Abstract
Any multicellular organism may be considered a metaorganism or holobiont—comprised of the macroscopic host and synergistic interdependence with bacteria, archaea, fungi, viruses, and numerous other microbial and eukaryotic species including algal symbionts. Defining the individual microbe-host conversations in these consortia is a challenging but necessary step on the path to understanding the function of the associations as a whole. Dissecting the fundamental principles that underlie all host-microbe interactions requires simple animal models with only a few specific bacterial species. Here I present Hydra as such a model with one of the simplest epithelia in the animal kingdom, with the availability of a fully sequenced genome and numerous genomic tools, and with few associated bacterial species.
Keywords: cnidaria, innate immunity, commensal microbiota, epithelial cells
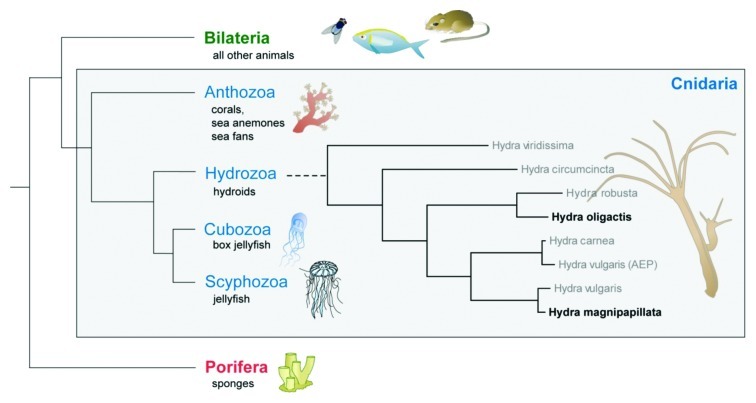
As member of the phylum Cnidaria, Hydra bears a deep metazoan ancestry (Fig. 1). Paleontological findings of fossils from basal Cambrian rocks in China and Siberia containing exceptionally well preserved cnidarian embryos1 suggest that cnidarians were members of the pre-Ediacaran fauna between 1,200 and 600 million years ago and diverged from the main line of metazoan evolution long before the pre-Cambrian radiation. Cnidarians not only are among the earliest known phyletic lineages to form natural symbiotic relationships but also possess most of the gene families found in bilaterians and have retained many genes that have been lost in Drosophila and Caenorabditis elegans.2-4 Since the genome organization and genome content of cnidarians is remarkably similar to that of morphologically much more complex bilaterians,5-7 these animals offer unique insights into the content of the “genetic tool kit” present in the cnidarian-bilaterian ancestor. Surprising in light of the simple body plan is the relatively large genome of Hydra magnipapillata of 1300 Mb.8 Up to 40% of the genome are transposable elements.8 Whether this, in combination with horizontal gene transfer and trans-splicing, allows the constantly regenerating and asexually proliferating polyps to quickly adapt to changing environmental conditions, remains a matter of debate.
Figure 1. The early occurrence of Cnidaria on Earth. Cnidarians serve as models for studying pattern formation, the origin of stem cell systems, and the evolution and function of host-microbe interactions (modified from ref. 44).
For analytical purposes, Hydra is an almost perfect model. Clonally growing mass cultures can be kept easily in plastic or glass dishes for decades. Additionally, the efficient generation of transgenic Hydra lines by embryo microinjection9 allows functional analysis of genes controlling development and immune reactions as well as in vivo monitoring of host-microbe interactions. Last, but not least, to facilitate access and analysis of genomic and transcriptomic data of the members of the Hydra holobiont, a local bio-computational platform termed “compagen”5 has been established. The database at http://www.compagen.org, containing selected raw genomic and expressed sequence tag (EST) data sets from sponges and cnidarians up to the lower vertebrates, offers convincing proof for the above mentioned view that cnidarians share most of their genes with the Bilateria and that many human disease genes already evolved in the common ancestor of the Cnidaria and Bilateria2,4,10-12.
Hydra, a model system in evolutionary developmental biology since the 1980´s,13-15 is organized as a gastric tube with a mouth and tentacle ring at the oral end of the single body axis, and a peduncle and mucous cell disc at the aboral pole (Fig. 2A). Its body wall along the entire longitudinal axis is structurally reduced to an epithelio-muscular bilayer with an intervening extracellular matrix and a few cell types derived from three distinct stem cell lineages (Fig. 2B and D). The tube-like body resembles in several aspects the anatomy of the vertebrate intestine with the endodermal epithelium lining the gastric cavity and the ectodermal epithelium providing a permanent protection barrier to the environment (Fig. 2B, C). Similar to Hydra´s endothelium, the mammalian intestinal tract is essentially a tube16 with the epithelium, here termed the mucosa, being responsible for the processing and absorption of nutrients. The intestinal epithelium renews every 5 days and is fueled by proliferative stem cells that differentiate into absorptive enterocytes and three classes of secretory cells.17-19 The endothelium in Hydra is considered the simplest epithelia in the animal kingdom. Because of the reduced number of cell types, differentiated cells in Hydra are multifunctional. For example, epithelial cells in both layers have both secretory and phagocytic activity20,21 and also function in other processes such as fluid and electrolyte transport. Although there are no motile immune effector cells or phagocytes present in sensu strictu, endodermal epithelial cells not only contribute to digestion and uptake of food, but also are capable of phagocytosing bacteria present in the gastric cavity.21 Similar to the cells of the mammalian intestinal tract, the cells in Hydra are also in a dynamic state of proliferation and turnover. Epithelial cells continuously proliferate along the body column leading to an epithelium renewal every 3.5 days.22-24 As a result of this extensive cell turnover, Hydra has an extensive regenerative capacity.15
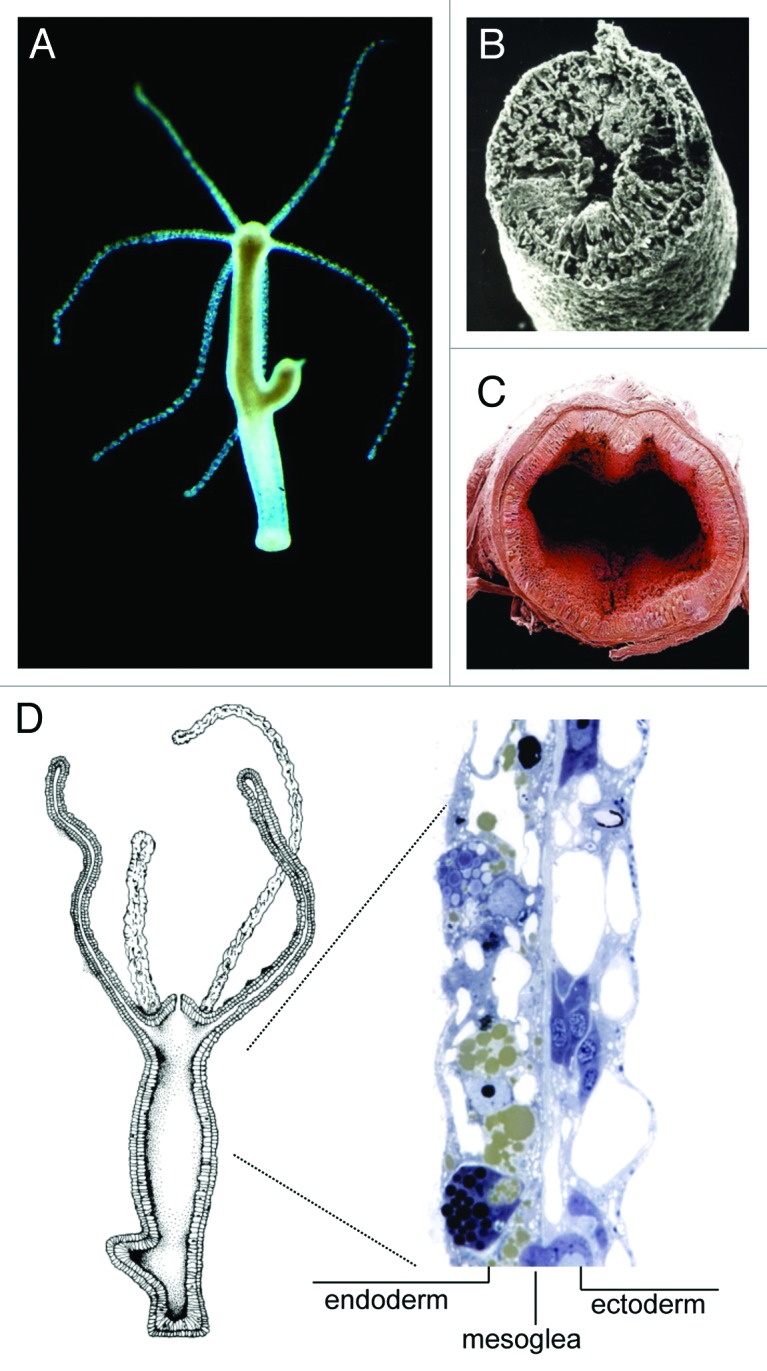
Figure 2. Hydra’s body anatomy resembles the mammalian intestinal epithelium. (A) Hydra oligactis polyp with a bud. (B) Scanning electron microscope (SEM) image of a cross section of Hydra. Taken from Bosch, BIUZ (2009). (C) View into human intestinal tract (from ref. 50). (D) The Hydra body is an epithelio-muscular bilayer with an intervening extracellular matrix and a few cell types derived from three distinct stem cell lineages.
Hydra Actively Shapes the Composition of its Colonizing Microbiota
For decades a number of Hydra species have been cultivated under standard conditions at constant temperature and identical food. It came as a complete surprise, therefore, that examining the microbiota in different Hydra species kept in the laboratory for more than 20 years under controlled conditions revealed an epithelium colonized by a complex community of microbes, and that individuals from different species differed greatly in their microbiota.25 Even more astonishing was the finding that individuals living in the wild were colonized by a group of microbes that is similar to that in polyps grown in the lab, pointing to the maintenance of specific microbial communities over long periods of time.25 This observation strongly indicates that distinct selective pressures are imposed on and within the Hydra epithelium, suggesting that the epithelium actively selects and shapes its microbial community, and that there is a direct interaction between epithelia and microbiota. In the meantime it is increasingly recognized that all animals, ranging from Hydra to primates, are host to complex microbial ecosystems, engaged in a symbiotic relationship that is essential for host physiology and homeostasis.26 Deep sea corals, for example, have complex microbial assemblages, just like shallow corals, and each coral species appears to differ in their bacterial microbiota.27,28 For that reason we consider the host and its associated microbiota an holobiont or metaorganism which is considered the unit of natural selection.29 The homeostasis within the metaorganism is critical for its health and ecological performance. In cnidarians as in other animals, disturbance or shifts in any of these interacting partners can affect the fitness of the collective holobiont, compromise the health of the whole animal, and lead to disease.30,31 Thus, to understand disease, be it bleaching in corals or inflammatory bowel disease in man, an in-depth knowledge of the basic biology of each holobiont member is required. It comes, therefore, as no surprise that the forces that shape the colonizing microbial community are the focus of much current investigation.
How intimate is the relationship between microbes and this epithelium? Do microbes react to changes in the Hydra epithelium? To decipher putative links between epithelial homeostasis and species-specific bacterial phylotypes, we removed the cells of the interstitial cell lineage from the Hydra tissue while leaving both epithelial stem cell lineages untouched. These changes in the epithelia cell composition led to significant modulation of the microbial community.32 Especially two bacterial phylotypes changed drastically when one host-cell lineage was removed. The dominant bacterial phylotype in control polyps belonging to the β-Proteobacteria was decreased in polyps lacking the interstitial cell lineage. In contrast, a bacterial phylotype belonging to the Bacteroidetes increased in abundance in polyps lacking interstitial cell lineage compared with control polyps.32 Thus, changes in epithelial homeostasis cause significant changes in the microbial community, implying a direct interaction between epithelia and microbiota.
Where Do the Hydra-associated Microbes Live?
The Hydra polyp, as any other animal body, may be considered an “ecological landscape” that harbors different ecosystems and meta-communities33 with environmental characteristics such as dispersal barriers likely to affect the localization of host-associated microbial communities. One such dispersal barrier appears to be the endodermal epithelium. Most likely due to the activity of numerous endodermally produced antimicrobial substances,4,20 up to now we cannot identify a stable microbiota within Hydra´s gastric cavity. In contrast to the endoderm, the ectoderm in Hydra is coated by a glycocalyx, a complex and highly dynamic polymeric meshwork that consists mostly of proteoglycans, glycosaminoglycans, and glycoproteins. Unexpectedly, and made possible only through an improved cryofixation method, we recently identified Hydra-associated bacteria outside the ectoderm on or within the glycocalyx (Anton-Erxleben; Augustin, Fraune and Bosch, unpubl.). In microbial biogeography, a predominant theory is that “everything is everywhere, but the environment selects”34. Whether Hydra’s sugar-rich glycocalyx provides such environmental determinants that facilitate microbial localization remains to be shown. However, it is clear that in both ectoderm and endoderm, multifunctional epithelial cells are points of microbe recognition. Their unique properties allow each epithelial barrier to deal with specific microenvironments and to control the microbiota of each milieu. Neither epithelium functions alone, but instead acts within the context of the organism, and thus, the function of each epithelial barrier is likely to influence the others.34,35
How Does Hydra Assemble the Specific Set of Microbes?
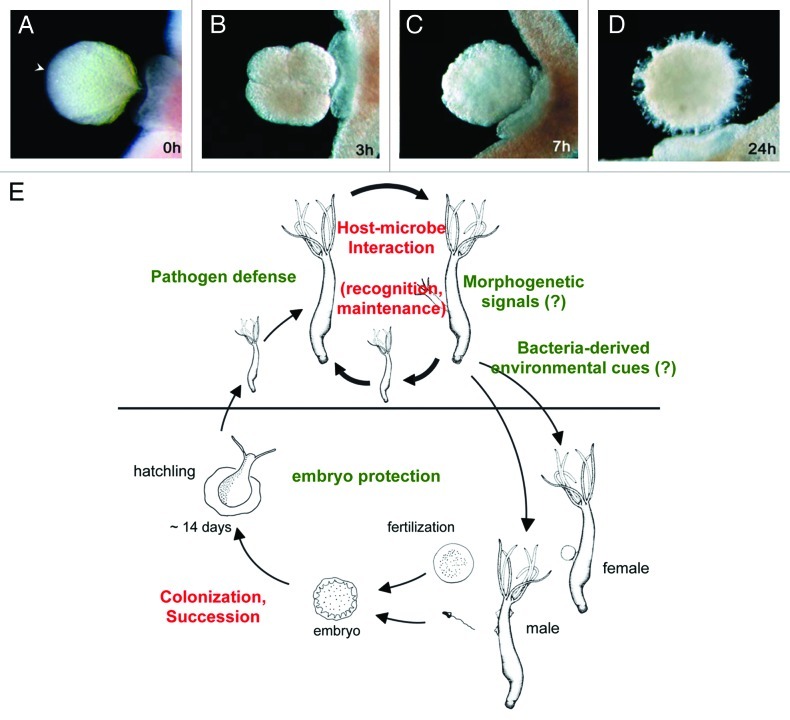
In the same way that microbial communities are expected to change in different parts of a body, they are also dynamic in time. For a first understanding of the temporal dynamics in Hydra-microbe interactions we investigated the establishment of the microbiota during oogenesis and embryogenesis.36,37 Early embryonic stages in Hydra (Fig. 3A) are colonized by a limited number of microbes.37 During embryogenesis the number of bacterial colonizers changes in number and composition. For example, Curvibacter-related Betaproteobacteria (phylum Chloroflexi) are present only in late developmental stages while they appear to be absent in the early embryo.37 Thus, early developmental stages have a microbiota that is clearly distinct from later developmental stages. Interestingly, the differential colonization is reflected in differences in antimicrobial activity.36,37 Hydra embryos are protected by a maternally produced antimicrobial peptide (AMP) of the periculin peptide family, which controls the establishment of the microbiota during embryogenesis. Beginning with the gastrula stage, i.e., after the “Maternal to Zygotic Transition” (MZT) stage, Hydra embryos express a set of periculin peptides (periculin 2a and 2b), which replaces the maternal produced periculin peptides 1a and 1b. This shift in the expression within the periculin peptide family represents a shift from maternal to zygotic protection of the embryo.36,37 In adult Hydra polyps, additional AMPs including Hydramacin38 and Arminin39 contribute to the host-derived control of bacterial colonization.
Figure 3.Hydra, an epithelial model for dissecting the fundamental principles that underlie host-microbe interactions in embryos and adults. (A) Oocyte of Hydra vulgaris. Arrowhead marks the oral end of the body axis corresponding to the sperm entry point. (B) 4-cell stage embryo. (C) Late cleavage stage. (D) Postgastrula cuticle stage embryo. (A) to (D) modified from ref. 51. (E) Hydra may provide one of the simplest possible systems to address key questions of how a stable host-microbe community is established and remains in balance over time (red letters); and in which ways bacteria exert influence on the hydra life cycle (green letters).
How Does Hydra Control the Composition of the Microbiota in the Context of Specific Environmental Conditions?
Hydra recognizes “Microbial Associated Molecular Patterns” (MAMPs) with the help of the Toll-Like-Receptor (TLR) signaling pathway.20 The Toll-like receptor in Hydra functions as a co-receptor with MAMP-recognizing leucine-rich repeats and a signal transmitting TIR domain on two separated but interacting proteins.20,40 Homologous sequences to nearly all other components of the TLR pathway have been identified.20,41 RNAi knock-down experiments with Hydra TLR showed a drastic reduction of antimicrobial activity in the knock down tissue compared with the wild type, which makes it apparent that antimicrobial activity relies directly on the activation of the TLR cascade.20 In addition to recognizing MAMPs at the cell membrane, intracellular recognition of bacteria in Hydra is mediated by an unexpectedly large number of cytosolic NOD-like receptors.42 We have proposed elsewhere42 that apoptosis as an evolutionary ancient mechanism in immune defense might be initiated upon activation and dimerization of Hydra´s NOD-like receptors.
Prominent effector molecules downstream of the conserved TLR cascade are antimicrobial peptides (AMPs). Up to now we have isolated four families of antimicrobial peptides including the hydramacins, arminins, periculins and serine protease inhibitors of the Kazal type.20,38,39,43 Interestingly, all antimicrobial peptides isolated so far are synthesized in endodermal tissue only. This confined location supports the view that the endodermal epithelium surrounding the gastric cavity is especially endangered by the regular uptake of food, and that Hydra´s AMPs contribute to the chemical defense properties of this layer similar to AMPs in the human small intestine. Periculins and arminins are made as precursors. To activate them, a negatively charged N-terminal part is cleaved releasing a highly positively charged C-terminal portion.20,39 In the periculin family, this cationic C-terminal region is rich in cysteines indicating that this domain requires a distinct three dimensional structure for activity. In addition to endodermal epithelial cells, periculin is also expressed in female germ cells and used for maternal protection of the embryo.36,37 Arminins are characterized by a positively charged and variable C-terminus that includes 31 amino acids.39 When used in liquid growth inhibition assays, the C-terminal part of arminin 1a is capable of killing a large number of antibiotic resistant bacteria including methicillin resistant S. aureus and vancomycin resistant strains of Enterococcus faecalis and E. faecium.39 In contrast to Periculins and Arminins, Hydramacins have no precursor form. They are made as single peptides with an N-terminal signal peptide that is followed by a cationic C-terminal part containing 8 cysteins. The peptide seems to be highly active mostly against gram-negative bacteria also including human pathogenic multi-drug resistant bacteria.20 The three-dimensional (3D) structure38 is characterized by an unusual arrangement of cationic and hydrophobic amino acid residues. Based on a 3D model we hypothesize that bacteria attached to hydramacin from large aggregates.38 The model is supported by the observation that after application of Hydramacin-1 bacteria aggregate, settle-out and finally die. Electron microscopic pictures show that these bacteria die within intact membranes. We assume, therefore, that the aggregation processes may induce some kind of programmed cell death in bacteria.38
Taken together, these data indicate that Hydra has an effective innate immune system to interact with bacteria at the epithelial interface, and that antimicrobial peptides (AMPs) have key regulatory functions in host-microbe homeostasis. The crucial question now is what is the driving force that leads to changes in microbiota composition? Because of their obvious ability to affect bacterial life, promising candidate molecules are AMPs. To investigate whether the ectotopic expression of an AMP may affect the number and composition of the colonizing microbiota at the ectodermal epithelial surface, we generated transgenic Hydra expressing periculin1a in ectoderm epithelial cells. Comparing the bacterial load of these transgenic polyps with that of wild-type control polyps revealed not only a significantly lower bacterial load in transgenic polyps overexpressing Periculin1a but also, unexpectedly, drastic changes in the bacterial community structure.36 Analyzing the identity of the colonizing bacteria showed that the dominant β-Proteobacteria decreased in number, whereas α-Proteobacteria were more prevalent. Thus, overexpression of Periculin causes not only a decrease in the number of associated bacteria but also a changed bacterial composition.36 With the transgenic polyps overexpressing periculin we apparently have created a new holobiont that is different from all investigated Hydra species. Future efforts will be directed toward analyzing the performance of this new phenotype under different environmental conditions.
Intriguingly, most of the antimicrobial peptide genes identified in Hydra as well as in other animals such as Drosophila or C. elegans, have no homology to sequences in other species and therefore are classified as “taxonomically restricted genes” (TRGs).44 An informative example in Hydra is periculin-1, termed due to its rapid response to a wide variety of bacterial and tissue “danger” signals.20 Analysis of the deduced amino acid sequence of periculin-1 and the charge distribution within the molecule revealed an anionic N-terminal region and an 8-cysteine residues containing cationic C-terminal region.20 No identifiable orthologs were found in any sequence database. Periculin-1 has a strong bactericidal activity and is expressed in the endodermal epithelium as well as in a subpopulation of ectodermal interstitial cells. As discussed elsewhere,44 each animal species contains a significant number of such “orphan” genes encoding potent antimicrobial peptides. For example, Aurelia aurita, one of the most common jellyfish, contains the novel 40 amino-acid antimicrobial peptide aurelin.45 Similarily, the antibacterial immune response gene encoding diptericin is restricted to insects of the Diptera group46; and the 11 kDa metal ion−binding S100 protein psoriasin protects epithelia in mammals but not in other animals from infection.47 Taxonomically restricted host defense molecules appear to represent an extremely effective chemical warfare system to shape the colonizing microbiota and at the same time to cope with specific environmental challenges.
Are Bacteria-deprived Hydra at a Disadvantage?
The intimacy of the interaction between host and microbiota, as well as the high evolutionary pressure48 to maintain a specific microbiota, points to the significance of the interkingdom association and implies that hosts deprived of their microbiota should be at a disadvantage. To investigate the effect of absence of microbiota in Hydra we have produced gnotobiotic Hydra polyps that are devoid of any bacteria. While morphologically no differences could be observed to control polyps, we are currently finding evidence that Hydra lacking bacteria suffer from fungal infections unknown in normally cultured polyps (Augustin, Franzenburg, Fraune and Bosch). Thus, do beneficial microbes associated with Hydra produce anti-fungal compounds? Future efforts are directed toward isolating the active substances from these bacteria that eventually may lead to the development of novel antimycotics.
Conclusion and Perspectives
Discovering that individuals from Hydra to man are not solitary, homogenous entities but consist of complex communities of many species that likely evolved during a billion years of coexistence48 led to the hologenome theory of evolution49 which considers the holobiont with its hologenome as the unit of selection in evolution. Here I have introduced Hydra as a powerful and intellectually attractive model system allowing profound insights into understanding a microbe-dependent life style and its evolutionary consequences (Fig. 3B). Organisms become models when they support sustainable opportunities with uncompromising experimental rigor and ease of use. With the molecular dissecting of the components controlling host-microbe interactions in Hydra, the stage is now set for biologists to uncover the secrets of the cnidarian holobiont. Findings derived from the in vivo context of the Hydra model may provide one of the simplest possible systems to address questions of how a stable host-microbe community is established and remains in balance over time. The uncovered basic molecular machinery can be transliterated to more complex organisms, providing conceptual insights into the complexity of host-microbe interactions.
Although important first steps have been made in describing host-microbe interactions in Hydra, much needs to be learned about the underlying controlling mechanisms and the role of the individual members of the holobiont. Questions to be addressed in the future include: What are the functional roles of all members of the Hydra holobiont? What is the function of the main colonizing β-Proteobacteria? How are the Hydra–microbe interactions regulated in the healthy holobiont, and how do shifts in holobiont members lead to a transition into a disease state? What effect does a changing environment have on microbial associates and the fitness of the holobiont? Elucidating these issues will not only contribute to the understanding of the interactions of Hydra epithelia with microbial communities, but also may be key to dissecting the fundamental principles that underlie all host-microbe interactions.
Acknowledgments
My sincere thanks are to the members of my laboratory: Friederike Anton-Erxleben, René Augustin, Sören Franzenburg, Sebastian Fraune, Konstantin Khalturin, Alexander Klimovich, and Jörg Wittlieb. Judith Ittner-Bosch read the manuscript, and her support has been invaluable. I am also grateful to two anonymous referees for their constructive criticism. The work described in this review was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) and grants from the DFG Cluster of Excellence programs “The Future Ocean” and “Inflammation at Interfaces.”
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20660
References
- 1.Morris SC. Eggs and embryos from the Cambrian. Bioessays. 1998;20:676–82. doi: 10.1002/(SICI)1521-1878(199808)20:8<676::AID-BIES11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–5. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 3.Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21:536–9. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–9. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Hemmrich G, Bosch TC. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays. 2008;30:1010–8. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- 6.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 7.Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–60. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 8.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The dynamic genome of Hydra. Nature. 2010;464:592–6. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A. 2006;103:6208–11. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemmrich G, Miller DJ, Bosch TCG. The evolution of immunity: a low-life perspective. Trends Immunol. 2007;28:449–54. doi: 10.1016/j.it.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domazet-Loso T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol Biol Evol. 2008;25:2699–707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [Review] [DOI] [PubMed] [Google Scholar]
- 14.Bode HR. Axial patterning in hydra. Cold Spring Harb Perspect Biol. 2009;1:a000463. doi: 10.1101/cshperspect.a000463. [Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch TCG. Hydra and the evolution of stem cells. Bioessays. 2009;31:478–86. doi: 10.1002/bies.200800183. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Hou SX. Regulation of intestinal stem cells in mammals and Drosophila. J Cell Physiol. 2010;222:33–7. doi: 10.1002/jcp.21928. [DOI] [PubMed] [Google Scholar]
- 17.Sancho E, Batlle E, Clevers H. Live and let die in the intestinal epithelium. Curr Opin Cell Biol. 2003;15:763–70. doi: 10.1016/j.ceb.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–9. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 19.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–64. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–69. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Bosch TCG, David CN. Immunocompetence in Hydra: Epithelial cells recognize self-nonself and react against it. J Exp Biol. 1986;238:225–34. [Google Scholar]
- 22.Bosch TCG, David CN. Growth regulation in Hydra: relationship between epithelial cell cycle length and growth rate. Dev Biol. 1984;104:161–71. doi: 10.1016/0012-1606(84)90045-9. [DOI] [PubMed] [Google Scholar]
- 23.Bode H, Dunne J, Heimfeld S, Huang L, Javois L, Koizumi O, et al. Transdifferentiation occurs continuously in adult hydra. Curr Top Dev Biol. 1986;20:257–80. doi: 10.1016/S0070-2153(08)60668-7. [DOI] [PubMed] [Google Scholar]
- 24.Bosch TCG, Anton-Erxleben F, Hemmrich G, Khalturin K. The Hydra polyp: nothing but an active stem cell community. Dev Growth Differ. 2010;52:15–25. doi: 10.1111/j.1440-169X.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- 25.Fraune S, Bosch TCG. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A. 2007;104:13146–51. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bevins CL, Salzman NH. The potter’s wheel: the host’s role in sculpting its microbiota. Cell Mol Life Sci. 2011;68:3675–85. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–62. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 28.Rohwer F, et al. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 29.Bosch TCG, McFall-Ngai MJ. Metaorganisms as the new frontier. Zoology (Jena) 2011;114:185–90. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiser DM, et al. Cause of sea fan death in the West Indies. Nature. 1998;394:137–8. doi: 10.1038/28079. [DOI] [Google Scholar]
- 31.Rosenberg E, Falkovitz L. The Vibrio shiloi/Oculina patagonica model system of coral bleaching. Annu Rev Microbiol. 2004;58:143–59. doi: 10.1146/annurev.micro.58.030603.123610. [DOI] [PubMed] [Google Scholar]
- 32.Fraune S, Abe Y, Bosch TCG. Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ Microbiol. 2009;11:2361–9. doi: 10.1111/j.1462-2920.2009.01963.x. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez A, Clemente JC, Shade A, Metcalf JL, Song S, Prithiviraj B, et al. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12:775–84. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Malley MA. The nineteenth century roots of ‘everything is everywhere’. Nat Rev Microbiol. 2007;5:647–51. doi: 10.1038/nrmicro1711. [DOI] [PubMed] [Google Scholar]
- 35.Augustin R, Bosch TCG. Cnidarian immunity: a tale of two barriers. Adv Exp Med Biol. 2010;708:1–16. doi: 10.1007/978-1-4419-8059-5_1. [DOI] [PubMed] [Google Scholar]
- 36.Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–72. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraune S, Augustin R, Bosch TCG. Embryo protection in contemporary immunology: Why bacteria matter. Commun Integr Biol. 2011;4:369–72. doi: 10.4161/cib.4.4.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung S, Dingley AJ, Augustin R, Anton-Erxleben F, Stanisak M, Gelhaus C, et al. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J Biol Chem. 2009;284:1896–905. doi: 10.1074/jbc.M804713200. [DOI] [PubMed] [Google Scholar]
- 39.Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother. 2009;53:5245–50. doi: 10.1128/AAC.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Augustin R, Fraune S, Bosch TCG. How Hydra senses and destroys microbes. Semin Immunol. 2010;22:54–8. doi: 10.1016/j.smim.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange C, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Miller DJ, Rahn T, Weiss Y, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28:1687–702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- 43.Augustin R, Siebert S, Bosch TCG. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev Comp Immunol. 2009;33:830–7. doi: 10.1016/j.dci.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–13. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Ovchinnikova TV, Balandin SV, Aleshina GM, Tagaev AA, Leonova YF, Krasnodembsky ED, et al. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem Biophys Res Commun. 2006;348:514–23. doi: 10.1016/j.bbrc.2006.07.078. [DOI] [PubMed] [Google Scholar]
- 46.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 47.Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schröder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 48.Fraune S, Bosch TCG. Why bacteria matter in animal development and evolution. Bioessays. 2010;32:571–80. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 49.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–35. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 50.Bosch TCG. Evolutionäres Vermächtnis: Stammzellen in Hydra. Biol Unserer Zeit. 2009;2/2009:114–22. doi: 10.1002/biuz.200910390. [DOI] [Google Scholar]
- 51.Fröbius AC, Genikhovich G, Kürn U, Anton-Erxleben F, Bosch TCG. Expression of developmental genes during early embryogenesis of Hydra. Dev Genes Evol. 2003;213:445–55. doi: 10.1007/s00427-003-0344-6. [DOI] [PubMed] [Google Scholar]