Abstract
With the rapid advances in sequencing technologies in recent years, the human genome is now considered incomplete without the complementing microbiome, which outnumbers human genes by a factor of one hundred. The human microbiome, and more specifically the gut microbiome, has received considerable attention and research efforts over the past decade. Many studies have identified and quantified “who is there?,” while others have determined some of their functional capacity, or “what are they doing?” In a recent study, we identified novel salt-tolerance loci from the human gut microbiome using combined functional metagenomic and bioinformatics based approaches. Herein, we discuss the identified loci, their role in salt-tolerance and their importance in the context of the gut environment. We also consider the utility and power of functional metagenomics for mining such environments for novel genes and proteins, as well as the implications and possible applications for future research.
Keywords: functional metagenomics, human gut microbiome, salt tolerance, meta-biotechnology
Introduction
Bacteria encounter numerous environmental stresses in various environments and the gastrointestinal tract is no exception.1 This dynamic environment poses a set of challenges that both transient and symbiotic microorganisms must overcome in order to colonise and proliferate.2 Low pH, bile acids, elevated osmolarity, iron limitation, intermittent nutrient availability and host immune factors are just some of the challenges faced in the gastrointestinal tract.3 The ability to cope with rapid changes in external osmolarity is an important mechanism that allows microorganisms adapt to and colonise a given environmental niche.4 The cellular response to hyper-osmotic stress is broad and involves a number of different processes such as potassium (K+) uptake,5 compatible solute accumulation6 and numerous ancillary systems.7,8
The emergence of metagenomics as a key area of scientific research in recent years has transformed how we view ourselves as living organisms.9,10 Working with microbes in pure cultures is very reductive in terms of understanding microbial behavior in complex ecological niches. Metagenomics can, in principle, allow us access the entire genetic complement of our associated microbiome without the need for classic microbiological culturing techniques. Despite recent advances in high-throughput anaerobic culturing techniques with gnotobiotic animal husbandry; which suggests that the human faecal microbiota consists largely of taxa and predicted functions that are represented in its cultured members;11 functional metagenomics allows us to rapidly separate the ‘wheat from the chaff’. Gaining insights about the functional capacity of the human gut microbiome was a key aim in our efforts to elucidate novel genetic loci conferring a salt tolerance phenotype. We employed a functional metagenomic screen of a human gut metagenomic library which resulted in the identification of five novel genes involved in salt tolerance.12 An advantage of functional metagenomics is its ability to uncover completely novel functions for new or known genes without the need for any previous sequence information.
Novel Salt Tolerance Loci Identified
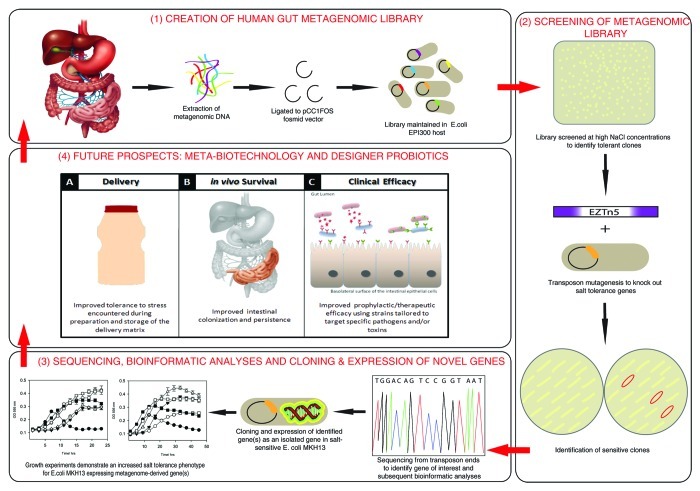
From our initial screen (the overall scheme is presented in Figure 1) of over 20,000 metagenomic clones, we identified 53 that could tolerate high sodium chloride (NaCl) concentrations. We termed these clones SMG 1–53 (salt metagenome). Through a combined transposon mutagenesis and bioinformatic strategy we identified five novel salt tolerance genes from clone SMG 3 (namely galE, murB and mazG) as well as additional mazG and galE genes from clone SMG 5 and SMG 25, respectively. Phylogenetic assignments revealed SMG 3 had the highest genetic identity to Collinsella aerofaciens, while SMG 5 and SMG 25 corresponded to Eggerthella spp YY7918 and Akkermansia muciniphila respectively. Each of the five genes were cloned separately and expressed in the osmosensitive strain Escherichia coli MKH13, which resulted in an increased tolerance to the ionic osmotic stressors NaCl and KCl (potassium chloride), but not to non-ionic stressors such as sucrose and glycerol, a finding which seems to suggest that these genes confer a salt-specific protective effect. While E. coli has been shown to upregulate a set of genes in response to both ionic and non-ionic osmotic stress, it also regulates genes specific to each type of stress.13 Furthermore each of the three genes was also found to be over-represented in the human gut metagenome and abundant among healthy subjects from the MetaHit data set.12,14
Figure 1. An overview of novel gene discovery using functional metagenomics; from metagenomic library creation to novel therapeutics.
galE
The galE gene product (UDP glucose 4-epimerase) catalyzes the inter-conversion of UDP glucose and UDP galactose and has been previously linked to the osmotic stress response through different mechanisms such as the production of the osmoprotectant trehalose or through cellular signaling.15,16 We theorise that galE may be important in maintaining the integrity of the lipopolysaccharide (LPS) in Gram negative or lipoteichoic acid (LTA) layers in Gram positive bacteria, making the cell more resistant to salt-induced osmotic stress. A recent functional metagenomic study identified a galE gene, that when cloned and expressed in E. coli conferred resistance to menadione, which can cause membrane damage through the generation of reactive oxygen species.17 The authors believe the resistance is mediated through galE by increasing the permeability barrier of the cell. Such menaquinones are also found at significant concentrations in the human gastrointestinal tract.18 The ability to form biofilms is likely to be critically important in the gut environment and for homeostasis within the community.19 Furthermore, it has been shown that galE mutants have reduced ability to form biofilms, while gal mutants are defective in intestinal colonisation, both of which could be an important factor in the gastrointestinal tract.20,21
murB
The murB gene is involved in the biosynthesis of peptidoglycan and the bacterial cell wall, which itself plays an important role in withstanding osmotic stress.22 Disruption or deletion of the murB gene could make cells acutely sensitive to osmotic stress due to a reduction in cell wall integrity as well as causing a reduction in turgor pressure which is a driving force for cellular growth and division. Bacteria remodel the structure of their peptidoglycan in response to changes in environmental conditions,23 which could be important in the gut by allowing for varying levels of rigidity or elasticity depending on the conditions in the immediate environment. One of the more interesting functions of peptidoglycan, and a possible reason why genes for its synthesis are enriched among the human gut microbiota, is its stimulation of host immunity. Clarke et al. (2010), have demonstrated peptidoglycan from the commensal microbiota modulate the innate immune system by improving neutrophil function even in the absence of infection.24 The authors note that peptidoglycan can be translocated to the bloodstream, with concentrations at similar levels to those in faeces, indicating that there is constant peptidoglycan turnover among the microbiota and that immune stimulation by the microbiota can affect sites distal from the GI tract. Stimulated neutrophils demonstrated increased killing of the pathogenic bacteria Streptococcus pneumoniae and S. aureus.24 This may indicate a co-evolution of a mutually beneficial arrangement by removing potentially harmful host pathogens and competitors to our symbiotic gut microbiota.
mazG
The mazG gene represents the most interesting of the identified genes, in that its possible mode of action in response to salt stress is not as immediately clear as galE or murB. As the latter two genes are related to outer membrane or cell wall functions, one can envisage how they could mediate resistance to external environmental stresses. MazG has been shown to play a role in different cellular processes, such as the removal of aberrant dNTP’s from DNA strands,25 as well as the oxidative26 and nutritional stress responses.27 Often found in association with the mazEF toxin-antitoxin (TA) system, the mazG gene product can delay programmed cell death and allow the cell to survive for longer periods under stress in the event that additional nutrients become available.27 TA systems such as mazEF can induce cell death or arrest in response to various cellular stresses,28 particularly those which induce DNA damage. MazG may delay apoptosis in salt-stressed cells as well as providing a mechanism to reduce or repair salt-induced damage to DNA. It has been speculated that individual TA systems may respond to specific stresses29 and play a role in biofilm and persister cell formation, as well as having numerous other putative functions.30 The development of persister cells allows for the survival of a small subpopulation through the death of the majority of the population.31 Such an altruistic characteristic benefits long-term survival and it seems, at the microscopic level, that the sacrifice of many for the good of a few may be a tenet of bacterial survival. Although the mazG genes identified in our study12 are not located in the genomic neighborhood of any obvious TA system, it is possible their encoded proteins could still regulate such TA systems at distant chromosomal locations or indeed regulate as yet unidentified, stress responsive genes or function as a general stress responsive protein itself.
Future Perspectives
While expanding our current knowledge on the diverse mechanisms employed by bacteria to overcome salt stress, the identification of novel genes may also assist in the development of novel drugs and drug targets as well as novel strategies to control some of the resident gut microbiota.32,33 Interestingly murB has been investigated as a possible target for novel antibiotics and antibacterial compounds34 as it is exclusively found in bacteria and it has an important role in maintaining cellular integrity and viability. Furthermore, galE has been investigated as target for novel therapeutics against African sleeping sickness35 and a galE mutant of Salmonella enterica serovar Typhi has been used to create an oral live attenuated vaccine for typhoid.36 TA systems have also received attention as possible novel antibacterial drug targets.37 MazG could also be a putative target, if disruption of its function could allow the toxin component of the TA system to cause cell death to certain bacterial populations.
Ultimately we would envisage that some of these novel salt tolerance genes could be used as part of a “meta-biotechnology”38 strategy for the development of technologically more robust probiotic cultures and with greater ability to survive gastrointestinal transit and colonise the gut. Meta-biotechnology describes the “mining” of the human gut metagenome for novel genes for use in medicine, science and industry for the development of novel therapeutics38 and is an extension of the patho-biotechnology39,40 concept. Patho-biotechnology has been used with success to engineer probiotic strains with increased stress resistance, improved gastrointestinal persistence and colonisation and therapeutic efficacy.41
Mining the metagenome is not limited to the identification of novel stress tolerance genes but may be used for the identification of novel antimicrobial compounds or genes that may be used for the development of designer probiotics which can be used to target specific pathogens or toxins.42 New ways of thinking and alternative therapies are needed to control and combat pathogenic microorganisms in the era of increasing antibiotic resistance and emerging superbugs. The identification of such novel genes will broaden our understanding of salt tolerance in bacteria and uncover novel mechanisms employed for survival in the gastrointestinal tract as well as providing a platform for the development of novel biological therapeutics or novel drug targets.
Acknowledgments
EPC is funded by Science Foundation Ireland under the CSET Uplift Grant. We acknowledge the continued financial assistance of the Alimentary Pharmabiotic Centre, funded by Science Foundation Ireland. JRM acknowledges funding from The Royal Society which supports the bioinformatic cluster (Hive) at Cardiff University, School of Biosciences. RDS is an ESCMID Research Fellow.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20984
References
- 1.Sleator RD, Hill C. Engineered pharmabiotics with improved therapeutic potential. Hum Vaccin. 2008;4:271–4. doi: 10.4161/hv.4.4.6315. [DOI] [PubMed] [Google Scholar]
- 2.Sleator RD, Hill C. New frontiers in probiotic research. Lett Appl Microbiol. 2008;46:143–7. doi: 10.1111/j.1472-765X.2007.02293.x. [DOI] [PubMed] [Google Scholar]
- 3.Louis P, O’Byrne CP. Life in the gut: microbial responses to stress in the gastrointestinal tract. Sci Prog. 2010;93:7–36. doi: 10.3184/003685009X12605525292307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 5.Epstein W. The roles and regulation of potassium in bacteria. Prog Nucleic Acid Res Mol Biol. 2003;75:293–320. doi: 10.1016/S0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 6.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–30. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 7.Piuri M, Sanchez-Rivas C, Ruzal SM. Adaptation to high salt in Lactobacillus: role of peptides and proteolytic enzymes. J Appl Microbiol. 2003;95:372–9. doi: 10.1046/j.1365-2672.2003.01971.x. [DOI] [PubMed] [Google Scholar]
- 8.Sleator RD, Hill C. A novel role for the LisRK two-component regulatory system in listerial osmotolerance. Clin Microbiol Infect. 2005;11:599–601. doi: 10.1111/j.1469-0691.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- 9.Sleator RD, Shortall C, Hill C. Metagenomics. Lett Appl Microbiol. 2008;47(c):361–6. doi: 10.1111/j.1472-765X.2008.02444.x. [DOI] [PubMed] [Google Scholar]
- 10.Sleator RD. The human superorganism—of microbes and men. 2010;Med Hypotheses74:214–5. doi: 10.1016/j.mehy.2009.08.047. [DOI] [PubMed] [Google Scholar]
- 11.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–7. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culligan EP, Sleator RD, Marchesi JR, Hill C. Functional metagenomics reveals novel salt tolerance loci from the human gut microbiome. ISME J. 2012 doi: 10.1038/ismej.2012.38. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shabala L, Bowman J, Brown J, Ross T, McMeekin T, Shabala S. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ Microbiol. 2009;11:137–48. doi: 10.1111/j.1462-2920.2008.01748.x. [DOI] [PubMed] [Google Scholar]
- 14.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böhringer J, Fischer D, Mosler G, Hengge-Aronis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of sigma S and sigma S-dependent genes in Escherichia coli. J Bacteriol. 1995;177:413–22. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leloir LF. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch Biochem Biophys. 1951;33:186–90. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- 17.Mori T, Suenaga H, Miyazaki K. A metagenomic approach to the identification of UDP-glucose 4-epimerase as a menadione resistance protein. Biosci Biotechnol Biochem. 2008;72:1611–4. doi: 10.1271/bbb.70815. [DOI] [PubMed] [Google Scholar]
- 18.Conly JM, Stein K. Quantitative and qualitative measurements of K vitamins in human intestinal contents. Am J Gastroenterol. 1992;87:311–6. [PubMed] [Google Scholar]
- 19.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–96. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 20.Ho TD, Waldor MK. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect Immun. 2007;75:1661–6. doi: 10.1128/IAI.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakao R, Senpuku H, Watanabe H. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun. 2006;74:6145–53. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Heijenoort J. Murein synthesis. In: Neidhardt FC, Curtis, R., III., Ingraham, J.L., Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaecter, M. and Umbarger, H.E, ed. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology, 1996:1025-34. [Google Scholar]
- 23.Vijaranakul U, Nadakavukaren MJ, de Jonge BL, Wilkinson BJ, Jayaswal RK. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J Bacteriol. 1995;177:5116–21. doi: 10.1128/jb.177.17.5116-5121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galperin MY, Moroz OV, Wilson KS, Murzin AG. House cleaning, a part of good housekeeping. Mol Microbiol. 2006;59:5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- 26.Lu LD, Sun Q, Fan XY, Zhong Y, Yao YF, Zhao GP. Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J Biol Chem. 2010;285:28076–85. doi: 10.1074/jbc.M109.088872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross M, Marianovsky I, Glaser G. MazG -- a regulator of programmed cell death in Escherichia coli. Mol Microbiol. 2006;59:590–601. doi: 10.1111/j.1365-2958.2005.04956.x. [DOI] [PubMed] [Google Scholar]
- 28.Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186:3663–9. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–83. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 31.Erental A, Sharon I, Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10:e1001281. doi: 10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sleator RD. Probiotic therapy - recruiting old friends to fight new foes. Gut Pathog. 2010;2:5. doi: 10.1186/1757-4749-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleator RD. Probiotics -- a viable therapeutic alternative for enteric infections especially in the developing world. Discov Med. 2010;10(c):119–24. [PubMed] [Google Scholar]
- 34.Shapiro AB, Livchak S, Gao N, Whiteaker J, Thresher J, Jahić H, et al. A homogeneous, high-throughput-compatible, fluorescence intensity-based assay for UDP-N-acetylenolpyruvylglucosamine reductase (MurB) with nanomolar product detection. J Biomol Screen. 2012;17:327–38. doi: 10.1177/1087057111425188. [DOI] [PubMed] [Google Scholar]
- 35.Urbaniak MD, Tabudravu JN, Msaki A, Matera KM, Brenk R, Jaspars M, et al. Identification of novel inhibitors of UDP-Glc 4′-epimerase, a validated drug target for african sleeping sickness. Bioorg Med Chem Lett. 2006;16:5744–7. doi: 10.1016/j.bmcl.2006.08.091. [DOI] [PubMed] [Google Scholar]
- 36.Germanier R, Füer E. Isolation and characterization of Gal E mutant Ty 21a of Salmonella typhi: a candidate strain for a live, oral typhoid vaccine. J Infect Dis. 1975;131:553–8. doi: 10.1093/infdis/131.5.553. [DOI] [PubMed] [Google Scholar]
- 37.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog. 2009;1:19. doi: 10.1186/1757-4749-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleator RD, Hill C. Patho-biotechnology: using bad bugs to do good things. Curr Opin Biotechnol. 2006;17:211–6. doi: 10.1016/j.copbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Sleator RD, Hill C. Patho-biotechnology; using bad bugs to make good bugs better. Sci Prog. 2007;90:1–14. doi: 10.3184/003685007780440530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehan VM, Sleator RD, Hill C, Fitzgerald GF. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003. Microbiology. 2007;153:3563–71. doi: 10.1099/mic.0.2007/006510-0. [DOI] [PubMed] [Google Scholar]
- 42.Focareta A, Paton JC, Morona R, Cook J, Paton AW. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology. 2006;130:1688–95. doi: 10.1053/j.gastro.2006.02.005. [DOI] [PubMed] [Google Scholar]