Abstract
The interaction of the host with its abundant intestinal microbiota is complex and engages most of the cells in the intestinal mucosa. The inflammatory bowel diseases appear to be disorders of the host immune response to the microbiota. This is supported by data from induced gene mutations in mice and more recently by the identification of gene variants in humans that result in IBD or IBD susceptibility. These genetic studies have provided insights into the cells and molecular pathways involved in the host-microbiota dialog. This review discusses the innate, adaptive, and regulatory immune response to the microbiota in the context of the mouse and human genes that are involved in maintaining intestinal homeostasis and preventing inflammation. These data continue to support the hypothesis that inflammatory bowel disease results from a dysregulated adaptive immune response, particularly a CD4 T-cell response, to the microbiota. The microbiota itself is an active participant in these homeostatic processes. The microbiota composition is perturbed during inflammation, resulting in a dysbiosis that may induce or perpetuate inflammation. However, host genotype and the environment have a major impact on the shape of such dysbiosis, as well as upon which members of the microbiota stimulate pathogenic immune responses.
Keywords: B cell, T cell, T regulatory cell, Th1, Th17, adaptive immunity, dendritic cell, dysbiosis, epithelial cell, innate immunity, macrophage, microbiota, mucin
Introduction
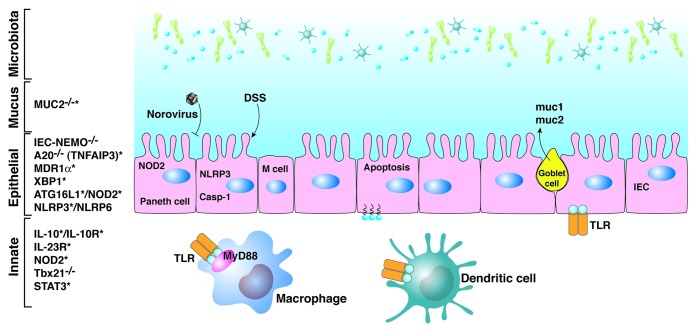
Over the past decade genetic technologies have opened new avenues of investigation in inflammatory bowel disease. The first of these was the ability to manipulate the genome of experimental animals, mostly mice, by deleting (knockout) or overexpressing (transgenic) individual genes collectively termed “induced mutant” mice. Virtually every immune gene has either been deleted or expressed as a transgene and a small subset of such mutants were found to develop inflammatory bowel disease. In virtually every instance, induced mutants made germ-free no longer developed IBD, indicating that the microbiota played a major role in disease pathogenesis. These models have also taught us that multiple pathways are involved, that CD4 T cells were the major effector cells mediating inflammation in most models, and that multiple hits were required in the innate, adaptive, and regulatory compartments for disease expression.1,2 A second important genetic technology has been the ability to identify IBD susceptibility genes in humans. The most recent version of these studies has been the genome-wide association studies, which have identified some 100 genes involved in IBD susceptibility.3 Interestingly, given that these studies were completely independent of the experimental models, these two scientific approaches have been largely convergent. Thus, many of the genes that have been identified by GWAS in humans fit into the current paradigm generated from experimental models that inflammatory bowel disease is due to a dysregulated adaptive immune response, particularly a CD4+ T-cell response to the intestinal microbiota. A third genetic technology has been culture-independent techniques allowing delineation of the intestinal microbiome, including PCR of the 16S rDNA gene, next generation sequencing, and the computational resources that are required to analyze such data.4 Together, these powerful tools allow experiments that were only dreamt about just 10 y or so ago. The cost of these advances, however, is data that is streaming out so fast that it is like drinking from the proverbial fire hose. In this review, we will start with the microbiota in the intestinal lumen and move into the mucosa discussing various cells and cell products that are involved in homeostasis with the microbiota (Fig. 1). The focus will be on gene variants or mutations that confer susceptibility to IBD as a framework for understanding the pathogenesis of IBD. For most of the gene mutations or variants, it’s unclear which cell or cells are most affected, and we will arbitrarily put them in the cellular context that seems most likely.
Figure 1. Innate immune interactions with the microbiota. Genes resulting in or conferring susceptibility to IBD are shown on the left. Genes targeted for deletion in mice that result in IBD and that have also been identified in humans as susceptibility loci are marked with an asterisk, and illustrate the convergence between experimental models and genome-wide association studies.
Microbiota Composition and Impact on the Host
The Human Microbiome Project and related studies have allowed a catalog to be assembled of the intestinal microbiota.5 There is high interpersonal variation in species composition of the gut microbiota in humans and no single species is present in all humans.6 However, when the metagenome of the human intestinal microbiota has been analyzed, a core composition of genes is evident and comparable among humans.7,8 There may be a limited number of enterotypes, which are marked by predominance of a particular family of microbes.9 The microbiota effects on the host are profound and occur at multiple levels. The microbiota clearly has a major impact on host metabolism, and can contribute to obesity, insulin resistance, and even metabolic syndrome.10 The microbiota also has effects on the vascular system, nervous system, and, of course, on intestinal development and function. The microbiota educates and causes the maturation of the immune system, both in the intestine and systemically. For example, mice with a limited microbiota have reduced populations of certain lymphoid subsets.11 Microbiota products prime neutrophils for effector function via NOD1 signaling.12 The intestinal microbiota affect immune responses in the lung to influenza virus13 and also can play a role in systemic autoimmunity in certain experimental models. However, the most profound effects are on the mucosal immune system itself, which contains most of the lymphocytes and immunoglobulin in the body. Not all members of the microbiota are equal in these various activities and it’s clear that low abundance members of the microbiota can have a major impact on the immune system.
Microbiota Changes during Intestinal Inflammation
Numerous studies have demonstrated that the microbiota shifts during inflammatory bowel disease, with a decrease in strict anaerobes, particular Firmicutes, and a bloom of proteobacteria14 (Table 1). There is also loss of some potentially anti-inflammatory species such as Faecalibacterium prausnitzii.15 Interestingly, similar changes are induced during infections by enteric pathogens such as Salmonella,16 Citrobacter, and Campylobacter.17 Induction of non-specific inflammation with oral dextran sodium sulfate (DSS) also induces similar changes. The consequences of these shifts in microbiota are unclear, particularly whether they are cause or effect. In favor of the latter, expansion of a particular strain of E. coli, adherent invasive E. coli, has been demonstrated in a subset of patients with Crohn’s ileitis.18 Tissue invasion by such AIEC may be due to defects in autophagy, which has been shown to be an important defense mechanism against this microbe.19 An expansion of proteobacteria has also been demonstrated as a feature of dysbiosis in the TRUC mouse, which is deficient in both T-bet and RAG genes and thus has no adaptive immune system and significant defects in innate immunity.20 This dysbiosis is marked by expansion of enterobacteriaceae, particularly of Klebsiella pneumonia and Proteus mirabalis, and this dysbiotic microbiota can transmit colitis even to wild type hosts.21 Although there is a similar expansion of proteobacteria, mostly Enterobacteriaceae, in a mouse line deficient in both IL-10 and TGFß signaling, colonization experiments have identified Bacteroides species, rather than Enterobacteriaceae, as important in triggering the colitis.22 Monocolonization of germ-free 129.IL10-deficient mice has demonstrated, in contrast to the above, that colonization with E. coli and Enterococcus faecalis induced colitis, whereas colonization with Bacteroides vulgatus and Pseudomonas fluorescens did not.23 In C3H/HeJBir mice that develop spontaneous colitis, serologic expression cloning has identified a limited set of some 60 microbiota antigens, among which flagellins from Clostridia, particularly Lachnospiraceae, were the largest cluster.24 Half of patients with Crohn’s disease have serum IgG to this cluster of flagellins. Based on BLAST matches of the remaining non-flagellin antigens, a diverse assortment of bacteria of origin are likely. However, no E. coli, Klebsiella, or Proteus antigens were detected. In these reports, mice are housed in different locations and have different genotypes that could impact the microbes and antigens that are inducing or being targeted by the immune system during IBD. The microbiota composition is clearly modulated by the host genotype25 and a strain that is detrimental for one genotype may be beneficial or neutral for another. There does not appear to be a single organism responsible for inflammatory bowel disease. Rather, these data are consistent with the hypothesis that a dysregulated host immune response to components of the microbiota is central to inflammatory bowel disease, but host genotype and the environment exert a major influence on which members and antigens of the microbiota are targeted by effector T cells.
Table 1. Microbiota changes during or contributing to gut inflammation.
|
Crohn’s disease |
↓Firmicutes ↑Proteobacteria (AIEC) Loss of F. prausnitzii |
|
Salmonella/Citrobacter infection |
↓Firmicutes ↑Proteobacteria (Salmonella/Citrobacter) |
|
TRUC mice |
↑Proteobacteria – Klebsiella, Proteus |
|
B6.L-10R2−/−.TGFßRIIDN |
↑Enterobactriaceae but Bacteroides → colitis |
|
129.IL-10−/− |
E. coli, E. faecalis → colitis |
|
C3H/HeJBir |
Clostridia/Lachnospiraceae |
| Normal C57Bl/6 | Enterotoxigenic B. fragilis |
Innate Immune Response to the Microbiota
Mice with innate immunity but lacking adaptive immune responses such as RAG−/− and SCID mice co-exist with the microbiota and do not develop inflammatory bowel disease spontaneously. However, various perturbations of such mice can result in inflammatory bowel disease, even in the absence of adaptive immunity. These include DSS-induced colitis,26 administration of an agonist anti-CD40 monoclonal antibody,27 T-bet−/−RAG−/− (TRUC) mice,20 and RAG mice infected with Helicobacter hepaticus.28 These model systems have allowed investigators to dissect mechanisms of innate inflammation and its regulation independent of adaptive immune cells. However, it should be noted that most induced mutant mice that develop IBD do not do so when crossed onto a RAG background, i.e., in the absence of adaptive immune cells, particularly CD4 T cells.1 Thus, these innate models are the exception rather than the rule and in most instances abnormalities of the innate immune response is pathogenic by resulting in a colitogenic T-cell effector response. That being said, many of the gene variants identified by GWAS studies involve innate pathways and these will be considered in the section below.
Mucin
The surface of the intestine is protected by an overlying layer of mucin that is generated by goblet cells in the epithelial layer. In addition to mucin proteins, the mucus layer contains antimicrobial peptides and IgA, which serve to select and limit access to this space closest to the epithelial cells and protect against pathogens such as attaching effacing E. coli.29 Goblet cells also product trefoil factor, which is important in epithelial restitution and repair. In the colon, a very tight mucin layer is present in mice, which excludes all bacteria.30 Above this layer is a loose layer of mucin in which certain organisms reside. The tight layer is formed by Muc2, the major intestinal secretory mucin. In the mouse, this tight layer increases in thickness along the length of the colon, being thickest in the distal colon. A variant in the Muc2 gene confers susceptibility for human IBD and Muc2-deficient mice develop colitis spontaneously,31 illustrating the importance of the mucus layer for mucosal homeostasis. Goblet cell depletion and an impaired mucus layer occur in human IBD and in Crohn’s disease; this allows bacteria to adhere directly to epithelial cells, which may contribute to disease progression.32 The mechanism by which mucosa-associated bacteria might cause more inflammation is unknown. It’s often postulated that bacteria closest to the mucosa would be more involved in disease pathogenesis. However, co-evolution of microbiota with host over the millennia makes it more likely that bacteria that live in the mucus layer are beneficial to the host. Mutations of the Muc2 gene, rather than its total absence, in mice can also cause colitis. These mutants cause endoplasmic reticulum stress in goblet cells. Genetic deletion of XBP1, a stress response protein, results also in depletion of goblet and Paneth cells and susceptibility to colitis in mice.33 Variants in XBP1 are also linked to human IBD.3
The Epithelium
The epithelium has many functions, not least of which are digestion, secretion, and absorption. The intestinal epithelium forms tight junctions between enterocytes that can exclude small molecules, thus forming a mucosal barrier to entry of most substances. Tight junctions are regulated by cytokines and other stimuli and dynamically respond to the local microenvironment. This exclusion of nutrients and small molecules is usually referred to as barrier function, however epithelial barrier function subsumes a broader role in that the epithelium also forms a barrier to the microbiota in the lumen. Positioned between the microbiota in the lumen and the immune cells in the intestinal lamina propria, the epithelium communicates with and is signaled by both. For example, the microbiota signals enterocytes as well as innate cells in the lamina propria via pattern recognition molecules via receptors such as the TLR, NOD, etc. In fact, such microbiota signaling via TLRs is required for normal epithelial organization and resistance to injury.34 Immune cytokines such as IFNγ, IL-17, and IL-22 have substantial effects on the epithelium. In turn, the epithelium secretes certain mediators, such as TSLP, IL-33, and IL-25, that “instruct” intestinal dendritic cells to become tolerogenic, i.e., to preferentially induce lamina propria Treg cells and thus maintain intestinal homeostasis.35 It should not be too surprising that genes linked to epithelial renewal, repair, stress response, and to antimicrobial peptide secretion have all been linked to IBD susceptibility in humans or mice.
The epithelial layer is renewed every 2–3 d by a process involving proliferation of epithelial cells in the crypts with migration up on the villi in the small intestine or onto the surface epithelium in the colon. The proliferation is, in turn, balanced by enterocyte apoptosis and shedding of the epithelium. Disruption of these processes impairs the epithelial barrier and results in chronic inflammation. NF-κB signaling in intestinal epithelial cells stimulates important anti-apoptotic molecules. Intestinal epithelial cell-specific deletion of NEMO (IKKγ) or of both IKKα and IKKß in mice results in apoptosis of colon epithelial cells, impaired expression of antimicrobial peptides, translocation of intestinal bacteria, and chronic colitis.36 Concomitant deficiency of MYD88 prevented the colitis due to a reduction in TNFα-induced apoptosis.36 A second molecule regulating epithelial apoptosis is A20 (TNFAIP3), an E3 ligase-editing enzyme required for inhibition of TLR and TNFα signaling. A20 gene variants have been linked to Crohn’s disease. Epithelial-specific deletion of A20 did not result in spontaneous colitis, but rendered the epithelial cells highly sensitive to TNFα-induced apoptosis and thus a high mortality upon challenge with DSS.37 Generalized deletion of A20 in mice results in spontaneous diffuse inflammation, including the intestine, resulting in cachexia and early death. This inflammation is dependent on microbiota-mediated homeostatic TLR-MYD88 signaling in myeloid cells.38
A SNP variant of the multi-drug resistance I gene (MDR1) has been linked to human IBD susceptibility.3 The homologous gene in mice, MDR1α, is expressed in intestinal epithelial cells and in some lamina propria myeloid cells. MDR1α, or p-glycoprotein, is an ATP binding cassette transporter that is postulated to pump toxic xenobiotic molecules out of epithelial cells and other cells, but its exact role in intestinal epithelial cells is not known. Mice deficient in MDR1α develop colitis spontaneously and antibiotic treatment prevents this colitis.39 T cells from MDR1 α colitic mice demonstrate increased responsiveness to the microbiota. Based on bone marrow chimera experiments, the disease is caused by MDR1α deficiency in non-hemopoietic, likely epithelial, cells making this an interesting example on which an epithelial cell defect results in an abnormal adaptive T-cell response to the microbiota.39 Colitis onset is accelerated by colonization with Helicobacter bilus. Interestingly, co-infection with H. bilus and murine norovirus 4 increased severity of the colitis, demonstrating an environment-microbiota-immune interaction leading to severe IBD.40
Variants in the gene encoding the transcription factor XBP1 have been linked to susceptibility to IBD in humans.3,33 XBP1 is part of the unfolded protein response (UPR), which, in turn, is important for the maintenance of cellular integrity, particularly of secretory epithelial cells. The UPR is activated by accumulation of unfolded or misfolded proteins in the endoplasmic reticulum. Conditional deletion of XBP1 in intestinal epithelial cells resulted in apoptosis of Paneth cells, reduced numbers of small intestinal goblet cells, and small intestinal inflammation.33 XBP1-deficient mice were more susceptible to DSS colitis, an effect that was dependent on the colonic microbiota.33 Human ileal and colonic biopsies from patients with IBD have signs of ER stress, demonstrating the relevance of this mouse model data for human IBD.
Another pathway that is activated by cell stress is autophagy.41 Autophagy is best known as a cellular process whereby a cell can recycle its components via the lysosomes, a process that is particularly important during starvation. However, autophagy is present widely in nature and provides host resistance to intracellular pathogens such as mycobacteria. Two autophagy genes, ATG16L1 and IRGM, are linked to Crohn’s disease.3 An ATG16L1 hypomorphic mouse line that expresses about 1% of the normal level of ATG16L1 demonstrates Paneth cell granule abnormalities that are very similar to those found in ileal resections in patients with Crohn’s disease who carry the relevant ATG16L1 gene variant.42 Although these ATG16L1HM do not develop IBD themselves, they are more susceptible to DSS-induced injury. When ATG16L1HM mice were re-derived virus-free, they lost the Paneth cell phenotype and the increased susceptibility to DSS-induced colitis. Infection with norovirus restored both the Paneth cell phenotype and the DSS colitis susceptibility, although the latter was prevented with treatment with broad spectrum antibiotics.43 This is another example of a virus-immune-microbiota interaction having a profound effect on disease expression. The pattern recognition receptor, NOD2, is highly expressed in Paneth cells and regulates their secretion of antimicrobial peptides.44 NOD2 has recently been found to regulate autophagy, including the autophagy pathway involving ATG16L145 linking these two IBD susceptibility loci. Moreover, ATG16L1 deficiency in mice has been found to increase endotoxin-induced inflammasome activation, resulting in high production of IL-1β and IL-18.46 In these studies, mice lacking ATG16L1 on in hematopoietic cells were highly susceptible to DSS colitis, which was mediated by IL-1β and IL-18. Thus, autophagy also appears to regulate also inflammasome-mediated inflammation.
Recent studies have identified the inflammasome as critical to resistance to innate injury with DSS and maintenance of homeostasis. Inflammasomes are multimeric proteins in the cytoplasm that form complexes that act as sensors of damage-associated molecular patterns and regulate the maturation and secretion of IL-1β and IL-18. NLRP3 inflammasomes, which include apoptosis-associated spec-like protein (ASC), activate caspase-1, which, in turn, cleaves pro-IL-1β and pro-IL-18 into their active forms. NLRP3 inflammasomes have been found to play an important role in resistance to DSS colitis. Deletion of NLRP3, ASC, caspase-1, or IL-18 all result in an increased severity and mortality to DSS colitis.47,48 NLRP3 gene variants have been linked to human IBD susceptibility, which may involve a similar mechanism.3
NLRP6-containing inflammasomes in colon enterocytes also provide resistance to DSS colitis, but surprisingly appear to regulate the composition of the colonic microbiota. Mice deficient in NLRP6 in colon enterocytes had reduced IL-18 levels and an altered fecal microbiota characterized by increases in the Provotellaceae of the Bacteroidetes phylum and increases in TM7.49 This microbiota conferred increased susceptibility to DSS colitis, which could be transferred to neonatal or adult wild type mice by co-housing. The mechanism for this shift in the microbiota and which members of the altered microbiota account for the increased DSS-colitogenicity are unknown. This highlights an important feature of DSS-induced injury, namely that DSS acts in concert with the microbiota to induce injury to the epithelial layer. The mechanism of such injury and the specific role of the microbiota in this process are unknown.
Innate Lymphoid Cells
Recent studies have identified a number of innate lymphoid cells that respond to the microbiota and resemble lymphoid tissue inducer (LTi) cells (Table 2). LTi cells are required for development of Peyer’s patches, lymph nodes, and intestinal lymphoid follicles. LTi cells do not have antigen receptors, but are identified by being lineage-negative and c-Kit+, CD127+ (IL-7 receptor-α), and RORγτ+.50 A LTi-like set of cells has now been identified in the intestinal mucosa, and are referred to as innate lymphoid cells (ILC). These also are RORγτ-positive and produce IL-22 and/or IL-17 rapidly upon either TLR or IL-23 signaling. These have been shown to play a role during innate colitis during which they produce IFNγ, IL-17A, and IL-22,27 because ablation of these innate lymphoid cells ameliorated colitis in this model. A CD4+ LTi-like cell appears to be a dominant source of IL-22 early during Citrobacter infection in mice and adoptive transfer of these cells could rescue normal lymphoreplete mice from death from Citrobacter infection.50 These innate lymphoid cells also express the aryl hydrocarbon receptor (AHR) and are dependent on AHR signaling for IL-22 production and survival.51 LTß receptor signaling in dendritic cells also regulates IL-22 production by innate lymphoid cells.52 A similar cell, but with Natural Killer cell (NK)-like markers, has also been found to be present in both human and mouse intestine.53 This cell has the NK marker CD56 and expresses either NK-p44 in humans or NK-p46 in mice. Both in human and mouse, these cells express the RORγτ transcription factor and are CD127 (IL-7Rα) positive and produce IL-22. The microbiota appears to drive their generation. This cell type has been found to be decreased in Crohn’s disease mucosa.54 Yet another cell type that has similar function to ILCs are a subset of Tγδ T cells, which are also RORγτ+, AHR+, CC6+, and IL-23R+. These Tγδ cells secrete IL-17, IL-21, and IL-22 in response to either IL-23 or IL1β without any TCR engagement.55 These cells express TLR1, TLR2, and Dectin-1 and can rapidly produce IL-17, amplifying IL-17 production by CD4+ T cells. All of these cells appear to serve as microbe sensors in the lamina propria via their TLR and AHR receptors, and are able to rapidly produce cytokines such as IL-17, IL-22, and IL-21 when triggered.56 IL-17 and IL-22 both act on the epithelium to maintain barrier function and thus these cells could be viewed as a way of tuning up or tuning down barrier function in normal homeostasis with the microbiota, as well as providing rapid protection during infection with enteric pathogens. Given that IL-23 signaling is important in at least some of these cells, this is another potential cell type that may be affected by variants in the IL-23 receptor gene that have been linked to IBD.
Table 2. Innate lymphoid cells.
| Cell | Markers | Cytokines | Reference |
|---|---|---|---|
| |
|
|
|
| LTi |
Lin- ckithi, CD127hi RORγt+ |
|
|
| LTi-like |
CD90+, CD4- or +, IL-23R+, RORγt+ |
IL-17, IL-22, IFNγ |
27 |
| |
Lin-, ckit+, CD4+, CD44+, CD127+, CD90+, CCR6+, IL-23R |
IL-22 |
50, 132 |
| |
CD3-, CD127+, CD4+ |
IL-22, IL-17 |
133 |
| |
AHR |
|
51 |
| NK-like |
CD56+, NKp44+, CCR6+ (Human), CD96+, CD103+, IL-23R, RORα+, RORγ+, AHR+, IRF4 |
IL-22, IL-26, LIF (without IL-23 stimulation) |
53 |
| |
NKp46+, NK1.1+/− (mouse) |
IL-22 |
46, 53 |
| γδ T cells | CCR6+, IL-23R+, RORγ+, AHR TLR1, 2, Dectin-1 |
IL-17, IL-22 IL-17, Il-21, IL-22 |
55, 56 |
LTi, lymphoid tissue inducer; LTi-like, innate lymphoid cells with features in common with LTi cells; NK, Natural Killer
Intestinal Macrophages
A substantial proportion of all innate myeloid cells in the intestine are macrophages. Intestinal macrophages have a distinctive phenotype compared with macrophages in other tissues or compared with blood monocytes from which they are derived. Human intestinal macrophages are strongly phagocytic and bactericidal, but do not produce cytokines and do not express a variety of innate immune response receptors, including CD14, which is required for LPS recognition.57 This state of “inflammatory anergy” is induced by TGFß derived from the stroma, which blocks TLR and NF-κB signaling.58 Mouse intestinal macrophages have had limited study, but a CD11b+F4/80+CD11c- cell that produces IL-10, but not inflammatory cytokines, and that induces Treg cells, has been identified.59 Intestinal macrophages thus serve as non-inflammatory scavengers that clear translocating microbiota and their products, thus preventing activation of potentially detrimental adaptive immune responses.
During infection or inflammation, monocytes move into the intestine and convert into macrophages and dendritic cells and retain the ability to produce inflammatory cytokines.60 For example, during Citrobacter infection in mice, recruitment of myeloid cells and T cells due to NOD2-dependent CCL2 production by stromal cells is critical to resolving the infection.61 This appears to be a general mechanism of host defense to enteric pathogens but occurs aberrantly, i.e., in the absence of a pathogen, in inflammatory bowel disease. Interestingly, recruitment of innate cells to sites of acute bacterial challenge in patients with Crohn’s disease appears to be defective, and this is postulated to contribute to the pathogenesis of IBD.62
Signal transducer and activation of transcription 3 (STAT3) mediates signaling of a number of cytokines, including IL-10. STAT3 has been identified as a susceptibility gene for human IBD. Conditional STAT3 deletion in macrophages and neutrophils results in colitis in mice, a phenotype that is quite similar to IL-10 deficiency. Macrophages from these mice do not respond to IL-10 and produce high amounts of TNFα, IL-1, IL-6, and IFNγ.63 Similarly, conditional deletion of STAT3 in bone marrow myeloid cells results in a severe enterocolitis in mice.64 Thus, IBD susceptibility due to STAT3 gene variants might be macrophage-mediated. However, STAT3 is expressed widely in multiple cells and the effects of its deletion appear to be cell intrinsic. For example, intestinal epithelial STAT3 is important for cell survival65 and epithelial repair,66 and T-cell STAT3 is required for induction of colitis in the CD45RBhi transfer model of colitis.67
Dendritic Cells
A number of genes affecting dendritic cell (DC) function cause or contribute to IBD. The intestinal lamina propria contains numerous DCs of different subsets. Mucosal DCs have distinctive properties and cell markers compared with DCs in lymphoid or other tissues.68 Among these are the preferential induction of Treg cells, induction of gut homing receptors on B cells and T cells, and the selective induction of IgA antibodies, all of which contribute to maintenance of gut homeostasis.69,70 Thus, like intestinal macrophages, under homeostatic conditions mucosal DCs are anti-inflammatory. Many of these features are due to DC conditioning by epithelial cells,35 and interestingly, these tolerogenic properties of mucosal DC require signaling via the β catenin-WNT pathway, which is critical in epithelial cell turnover.71 This situation changes during inflammatory conditions, with chemokines recruiting inflammatory DCs into the lamina propria from the circulation.60,72 The transition from homeostatic to inflammatory DCs occurs during pathogen invasion of the gut, but what triggers this shift in chronic relapsing IBD is unclear.
IL-10 is a critical cytokine for intestinal homeostasis. IL-10 deficiency in mice results in spontaneous colitis as it does also in humans. In the gut, IL-10 is produced by multiple cell types, including DCs. In DCs it provides feedback inhibition of IL-12 and TNFα. IL-10 signals via the transcription factor STAT3, genetic variants of which have also been linked to human IBD. STAT3 deficiency in mice results in chronic colitis with a phenotype similar to IL-10 deficiency. Myeloid cell-specific STAT3 deficiency results in a Crohn’s disease-like IBD syndrome, indicating that myeloid DCs are the critical cell in STAT3 deficiency-IBD.63 IL-10 deficient mice made germ-free do not develop colitis unless colonized by microbiota.23 Mono-association studies of germ-free IL-10 deficient mice have shown that E. faecalis and E. coli can induce colitis, whereas neither B. vulgatus nor P. fluoresceins did so.23 In this mono-association system, E. faecalis induced more distal disease and E. coli more proximal disease, suggesting that disease localization in IBD may be due to the biogeography of the microbiota in the lumen.
IL-23R SNPs have been linked to IBD in humans.3 IL-23 is produced by lamina propria dendritic cells and acts on both innate and adaptive immune cells. IL-23 is critical for induction of IL-22, a cytokine that enhances epithelial barrier function and antimicrobial peptide production and is absolutely required for resistance to infection with Citrobacter rodentium, a murine enteropathogen.73 IL-23 is also critical for the survival of CD4 Th17 cells, an effector subset discussed below, which is pathogenic in IBD and in several experimental autoimmune models. Monoclonal antibody neutralization of IL-23 in a microbiota-specific CD4+ T-cell transfer model of colitis prevented and treated disease.74 Perhaps related, an IL-23R SNP in humans is protective for Crohn’s disease, and this SNP has been shown to encode a truncated splice variant resulting in a soluble receptor inhibitor of IL-23.75
NOD2 is an intracellular pattern recognition receptor that binds muramyl dipeptide, a component of bacterial peptidoglycan. NOD2 gene polymorphisms are risk factors for Crohn’s disease. NOD2 is present in many cell types and has a number of functions. For example, acute muramyl dipeptide stimulation of DCs results in NF-κB and MAP kinase activation and inflammatory cytokine production. However, chronic stimulation of DCs with MDP reduces TLR- and NOD-induced cytokine production. Both of these effects are impaired in DCs from individuals with Crohn’s disease-associated polymorphisms.76 NOD2 has more recently been found to stimulate autophagy in DCs, which is required for resistance to intracellular bacteria and for antigen-specific MHC class II stimulation of CD4+ T cells.77 NOD2 was found to recruit ATG 16L1 to the plasma membrane at the bacteria (Salmonella) entry site.45 Crohn’s disease-related polymorphisms in either NOD2 or ATG 16L1 impaired this bacterial handling function.78 These data link two major susceptibility loci for Crohn’s disease into a single pathway of response to bacteria.
Adaptive Response to Microbiota in IBD
Although the host innate response to the microbiota plays a crucial role in the maintenance of intestinal immune homeostasis and in the pathogenesis of IBD, deficiencies in innate pathways alone, in most model systems, do not result in the disruption of homeostasis and development of intestinal inflammation. The latter requires an adaptive immune response to the microbiota. Host and microbiota interact with each other in an interdependent manner: colonization of intestinal tract with a diverse microbiota profoundly influences the development and function of both innate and adaptive arms of the host immune system, and educates local immune cells to generate a homeostatic response characterized by active immune readiness against pathogens and hyporesponsiveness against microbiota. Conversely, the host immune system shapes the composition of the gut microbiota both through innate responses and the production of bacteria-reactive IgA by B cells.79 The effects of the microbiota on the mucosal immune system are best demonstrated by germ-free (GF) mice in whom mucosal immunity is impaired. Reconstitution of GF mice with microbiota results in the expansion of innate cells, B cells and T cells in all mucosal compartments in the intestine to a level comparable to that of specific pathogen-free (SPF) mice. Both B cells and T cells, including T helper 1 (Th1), Th17, and Foxp3+ regulatory T (Treg) cells, are important for intestinal immune homeostasis in the presence of a nonpathogenic microbiota.
Influence of Microbiota Components on Adaptive Cells
Several recent studies have shown that members of the microbiota may act together or may act individually to affect mucosal adaptive responses.80-84 Some specific organisms have defined effects on mucosal immune function. Intestinal lamina propria has been identified as a “natural site” for Th17 cell development, in that Th17 cells are enriched in the intestinal lamina propria (LP) of SFP mice whereas they are absent in GF mice, indicating that microbiota induce Th17 cells in the lamina propria. Monocolonization of GF mice with segmented filamentous bacteria (SFB) restores mucosal Th17 cells, indicating that SFB stimulate mucosal Th17 cell development. Interestingly, SFB have also been shown to induce intestinal IgA. How SFB promote intestinal Th17 cell development and IgA production is still unknown, although the close interaction of SFB with epithelial cells and Peyer’s patches could be involved in such effects. A toxigenic form of human B. fragilis is also able to induce mucosal Th17 cells and causes colitis, which is associated with toxin-induced signal transducer and activator of transcription (STAT)-3 activation in epithelial cells and immune cells.85 A recent study demonstrated that, in the absence of a mutualistic Treg cell response, intestinal altered Schaedler flora (ASF), which contains 8 defined bacteria but no SFB or B. fragilis, could also induce intestinal Th17 cells, indicating that combinations of microbial species can induce Th17 cells in the intestine.86 Although induction of Th17 cells by SFB has been implicated in the pathogenesis of arthritis and experimental autoimmune encephalomyelitis (EAE),85,87 the role of SFB induction of Th17 cells in the pathogenesis of IBD is unknown. SFB together with multiple defined SPF species was effective in triggering intestinal inflammation in reconstituted SCID mice whereas there was no colitis in GF mice colonized either with the defined SPF species or monoassociated only with SFB.88
Treg cells are also constitutively present and enriched in the intestinal lamina propria. There is evidence that indigenous microflora are potent inducers of mucosal Treg cells.80,83,84,86 Colonization of GF mice with commensal Bacteroides fragilis (B. fragilis) was able to induce the development of CD4+Foxp3+ regulatory T (Treg) cells that produced interleukin (IL)-10. This effect was mediated through production of polysaccharide A (PSA) as it did not occur after colonization with a B. fragilis PSA mutant. Importantly, B. fragilis-induced Treg cells protected mice from colitis induced by Helicobacter hepaticus infection via Treg production of IL-10.83 The colonization of GF mice with a defined mixture of 46 strains of commensal Clostridia was able to induce Treg cells.80 These Clostridia species, mostly members of class XIVa, preferentially colonize and form a thick layer on the cecal and colon mucosa. Interestingly, colonization with Clostridia also induced epithelial cell expression of indoleamine 2,3-dioxygenase (IDO), which could induce Treg cells locally. In a more truly mutualistic immunological adaptation model, colonization of GF mice with ASF was also shown to be able to induce Treg cells in the intestinal mucosa.84 Thus, the ability of certain microbiota species to induce either Th17 cells or Treg cells reflects a differential regulation of the mucosal immune system by different members of the microbiota resulting in maintenance of immune homeostasis with the host.
Effector T cells in IBD
Both Th1 and Th17 cells have been implicated as important mediators of inflammation in IBD.74,89-92 There is an increased production of IL-12 and IL-23 in Crohn’s disease, and MLN DC from patients with Crohn’s diseases induce both Th1 and Th17 immune responses.93-97 T cells of inflamed Crohn’s disease lesions contain high levels of activated STAT4 and T-bet, the Th1-associated transcription factors indicative of IL-12 signaling.98,99 The important role of Th17 cells, which express the IL-23 receptor (IL-23R) on their surface, in the pathogenesis of IBD is supported by recent genome-wide association studies indicating that IL23R and other genes involved in Th17 differentiation (IL12B, JAK2, STAT3, CCR6 and TNFSF15) are associated with susceptibility to Crohn’s disease and partly also to ulcerative colitis100-102 ( Fig. 2). Anti-IL-12/IL-23p40 antibody therapy, which targets both Th1 and Th17 cells, is effective in Crohn’s disease.103,104 Anti-IL-23p19 mAb prevents as well as treats colitis in a colitis model induced by microbiota-specific T cells, further confirming a role of the IL-23/Th17 pathway in the pathogenesis of chronic intestinal inflammation.74 However, in patients with Crohn’s disease, a subset of CD14+ macrophages producing IL-23 and TNFα has been identified that contributes to the pathogenesis of Crohn’s disease by promoting IFNγ production rather than IL-17 production by lamina propria mononuclear cells.105 Significant IL-17 mRNA upregulation is found in lamina propria CD4+ cells from patients with ulcerative colitis, whereas IFNγ is increased in Crohn’s disease, arguing somewhat against the concept that IL-23 contributes only to Th17 cytokine production,94 because IL-23 can well promote Th1 cell IFNγ production. A number of reports have identified a subset of Th17 cells that co-produce the Th1 cytokine IFNγ.106,107 This is particularly prominent at sites of inflammation such as those found in active Crohn’s disease.107
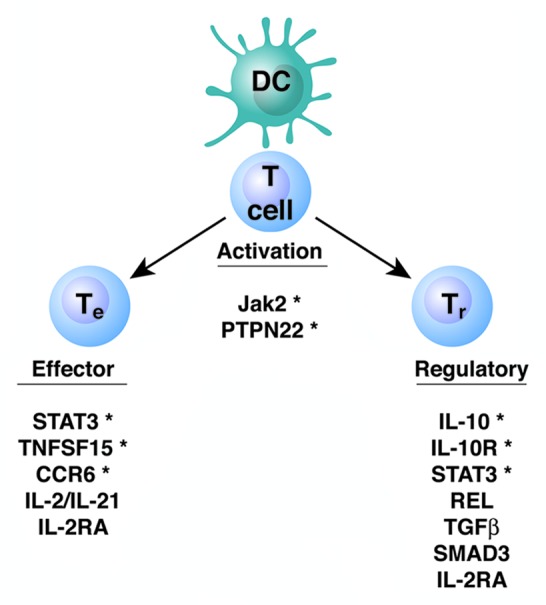
Figure 2. Genes involved in the adaptive immune response and linked to IBD susceptibility. The asterisk denotes genes linked to IBD in both experimental models and in human genome-wide association studies.
Microbiota antigen stimulation of T cells plays a crucial role in the pathogenesis of colitis. T cells respond to specific antigens but little is known about which microbiota antigens are driving the pathogenic T-cell response in IBD. Using serologic expression cloning, microbiota flagellins have been identified as immunodominant antigens in patients with Crohn’s disease and in multiple models of colitis.24 Flagellin possesses the properties of both a potent antigen for adaptive responses and is a ligand for Toll-like receptor (TLR)-5 ligand that stimulates innate responses. Thus, flagellin can bridge innate and adaptive responses, and provides a window into the normal host immune response to its microbiota and abnormal responses resulting in colitis development. In an adoptive transfer model, Th17 cells specific for CBir1 flagellin were more potent than CBir1 flagellin cells in the induction of colitis. However, Th17 cells are not stable and some Th17 cells produce IFNγ and convert into Th1 cells during intestinal inflammation, which might contribute to their increased potency. Interestingly, IL-17 produced by Th17 cells promotes T-cell IFNγ production via induction of local mucosal DC IL-12 and IL-23 production in the inflamed lesions.108 However, the complex relationship between Th1 and Th17 cell-mediated pathophysiology has not been completely analyzed due to a lack of appropriate reporter mice and suitable colitis models. This will be of great importance for delineating the specific contributions of these cells to chronic intestinal inflammation, especially to the persistence and progression of colitis.
Adaptive Regulatory Cells
The role of Treg in intestinal homeostasis
Among the multiple mechanisms that regulate intestinal immune homeostasis and control the compartmentation of immune responses to microbiota antigens, regulatory T cells (Treg), mainly including CD25+CD4+Foxp3+ Tregs and Tr1 cells, play an essential role. Treg cells constitutively express their signature transcription factor Foxp3, which is required for their development as well as their regulatory activity. CD25+CD4+Foxp3+ Tregs can be generated in the thymus (natural Treg cells) as well as in periphery (induced Treg cells) through antigen stimulation in the presence of TGF-β. Foxp3+ Treg cells distribute to essentially all lymphoid organs with the frequencies of approximately 10% within the CD4+ cell subset, but are enriched in the intestinal mucosa. Although both natural and induced Treg cells are present in intestinal lamina propria, the amino-acid sequences of TCRs on Treg cells from the mouse colon are different from those of receptors on T cells from other organs.109 Many of these TCRs on colonic Treg cells recognize antigens derived from the intestinal microbiota. In addition, colonic mucosal Treg cell numbers are decreased in GF mice and antibiotic-treated mice.80,86 It is thus likely that most Tregs in the intestine are induced by the microbiota, which plays a critical role in the accumulation and functional maturation of intestinal Treg cells. It has been shown that colonization of GF mice with commensal B. fragilis, Clostridium spp, or ASF induces Treg cells in intestinal mucosa, although the mechanisms involved could be different.80,83,84,86
Germline mutation in Foxp3 in humans results in multiple organ inflammation (X-linked IPEX syndrome),110 indicating the importance of Treg cells in maintaining intestinal homeostasis in humans. Mice with Foxp3 deficiency also develop fatal multiorgan inflammation including chronic intestinal inflammation that can be suppressed by adoptive transfer of Foxp3+ Treg cells. Treg cells also inhibit experimental colitis induced by adoptive transfer of CD4+CD45RBhi naive T cells to immunodeficient mice, and Treg can cure established transfer colitis in an IL-10 dependent manner.111 Among several independent mechanisms mediating the regulatory activity of Treg cells, mice lacking anti-inflammatory cytokine IL-10 or TGF-β in T cells develop wasting disease and colitis when they are housed under SPF conditions,112,113 highlighting the importance of IL-10 and TGF-β in intestinal homeostasis. Foxp3+ and Foxp3- Treg cells in the colonic LP CD4+ T-cell population produce IL-10 with about one-third of the IL-10-producing colon CD4+ T cells being Foxp3+.114 CD4+ T cell- or Treg-specific deletion of IL-10 in mice results in spontaneous colitis. Thus, Treg production of IL-10 is indispensable for the maintenance of intestinal immune homeostasis. Recent GWAS studies have identified IL-10 as a susceptibility locus for the development of IBD in humans and the patients lacking functional IL10 receptor (IL10R) develop early onset colitis,115,116 implicating IL-10 in the regulation of human intestinal immune homeostasis and its impairment in the pathogenesis of IBD in humans. It is currently unknown, however, why T-cell-produced TGF-β and IL-10 play such a decisive role when TGF-β and IL-10 produced by other cells are readily available in the intestinal environment.
T regulatory cells in IBD
Paradoxically, in patients with UC and CD, as well as in animal models of colitis, there are increased numbers of Foxp3+ Tregs and high levels of their signature cytokines TGFβ and IL-10 in inflamed intestinal lesions.111,117-120 Tregs can constitute as much as 20–30% of total CD4+ T cells in inflamed lamina propria (LP), which is much higher than the percentage of Tregs in the steady-state (5–10%). Tregs in inflamed lesions express TNFR2, and anti-TNFα treatment, effective in a majority of IBD patients to inhibit intestinal inflammation, results in a decrease of local mucosal Tregs with a concomitant increase of blood Tregs,119 suggesting that high levels of inflammation and inflammatory cytokines can also promote Treg expansion and accumulation in inflamed lesions. It is still hotly debated whether Tregs in inflamed lesions remain functional, as some report functional Tregs while others describe resistance of effector T cells to Tregs in the inflamed lesions in IBD. The presence of increased Tregs in the affected tissues of patients with IBD and animal models may mean that failed regulation in the inflamed tissues is due to an insufficient or defective Treg function as a result of intrinsic defects in Tregs or Teff or extrinsic factors present at the site of intestinal inflammation. However, due to a lack of suitable models, most studies have been done either in vitro or ex vivo, and these did not completely reproduce the physiological conditions in inflamed intestinal lesions. Investigating Treg and Teff functions during the course of chronic intestinal inflammation and mechanisms of Treg expansion during inflammation will be essential to allow us to achieve the goal of restoring intestinal homeostasis in IBD patients.
The T regulatory-IgA axis
A key component of intestinal homeostasis is IgA, the most abundant antibody isotype produced in vivo, which accounts for over 75% of the total antibody synthesized. Unlike the other isotypes, which are mainly found in serum and other lymphoid organs, IgA is a mucosal immunoglobulin and is secreted across the mucosal epithelium. Intestinal IgA plays a key role in intestinal immune protection in a non-inflammatory manner. Secreted IgA limits bacterial association with the intestinal epithelial cell surface and restricts the penetration of symbiotic bacteria across the gut epithelium by inducing bacterial agglutination, masking of bacterial proteins involved in epithelial attachment, and anchoring bacterial cells to mucus. IgA also shapes the microbiota bacterial communities in the intestine and decreases the inflammatory tone of the intestines. Intestinal IgA has also been shown to shape the metabolic features of intestinal epithelial cells and such function depends on the presence of intestinal microbiota.121 Lack of IgA changes the composition of the intestinal microbiota in a manner that can cause hyperactivation of the immune system and inflammation.79,122,123 IgA has long been shown for a long time to participate in host responses against infection, however, the major role of IgA probably is to maintain a balance between the host and its microbiota, as evidenced by SPF mice that have no pathogen exposure but have abundant IgA, and the greatly reduced intestinal IgA levels in germ-free mice.
IgA also regulates systemic T-cell responses to microbiota.124 Large amounts of IgA against microbiota antigens are present in the intestinal lumen. For example, high levels of intestinal IgA but not systemic IgG responses against commensal CBir1 flagellin are detected in wild-type mice. CBir1 Tg T cells do not respond to CBir1 flagellin delivered mucosally into wild-type mice but do proliferate well to mucosal CBir1 challenge when transferred into IgA−/− mice. Collectively, these studies indicate that antigen-specific intestinal IgA plays a critical role in regulating systemic CD4+ T-cell responses to commensal bacterial antigens by limiting microbiota antigen uptake.
IgA induction against commensal bacteria occurs in the intestine in both a T-cell-dependent and independent manner. Although some studies suggested that the production of low affinity IgA response for microbiota is mainly T-cell-independent, in TCRβxδ−/− mice lacking both αβ and γδ T cells, the total levels of secretory IgA, as well as commensal bacterial antigen-specific intestinal IgA, are reduced to about one-fourth of the wild-type levels, even though the same specificities are generated.124,125 These data indicate that the T-cell-dependent IgA response is predominant in the intestine. Among multiple mechanisms regulating IgA production, TGF-β is probably the most important cytokine that promotes IgA induction, as TGF-β−/− mice have low levels of IgA and there is almost a complete absence of IgA in mice deficient for TGF-β receptor II.126 Acute depletion of Treg cells results in a decrease of lamina propria IgA+ B cells, with a substantial reduction in luminal total and anti-CBir1 flagellin-specific IgA. Adoptive transfer of Tregs restores total and CBir1-specific intestinal IgA production in TCRβxδ−/− mice. Further studies indicate that Treg cells promote B cell AID expression and IgA production through production of TGF-β.124 Collectively, these data demonstrate that Tregs play a dominant role in regulating intestinal IgA production to microbiota antigens, and in preventing systemic T-cell activation to them in mice.
Regulatory B cells
In addition to producing antibody and presenting antigens to T cells, some murine B cells negatively regulate immune responses. Regulatory B cells (Breg) have been shown in experimental autoimmune diseases, such as EAE, as well as in IBD. Breg were first described in IBD in TCRα−/− mice, which develop a UC-like colitis spontaneously; B cell-deficient TCRα−/− double KO (DKO) mice developed much more severe intestinal inflammation.127 Breg have been identified also in some Crohn’s disease-like disease models. A potent subset of regulatory B cells, B10 cells, with a phenotype of CD1dhiCD5+, has been shown to regulate DSS-induced colitis in an IL-10-dependent manner.128 B10 cells are the predominant source of B-cell IL-10 production. Adoptive transfer of antigen-primed B10 cells ameliorates the severity of colitis in TCRα−/− mice as well as in the DSS-model.128,129 Interestingly, Breg numbers are increased in autoimmune diseases as well as in IBD, thus Breg are possibly induced under inflammatory conditions and are capable of suppressing the exacerbation of inflammation and/or enhancing the recovery process.
Unanswered Questions and Future Investigation
The relationship between innate and adaptive immunity is poorly understood, particularly how defects of innate immunity result in disease. A common assumption is that innate immune stimulation, for instance via TLR ligands, in turn activates adaptive immunity. This is thought to be the basis of adjuvant function for vaccine immunization. However, surprisingly, TLR-activating adjuvants were able to immunize mice in which all TLR signaling was ablated. The microbiota stimulation of innate immunity appears to be required for induction of colitis, even colitis induced by colitogenic CD4 T cells reactive to a microbiota flagellin.130 That being said, decreased innate immune reactivity has been found to be associated with increased T-cell responses to microbiota antigens such as flagellin.131 Thus, how genetic variants or defects in innate immunity result in pathogenic CD4 T cell responses to microbiota antigens remains a topic that needs further investigation. Norovirus infection exacerbates the Mdr1α−/− colitis and is required for the Paneth cell abnormality and DSS sensitivity seen in ATG16L1HM mice. The mechanisms of these viral effects are undefined. Perturbation of mucosal immunity by other pathogens disturbs homeostasis with the microbiota, but the microbiota is restored in normal animals, much less how pathogens impact an IBD susceptibility gene variant, is unknown. GWAS studies have identified over 100 genes whose variants result in susceptibility to IBD. These are likely to affect expression of the gene products, but are unlikely to cause complete deletion such as exists in knockout mice. How partial reductions in gene expression results in IBD susceptibility remains to be defined. It seems likely that multiple genes will need to be defective to result in IBD, the “multi-hit” hypothesis.1,28 Turning to the microbiota, is dysbiosis a consequence or cause of IBD? Will restoring “normobiosis” be beneficial or detrimental in IBD in that resolving the dysbiosis may provide more antigens to drive a pathogenic T-cell response. Do rare strains with potent biologic effects have more of an impact in IBD compared with an altered community composition of the microbiota? Lastly, what are the antigens driving pathogenic CD4 T-cell responses? Clearly there is selectivity and immunodominance among certain classes of antigen. How this immunodominance is determined is at present unknown but ripe for future investigation. This is an exciting time for this field of research because we now have the tools to answer these and other important questions in IBD.
Acknowledgments
This work was supported by research grants from NIH grants DK071176, DK079918, Digestive Diseases Research Development Center grant DK064400, and NIH RR20136.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20228
References
- 1.Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–76. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 2.Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 3.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, et al. Human Microbiome Jumpstart Reference Strains Consortium A catalog of reference genomes from the human microbiome. Science. 2010;328:994–9. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara D, Wei B, Presley LL, Brewer S, McPherson M, Lewinski MA, et al. Systemic control of plasmacytoid dendritic cells by CD8+ T cells and commensal microbiota. J Immunol. 2008;180:5843–52. doi: 10.4049/jimmunol.180.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 19.Lapaquette P, Glasser A-L, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieleman LA, Ridwan BU, Tennyson GS, Beagley KW, Bucy RP, Elson CO. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. 1994;107:1643–52. doi: 10.1016/0016-5085(94)90803-6. [DOI] [PubMed] [Google Scholar]
- 27.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–5. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 29.Bergstrom KSB, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjövall H, et al. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 33.Kaser A, Lee A-H, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Iliev ID, Spadoni I, Mileti E, Matteoli G, Sonzogni A, Sampietro GM, et al. Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut. 2009;58:1481–9. doi: 10.1136/gut.2008.175166. [DOI] [PubMed] [Google Scholar]
- 36.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–61. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 37.Vereecke L, Sze M, Mc Guire C, Rogiers B, Chu Y, Schmidt-Supprian M, et al. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–23. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, et al. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J Exp Med. 2008;205:451–64. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol. 1998;161:5733–44. [PubMed] [Google Scholar]
- 40.Lencioni KC, Seamons A, Treuting PM, Maggio-Price L, Brabb T. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp Med. 2008;58:522–33. [PMC free article] [PubMed] [Google Scholar]
- 41.Huett A, Xavier RJ. Autophagy at the gut interface: mucosal responses to stress and the consequences for inflammatory bowel diseases. Inflamm Bowel Dis. 2010;16:152–74. doi: 10.1002/ibd.20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cadwell K, Patel KK, Maloney NS, Liu T-C, Ng ACY, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J. Defensins and Paneth cells in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1284–92. doi: 10.1002/ibd.20197. [DOI] [PubMed] [Google Scholar]
- 45.Travassos LH, Carneiro LAM, Ramjeet M, Hussey S, Kim Y-G, Magalhães JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 47.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti T-D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–91. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KSB, McIntire CR, LeBlanc PM, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–78. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–79. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–34. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JKM, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–5. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, et al. Imbalance of NKp44(+)NKp46(-) and NKp44(-)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139:882–92, 892, e1-3. doi: 10.1053/j.gastro.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 55.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KHG. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–41. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–30. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem. 2010;285:19593–604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–94. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 60.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–55. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y-G, Kamada N, Shaw MH, Warner N, Chen GY, Franchi L, et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34:769–80. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith AM, Rahman FZ, Hayee B, Graham SJ, Marks DJ, Sewell GW, et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn’s disease. J Exp Med. 2009;206:1883–97. doi: 10.1084/jem.20091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Förster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/S1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 64.Welte T, Zhang SSM, Wang T, Zhang Z, Hesslein DGT, Yin Z, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100:1879–84. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grivennikov S, Karin E, Terzic J, Mucida D, Yu G-Y, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–13. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–72. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–15. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rescigno M, Di Sabatino A. Dendritic cells in intestinal homeostasis and disease. J Clin Invest. 2009;119:2441–50. doi: 10.1172/JCI39134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205:2139–49. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang YC, et al. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 2010;329:849–53. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siddiqui KRR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32:557–67. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 74.Elson CO, Cong Y, Weaver CT, Schoeb TR, McClanahan TK, Fick RB, et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–70. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 75.Kan SH, Mancini G, Gallagher G. Identification and characterization of multiple splice forms of the human interleukin-23 receptor alpha chain in mitogen-activated leukocytes. Genes Immun. 2008;9:631–9. doi: 10.1038/gene.2008.64. [DOI] [PubMed] [Google Scholar]
- 76.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–5. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 78.Homer CR, Richmond AL, Rebert NA, Achkar J-P, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–41, e1-2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–7. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 80.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geuking MB, Cahenzli J, Lawson MAE, Ng DCK, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 82.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 83.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 84.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 87.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–11. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 89.Berg DJ, Davidson N, Kühn R, Müller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–20. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157:1261–70. [PubMed] [Google Scholar]
- 91.Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–62. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 92.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fuss IJ, Becker C, Yang Z, Groden C, Hornung RL, Heller F, et al. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis. 2006;12:9–15. doi: 10.1097/01.MIB.0000194183.92671.b6. [DOI] [PubMed] [Google Scholar]
- 94.Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut. 2008;57:1682–9. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 95.Monteleone G, Biancone L, Marasco R, Morrone G, Marasco O, Luzza F, et al. Interleukin 12 is expressed and actively released by Crohn’s disease intestinal lamina propria mononuclear cells. Gastroenterology. 1997;112:1169–78. doi: 10.1016/S0016-5085(97)70128-8. [DOI] [PubMed] [Google Scholar]
- 96.Sakuraba A, Sato T, Kamada N, Kitazume M, Sugita A, Hibi T. Th1/Th17 immune response is induced by mesenteric lymph node dendritic cells in Crohn’s disease. Gastroenterology. 2009;137:1736–45. doi: 10.1053/j.gastro.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 97.Schmidt C, Giese T, Ludwig B, Mueller-Molaian I, Marth T, Zeuzem S, et al. Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-27p28 in Crohn’s disease but not in ulcerative colitis. Inflamm Bowel Dis. 2005;11:16–23. doi: 10.1097/00054725-200501000-00003. [DOI] [PubMed] [Google Scholar]
- 98.Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J Exp Med. 2002;195:1129–43. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wirtz S, Finotto S, Kanzler S, Lohse AW, Blessing M, Lehr HA, et al. Cutting edge: chronic intestinal inflammation in STAT-4 transgenic mice: characterization of disease and adoptive transfer by TNF- plus IFN-gamma-producing CD4+ T cells that respond to bacterial antigens. J Immunol. 1999;162:1884–8. [PubMed] [Google Scholar]
- 100.Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, Fisher SA, et al. Wellcome Trust Case Control Consortium Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–9. e3. doi: 10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 102.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mannon PJ, Fuss IJ, Mayer L, Elson CO, Sandborn WJ, Present D, et al. Anti-IL-12 Crohn’s Disease Study Group Anti-interleukin-12 antibody for active Crohn’s disease. N Engl J Med. 2004;351:2069–79. doi: 10.1056/NEJMoa033402. [DOI] [PubMed] [Google Scholar]
- 104.Sandborn WJ, Feagan BG, Fedorak RN, Scherl E, Fleisher MR, Katz S, et al. Ustekinumab Crohn’s Disease Study Group A randomized trial of Ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology. 2008;135:1130–41. doi: 10.1053/j.gastro.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 105.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. 2008;118:2269–80. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–46. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 107.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–61. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feng T, Qin H, Wang L, Benveniste EN, Elson CO, Cong Y. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J Immunol. 2011;186:6313–8. doi: 10.4049/jimmunol.1001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 111.Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, et al. Characterization of Foxp3+CD4+CD25+ and IL-10-secreting CD4+CD25+ T cells during cure of colitis. J Immunol. 2006;177:5852–60. doi: 10.4049/jimmunol.177.9.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–91. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 113.Roers A, Siewe L, Strittmatter E, Deckert M, Schlüter D, Stenzel W, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–97. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–41. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 115.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. IBSEN study group Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 116.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eastaff-Leung N, Mabarrack N, Barbour A, Cummins A, Barry S. Foxp3+ regulatory T cells, Th17 effector cells, and cytokine environment in inflammatory bowel disease. J Clin Immunol. 2010;30:80–9. doi: 10.1007/s10875-009-9345-1. [DOI] [PubMed] [Google Scholar]
- 118.Holmén N, Lundgren A, Lundin S, Bergin AM, Rudin A, Sjövall H, et al. Functional CD4+CD25high regulatory T cells are enriched in the colonic mucosa of patients with active ulcerative colitis and increase with disease activity. Inflamm Bowel Dis. 2006;12:447–56. doi: 10.1097/00054725-200606000-00003. [DOI] [PubMed] [Google Scholar]
- 119.Li Z, Arijs I, De Hertogh G, Vermeire S, Noman M, Bullens D, et al. Reciprocal changes of Foxp3 expression in blood and intestinal mucosa in IBD patients responding to infliximab. Inflamm Bowel Dis. 2010;16:1299–310. doi: 10.1002/ibd.21229. [DOI] [PubMed] [Google Scholar]
- 120.Maul J, Loddenkemper C, Mundt P, Berg E, Giese T, Stallmach A, et al. Peripheral and intestinal regulatory CD4+ CD25(high) T cells in inflammatory bowel disease. Gastroenterology. 2005;128:1868–78. doi: 10.1053/j.gastro.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 121.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–93. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 123.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–70. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 124.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–61. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–5. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 126.Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–51. doi: 10.1016/S1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 127.Mizoguchi A, Mizoguchi E, Smith RN, Preffer FI, Bhan AK. Suppressive role of B cells in chronic colitis of T cell receptor alpha mutant mice. J Exp Med. 1997;186:1749–56. doi: 10.1084/jem.186.10.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yanaba K, Yoshizaki A, Asano Y, Kadono T, Tedder TF, Sato S. IL-10-producing regulatory B10 cells inhibit intestinal injury in a mouse model. Am J Pathol. 2011;178:735–43. doi: 10.1016/j.ajpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16:219–30. doi: 10.1016/S1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 130.Feng T, Elson CO, Cong Y. Microbiota: dual-faceted player in experimental colitis. Gut Microbes. 2010;1:388–91. doi: 10.4161/gmic.1.6.13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beckwith J, Cong Y, Sundberg JP, Elson CO, Leiter EH. Cdcs1, a major colitogenic locus in mice, regulates innate and adaptive immune response to enteric bacterial antigens. Gastroenterology. 2005;129:1473–84. doi: 10.1053/j.gastro.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 132.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Bérard M, Kleinschek M, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–6. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 133.Van Maele L, Carnoy C, Cayet D, Songhet P, Dumoutier L, Ferrero I, et al. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J Immunol. 2010;185:1177–85. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]