Abstract
Daphnia pulex is quickly becoming an attractive model species in the field of ecological genomics due to the recent release of its complete genome sequence, a wide variety of new genomic resources, and a rich history of ecological data. Sequences of the mitochondrial NADH dehydrogenase subunit 5 and cytochrome c oxidase subunit 1 genes were used to assess the global phylogeography of this species, and to further elucidate its phylogenetic relationship to other members of the Daphnia pulex species complex. Using both newly acquired and previously published data, we analyzed 398 individuals from collections spanning five continents. Eleven strongly supported lineages were found within the D. pulex complex, and one lineage in particular, panarctic D. pulex, has very little phylogeographical structure and a near worldwide distribution. Mismatch distribution, haplotype network, and population genetic analyses are compatible with a North American origin for this lineage and subsequent spatial expansion in the Late Pleistocene. In addition, our analyses suggest that dispersal between North and South America of this and other species in the D. pulex complex has occurred multiple times, and is predominantly from north to south. Our results provide additional support for the evolutionary relationships of the eleven main mitochondrial lineages of the D. pulex complex. We found that the well-studied panarctic D. pulex is present on every continent except Australia and Antarctica. Despite being geographically very widespread, there is a lack of strong regionalism in the mitochondrial genomes of panarctic D. pulex – a pattern that differs from that of most studied cladocerans. Moreover, our analyses suggest recent expansion of the panarctic D. pulex lineage, with some continents sharing haplotypes. The hypothesis that hybrid asexuality has contributed to the recent and unusual geographic success of the panarctic D. pulex lineage warrants further study.
Introduction
Scientific understanding of the freshwater crustacean Daphnia is unusually deep in ecology and toxicology [1], [2], and with the release of a Daphnia genome sequence [3], this organism has now become a promising model system with which to investigate a wide range of biological phenomena [4]–[7]. The Daphnia genome sequence was derived from a member of the Daphnia pulex species complex, which is common throughout the Holarctic, but is also found in South America and Africa [8], [9]. Daphnia typically reproduce by cyclic parthenogenesis, in which production of direct-developing embryos by apomixis alternates with production of diapausing eggs via meiosis and fertilization. However, some lineages in the D. pulex species complex also produce their diapausing eggs apomictically (obligate parthenogenesis), and have therefore lost the capacity for sexual reproduction. To date, obligate parthenogenesis has not been identified in any other lineages in the subgenus Daphnia [10].
Based on morphology, the D. pulex species complex was originally thought to include few species with extremely broad geographic distributions, but studies of mitochondrial DNA (mtDNA) variation have revealed three major groups (pulex, pulicaria, and tenebrosa) and at least 12 named lineages (1 in the pulex group, 9 in the pulicaria group, and 2 in the tenebrosa group) of which all but four are found only on one continent [8]. Even so, the geographic limits of some lineages are unknown because of limited genetic analysis and sampling in many parts of the world [9], [11], [12], [13], [14], [15], [16]. In addition, the origin of the most speciose pulicaria group is controversial. Brooks [17] proposed either a North American or Eurasian origin based on greater abundance on these continents compared to southern hemisphere continents. The absence of sexual lineages outside the Holarctic, and higher levels of genetic diversity in the northern hemisphere also provides evidence for a Holarctic origin [9], [18]. However, Mergeay et al. [13] recently proposed a South American origin for the pulicaria group based on phylogenetic evidence showing that its two most divergent lineages are restricted to South America.
One mtDNA lineage in the pulicaria group, panarctic D. pulex, has been reported from temperate and arctic North America, the northeastern Palearctic, and Africa [9], [12], [16]. However, this lineage is unknown from most of the Palearctic and Alaska [15]. Furthermore, the basic phylogeographic structure of the panarctic D. pulex lineage is unclear. The formation of sublineages has been attributed to Pleistocene or older vicariance events [12]. Hebert et al. [19] first proposed two separate glacial refugia for panarctic D. pulex based on the geographic pattern of breeding system, with obligate parthenogenesis having arisen in this taxon in an eastern North America refugium. Later, three distinct glacial refugia were proposed based on regional phylogenetic lineages and patterns of mtDNA nucleotide diversity in relation to refugial locations [14]. Lynch et al. [20] also found well-supported geographic sublineages in North America, and estimated the age of the oldest obligately parthenogenetic lineage of panarctic D. pulex (Quebec and New Brunswick) to be only 172,000 generations (Late Glacial).
In the present study, we employ new mtDNA data from expanded geographic surveys to further examine the phylogeographic history of the D. pulex species complex. We aimed to address conflicting hypotheses on the phylogeny of this group by combining data from previous studies with new data from vast geographic regions including those that were not well represented previously. Specifically, we aim to investigate (1) the geographic distribution of the mtDNA lineages within this complex, (2) the origin of the pulicaria group, and (3) the phylogeographic history of panarctic D. pulex.
Results
Phylogenetic Analyses
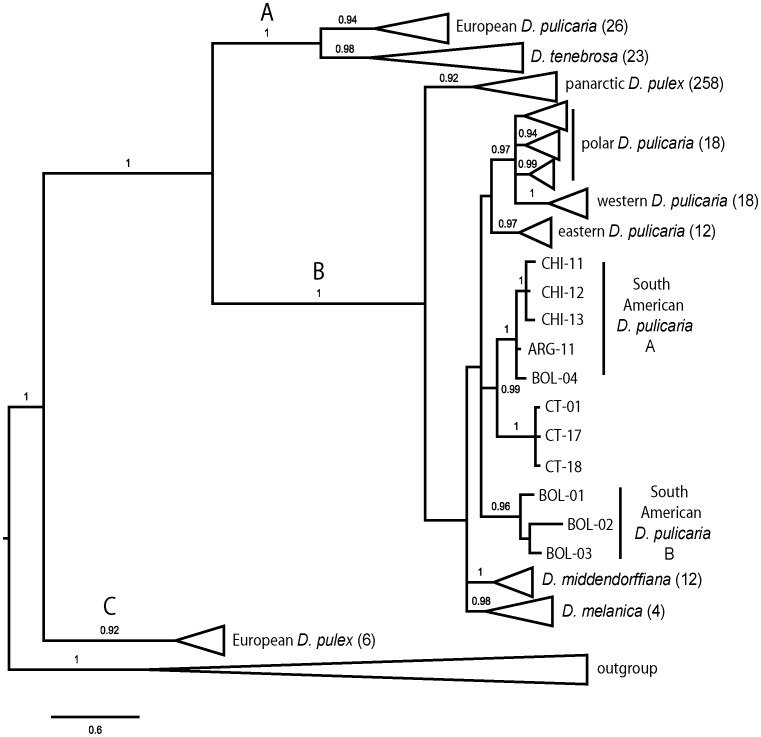
Bayesian phylogenetic analysis of a dataset containing 398 partial sequences of the mitochondrial NADH dehydrogenase subunit 5 (ND5) gene (Figure 1) reveals strong support for the three major groups within the D. pulex species complex [12]: pulex (posterior probability [PP] = 0.92), pulicaria (PP = 1), and tenebrosa (PP = 1). In addition, previously described lineages [12], [13] within these major groups are identified with high posterior probability values, although relationships among lineages are weakly supported. Synonymous site diversity ranges from 0.00 to 0.04 in all lineages except D. tenebrosa, which has extremely high diversity at 0.106 (Table 1).
Figure 1. Consensus Bayesian phylogeny of the Daphnia pulex species complex based on the mitochondrial ND5 gene.
The alignment contains 398 sequences of length 496 nt with 241 polymorphic nt positions of which 204 are phylogenetically informative (excluding the outgroup). Posterior probabilities are indicated on the nodes of the tree and are not shown if less than 0.80. The tree is rooted using an ND5 sequence from Daphnia obtusa. Triangles represent clusters that are collapsed to save space and the number of individuals included is shown in parentheses. The large letters indicate the three major groups within the D. pulex species complex: A = tenebrosa; B = pulicaria; C = pulex. The two or three-letter code names of some individuals correspond to sampling locations as follows: ARG = Argentina, BOL = Bolivia, CHI = Chile, CT = Connecticut, USA. The expanded version of this tree showing all individuals is available in Figure S1.
Table 1. Polymorphism statistics for ND5 and COX1 (panarctic D. pulex) in lineages of the Daphnia pulex species complex.
| Lineage | N1 | K2 | H3 | S4 | πs 5 | πn |
| panarctic D. pulex – COX1 | 68 | 59 | 0.99 | 128 | 0.0248 | 0.0008 |
| panarctic D. pulex – ND5 | 171 | 76 | 0.95 | 96 | 0.0396 | 0.0021 |
| European D. pulex | 3 | 3 | 1.00 | 8 | 0.0412 | 0.0018 |
| European D. pulicaria | 23 | 14 | 0.93 | 31 | 0.0306 | 0.0041 |
| eastern D. pulicaria | 10 | 9 | 0.98 | 12 | 0.0257 | 0.0015 |
| western D. pulicaria | 12 | 7 | 0.83 | 12 | 0.0179 | 0.0014 |
| polar D. pulicaria | 10 | 10 | 1.00 | 19 | 0.0227 | 0.0063 |
| S. American D. pulicaria A | 5 | 4 | 0.90 | 3 | 0.0000 | 0.0038 |
| S. American D. pulicaria B | 3 | 3 | 1.00 | 9 | 0.0281 | 0.0081 |
| D. middendorffiana | 6 | 6 | 1.00 | 12 | 0.0305 | 0.0033 |
| D. tenebrosa | 11 | 11 | 1.00 | 49 | 0.1055 | 0.0076 |
The number of individuals analyzed. Multiple individuals collected from the same location were removed.
The number of haplotypes.
The haplotype diversity, defined as the probability that two alleles randomly chosen from the sample are different.
The number of segregating sites.
π was estimated for nonsynonymous (πn) and synonymous (πs) sites using the correction of Jukes and Cantor.
In addition to panarctic D. pulex and the other eight lineages previously described by Colbourne et al. [12], two distinct lineages from South America are evident; South American D. pulicaria A (SA-A), and South American D. pulicaria B (SA-B) [13]. In our analysis, SA-B clusters within the pulicaria group, and panarctic D. pulex is the sister taxon to all the other pulicaria lineages (Figure 1). Mergeay et al. [13] identified a third South American lineage (SA-C), but it is not represented in our analysis as repeated attempts to amplify ND5 from individuals of this lineage have been unsuccessful (this study, Mergeay et al. [13]).
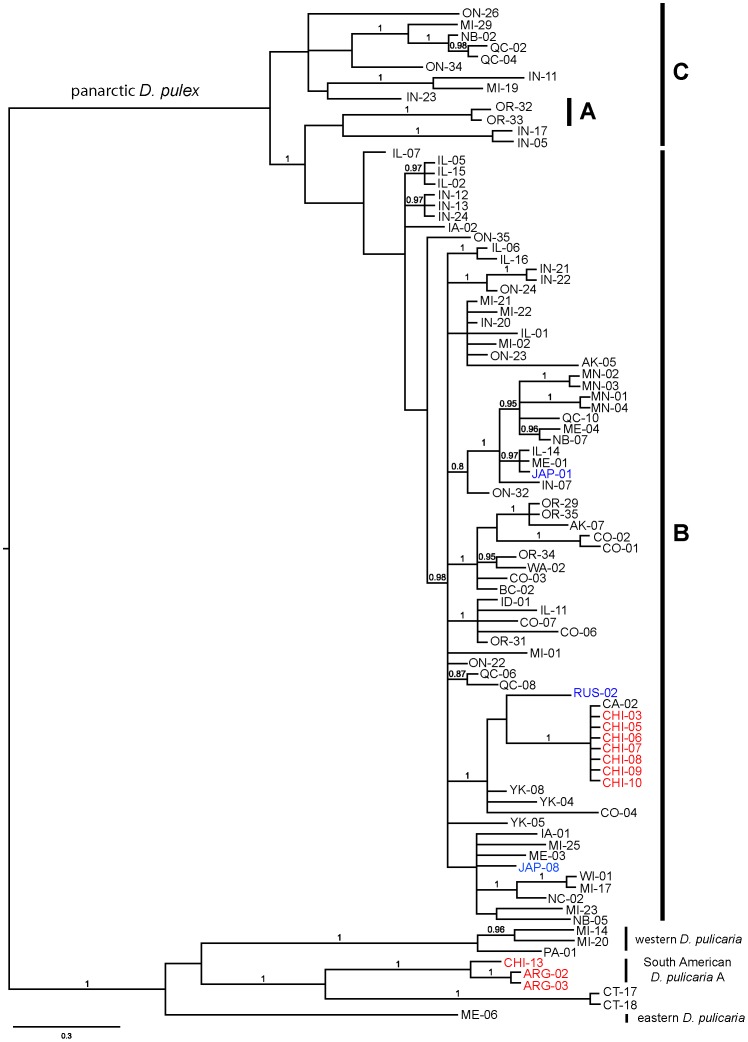
Bayesian phylogenetic analysis was also conducted on a dataset consisting of 98 sequences of ND5 combined with partial sequences of the mitochondrial cytochrome c oxidase subunit 1 (COX1) gene. This dataset includes 8 of the 12 lineages in the species complex, including SA-A and SA-B. After examining a variety of partitioning schemes, we obtained the lowest average standard deviation of split frequencies with unpartitioned data. The results from the different partitioning schemes are largely consistent; incongruence appeared only at branches with low posterior probability values. Both the ND5 (Figure 1) and the ND5+COX1 tree (Figure 2) suggest that SA-A and SA-B are derived from North American ancestors, and that there were at least two, and likely three introductions into South America from North America. For example, SA-A forms a strongly supported sister group to isolates CT-17 and CT-18 from Connecticut, USA.
Figure 2. Consensus Bayesian phylogeny of the Daphnia pulex species complex.
based on the mitochondrial ND5 and COX1 genes. The alignment contains 98 sequences consisting of 496 nt of ND5 and 552 nt of COX1 with 295 polymorphic positions of which 258 are phylogenetically informative. The tree is rooted through the midpoint. Posterior probabilities are indicated on the nodes of the tree and are not shown if less than 0.80. Isolates CT-17 and CT-18 were collected from Connecticut, USA.
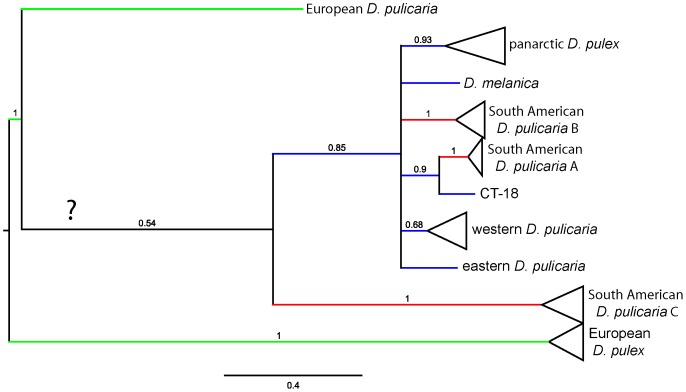
Analysis of 95 longer ND5+COX1 sequences representing four lineages (panarctic D. pulex, eastern and western D. pulicaria, SA-A, Figure 3), identified two of the three subgroups of panarctic D. pulex (A, B and C) identified by Paland et al. [14]. However, in contrast to their results, group A is not the sister taxon to the other two; instead, individuals from Paland et al.’s group C occupy this position. Moreover, their group C is paraphyletic to both group A and group B in our tree.
Figure 3. Consensus Bayesian phylogeny of the Daphnia pulex species complex based on the mitochondrial ND5 and COX1 genes.
The alignment contains 95 sequences consisting of 762 nt of ND5 and 1386 nt of COX1 with 321 polymorphic positions of which 204 are phylogenetically informative. The tree is rooted through the midpoint. Posterior probabilities are indicated on the nodes of the tree and are not shown if less than 0.80. Taxon colors represent geographic locations as follows: black = North America, blue = east Asia, red = South America.
Panarctic D. pulex is by far the most geographically widespread and well represented lineage in our analysis (65% of individuals analyzed), being observed on every continent except Australia and Antarctica (Figure 4, Table S1). Our new records from Japan, eastern Russia, western Alaska, and Chile provide large range extensions for this lineage. Even so, there is little obvious regional association of haplotypes in this lineage despite its large geographic range, with the exception of group A, which is restricted to western Oregon [14], [21]. Indeed, many of the individuals collected outside of North America share a very similar or even identical mitochondrial haplotype with individuals from North America (Figure 3, Figure S1). For example, individuals from two populations in Chile (Llanquihue Lake and Lake Riñuhue, Table S1) have the same panarctic D. pulex haplotype as individuals from California, USA (CA-02). In addition, haplotypes from Russia cluster with haplotypes from western North America (Colorado, California, Yukon) and those from Japan cluster with haplotypes from eastern (Maine) and midwestern USA (Iowa, Michigan).
Figure 4. Location of collection sites for individuals in the panarctic Daphnia pulex lineage.
(a) Red circles indicate collection sites for all individuals in the panarctic D. pulex lineage. (b) Blue circles indicate collection sites for individuals with the predicted ancestral haplotype for panarctic D. pulex. Sampling locations for all other individuals included in this study are provided in Figure S2.
Because ND5 data are not available for SA-C, we constructed a tree from partial sequences of the COX1 gene from 52 individuals representing this lineage and seven of the other lineages. SA-C is the sister taxon to all the other pulicaria group lineages in this tree, as reported by Mergeay et al. [13], but relationships among these lineages are not well resolved with these data (Figure 5).
Figure 5. Consensus Bayesian phylogeny of the Daphnia species complex based on the mitochondrial COX1 gene.
The alignment contains 52 sequences of length 552 nt with 151 polymorphic positions of which 112 are phylogenetically informative. Nine of the 12 lineages in the Daphnia pulex species complex, including all three South American lineages, are represented. The tree is rooted through the midpoint. Numbers at the nodes are Bayesian posterior probabilities and are not shown if less than 0.80. Branch colors correspond to continents as follows: green = Europe, blue = North America, red = South America. Individual CT-18 was collected from Connecticut, USA.
Demographic History of Panarctic D. pulex
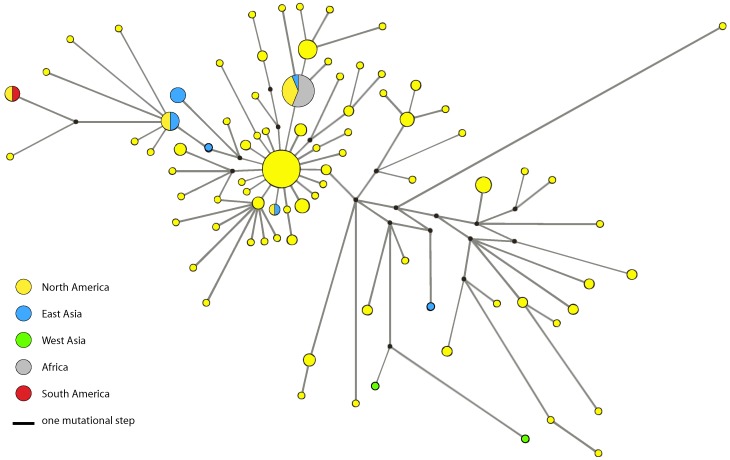
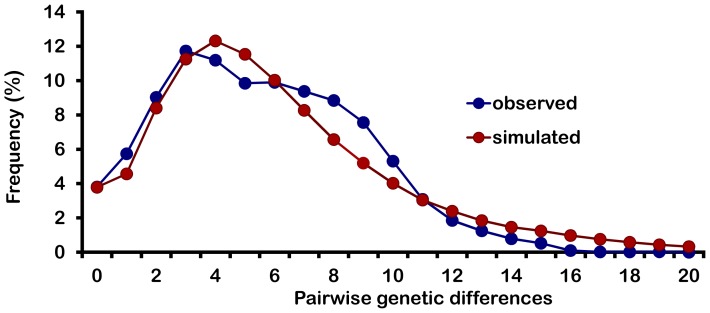
Network analysis of 173 panarctic D. pulex using the ND5 alignment revealed 84 haplotypes. One haplotype was sampled from 27 sites throughout North America (Figure 4b) and is located in the center of the network (Figure 6), which shows the characteristic star-like pattern commonly associated with recent demographic or spatial expansion. A second widespread haplotype was observed in 16 sites from North America, Japan, and throughout Africa. A mismatch distribution plot based on this dataset is not significantly different from the expected distribution under an expansion model despite the appearance of a slightly jagged curve (P = 0.738, Figure 7). The raggedness index is low at 0.004 (P = 0.988) and values for Tajima’s D and Fu’s Fs (D = −2.212, Fs = −25.058, P<0.001) are significantly negative providing further support for recent population expansion. The τ parameter, which dates the initiation of the expansion in mutational time [22], was estimated as τ = 2.88 (95th percentile confidence interval 1.45–7.38). Adopting Paland’s [23] direct estimate of the mitochondrial mutation rate in D. pulex (6.6×10−8), spatial expansion of panarctic D. pulex is estimated to have occurred 44,000 generations ago (95th percentile confidence interval ranges from 22,000 to 113,000 generations ago).
Figure 6. Median-joining haplotype network (ε = 0) for the panarctic D. pulex lineage.
The network is based on the mitochondrial ND5 gene (496 nt, 173 individuals). Multiple individuals from the same location were removed. Each haplotype is represented by a circle whose area is proportional to the number of times the haplotype was observed. Median vectors, which represent either extant unsampled sequences or extinct ancestral sequences, are indicated by small black circles.
Figure 7. Mismatch distribution plot for the panarctic D. pulex lineage.
This plot is based on the mitochondrial ND5 gene (496 nt, 173 individuals). Multiple individuals collected from the same location were removed.
Discussion
Phylogeny and Origin of the Pulicaria Group
Our phylogenetic analyses confirm the existence of at least 12 lineages in the D. pulex complex, and extend the geographic range of SA-A from one to two continents, and the range of panarctic D. pulex from three to five continents. SA-C is the sister taxon to the rest of the pulicaria group in our analysis (Figure 5), which is concordant with previous results [13], but we found that both SA-A and SA-B cluster within the pulicaria group, and that panarctic D. pulex is its sister taxon (Figure 2). In contrast, Mergeay et al. [13] found that both SA-B and SA-C cluster outside the rest of the pulicaria group, and consequently proposed that the pulicaria ancestor came from South America. All three SA lineages are as yet associated only with obligately parthenogenetic, polyploid individuals from high-altitude lakes and appear to be interspecific hybrids [13], [18], a situation that closely parallels that observed in arctic North America [16], [24]. Moreover, we found close relatives of SA-A in temperate North America (CT-17 and CT-18 from Connecticut), which suggests that both SA-A and SA-B are derived from a North American ancestor, consistent with the Holarctic origin proposed for this group [9], [17], [19]. Thus, SA-C is currently the only lineage in the pulicaria group without a close North American relative.
The Panama land bridge formed during the Pliocene and facilitated the movement, in both directions, of fauna between North and South America [25]. Successive expansion of Daphnia through Central and into South America (which were cooler during the Pliocene) could have occurred during this period, with subsequent extinction of the intervening lineages in regions that are now tropical. Indeed, Adamowicz et al. [8] noted a propensity for dispersal between these two continents throughout the genus, Daphnia. Discovery of the haplotypes from Connecticut (CT), and of panarctic D. pulex haplotypes in Chile that are identical to haplotypes in western USA, provide support for the hypothesis that members of the pulicaria group have moved from North to South America several times. Thus, the possibility remains that SA-C also exists but is yet undetected in North America, or has gone extinct there.
Adamowicz et al. [8] mapped continental regions onto the phylogeny of the entire genus Daphnia in order to explore the frequency and orientation of intercontinental speciation. Based on their analysis, it was not possible to determine the region from which the ancestor of the pulicaria group came (Figure 5). However, based on the Holarctic distribution of the D. pulex species complex, it seems unlikely that it came from South America. A more parsimonious hypothesis is that it colonized the Nearctic from Asia and spread to South America soon thereafter. Testing hypotheses about the origin of the pulicaria group will require even more extensive sampling in the temperate regions of South America, as well as the temperate and high-altitude regions of Central America.
Demographic History of Panarctic D. pulex
Despite a near worldwide sampling effort and our discovery of panarctic D. pulex from Alaska, Central Asia, Japan, and South America, the only evidence of regionalism in this lineage is the A subgroup, which is restricted to Oregon, USA and is thought to have diverged during the Pleistocene in a refugium located south of the Cordilleran ice sheet [14], [21]. This lineage, to which the isolate whose full genome was sequenced [26] belongs, is also divergent at nuclear loci [21], [27], [28]. Otherwise, panarctic D. pulex has apparently undergone a Late Quaternary expansion and recently emigrated from North America to other continents. Even so, this lineage is primarily restricted to temperate regions (Figure 4) while other members of the D. pulex complex (D. pulicaria, D. middendorffiana, D. tenebrosa) dominate in the Arctic [15], and other species in the subgenus Daphnia dominate in the desert and subtropical regions of North America [29].
Panarctic D. pulex is by far the most prevalent lineage in our samples. Given that it has been studied more rigorously than other lineages in North America, and we incorporated a substantial amount of previously published data, its overrepresentation in our dataset may be partly due to sampling bias. Nevertheless, it is clear that panarctic D. pulex has a more extensive geographical range than any other lineage in the D. pulex complex with individuals sampled from five continents.
Past bottlenecks and subsequent range expansions are commonly documented for taxa that currently inhabit geographic regions that were glaciated during the Pleistocene [30]. Genetic signatures of such expansion include star-like haplotype networks with a common, presumably ancestral haplotype being central to other rarer haplotypes [31], [32], and unimodal plots of the number of differences between pairs of haplotypes (mismatch distribution, [22], [32]). In addition, significantly negative values of Tajima’s D and Fu’s Fs indicate an excess of low frequency variants, which can be attributed to expanding population size or range. Our results suggest that the panarctic D. pulex lineage is likely to have experienced spatial expansion in North America approximately 44,000 (22,000 to 113,000) generations ago based on the mitochondrial haplotype network (Figure 6), mismatch distribution (Figure 7), Tajima’s D and Fu’s Fs analyses. Assuming that Daphnia undergo between two and five generations per year depending on location, this would suggest that expansion occurred between 8,800 and 22,000 years ago. Although this estimate is more recent than others reported for this group [14], [20], it is not unreasonable given that the maximum extent of the last glacial advance in North America occurred about 21,000 years ago and ended about 10,000 years ago [33]. Moreover, it overlaps with the Hypsithermal or Holocene thermal maximum period of warming that occurred from about 9,000 to 4,000 years ago [34], and during which range expansion of other temperate flora and fauna is thought to have occurred [35].
There are some caveats to our finding of recent range expansion as other scenarios, such as ancient range expansion with high migration between neighboring demes [36], [37], can also mimic the signature of recent expansion. However, this scenario seems unlikely in the case of panarctic D. pulex as most of its range in North America was glaciated during the last glacial maximum. In addition, regional studies of genetic variation indicate that gene flow among populations on a local scale is typically very low [38], [39], [40], [41], although some obligately parthenogenetic clones have spread over very large geographic areas [9], [16]. Heterogeneity of mutation rates among sites [42], [43], and/or selective sweeps [32], [44] can also mimic the signature of population expansion, and we cannot exclude the possibility that one or both of these factors has contributed to the distribution of mitochondrial variation that we observed.
Morphological similarity across wide geographic areas coupled with high potential for long-distance dispersal via diapausing eggs originally led to the conclusion that many Daphnia species are cosmopolitan [17], [45]. However, subsequent morphological [17], [46] and genetic analyses showed that regionalism is more common [8], [15], [21], [47], [48], [49] and that many species are restricted to a single continent. Even so, cases of transcontinental establishment in cladocerans have been documented [8], [50], [51], [52], [53]. For example, eastern D. pulicaria, which was thought to occur only in North America, has now been reported in the High Tatra and Pyrenees mountains in Europe [54]. Moreover, we found that the clone currently spreading in Africa [9] has a haplotype that is widespread in North America (Figure 6, Figure S1), that a haplotype from Chile is identical to one from California, USA, and that haplotypes from Japan are very similar (but not identical) to the putative ancestral haplotype, suggesting that panarctic D. pulex has repeatedly traversed great distances to colonize new regions.
It has long been suggested that migratory birds act as long-distance dispersal vectors of invertebrate resting eggs [55], and a recent study has shown that the genetic structure of Daphnia ambigua and Daphnia laevis in North America is consistent with the movement of water fowl [56]. However, recent intercontinental dispersal of cladocerans has also been attributed to human activity [55] and indeed, Mergeay et al. [9] suggested that the panarctic D. pulex clone in Africa was introduced into Lake Naivasha (Kenya) from the USA during fish stocking programs in the late 1920s. However, they also suggested that dispersal by migrant waterfowl has helped to spread the clone throughout the continent.
There are four major migratory bird flyways between North and South America (Atlantic, Central, Mississippian and Pacific), and the occurrence of the same panarctic D. pulex haplotype in California and Chile is consistent with movement of resting eggs along the Pacific flyway. In addition, Weider et al. [15] suggested that panarctic D. pulex could have invaded northern Europe via waterfowl that migrate between wintering grounds in Europe and summer breeding grounds in arctic Canada, which could also explain the occurrence of eastern D. pulicaria in Europe [54]. Waterfowl are also known to migrate between eastern Asia and Alaska [57], which could account for the presence of panarctic D. pulex in Japan and Russia.
One hypothesis to explain the success of the ubiquitous panarctic D. pulex lineage outside its typical range in temperate North America is its frequent hybridization with other members of the pulicaria group, and the maintenance of these hybrid genotypes via obligate parthenogenesis. For example, no sexual populations of the pulicaria group have been detected in South America, and all the clones appear to be of hybrid origin [13], [18]. Moreover, when pulicaria-group populations are detected in desert or subtropical regions of North America they are typically D. pulex × D. pulicaria hybrids that reproduce by obligate parthenogenesis [29], as is the North American clone that is invading Africa [9]. Previous work has suggested that the lack of gene flow between regional populations of Daphnia is likely the result of priority effects and local adaptation, which makes it difficult for immigrant genotypes to integrate into well-established Daphnia communities [58]. In contrast, Mergeay et al. [9] highlighted the fact that the clone introduced to Africa was able to displace a genetically diverse, sexually reproducing indigenous species that was supposedly well-adapted to its environment in only 75 years. Further study of such Daphnia clones should provide important insights into the characteristics that make them such successful invaders of established Daphnia communities.
Conclusions
Our phylogenetic analyses support the evolutionary relationships among eleven of the major mitochondrial lineages of the D. pulex species complex. However, we find little support for the hypothesis that the pulicaria group originated in South America. While most lineages within the species complex are restricted to one or two continents, the panarctic D. pulex lineage was detected on every continent except Australia and Antarctica. Despite this wide geographic range, no strong regionalism was detected, a pattern that differs from that observed in other cladocerans. In addition, our results suggest that this lineage has undergone recent expansion with some haplotypes spreading to more than one continent. The hypothesis that hybridization and maintenance of the resulting genotypes by obligate parthenogenesis has contributed to the recent and unusual geographic success of the panarctic D. pulex lineage warrants further study.
Materials and Methods
Taxonomic Sampling
We sought to examine individuals of the D. pulex complex from regions that had not been sampled extensively in the past, i.e., western Alaska, Japan, Russia, the Canadian prairies, South America, and the Southwestern United States. Species delimitation and nomenclature within this complex, and especially the pulicaria group, is problematic (reviewed in Mergeay et al. [13]) and we used the nomenclature based on phylogenetic analysis of mtDNA described by Colbourne et al. [12] and Mergeay et al. [13] for our samples.
We generated new sequences of 762 nt of the ND5 gene from 195 individuals and combined these with 203 published sequences [9], [12], [13], [14], [18], [54], [59], [60] to generate an ND5 dataset of 398 individuals representing all but one lineage in this species complex (Table S1). Sequences from previous studies with a substantial amount of missing nucleotide data (greater than 30 Ns in a single read) were not included in our analyses. We used Daphnia cf. obtusa from North America as an outgroup in the phylogenetic analysis of this dataset as it has been identified as a close relative of the D. pulex species complex with independent evidence [61].
We also amplified the COX1 gene from 66 of our DNA samples (mainly panarctic D. pulex), and added them to previously published sequences [59], [60] for a total of 95 COX1 sequences of 1386 nt. We added these sequences to our longer ND5 sequences (762 nt) to create a dataset representing four lineages with an alignment length of 2148 nt. We also generated a larger dataset representing eight lineages by combining 496-nt sequences of ND5 with 552-nt sequences of COX1 from our new and previously published sequences. New sequence data have been deposited in GenBank under accession numbers JX532724 - JX532790 (1386 nt COX1), JX532791 - JX532857 (762 nt ND5) and JX532858 - JX532985 (496 nt ND5).
DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA was extracted from Daphnia using QuickExtract (Epicentre). Samples were homogenized in 50 µL of QuickExtract solution, incubated at 65°C for four hours, and 98°C for two min. PCR reactions were 50 µl total volume and contained 35 µl water, 5 µl 10X PCR buffer (1.5 mM MgCl2), 10 nmoles dNTPs, 15 pmoles of each primer, 1 U of Taq polymerase, and 25–50 ng of DNA template. PCR was conducted on an MJ Thermocycler with the following conditions: 40 cycles of 30 sec at 94°C, 30 sec at 48°C, and one min at 72°C; followed by one cycle of six min at 72°C. PCR amplicons were sequenced in both directions by High-Throughput Sequencing Solutions at the University of Washington, Department of Genome Sciences (Seattle, WA).
The primers ND5pulexF (5′GGGGTGTATCTATTAATTCG) and ND5pulexR (5′ATAAAACTCCAATCAACCTTG) were used to generate an amplicon of 897 nt from the ND5 gene. These primers were used for both PCR and sequencing reactions. The primers COX1pulexF (5′CCTACTCCTCGGCCATTTG) and COX1pulexR (5′GGGGATGCTCTATTTTGGAA) were used to generate an amplicon of 1679 nt containing the complete COX 1 gene. Internal sequencing primers were used for this locus and are as follows: COX1seqintR (5′TGAATCTTTAACCAACGGG) and COX1seqint_B (5′ CGTGAAGTGTGCCAAGTCAT).
Phylogenetic Analyses, Sequence Divergence, and Diversity
DNA sequence electropherograms were edited and assembled with Sequencher 4.7 (Gene Codes Corp.) and CodonCode Aligner, and aligned with ClustalW in MEGA 4 [62]. The alignment for these protein coding regions was unambiguous and no indels were observed. We constructed phylogenetic trees using Bayesian inference in MrBayes 3.1.2 [63]. The effects of data partitioning were explored by conducting an analysis without partitioning, an analysis with the data partitioned by locus, and an analysis with the data partitioned by one noncoding and three codon positions (1st, 2nd, and 3rd codon positions). Model selection for each partition was based on the Akaike information criterion in the program Modeltest 3.7 [64], [65]. The best-fitting model for each partition was employed in subsequent phylogenetic analysis. Default prior settings were used with the exception of the ‘ratepr’ parameter, which was set to ‘variable’ so that partitions could evolve at different rates. Branch lengths and topology were shared among partitions, but the substitution rate matrix, state frequency, and shape parameter of the gamma distribution were unlinked to allow separate parameter estimates.
Two independent and simultaneous Markov chain Monte Carlo (MCMC) analyses of fifteen heated and one cold chain were run for six million generations with sampling from the chain every 100 generations. After verifying chain convergence (average standard deviation of split frequencies <0.01), we discarded the initial 25% of trees as ‘burn-in’. A 50% majority-rule consensus tree with posterior probability (PP) values for each node was constructed from the remaining Bayesian trees. Branch support was the proportion of trees that contained a lineage, which represents the posterior probability of lineage existence, given the data and the model of evolution. Each phylogenetic analysis was repeated at least twice and the results were inspected with Tracer 1.4 [66].
Nucleotide diversity (π), defined as the average number of pairwise differences among DNA sequences [67], was estimated for each lineage and for the following categories of nucleotide sites in the ND5 gene: synonymous (πs), nonsynonymous (πn), and total (πT) using DNASP [68].
Genealogical relationships in panarctic D. pulex were further examined with median-joining haplotype networks using the program Network 4.5 [69]. The ε parameter was set to 0. We used the MP post-processing option, which removes all superfluous median vectors and links that are not contained in the shortest trees of the network. For network and subsequent demographic analyses, the panarctic D. pulex dataset was pruned so that one randomly chosen individual per collection site was included (Table S1). This pruning minimizes bias due to uneven sampling across geographic sites.
Demographic History of Panarctic D. pulex
To elucidate the demographic history of panarctic D. pulex, frequency (mismatch) distributions of the number of differences between pairs of haplotypes [29], [33], [70], [71] were constructed with Arlequin 3.11 [72]. In addition, the raggedness index [73] was used to measure the smoothness of the pairwise mismatch plots. The simulated model of spatial expansion was compared to the observed mismatch distribution and the sum of square deviations (SSD) between the observed and expected distributions was tested with 10,000 permutation replicates. P-values were obtained by calculating the proportion of simulations that had an SSD that was equal to or larger than the observed SSD. A generalized non-linear least-square approach was employed in Arlequin to estimate the τ parameter, which is time to expansion in mutational time (τ = 2µt), where µ is the mutation rate for the entire haplotype and t is time in generations. Confidence intervals for τ were obtained by parametric bootstrapping [71].
We used Arlequin 3.11 [72] to calculate Tajima’s D and Fu’s FS, which are commonly used to detect departures from the neutral model, from panarctic D. pulex sequences. The significance of Tajima’s D was determined by generating random samples under the hypothesis of selective neutrality and population equilibrium using coalescent simulations. The significance of Fu’s Fs was determined by generating random samples under the hypothesis of neutrality and then estimating P-values as the proportion of Fs statistics that were less than or equal to the observed value [72].
Supporting Information
Full Bayesian phylogeny of the Daphnia pulex species complex from Figure 1 . This is a PDF file. The tree is based on 496 nt of the mitochondrial ND5 gene and shows all 398 individuals included in this study, plus the outgroup. Taxa represented on branches followed by a letter are given below. A CZE-02, CZE-04, CZE-06, GER-02, SWI-01, wSIB-10 B SAF-01, BOT-01, BOT-02, BOT-03, ETH-01, ETH-02, JAP-01, KEN-01, KEN-02, KEN-03, ZIM-01, AB-01, IL-13, IL-14, ME-01, ME-02, OR-06, SK-07, SK-10, SK-13 C ME-04, MN-01, MN-04, NB-06, NB-07, NY-01, QC-10, QC-17, QC-18 D BC-01, BC-02, CO-01, CO-02, CO-03, IA-01, ID-01, IL-10, IL-12, IN-20, MI-03, MI-12, MI-13, MI-21, MI-22, NB-03, NC-01, NC-02, NU-01, NU-03, ON-07, ON-08, ON-10, ON-11, ON-17, ON-21, ON-22, ON-28, ON-29, ON-32, OR-28, OR-29, OR-30, OR-31, SK-05, YK-05, YK-06 E RUS-01, RUS-02, SK-04, wSIB-03, YK-01, YK-02, YK-07, YK-08 F CA-01, CA-02, CA-03, CHI-01, CHI-02, CHI-03, CHI-04, CHI-05, CHI-06, CHI-07, CHI-08, CHI-09, CHI-10 G JAP-02, JAP-04, JAP-05, JAP-06, JAP-07, JAP-10, JAP-11 H OR-03, OR-07, OR-08, OR-11, OR-18, OR-19 I NB-01, NB-02, NS-03, NS-04, QC-02, QC-04 J CHI-12, CHI-14, CHI-15, ARG-01, ARG-02, ARG-03, ARG-09.
(PDF)
Collection sites for lineages of the Daphnia pulex species complex included in this study, excluding panarctic D. pulex . This is a PDF file. Colors are used to indicate lineages as follows: black = European D. pulex orange = European D. pulicaria green = eastern D. pulicaria magenta = western D. pulicaria light blue = polar D. pulicaria brown = D. middendorffiana dark purple = D. tenebrosa light purple = S. American D. pulicaria A pink = S. American D. pulicaria B.
(PDF)
Individuals of the Daphnia pulex species complex included in this study. This Excel spreadsheet provides information on all individuals included in this study. The individuals that were included in each phylogenetic or network analysis are indicated in the last 5 columns. Latitude and longitude of collection sites for some previously published accessions were estimated with Google Earth.
(XLS)
Acknowledgments
We thank the following people who collected Daphnia specimens that were used in our study: Desiree Allen, Maria Belyaeva, Carla Caceres, Sandra Connelly, Jeffrey Dudycha, Seiji Ishida, Alexey Kotov, Michael Lynch, Sarah Schaack, and Larry Weider. We also thank two anonymous reviewers whose comments on an earlier draft improved the manuscript. Part of this work was carried out using the resources of the Computational Biology Service Unit at Cornell University.
Funding Statement
TJC was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (www.nserc.ca). DJT was funded by grants ARC1023334 and DEB0331095 from the National Science Foundation, United States of America (www.nsf.gov). Part of this work was carried out using the resources of the Computational Biology Service Unit from Cornell University that is partially funded by Microsoft Corporation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gewin V (2005) Functional genomics thickens the biological plot. PLoS Biol 3: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters RH, de Bernardi R (1987) Daphnia. Memorie dell’Instituto Italiano di Idrobiologia, 45. Instituto Italiano di Idrobidlogia Verbania Pallanza.
- 3. Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex. Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eads BD, Colbourne JK, Bohuski E, Andrews J (2007) Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex. BMC Genomics 8: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Omilian AR, Scofield DG, Lynch M (2008) Intron presence-absence polymorphisms in Daphnia. Mol Biol Evol 25: 2129–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papillon D, Telford MJ (2007) Evolution of Hox3 and ftz in arthropods : insights from the crustacean Daphnia pulex . Dev Genes Evol 217: 315–322. [DOI] [PubMed] [Google Scholar]
- 7. Schaack S (2008) Daphnia comes of age : an ecological model in the genomic era. Mol Ecol 17: 1634–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adamowicz S, Petrusek A, Colbourne J, Hebert P, Witt J (2009) The scale of divergence : A phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus. Mol Phylogenet Evol 50: 423–436. [DOI] [PubMed] [Google Scholar]
- 9. Mergeay J, Verschuren D, De Meester L (2006) Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc Biol Sci 273: 2839–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decaestecker E (2009) Cyclical parthenogenesis in Daphnia: sexual versus asexual reproduction. In: Schön I, Martens K, van Dijk P, editors. Lost Sex: The evolutionary biology of parthenogenesis. Dordrecht: Springer 295–316.
- 11. Cerny M, Hebert PDN (1999) Intercontinental allozyme differentiation among four holarctic Daphnia species. Limnol Oceanogr 44: 1381–1387. [Google Scholar]
- 12. Colbourne JK, Crease TJ, Weider LJ, Hebert PDN, Dufresne F, et al. (1998) Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex). Biol J Linnean Soc 65: 347–365. [Google Scholar]
- 13. Mergeay J, Aguilera X, Declerck S, Petrusek A, Huyse T, et al. (2008) The genetic legacy of polyploid Bolivian Daphnia: the tropical Andes as a source for the North and South American D. pulicaria complex. Mol Ecol 17: 1789–1800. [DOI] [PubMed] [Google Scholar]
- 14. Paland S, Colbourne J, Lynch M (2005) Evolutionary history of contagious asexuality in Daphnia pulex . Evolution 59: 800–813. [PubMed] [Google Scholar]
- 15. Weider LJ, Hobaek A, Colbourne JK, Crease TJ, Dufresne F, et al. (1999) Holarctic phylogeography of an asexual species complex. I. Mitochondrial DNA variation in arctic Daphnia. Evolution 53: 777–792. [DOI] [PubMed] [Google Scholar]
- 16. Weider LJ, Hobaek A, Hebert PDN, Crease TJ (1999) Holarctic phylogeography of an asexual species complex. II. Allozymic variation and clonal structure in Arctic Daphnia. Mol Ecol 8: 1–13. [Google Scholar]
- 17. Brooks JL (1957) The systematics of North American Daphnia. Mem Conn Acad Arts Sci 13: 1–180. [Google Scholar]
- 18. Adamowicz SJ, Gregory TR, Marinone MC, Hebert PDN (2002) New insights into the distribution of polyploid Daphnia: the Holarctic revisited and Argentina explored. Mol Ecol 11: 1209–1217. [DOI] [PubMed] [Google Scholar]
- 19. Hebert PDN, Schwartz SS, Ward RD, Finston TL (1993) Macrogeographic patterns of breeding system diversity in the Daphnia pulex group. 1. Breeding systems of Canadian populations. Heredity 70: 148–161. [DOI] [PubMed] [Google Scholar]
- 20. Lynch M, Seyfert A, Eads B, Williams E (2008) Localization of the genetic determinants of meiosis suppression in Daphnia pulex . Genetics 180: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crease TJ, Lee SK, Yu SL, Spitze K, Lehman N, et al. (1997) Allozyme and mtDNA variation in populations of the Daphnia pulex complex from both sides of the Rocky Mountains. Heredity 79: 242–251. [Google Scholar]
- 22. Rogers A, Harpending H (P1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 23.Paland S (2004) Investigations into mitochondrial genome evolution using Daphnia. PhD thesis Indiana University.
- 24. Dufresne F, Hebert PDN (1997) Pleistocene glaciations and polyphyletic origins of polyploidy in an arctic cladoceran. Proc R Soc Lond [Biol] 264: 201–206. [Google Scholar]
- 25.Stehli F, Webb S (1985) The great American biotic interchange. New York: Plenum Press. 532 p.
- 26. Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex . Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crease TJ, Floyd R, Cristescu M, Innes D (2011) Evolutionary factors affecting Lactate dehydrogenase A and B variation in the Daphnia pulex species complex. BMC Evol Biol 11: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vergilino R, Markova S, Ventura M, Manca M, Dufresne F (2011) Reticulate evolution of the Daphnia pulex complex as revealed by nuclear markers. Mol Ecol 20: 1191–1207. [DOI] [PubMed] [Google Scholar]
- 29. Hebert PDN, Finston TL (1996) A taxonomic reevaluation of North American Daphnia (Crustacea : Cladocera). II. New species in the Daphnia pulex group from the south-central United States and Mexico. Can J Zool 74: 632–653. [Google Scholar]
- 30. Hewitt GM (2004) The structure of biodiversity - insights from molecular phylogeography. Front Zool 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avise JC : Phylogeography (2000) The History and Formation of Species. Cambridge: Harvard University Press.
- 32. Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dyke AS, Andrews JT, Clark PU, England JH, Miller GH, et al. (2002) The Laurentide and Innuitian ice sheets during the Last Glacial Maximum. Quat Sci Rev 21: 9–31. [Google Scholar]
- 34. Kaufman DS, Ager TA, Anderson NJ, Anderson PM, Andrews JT, et al. (2004) Holocene thermal maximum in the western Arctic (0–180°W). Quat Sci Rev 23: 529–560. [Google Scholar]
- 35. Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- 36. Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13: 853–864. [DOI] [PubMed] [Google Scholar]
- 37. Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20: 76–86. [DOI] [PubMed] [Google Scholar]
- 38. Allen MR, Thum RA, Caceres CE (2010) Does local adaptation to resources explain genetic differentiation among Daphnia populations? Mol Ecol 19: 3076–3087. [DOI] [PubMed] [Google Scholar]
- 39. Crease TJ, Lynch M, Spitze K (1990) Hierarchical analysis of population genetic variation in mitochondrial and nuclear genes of Daphnia pulex . Mol Biol Evol 7: 444–458. [DOI] [PubMed] [Google Scholar]
- 40. Miner BE, Kerr B (2011) Adaptation to local ultraviolet radiation conditions among neighbouring Daphnia populations. Proc Biol Sci 278: 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morgan KK, Hicks J, Spitze K, Latta L, Pfrender ME, et al. (2001) Patterns of genetic architecture for life-history traits and molecular markers in a subdivided species. Evolution 55: 1753–1761. [DOI] [PubMed] [Google Scholar]
- 42. Aris-Brosou S, Excoffier L (1996) The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol 13: 494–504. [DOI] [PubMed] [Google Scholar]
- 43. Tajima F (1996) The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaplan NL, Hudson RR, Langley CH (1989) The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frey DG (1982) Questions concerning cosmopolitanism in Cladocera. Arch Hydrobiol 93: 484–502. [Google Scholar]
- 46.Frey DG (1986) The non cosmopolitanism of chydorid Cladocera: implications for biogeography and evolution. In: Gore RH, Heck KL, editors. Crustacean Biogeography. Rotterdam: Balkema 237–256.
- 47. Ishida S, Taylor DJ (2007) Mature habitats associated with genetic divergence despite strong dispersal ability in an arthropod. BMC Evol Biol 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kotov AA, Taylor DJ (2010) A new African lineage of the Daphnia obtusa group (Cladocera: Daphniidae) disrupts continental vicariance patterns. J Plankton Res 32: 937–949. [Google Scholar]
- 49. Penton EH, Hebert PDN, Crease TJ (2004) Mitochondrial DNA variation in North American populations of Daphnia obtusa: continentalism or cryptic endemism? Mol Ecol 13: 97–107. [DOI] [PubMed] [Google Scholar]
- 50. De Melo R, Hebert PDN (1994) Founder effects and geographical variation in the invading cladoceran Bosmina (Eubosmina) coregoni Baird 1857 in North America. Heredity 73: 490–499. [Google Scholar]
- 51. Havel JE, Colbourne JK, Hebert PDN (2000) Reconstructing the history of intercontinental dispersal in Daphnia lumholtzi by use of genetic markers. Limnol Oceanogr 45: 1414–1419. [Google Scholar]
- 52. Ishida S, Taylor DJ (2007) Quaternary diversification in a sexual Holarctic zooplankter, Daphnia galeata . Mol Ecol 16: 569–582. [DOI] [PubMed] [Google Scholar]
- 53. Taylor DJ, Hebert PDN (1993) Cryptic intercontinental hybridization in Daphnia (Crustacea) : the ghost of introductions past. Proc R Soc Lond [Biol] 254: 163–168. [Google Scholar]
- 54. Markova S, Dufresne F, Rees DJ, Cerny M, Kotlik P (2007) Cryptic intercontinental colonization in water fleas Daphnia pulicaria inferred from phylogenetic analysis of mitochondrial DNA variation. Mol Phylogenet Evol 44: 42–52. [DOI] [PubMed] [Google Scholar]
- 55. Havel JE, Medley KA (2006) Biological invasions across spatial scales: Intercontinental, regional, and local dispersal of Cladoceran zooplankton. Biol Invasions 8: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Figuerola J, Green AJ, Michot TC (2005) Invertebrate eggs can fly: evidence of waterfowl-mediated gene flow in aquatic invertebrates. Amer Nat 165: 274–280. [DOI] [PubMed] [Google Scholar]
- 57. Koehler AV, Pearce JM, Flint PL, Franson JC, Ip HS (2008) Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta). Mol Ecol 17: 4754–4762. [DOI] [PubMed] [Google Scholar]
- 58. De Meester L, Gomez A, Okamura B, Schwenk K (2002) The Monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecol 23: 121. [Google Scholar]
- 59. Paland S, Lynch M (2006) Transitions to asexuality result in excess amino acid substitutions. Science 311: 990–992. [DOI] [PubMed] [Google Scholar]
- 60. Crease TJ (1999) The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea). Gene 233: 89–99. [DOI] [PubMed] [Google Scholar]
- 61. Colbourne JK, Hebert PDN (1996) The systematics of North American Daphnia (Crustacea: Anomopoda): a molecular phylogenetic approach. Philos Trans R Soc London [Biol] 351: 349–360. [DOI] [PubMed] [Google Scholar]
- 62. Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 63. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 64. Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53: 793–808. [DOI] [PubMed] [Google Scholar]
- 65. Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 66.Rambaut A, Drummond AJ (2007) Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer.
- 67. Tajima F (1993) Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 69. Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 70. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 71. Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites application to human mitochondrial DNA. Genetics 152: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Excoffier L, Laval G, Schneider S (2005) Arlequin 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47. [PMC free article] [PubMed] [Google Scholar]
- 73. Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66: 591–600. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Bayesian phylogeny of the Daphnia pulex species complex from Figure 1 . This is a PDF file. The tree is based on 496 nt of the mitochondrial ND5 gene and shows all 398 individuals included in this study, plus the outgroup. Taxa represented on branches followed by a letter are given below. A CZE-02, CZE-04, CZE-06, GER-02, SWI-01, wSIB-10 B SAF-01, BOT-01, BOT-02, BOT-03, ETH-01, ETH-02, JAP-01, KEN-01, KEN-02, KEN-03, ZIM-01, AB-01, IL-13, IL-14, ME-01, ME-02, OR-06, SK-07, SK-10, SK-13 C ME-04, MN-01, MN-04, NB-06, NB-07, NY-01, QC-10, QC-17, QC-18 D BC-01, BC-02, CO-01, CO-02, CO-03, IA-01, ID-01, IL-10, IL-12, IN-20, MI-03, MI-12, MI-13, MI-21, MI-22, NB-03, NC-01, NC-02, NU-01, NU-03, ON-07, ON-08, ON-10, ON-11, ON-17, ON-21, ON-22, ON-28, ON-29, ON-32, OR-28, OR-29, OR-30, OR-31, SK-05, YK-05, YK-06 E RUS-01, RUS-02, SK-04, wSIB-03, YK-01, YK-02, YK-07, YK-08 F CA-01, CA-02, CA-03, CHI-01, CHI-02, CHI-03, CHI-04, CHI-05, CHI-06, CHI-07, CHI-08, CHI-09, CHI-10 G JAP-02, JAP-04, JAP-05, JAP-06, JAP-07, JAP-10, JAP-11 H OR-03, OR-07, OR-08, OR-11, OR-18, OR-19 I NB-01, NB-02, NS-03, NS-04, QC-02, QC-04 J CHI-12, CHI-14, CHI-15, ARG-01, ARG-02, ARG-03, ARG-09.
(PDF)
Collection sites for lineages of the Daphnia pulex species complex included in this study, excluding panarctic D. pulex . This is a PDF file. Colors are used to indicate lineages as follows: black = European D. pulex orange = European D. pulicaria green = eastern D. pulicaria magenta = western D. pulicaria light blue = polar D. pulicaria brown = D. middendorffiana dark purple = D. tenebrosa light purple = S. American D. pulicaria A pink = S. American D. pulicaria B.
(PDF)
Individuals of the Daphnia pulex species complex included in this study. This Excel spreadsheet provides information on all individuals included in this study. The individuals that were included in each phylogenetic or network analysis are indicated in the last 5 columns. Latitude and longitude of collection sites for some previously published accessions were estimated with Google Earth.
(XLS)