it possesses a naturally limited microbiome that is dominated by two microbial constituents of the alimentary canal.23 One of these symbionts, an Aeromonas sp can serve as an opportunistic pathogen in humans and other animals and is amenable to genetic manipulation.24 The second primary symbiont, a Rikenella-like bacterium, is related to known bacterial members of other gastrointestinal microbiomes and was recently isolated in pure culture.25 In this review we discuss leech usage in medical practice and the role that the gut microbiome can play in infection following leech therapy, the composition and role of the leech microbiome, and how the transmission of symbionts is believed to occur. We also discuss colonization factors for the Aeromonas symbiont that have been identified from mutant screens and cover interactions between the symbionts themselves and with the leech host.
Leech Usage in Medicine
Records dating back to at least 200 BCE have been found that mention the use of leeches in a medicinal capacity. In Europe their use peaked around 1850. In 1851, France imported over 13 million leeches in order to supplement their local supply. Subsequently however, the use of leeches in medical practice fell into disfavor as the medical validity of blood-letting became justifiably questioned. In the past 30–40 years, the use of leeches has regained favor and interest within the medical community due to the powerful anticoagulants and vasodilators they produce. Leech therapy has become increasingly popular among plastic surgeons as part of the post-operative healing process.26 Widespread recognition and acceptance of leech usage within the medical community came in 2004 when the US Food and Drug Administration officially approved the marketing and sale of the medicinal leech as a medical device for relieving venous congestion. The European medical community has been actively investigating additional clinical applications for leech therapy, and recent studies have suggested that leech therapy is more effective than topical analgesics in treating symptoms associated with osteoarthritis.27,28
The European medicinal leech is the most commonly used leech in modern medical applications. While the species Hirudo medicinalis is historically believed to have served in this capacity, recent molecular analysis has shown that most leeches sold for medical use as H. medicinalis are usually Hirudo verbana and in some cases another species, Hirudo orientalis.29 As these species are difficult to visually differentiate, for medical purposes, they are used interchangeably as all three are hematophagus and produce anticoagulant proteins. However, it is possible that each species produces a different complement of secreted proteins, and greater attention should be given to correctly identify the species of leech used in medical procedures.26,30,31 Although standard practice dictates the administration of certain antibiotics prior to treatment, in ~5–15% of leech therapy cases for venous congestion treatment is followed by a bacterial infection of the bite wound, with higher rates of infection (~25–35%) occurring in patients that were not treated with antibiotics prophylactically.26,31,32 Culture-based recovery and biochemical identification of wound isolates indicated that Aeromonas species were typically the cause of these infections.31 Culture-based studies of the leech gut microbiome during the early 20th century identified an Aeromonas sp as a persistent and exclusive symbiont of the leech. This finding helped pave the way for use of the medicinal leech as a model organism for the investigation of host/symbiont relationships as well as host specific virulence mechanisms.24
Leech Gut Anatomy and Feeding Behavior
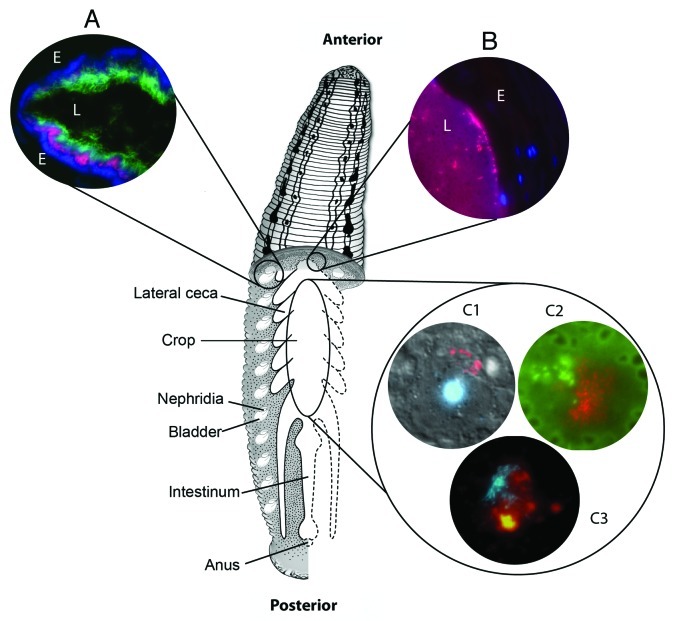
The medicinal leech is a freshwater member of the invertebrate phylum Annelida and its body is divided into 34 segments.33Figure 1 shows an overview of the alimentary tract and related structures of Hirudo verbana. The alimentary canal is divided into two primary sections, the crop which accounts for most of the overall capacity of the canal and intestinum, which is also referred to as the intestine in the literature.33 Lateral ceca extend off from the primary canal of the crop, which connects to the intestinum around the 19–20th body segment. During consumption of a blood meal, the leech uses a series of rhythmic muscle contractions to draw blood into the crop, where it is stored but not digested.33,34 Over the course of a single meal, the leech can consume over 5 times its own body weight in blood. After being satiated with the blood meal, the leech detaches from the host and the bite wound will continue to bleed until the effects of the powerful anticoagulants wear off.
Figure 1. Major alimentary structures and microbial interactions of Hirudo verbana. Drawing demonstrating the anatomy of Hirudo verbana showing the major structures of the alimentary system including the crop, lateral crop ceca, nephridia and bladders, intestinum, and anus. Inset images illustrate microbial interactions within the bladder (A), along the crop epithelium (B) and through the crop at large (C) and are as follows: A) FISH image showing the distribution of bacteria within the bladder lumen (colors correspond to probes targeted as follows: green = β-Proteobacteria, red = Bacteroidetes, blue = Bacteria); (B) FISH image of the Rikenella-like bacterium (red) along the crop epithelium, epithelial cell nuclei counter-stained with DAPI; (C1) composite image of Aeromonas (red) associated with the cell surface of a leech hemocyte, nucleus counter-stained with DAPI; (C2) FISH image of mixed microcolonies showing both Aeromonas (green) and the Rikenella-like bacterium (red), erythrocytes from a blood meal auto-fluoresce green and have darkened interiors; (C3) fluorescent image of lectin staining using succinylated wheat germ agglutinin (WGA-S, red) and DAPI counter-staining of bacterial colonies. For images A and B, the following indicators are used: E indicates the tissue epithelium of the bladder (A) or crop (B) and L indicates the lumen.
Within the first few hours after feeding, the leech is largely inactive while a series of modifications to the blood meal occur. Analogous to the human colon, water and select osmolytes of the blood meal are removed in the crop and excreted into the environment through a series of nephridia and bladders.33,35 This action removes nearly half of the ingested weight and effectively concentrates the remaining erythrocytes, which are stored in the crop predominantly intact. The crop hosts two numerically dominant symbiotic bacteria: an Aeromonas sp and a recently isolated bacterium related to the Bacteroidetes genus Rikenella.23 Analysis of the abundance of the Aeromonas symbiont in the crop before and after consuming a blood meal indicates that abundance is low (~5x105 cfu/ml of crop contents) immediately prior to consumption of the blood meal and increases roughly three orders of magnitude after feeding.24,36 After this period, the majority of host physiological changes to the blood meal have been completed and symbiont abundance in the crop gradually declines.
After consumption of a blood meal, the leech can go for a prolonged period of time, up to 6 mo, before feeding again.33 This prolonged feeding interval provides an opportunity to monitor nutrient preference and utilization by the gut microbiome in a temporal manner that may reflect similar patterns that occur along the length of longer alimentary tracts found in other organisms such as humans. During this “fasting” period, erythrocytes stored in the crop are slowly transported into the intestinum where they are lysed and the released nutrients are absorbed. Culture independent analyses have shown that similar to the crop, the Aeromonas and Rikenella-like symbionts are both present and numerically abundant in the intestinum, however the intestinum microbiome is also more diverse than that of the crop.23 In addition to the Aeromonas and Rikenella-like symbionts, Morganella morganii, members of the α, γ, and δ proteobacteria, Fusobacteria, and Firmicutes were all observed.23 This increased microbial diversity in the section of the gut where digestion and absorption of nutrients occurs is similar to that seen in other organisms such as humans and mice.37
Leech Endosymbionts
Nephridia/Bladder
Hirudo verbana possess 17 pairs of nephridia that lie along side the alimentary canal.33 The nephridia function in the removal of water and osmolytes from the leech hemolymph, especially following consumption of a blood meal.38,39 The bladders serve to store urine and nitrogenous wastes, primarily in the form of ammonia, prior to excretion.33 Büsing et al. in 1953 described two bacterial morphotypes closely associated with the nephridia and bladders and Wenning et al. in 1993 showed that these symbionts were present and associated both intra- and extra-cellularly with the bladder during leech embryogenesis.40,41 Further investigation into these relationships, however, was largely ignored until 2009 when they were investigated more thoroughly.
Using a combination of culture independent 16S rRNA sequencing, FISH probing, and transmission electron microscopy (TEM) Kikuchi et al. identified 6 bacterial symbionts and their physical arrangement within the leech bladder.42 An α-proteobacterium, phylogenetically clustered with members of the genus Ochrobactrum, was identified as living intracellularly within epithelial cells of the bladder. Two Bacteroidetes spp, phylogenetically related to Sphingobacterium and Niabella, were located along the luminal epithelium of the bladder while two β-Proteobacteria phylogenetically related to Comamonas and Sterolibacterium were located further into the bladder lumen. The sixth symbiont, phylogenetically related to Bdellovibrio, was located within the bladder lumen, typically in association with the two β-proteobacterial symbionts.
Determination of the functional roles that the nephridia and bladder symbionts fulfill in the leech is still a current area of exploration. It was generally postulated that the symbionts play an active role in the degradation and detoxification of nitrogenous wastes.40 Kikuchi et al., however, proposed an intriguing role of the symbionts in the recycling of nutrients during the prolonged “fasting” period in between blood meals.42 In the study by Wenning et al., antibiotics were injected into the cocoons of developing leech embryos. After hatching, the bacterial symbionts were not found to be present in adult leech bladders by TEM, suggesting that the bacterial symbionts do not play a critical role in the proper development of the nephridia and bladder.41
Digestive tract
The seemingly simple question of who are the digestive tract symbionts of the leech has proven to be tricky. Both the leech and particularly microbial taxonomy have changed in recent decades, while culture-independent analyses have revealed the presence of a more complex community than initially determined.
Aeromonas
Early culture based investigations of the microflora associated with the medicinal leech recovered a β-hemolytic and highly proteolytic species that was nearly always obtained in pure culture from the leech digestive tract.40 As chemotaxonomic identification methods improved, these isolates were ultimately classified as Aeromonas hydrophila. Aeromonas sp are Gram-negative bacteria that have been isolated from numerous aquatic habitants, and many Aeromonas spp have been identified as being opportunistic pathogens of both humans and various other animals including fish and amphibians.43 More recent molecular investigations into the identity of the Aeromonas symbiont in the medicinal leech, however, have suggested that the symbiont is in fact Aeromonas veronii and not A. hydrophila.24 Work performed by Siddall’s group has greatly clarified this confusion. It was initially assumed that leeches marketed commercially as H. medicinalis were indeed H. medicinalis, however using molecular taxonomy methods Siddall et al. showed that commercially distributed medicinal leeches were usually species of H. orientalis or H. verbana, with the latter being by far the most common species sold by medical suppliers for leech therapy in the UK and USA29
The issue of identifying the species of medicinal leech sold is not inconsequential as subsequent studies suggest that different species of medicinal leech carry a different complement of Aeromonas symbionts. The commonly sold medicinal leech, H. verbana, does not typically carry A. hydrophila but A. veronii instead.24,30 H. orientalis, meanwhile, carries A. veronii and/or Aeromonas jandaei while H. medicinalis carries A. hydrophila.30,44 A. jandaei has also been found to colonize the North American medicinal leech, Macrobdella decora.45 Of note, recent work by Siddall et al. has interestingly shown that the phylogenetic distribution of Aeromonas sp recovered from different species of leeches caught in the wild does not accurately reflect the established phylogeny of the leeches themselves.30 This apparent lack of co-evolution is intriguing, however only a few specimens have so far been analyzed, and these studies utilized sequenced PCR products, which often detect only the most abundant organisms. Considering that the taxonomy of the Aeromonads is still very much in the process of being refined, and that commercial phenotypic tests used in clinical settings often miss-identify some Aeromonas species as A. hydrophila, there is a concern that isolates previously identified as A. hydrophila from wound infections following leech therapy could be actually be A. veronii.46,47 Follow up studies to accurately identify the Aeromonas species isolated from wound infections have been challenging as these isolates are usually discarded after clinical identification; one recent study that isolated a ciprofloxacin resistant species saved the strain and our lab was able to independently confirm its identity as A.hydrophila (unplublished data).48 As A. hydrophila is generally regarded to be more pathogenic in humans than A. veronii, and different strains of each are susceptible to different types of prophylactic antibiotics administered during leech therapy, it will be important to better establish the diversity and distribution patterns of Aeromonas spp colonizing leeches supplied to the medical profession.
Rikenella-like bacterium
While it was widely stated in the literature that an Aeromonas sp (either A. hydrophila or A. veronii) was the sole microbial symbiont present in the leech gut, these studies were all based on the use of aerobic culturing methods.24 It is widely accepted, however, that the vast majority of all microbes are currently uncultivable for a variety of reasons and that culture-independent analyses provide a more accurate description of microbial populations. It was using culture-independent 16S rRNA sequencing methods, that Worthen et al. first discovered a second primary symbiont in the leech crop and intestinum, as well as confirming the presence of the previously identified Aeromonas veronii.23 Phylogenetic comparison of the newly identified symbiont to previously identified species suggested that it represented a novel genus that is related to the genus Rikenella, an obligate anaerobe and member of the Bacteroidetes.23 The closest phylogenetic match (89.7% identity) was to R. microfusus, which was isolated from a Japanese water fowl. Surveys of human fecal samples have found members of the family Rikenellaceae to be an abundant in the human gut microbiome, however few species from this family have been obtained in pure culture.49 Initial attempts to cultivate the Rikenella-like symbiont from the leech in the laboratory using previously described media proved to be unsuccessful until a directed culturing methodology was applied.
A fluorescent in situ hybridization (FISH) study revealed that both the Rikenella-like bacterium and Aeromonas proliferated within the leech crop after consumption of a blood meal, with the Aeromonas symbiont plateauing and subsequently decreasing in abundance earlier than the Rikenella-like bacterium.36 Using meta-transcriptomic data generated from the crop contents of leeches fed 42 h prior, Bomar et al. found elevated expression values for genes related to both glycan and mucin foraging and fermentative metabolism.25 Histological staining revealed the presence of mucin lining the crop epithelium. These data suggested that the Rikenella-like symbiont was foraging on mucins produced by the host and fermenting them to acetate.25 An anaerobic culture medium containing porcine mucin as the primary substrate for growth was designed and ultimately allowed for the pure isolation of the Rikenella-like symbiont.25 Having the second primary microbial leech symbiont in pure culture opens up exciting new avenues of research that were previously unavailable and may reveal the functions of these organisms in a variety of hosts.
Other known symbionts
In addition to the two dominant Aeromonas and Rikenella-like gut symbionts, a number of other bacterial species have been identified in lower abundance, particularly in the intestinum.23 One of these species, Morganella morganii, we have been able to isolate in the lab from H. verbana. M. morganii is a pervasive opportunistic pathogen of a number of animals and has been recovered from wound infections after leech therapy.26,50 Similar to other gastrointestinal microbiomes such as in mammals, a Desulfovibrio sp has also been observed in and isolated from the leech intestinum. In mammals, Desulfovibrio spp consume hydrogen and free sulfur end products of microbial fermentation and produce hydrogen sulfide, which can be toxic to the host gut epithelium.7 This toxicity has led to Desulfovibrio being implicated in bowel diseases, principally ulcerative colitis.51,52 Other bacterial species common to the gut microbiomes of other animal hosts have been identified using culture independent analyses and detailing the full scope of the intestinum microbiome is currently an active point of exploration.
Symbiont Transmission
One of the intriguing questions that arises from the study of host/microbe endosymbioses is where do the symbiotic bacteria come from and how do they become established within a host. In mammalian species such as mice and humans, the neonate gastrointestinal tract is sterile at birth and is subsequently colonized through interaction with the environment, particularly during feeding.3,53 This process of acquiring symbiotic microbes from the environment is known as horizontal transmission. Avian and presumably reptilian species are also born with sterile gastrointestinal tracts that are subsequently colonized post hatching through horizontal transmission.54 Leeches are hermaphroditic and reproduce through the deposition of multiple fertilized embryos in a cocoon.33 During cocoon deposition, the parent leech secretes a foamy liquid from specialized cells along the outer integument that is subsequently sloughed off over the anterior end.33 The leech then uses its mouth to form the cocoon’s final shape as the foamy liquid begins to harden. Contact with both bladder secretions and the mouth of the parental leech during cocoon deposition and formation could provide a simple and direct route for the vertical transmission of microbial symbionts from the parent crop to the cocoon.
This mode of direct vertical transmission from the parent to the embryo was explored by Rio et al. using diagnostic PCR. They showed that the Aeromonas symbiont was detectable within 24 h after cocoon deposition, while the Rikenella-like symbiont was first detected 2 weeks after the juveniles leeches hatched from the cocoon.55 While the Rikenella-like symbiont was not detectable using standard diagnostic PCR until two weeks after hatching, it is likely that this symbiont is present immediately following deposition but at a level below the limit of detection of the PCR assay used. Transmission of the nephridia and bladder symbionts to developing leeches is more complex considering the intracellular nature of one of the symbionts. In their same study, Rio et al. found that the intracellular Ochrobactrum symbiont was present in 100% of all cocoons examined from 24 h post-deposition onward while the Commamonas and Niabella symbionts were detected at all time points during development, although not in all cocoons sampled.55 Further, no transmission of bladder or nephridia symbionts was found to occur when housing juvenile leeches that had been exposed to antibiotics during development, and thus had no bladder symbionts, with adult leeches that were not exposed to antibiotics, suggesting that transmission and colonization of the bladder and nephridia symbionts must occur during cocoon deposition and embryogenesis.41
Proposed Role of the Leech Gut Microbiome
Microbial gut symbioses pose interesting questions regarding the functional interactions that occur between the microbiome and the host. The large microbial diversity and complexity of most digestive-tract microbiomes, such as in mice and humans, complicates studies attempting to determine the functional roles of individual species of the microbiome. The naturally occurring, low diversity microbiome within the leech gut, combined with our ability to culture both dominant symbionts and genetically manipulate one of them, makes the medicinal leech a powerful model for the study of microbe/host interactions. Since the initial discovery of the digestive-tract symbioses of the leech gut, three main hypotheses have been offered to explain why this relationship is maintained as well as its specificity.33,40,56 Two of these hypotheses are based on nutritional aspects and neither are mutually exclusive. The first hypothesis is that the microbial symbionts are responsible for the digestion of the blood meal (I) while the other is that the symbionts provide essential nutrients that the leech is unable to synthesize for itself and cannot derive in sufficient quantities from the blood meal directly (II). Early research suggested that the leech was incapable of lysing the consumed erythrocytes without the contribution of the Aeromonas symbiont due to an apparent lack of host proteases within the gut.33 Subsequent discovery of host produced proteases within the intestinum, which is the primary site of erythrocyte lysis and digestion of the blood meal, suggests that the microbial symbionts may not be essential for the digestion of the blood meal.57 However, recent transcriptome analysis has revealed which metabolic pathways of the microbial community are active within the leech crop, and suggests that the symbionts may be releasing nutrients such as acetate that can then be utilized by the host.25 Thus, while the host may be capable of digesting the blood meal itself, optimal nutritional benefits may require the presence of certain members of the gut microbiome. For example, the low concentration of B vitamins in blood, which the leech is incapable of synthesizing for itself, suggests that they would need to be supplied by the either the crop or intestinum microbiomes.56 Further transcriptome analysis of both the crop and intestinum microbiomes is currently under way and will greatly elucidate the role of each in the provision of nutrients to the host.
The third hypothesis for the role of the microbial symbionts is that they provide a form of either passive or active resistance to colonization of the leech gut by potentially pathogenic or detrimental species of bacteria.33 In humans and mice, the gut microbiome is though to perform an important function by preventing exogenous, potentially pathogenic microbes from colonizing, a process is termed colonization resistance. The specificity of the composition of the leech gut microbiome can be evaluated experimentally by utilizing a colonization assay that introduces different bacterial strains into the leech via the blood meal. This assay allows for the screening of factors that affect the introduced strains ability to colonize the leech crop and intestinum. Using this the assay, Indergrand and Graf first tested clinical isolates that were identified as Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa for their ability to colonize the leech crop when compared with growing in blood alone.58 Both S. aureus and P. aeruginosa were found to be significantly attenuated in their ability to proliferate within the leech crop compared with the in vitro blood control, suggesting either that their growth was inhibited or that cell proliferation was balanced by death. These data suggest that the leech and/or the gut microbiome creates conditions inside the leech gut that modulate the proliferation of these two species. E. coli, however, was found to have a significantly reduced ability to survive in both the leech and the in vitro blood control. This finding suggested that an innate immunity factor of the blood meal was responsible for inhibiting the proliferation of E. coli but not S. aureus and P. aeruginosa.58 Heat treatment of the blood prior to inoculation with the E. coli strain allowed for growth in the in vitro blood control and colonization of the leech crop, suggesting that the mammalian complement system is active within the leech crop and plays a role in preventing complement sensitive bacteria from colonizing the leech. This activity of the complement system is reinforced by findings that Aeromonas mutants that are unable to properly synthesize the lipopolysaccharide layer of the outer membrane are unable to colonize the crop or grow in blood where the complement system has not been inactivated.59
Factors Critical to Survival of Aeromonas in the Leech Gut
Insight into factors that are critical for the establishment and maintenance of the specificity of host/microbe symbioses is often gained through the creation of bacterial mutants and monitoring their ability to colonize and proliferate in the host.60 The Aeromonas symbiont is easily cultured in vitro and has a well-established system for genetic manipulation. Using random transposon mutagenesis screening, we have identified numerous colonization factors that are required for A. veronii to effectively colonize the leech crop. A brief overview of these factors is given in Table 1. Signature tagged mutagenesis (STM), a high-throughput transposon mutagenesis screening method has been used for a number of bacterial species to identify both virulence and symbiosis related genes.61-63 Using STM, Silver et al. identified 20 serum-resistant A. veronii mutants that showed a reduced ability to colonize the leech crop compared with the parent strain and 26 serum-sensitive mutants.59 Nine of the 20 serum-resistant mutants were observed as having a statistically lower ability to colonize the crop compared with the parent strain, however analysis of the transposon insertion site only revealed the disruption of hypothetical proteins or proteins of unknown function. The remaining 11 serum-resistant mutants, however, had insertions affecting identifiable genes, forming four groups based on the functional category of the disruption.
Table 1. List of Aeromonas veronii mutants.
| Functional Group | Strain ID(s) | Identified defect/mutation | Colonization deficiency* | Reference for strain |
|---|---|---|---|---|
| Parent/Wild-type Strains |
HM21R, HM21RS |
- |
- |
24 |
| Complement Resistance |
30+ isolates |
Multiple genetic loci |
+, ++, +++ |
59 |
| Oxidative Stress |
JG186 |
katA |
- |
72 |
| Surface Modification |
JG356 |
Glycosyltransferase |
+++ |
59 |
| JG736, JG730 |
lpp |
+++ |
59, Bomar and Graf unpublished |
|
| |
JG738 |
Methyltransferase type 11 |
++ |
59 |
| Regulatory |
JG574 |
Ribosomal operon |
+ |
59 |
| JG697 |
GTP-binding protein |
++ |
59 |
|
| |
JG741 |
Exoribonuclease II |
+ |
59 |
| Nutrition |
JG537 |
pstC homolog |
++ |
59 |
| JG698 |
GufA |
+ |
59 |
|
| |
JG750 |
Threonine/serine transporter |
++ |
59 |
| Host Interaction |
JG752 |
ascU - Type III secretion system |
++ |
70 |
| HE-1095 |
exeM - Type II secretion system |
+ |
69 |
|
| Unknown Role |
JG521, JG573, JG735, JG751 |
Hypothetical proteins |
+, ++, +++ |
59 |
| JG532 |
Conserved hypothetical protein |
+ |
59 |
|
| |
JG533 |
KAP family P-loop domain protein |
+ |
59 |
| |
JG538 |
Peptidase S15 |
+ |
59 |
| JG523, JG753 | No similar BLAST result | + | 59 |
Colonization deficiency of leech crop compared with parent/wild-type strain represented as follows: -, no colonization deficiency; +, 2–10 fold deficiency; ++, 10–100 fold deficiency; +++, > 100 fold deficiency
Resistance to blood complement system
Mammalian blood contains a number of innate immunity factors that serve to protect the host from bacterial infection. The complement system is one such innate immunity factor, and was found to remain active within the leech for up to two days after feeding.58 This prolonged activity limits the ability of foreign bacteria that are susceptible to the complement system to colonize the leech, such as was observed for E. coli. Of the 46 mutants identified by Silver et al., 56% were found to be sensitive to serum, an indicator for susceptibility to the complement system present in mammalian blood.59 This recovery of a large number of serum sensitive mutants reinforces the role that resistance to the complement system of the ingested blood meal provides as a colonization factor of A. veronii in the leech and highlights the large number of genes involved in protecting Aeromonas from the complement system.64
Regulatory control in response to host stimuli
Given that A. veronii is found not just in the leech gut, but also in many aquatic habitants and is a pathogen in both mammals and fish, one expects that tight regulatory control over cell proliferation, metabolic processes and colonization factors in response to the given environment would be crucial in allowing A. veronii to colonize and persist in the leech. Four mutants were initially reported by Silver et al. to have a disruption in regulatory genes which could be required for modulating activity in response to host stimuli.59 However, subsequent analysis of one of these mutants, JG730, revealed that the initial identification of the affected gene was most likely based on an annealing artifact during inverse PCR, and that the actual transposon insertion occurred in the lpp gene (Bomar and Graf, unpublished). The disruptions for the other three mutants occurred, respectively, in a ribosomal operon control region, a gene encoding a GTP-binding protein, and a gene encoding an exoribonuclease II.59
Cell membrane maintenance and modification
Studies examining other model symbioses, such as tsetse flies, have identified that certain cell membrane modifications serve to promote colonization of the host by only specific symbionts, and thus the ability to prevent or resist attack from membrane stress within the leech would be a critical colonization factor for Aeromonas.60 Three strains were reported in the STM study to have mutations in cell surface modification or stability proteins. Two strains have mutations in putative glycosyltransferase genes, which are known to play a role in the synthesis of the LPS layer that provides complement resistance in Aeromonas.59 However, as neither of these two mutants was found to be serum sensitive, it is unlikely that LPS synthesis alone was responsible for the colonization defect and that the affected genes may play a more subtle role in modifying the cell surface, the production of exopolysaccharides, or glycosylating certain surface proteins. The third mutant, JG736, was found to have a disruption of the lpp gene and is described in more detail in the STM study.59 Our subsequent re-identification of the transposon insertion site for JG730 as being in lpp would now make this four surface modification and stability mutants.
Nutrient acquisition
Competition for nutrients between the two primary symbionts, as well as with the host, provides a selective pressure that can prevent microbial species from effectively colonizing the leech crop. Three strains containing mutations in nutrient transporter genes were identified in the STM study. One of these mutants, JG537, was found to have a disruption of a pstC homolog gene.59 In E. coli, the classical documented activity of the Pst operon is to control phosphate transport into the cell under low phosphate conditions. Pst has also interestingly been implicated as being required for full pathogenicity of two pathogenic strains, O78 and O115, in animal models.65,66 Studies examining enteropathogenic E. coli in vitro and Citrobacter rodentium in vivo have found that the Pst operon is necessary for adhesion to host gastrointestinal epithelial cells and full virulence in murine models.67,68 Thus while Pst is a classical nutrient transporter system, it is possible that the failure of the Pst mutant to colonize the leech crop is not due to a defect in the acquisition of nutrients.
Host interaction
An interesting and clinically important feature of the Aeromonas symbiont is that it is also a pathogenic bacterium in both mammals and fish. Because of this dichotomous lifestyle, numerous investigations have been undertaken to elucidate the mechanisms that A. veronii uses to persist in the leech host without causing any apparent negative effects. Bacterial secretion systems have been identified as necessary virulence factors for a number of pathogenic bacteria including Aeromonas. Both the type 2 (T2SS) and type 3 (T3SS) secretion systems of A. veronii serve as virulence factors and were found to be required for colonization of the leech crop. The T2SS mutant was found during a screening of A. veronii transposon mutants that had lost the ability to lyse erythrocytes (β-hemolysis) when grown on blood agar plates.69 Sequencing of the insertion site found that the transposon disrupted transcription of two genes required for the synthesis of the inner membrane components of the T2SS, indicating that the T2SS is responsible for the export of hemolysins by A. veronii. The failure of the T2SS mutant to colonize the leech crop suggests that hemolysin export is a critical colonization factor for A. veronii, even though the majority of consumed erythrocytes remain visibly intact inside the leech crop. The T3SS mutant was identified in the above-mentioned STM study by Silver et al. and showed a diminished capacity to colonize the leech crop although no growth defect was witnessed in an in vitro blood control.59,70 Fluorescent in situ hybridization (FISH) microscopy determined that the T3SS mutant was phagocytosed by leech hemocytes, macrophage like cells that would serve to remove susceptible bacteria from the blood meal, while wild-type Aeromonas were only associated with the hemocyte cell surface. The T3SS mutant also demonstrated a significantly decreased ability to lyse murine macrophage cells when compared with the parent strain, and was subsequently less pathogenic when injected into mice.70 These finding suggest that A. veronii uses known virulence factors in a manner that allows it to colonize and persist in the leech crop.
Microbe/Microbe Interactions
The discovery and subsequent culture isolation of a second primary microbial symbiont in the leech gut provided new and exciting avenues of research not only into host/microbe interactions, but also into microbe/microbe interactions. The Rikenella-like symbiont, as with other members of the Bacteroidetes, is an obligate anaerobe, while the Aeromonas symbiont is a facultative anaerobe. As the leech epidermis is fairly permeable to oxygen and water, it was first questioned whether or not the leech gut was sufficiently anaerobic to support the proliferation of the Rikenella-like symbiont. This led to the hypothesis that the Aeromonas symbiont, a facultative anaerobe, could be responsible for the consumption of oxygen within the leech gut, keeping it sufficiently reduced to permit the growth of the Rikenella-like symbiont.23 This hypothesis is consistent with data that showed that the Aeromonas symbiont could be detected within 24 h after cocoon deposition, when the concentration of oxygen would be similar to the surrounding environment, while the Rikenella-like symbiont could not be detected until 2 weeks after emergence from the cocoon, at which point the gut would be fully formed and possibly anaerobic.55 Although the cocoon itself would not be anaerobic during embryogenesis, it is possible that during this period of development that the gut symbionts are in close proximity and that the Aeromonas symbiont has created an anaerobic microenvironment where the Rikenella-like bacterium can survive but not proliferate. This would explain how the obligately anaerobic Rikenella-like bacterium could be vertically transmitted from parent to offspring, but could not be detected until after emergence of the juvenile leech from the cocoon.55
Further evidence supporting this interaction between the Aeromonas and Rikenella-like symbionts can be found in the distribution of the symbionts within the crop of adult leeches. Using FISH microscopy, Kikuchi and Graf determined that the Aeromonas symbiont was found in one of two forms within the leech.36 The first form was as single, freely moving pelagic cells, while the second was in association with the Rikenella-like bacterium in what are described as mixed microcolonies. These microcolonies were found to be covered by a polysaccharide matrix, which could tightly bind the two species together. The close proximity of Aeromonas to the Rikenella-like bacterium would facilitate the maintenance of a sufficiently anaerobic environment for the Rikenella-like bacterium to proliferate within the microcolony, as well as allow for rapid nutrient transfer between the two species in a manner similar to that observed for bacteria in anaerobic granules present in certain waste treatment systems.36 An intriguing proposed function of these mixed microcolonies is that they aid the Rikenella-like bacterium in evading phagocytosis by leech hemocytes in the crop. This could occur by preventing recognition of the Rikenella-like bacterium due to the polysaccharide matrix or the clearing of hemocytes in the crop as a result of the activity of the T3SS of the Aeromonas symbiont as discussed above.36,70
Future Perspectives
Due to its naturally limited microbiome when compared with other common animal models, the ability to culture the two dominant symbionts, genetically manipulate one of them (Aeromonas), and modify the blood meal, the medicinal leech has proven to be a powerful model for examining the nature of microbial symbioses. While we discussed above a number of important findings regarding the nature of the leech microbiome, and have been able to apply these findings to other symbiosis models and areas of research, previous studies have only scratched the surface of what is occurring and the underlying mechanisms thereof. As the Aeromonas symbiont was the first isolate obtained from the leech, most of our knowledge is based on analyses of this symbiont only. Now that the Rikenlla-like bacterium has been obtained in pure culture, further characterization of the isolate and its role within the leech is an active point of exploration. An interesting finding in the python that Rikenella spp play a role in metabolizing host mucins to acetate, which could serve as a source of host nutrition during periods of fasting has interesting parallels to the leech, which also undergoes long periods between feeding.71
One of the important questions that we are currently investigating is how the symbiont/host dynamics change during the maturation of juveniles into adult leeches. Within the cocoon the embryonic leeches subsist on albumin, and do not consume their first blood meal until weeks after emerging from the cocoon. This represents a period in which a number of unique host and microbial interactions are believed to occur as the primary source of nutrients switches from albumin to blood. Using a combination of metagenomic and metatranscriptomic approaches, we are rapidly expanding our understanding of the host response in relation to changes in the physiology and composition of the gut microbiome, pointing out new research directions that we are just beginning to explore. Thus, despite its undeserved reputation and association with quackery, the medicinal leech can hold its head high as a powerful model for the study of microbial symbioses and as useful medical device in modern medicine.
Acknowledgments
Research was supported by NSF Career Award MCB 0448052 and NIH RO1 GM095390 to J.G. The authors thank Virge Kask for assistance with the artwork and members of the Graf lab for review and discussion of the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20227
References
- 1.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 2.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208–18. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 3.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–5. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol. 2010;1:363–95. doi: 10.1146/annurev.food.102308.124101. [DOI] [PubMed] [Google Scholar]
- 7.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–90. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchesi JR. Human distal gut microbiome. Environ Microbiol. 2011;13:3088–102. doi: 10.1111/j.1462-2920.2011.02574.x. [DOI] [PubMed] [Google Scholar]
- 11.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 13.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–89. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–6. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 2009;106:5859–64. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–8. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 22.Visick KL, McFall-Ngai MJ. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–87. doi: 10.1128/JB.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worthen PL, Gode CJ, Graf J. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol. 2006;72:4775–81. doi: 10.1128/AEM.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graf J. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect Immun. 1999;67:1–7. doi: 10.1128/iai.67.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bomar L, Maltz M, Colston S, Graf J. Directed culturing of microorganisms using metatranscriptomics. MBio. 2011;2:e00012–11. doi: 10.1128/mBio.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitaker IS, Josty IC, Hawkins S, Azzopardi E, Naderi N, Graf J, et al. Medicinal leeches and the microsurgeon: a four-year study, clinical series and risk benefit review. Microsurgery. 2011;31:281–7. doi: 10.1002/micr.20860. [DOI] [PubMed] [Google Scholar]
- 27.Andereya S, Stanzel S, Maus U, Mueller-Rath R, Mumme T, Siebert CH, et al. Assessment of leech therapy for knee osteoarthritis: a randomized study. Acta Orthop. 2008;79:235–43. doi: 10.1080/17453670710015030. [DOI] [PubMed] [Google Scholar]
- 28.Michalsen A, Lüdtke R, Cesur O, Afra D, Musial F, Baecker M, et al. Effectiveness of leech therapy in women with symptomatic arthrosis of the first carpometacarpal joint: a randomized controlled trial. Pain. 2008;137:452–9. doi: 10.1016/j.pain.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Siddall ME, Trontelj P, Utevsky SY, Nkamany M, Macdonald KS. Diverse molecular data demonstrate that commercially available medicinal leeches are not Hirudo medicinalis. Proc Biol Sci. 2007;274:1481–7. doi: 10.1098/rspb.2007.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddall ME, Min G-S, Fontanella FM, Phillips AJ, Watson SC. Bacterial symbiont and salivary peptide evolution in the context of leech phylogeny. Parasitology. 2011;138:1815–27. doi: 10.1017/S0031182011000539. [DOI] [PubMed] [Google Scholar]
- 31.Whitaker IS, Kamya C, Azzopardi EA, Graf J, Kon M, Lineaweaver WC. Preventing infective complications following leech therapy: is practice keeping pace with current research? Microsurgery. 2009;29:619–25. doi: 10.1002/micr.20666. [DOI] [PubMed] [Google Scholar]
- 32.Lineaweaver WC, Hill MK, Buncke GM, Follansbee S, Buncke HJ, Wong RK, et al. Aeromonas hydrophila infections following use of medicinal leeches in replantation and flap surgery. Ann Plast Surg. 1992;29:238–44. doi: 10.1097/00000637-199209000-00008. [DOI] [PubMed] [Google Scholar]
- 33.Sawyer R. Leech biology and behaviour.: Clarendon Press, 1986. [Google Scholar]
- 34.Lent CM, Fliegner KH, Freedman E, Dickinson MH. Ingestive behaviour and physiology of the medicinal leech. J Exp Biol. 1988;137:513–27. doi: 10.1242/jeb.137.1.513. [DOI] [PubMed] [Google Scholar]
- 35.Zebe E, Roters F, Kaiping B. Metabolic Changes in the Medical Leech Hirudo-Medicinalis Following Feeding. Comparative Biochemistry and Physiology a-Physiology. 1986;84:49–55. doi: 10.1016/0300-9629(86)90041-1. [DOI] [Google Scholar]
- 36.Kikuchi Y, Graf J. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl Environ Microbiol. 2007;73:1984–91. doi: 10.1128/AEM.01833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–29. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 38.Wenning A. Managing high salt loads: From neuron to urine in the leech. Physiol Zool. 1996;69:719–45. [Google Scholar]
- 39.Zerbst-Boroffka I, Bazin B, Wenning A. Chloride secretion drives urine formation in leech nephridia. J Exp Biol. 1997;200:2217–27. doi: 10.1242/jeb.200.16.2217. [DOI] [PubMed] [Google Scholar]
- 40.Büsing KH, Döll W, Freytag K. Die Bakterienflora der medizinischen Blutegel. Archiv fr Mikrobiologie. 1953;19:52–86. doi: 10.1007/BF00412315. [DOI] [PubMed] [Google Scholar]
- 41.Wenning A, Cahill M, Greisinger U, Kaltenhauser U. Organogenesis in the Leech - Development of Nephridia, Bladders and Their Innervation. Rouxs Arch Dev Biol. 1993;202:329–40. doi: 10.1007/BF00188732. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi Y, Bomar L, Graf J. Stratified bacterial community in the bladder of the medicinal leech, Hirudo verbana. Environ Microbiol. 2009;11:2758–70. doi: 10.1111/j.1462-2920.2009.02004.x. [DOI] [PubMed] [Google Scholar]
- 43.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laufer AS, Siddall ME, Graf J. Characterization of the digestive-tract microbiota of Hirudo orientalis, a european medicinal leech. Appl Environ Microbiol. 2008;74:6151–4. doi: 10.1128/AEM.00795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddall ME, Worthen PL, Johnson M, Graf J. Novel role for Aeromonas jandaei as a digestive tract symbiont of the North American medicinal leech. Appl Environ Microbiol. 2007;73:655–8. doi: 10.1128/AEM.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, Lara E, et al. Multilocus phylogenetic analysis of the genus Aeromonas. Syst Appl Microbiol. 2011;34:189–99. doi: 10.1016/j.syapm.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Martino ME, Fasolato L, Montemurro F, Rosteghin M, Manfrin A, Patarnello T, et al. Definition of microbial diversity in Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative genes of virulence. Appl Environ Microbiol. 2011;77:4986–5000. doi: 10.1128/AEM.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang EW, Warren DK, Ferris VM, Casabar E, Nussenbaum B. Leech-transmitted ciprofloxacin-resistant Aeromonas hydrophila. Arch Otolaryngol Head Neck Surg. 2011;137:190–3. doi: 10.1001/archoto.2010.257. [DOI] [PubMed] [Google Scholar]
- 49.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet J-P, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–84. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 50.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christophersen CT, Morrison M, Conlon MA. Overestimation of the abundance of sulfate-reducing bacteria in human feces by quantitative PCR targeting the Desulfovibrio 16S rRNA gene. Appl Environ Microbiol. 2011;77:3544–6. doi: 10.1128/AEM.02851-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46:64–72. doi: 10.1136/gut.46.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cressman MD, Yu Z, Nelson MC, Moeller SJ, Lilburn MS, Zerby HN. Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl Environ Microbiol. 2010;76:6572–82. doi: 10.1128/AEM.00180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rio RVM, Maltz M, McCormick B, Reiss A, Graf J. Symbiont succession during embryonic development of the European medicinal leech, Hirudo verbana. Appl Environ Microbiol. 2009;75:6890–5. doi: 10.1128/AEM.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graf J. The effect of symbionts on the physiology of Hirudo medicinalis, the medicinal leech. Invertebr Reprod Dev. 2002;41:269–75. doi: 10.1080/07924259.2002.9652760. [DOI] [Google Scholar]
- 57.Roters FJ, Zebe E. Proteinases of the medicinal leech, Hirudo medicinalis: purification and partial characterization of three enzymes from the digestive tract. Comp Biochem Physiol B. 1992;102:627–34. doi: 10.1016/0305-0491(92)90058-Y. [DOI] [PubMed] [Google Scholar]
- 58.Indergand S, Graf J. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech. Appl Environ Microbiol. 2000;66:4735–41. doi: 10.1128/AEM.66.11.4735-4741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silver AC, Rabinowitz NM, Küffer S, Graf J. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J Bacteriol. 2007;189:6763–72. doi: 10.1128/JB.00685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss BL, Wu Y, Schwank JJ, Tolwinski NS, Aksoy S. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc Natl Acad Sci U S A. 2008;105:15088–93. doi: 10.1073/pnas.0805666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heungens K, Cowles CE, Goodrich-Blair H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol Microbiol. 2002;45:1337–53. doi: 10.1046/j.1365-2958.2002.03100.x. [DOI] [PubMed] [Google Scholar]
- 62.Kizy AE, Neely MN. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect Immun. 2009;77:1854–65. doi: 10.1128/IAI.01306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menéndez A, Fernández L, Reimundo P, Guijarro JA. Genes required for Lactococcus garvieae survival in a fish host. Microbiology. 2007;153:3286–94. doi: 10.1099/mic.0.2007/007609-0. [DOI] [PubMed] [Google Scholar]
- 64.Braschler TR, Merino S, Tomás JM, Graf J. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl Environ Microbiol. 2003;69:4268–71. doi: 10.1128/AEM.69.7.4268-4271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daigle F, Fairbrother JM, Harel J. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect Immun. 1995;63:4924–7. doi: 10.1128/iai.63.12.4924-4927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamarche MG, Dozois CM, Daigle F, Caza M, Curtiss R, 3rd, Dubreuil JD, et al. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect Immun. 2005;73:4138–45. doi: 10.1128/IAI.73.7.4138-4145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng C, Tennant SM, Azzopardi KI, Bennett-Wood V, Hartland EL, Robins-Browne RM, et al. Contribution of the pst-phoU operon to cell adherence by atypical enteropathogenic Escherichia coli and virulence of Citrobacter rodentium. Infect Immun. 2009;77:1936–44. doi: 10.1128/IAI.01246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreira GM, Spira B. The pst operon of enteropathogenic Escherichia coli enhances bacterial adherence to epithelial cells. Microbiology. 2008;154:2025–36. doi: 10.1099/mic.0.2008/016634-0. [DOI] [PubMed] [Google Scholar]
- 69.Maltz M, Graf J. The type II secretion system is essential for erythrocyte lysis and gut colonization by the leech digestive tract symbiont Aeromonas veronii. Appl Environ Microbiol. 2011;77:597–603. doi: 10.1128/AEM.01621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silver AC, Kikuchi Y, Fadl AA, Sha J, Chopra AK, Graf J. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci U S A. 2007;104:9481–6. doi: 10.1073/pnas.0700286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–85. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rio RVM, Anderegg M, Graf J. Characterization of a catalase gene from Aeromonas veronii, the digestive-tract symbiont of the medicinal leech. Microbiology. 2007;153:1897–906. doi: 10.1099/mic.0.2006/003020-0. [DOI] [PubMed] [Google Scholar]