Abstract
Cartilage plays an essential role during skeletal development within the growth plate and in articular joint function. Interactions between the collagen fibrils and other extracellular matrix molecules maintain structural integrity of cartilage, orchestrate complex dynamic events during embryonic development, and help to regulate fibrillogenesis. To increase our understanding of these events, affinity chromatography and liquid chromatography/tandem mass spectrometry were used to identify proteins that interact with the collagen fibril surface via the amino terminal domain of collagen alpha 1(XI) a protein domain that is displayed at the surface of heterotypic collagen fibrils of cartilage. Proteins extracted from fetal bovine cartilage using homogenization in high ionic strength buffer were selected based on affinity for the amino terminal noncollagenous domain of collagen alpha 1(XI). Mass spectrometry was used to determine the amino acid sequence of tryptic fragments for protein identification. Extracellular matrix molecules and cellular proteins that were identified as interacting with the amino terminal domain of collagen alpha 1(XI) directly or indirectly, included proteoglycans, collagens, and matricellular molecules, some of which also play a role in fibrillogenesis, while others are known to function in the maintenance of tissue integrity. Characterization of these molecular interactions will provide a more thorough understanding of how the extracellular matrix molecules of cartilage interact and what role collagen XI plays in the process of fibrillogenesis and maintenance of tissue integrity. Such information will aid tissue engineering and cartilage regeneration efforts to treat cartilage tissue damage and degeneration.
Keywords: arthritis, cartilage, collagen fibril, extracellular matrix, interactions
Introduction
Microarray and proteomic studies of chondrocytes and cartilage have revealed greater than 2,400 gene products of which approximately ten percent are extracellular [1]. Cartilage tissue comprises chondrocytes surrounded by an extensive extracellular matrix (ECM) [2]. Composed primarily of water, the ECM also contains proteoglycans and collagens [3,4]. The physical entanglement and interactions between proteoglycan and collagen components contribute to the structural integrity of the cartilage matrix. Proteoglycans and glycosaminoglycans create a porous structure that provides compressive strength, while collagens form a framework for the ECM and cells and also provide tensile strength to the tissue. The collagenous framework of cartilage is formed by heterotypic fibril of collagen II crosslinked to collagen IX and collagen XI. Collagen XI, a heterotrimeric molecule of α1(XI), α2(XI) and α3(XI), is an essential, yet quantitatively minor component of the cartilage collagen fibrils [5]. The significance of collagen α1(XI) is demonstrated by cases of Fibrochondrogenesis, Stickler, and Marshall syndromes which are all attributed to mutations in the COL11A1 gene encoding the α1(XI) chain [6,7,8], and evidenced by the chondrodysplasia mouse (cho/cho) which carries a frameshift mutation resulting in a null allele of Col11a1 and neonatal lethality in homozygotes [9].
In exploring the relationship that collagen type XI may have with other components of the extracellular matrix, the amino terminal domain (NTD) of the α1(XI) chain is of specific interest. The NTD of collagen type α1(XI) is located at the surface of fibrils (Figure 1), is significantly larger than those of the major fibrillar collagens (types I, II and III) and is structurally related to α2(XI), α1(V) and α3(V) [10–17]. The NTDs share amino acid sequence homology (Table 1) and secondary structure with collagen α1(IX) NC3 domain, neurexin, sex hormone binding globulin, and thrombospondins as illustrated in Table 2 and Figure 2 [18–23]. ECM molecules with this domain have the potential to function as bridges between other ECM molecules, providing organization and stability [24–26]. The globular nature of collagen α1(XI) NTD leads to the restricted localization on the surface of collagen fibrils even though the triple helical domain of type XI collagen lies in the interior of the fibril [27–30]. The retained NTD may sterically hinder the further addition of collagen molecules and thereby limit the ultimate diameter of the collagen fibril [31–33]. Additionally, as the NTD of collagen α1(XI) is located on the exterior of the fibril, its association with other matrix components is plausible.
Figure 1.
Schematic diagram of collagen fibrils with collagen type XI (blue) located within the interior of type II collagen fibrils (orange). The globular nature of collagen α1(XI) NTD leads to the restricted localization on the surface of collagen fibrils even though the triple helical domain of type XI collagen lies in the interior of the fibril. The retained NTD may sterically hinder the further addition of collagen molecules and thereby limit the ultimate diameter of the collagen fibril. Additionally, the NTD of collagen α1(XI) on the surface of the fibril may interact with other matrix components.
Table 1.
MODELLER alignment based on amino acid sequence and secondary structure of the template.
 |
Table 2.
Template evaluation using BLAST PDB search (E value) and RMSD of homology models from PyMOL. Homology models were constructed in MODELLER. Secondary and tertiary structure comparison.
| Template | RMSD (Å) | E value |
|---|---|---|
| Collagen IX NC4 (2UUR) | 0.828 | 5.0 × 10−10 |
| Bovine Neurexin 1α LNS (21R6) | 2.506 | 0.65 |
| N-terminal Lg-domain Shbg (1LHN) | 8.619 | 1.3 |
| Thrombospondin (1Z78) | 7.482 | 1.5 |
Basic Alignment Search Tool (BLAST) Protein Database (PDB), root mean square deviation (RMSD). Smaller values indicate better match between template structure and generated model. Expected value (E value) refers to the number of hits expected when searching a database. Smaller value indicates a more accurate match.
Figure 2.
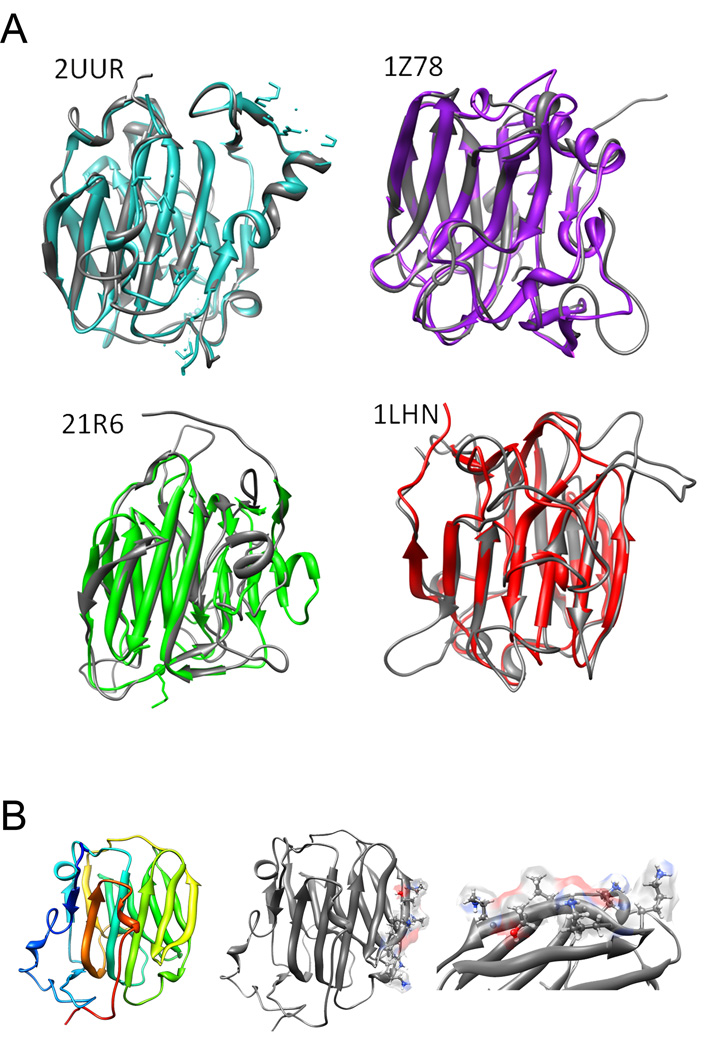
(A) Structural depictions of NTD of α1(XI) homology models (grey) aligned with respective templates [2UUR(blue), 21R6(green), 1LHN(red), and 1Z78(purple)]. (B) The α1(XI) NTD homology model from 2UUR (left), model with putative binding site (center) and zoom image of binding region (right).
While determining which components of the cartilage matrix associate with the NTD of collagen α1(XI) sets the stage for a number of interesting research questions, the premise that extracellular matrix components associate with the noncollagenous domain of collagens has been demonstrated for other collagens. There are a number of examples in the literature that illustrate FACIT collagens interacting with other extracellular matrix components. The NC4 region of the FACIT collagen α1(IX) is, like the NTD of collagen α1(XI), a globular domain that extends away from the surface of the fibril. Also, like collagen α1(XI), it is suggested that the NC4 domain serves as a potential dock between fibrils of the extracellular matrix [34]. Investigations of collagen α1(IX) demonstrate an association with collagen type II, its binding of matrilin-3 in a zinc-dependent manner, and binding of COMP in a manner inhibited by either calcium or zinc. In addition, both the NTD of collagen α1(XI)and the NC4 domain of collagen α1(IX) bind heparin [34–37]. The globular NTD domain of collagen α1(XI) adopts a thirteen anti-parallel beta sheet configuration similar to the NC4 domain of collagen α1(IX) and members of the LNS and thrombospondin family (Figure 2A). A glycosaminoglycan binding motif is present within beta strand 11 (Figure 2B). Interaction through this binding site may allow association with ECM constituents possessing glycosaminoglycan or those which would allow electrostatic interaction [20–22, 38]. Binding potential is not limited to the glycosaminoglycan binding site, and other as yet unidentified binding sites may exist on the NTD. Collagens XII and XIV are composed of three identical α1 chains, each with a large globular amino domain, NC3 which is homologous to NC4 of collagen α1(IX). The NC3 domains are thought to coordinate the interactions of tissues with banded collagen fibrils [37]. Collagen XII and the tenascin family of proteins, are both known to associate with collagen type I containing-fibrils as well as interacting with decorin [39–43]. More recently, it has been reported that tenascin-X interacts with the NC3 domain of collagen XII, demonstrating another physical link between the noncollagenous domains of collagens and other ECM constituents [43].
This study was carried out to identify molecular interactions between the collagen fibril and the other ECM components that are mediated by the NTD of collagen α1(XI) in cartilage. Candidates for interaction include collagenous proteins, proteoglycans and noncollagenous, nonproteoglycan matricellular protein components and peripherally associated membrane proteins [44]. Proteins extracted from cartilage under dissociating conditions were incubated with recombinant α1(XI) NTD under physiologically associative conditions. Associating constituents of the matrix were separated electrophoretically and identified by mass spectrometry. The globular NTD of collagen α1(XI), was found to associate with several components of the ECM including collagens type II, IX, XI, XII and XIV, the proteoglycans perlecan, fibromodulin, epiphycan, and biglycan, the thrombospondins 1 and 5 (cartilage oligomeric matrix protein), chondroadherin, and the matrilins 1 and 3, suggesting organizational, structural, and interactive roles of the NTD of α1(XI) in addition to its role in the restrictive growth of collagen fibrils by steric hindrance. Equally informative are those proteins found not to preferentially associate with the NTD of collagen α1(XI).
2. Materials and methods
2.1. Cloning, expression and purification of collagen α1(XI) NTD
The cDNA fragment encoding the NTD of type α1(XI) collagen was inserted into the expression vector pET11a (Stratagene), as previously described [32, 45, 46]. Briefly, recombinant collagen α1(XI) NTD was expressed in E. coli BL21 (DE3) cells (Novagen, Madison, WI), in a 100 L fermentator (Biostat 100D, Sartorius BBI, Bethlehem, PA) induced with 0.4mM IPTG at OD600 of 0.5–1.0. Cells were harvested with a continuous flow centrifuge (Sharples AS-14, Alfa Laval, Richmond, VA) five hours post-induction. Recombinant protein was extracted, isolated, purified and analyzed for primary and secondary structural composition as previously described [47].
2.2 Tissue preparation and extraction
Cartilage was obtained from the femoral head and condyles of an early third trimester (20–24 inches from crown to rump) fetal calf (Gem Meat, Boise, ID). Perichondrium and adhering tissues were removed. Approximately 4 grams of cartilage (wet weight) was harvested, minced and homogenized using a Polytron homogenizer (KINEMATICA Polytron, Brinkman Instruments, Westbury, NY). Cartilage proteins were extracted in buffered low salt solution (0.1 M NaCl) followed by buffered high salt solution (1 M NaCl), in 0.05 M Tris-HCl, pH 7.0, containing 0.01 M EDTA, and protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), pepstatinA, E-64, bestatin, leupeptin, and aprotinin. After an overnight extraction at 4°C with stirring, samples were centrifuged at 100,000 × g, at 4°C for 1 h using the TLA 110 rotor for the TL100 ultracentrifuge (Beckman Coulter, Brea, CA) to pellet insoluble material. The soluble proteins, extracted in the 1 M NaCl, 0.05 M Tris-HCl pH 7.5 were used for batch affinity chromatography after buffer exchange to adjust the NaCl concentration by ultrafiltration.
2.3 Affinity chromatography
Proteins extracted from cartilage were separated based on their ability to associate with the NTD of α1(XI) collagen. Briefly, 5 mg of recombinant collagen α1(XI) NTD, was immobilized on 1 mL Ni2+NTA Sepharose resin (Qiagen, Valencia, CA). Protein extracted from cartilage was applied to resin in batch at 4 °C for 16 h. Proteins that did not bind to the resin were collected as flow-through. After an initial wash equivalent to 4 × 10 resin volumes of 0.05 M Tris-HCl, pH 7.5, containing 0.14 M NaCl, bound proteins were eluted from the resin, by increasing NaCl concentration to 0.5 M. The eluate was evaluated under reducing conditions by SDS-PAGE stained with Bio-Safe Coomassie brilliant blue G250 dye (Bio-Rad, Hercules, CA). After evaluation, eluate was concentrated 10-fold using Centricon filters (Millipore, Billerica, MA) with a molecular mass cutoff of 10 kDa. Proteins were separated by SDS-PAGE for further analysis. As a control, extracted protein mix was applied to non-derivatized resin that was not carrying the recombinant α1(XI) NTD to assess nonspecific interaction.
2.4 Trypsin digestion and mass spectrometry
Prominent bands or regions of interest within the gel were manually excised, transferred to microcentrifuge tubes for two 30 minute destaining washes of 50% acetonitrile and 50mM ammonium bicarbonate, then dried in a vacuum concentrator (Eppendorf 5301). Digestion of proteins was performed by Trypsin Profile in-Gel Digestion (Sigma, Saint Louis, Missouri) with the addition of reduction and alkylation steps. Briefly, proteins were reduced with 10mM dithiothreitol/100mM ammonium bicarbonate for 45 minutes at 56°C and alkylated with 55mM iodoacetamide/100mM ammonium bicarbonate for 30 minutes in the dark. Following the tryptic digestion, proteins were dried and suspended in 5% formic acid to a final volume of 15µl.
Following acidification with formic acid, the released peptides were analyzed by mass spectrometry (LCQ Deca XP Plus, Thermo Fisher Scientific, Inc. Walthom, MA) with the Proteome X 1.0 operating system (Thermo Fisher Scientific, Inc. Walthom, MA). The digested peptides were loaded onto a BioBasic-18 reverse phase column (Thermo Hypersil-Keystone, Bellafonte, PA) and eluted using a linear gradient of 5–60% acetonitrile in 0.1% (v/v) HCOOH over 110 minutes followed by a two minute elution with 60–80% acetonitrile in 0.1% (v/v) HCOOH and injected into the mass spectrometer. The resulting mass spectral data were collected and analyzed for amino acid sequence identification.
2.5 Protein sequence database searches and analysis
The raw data (.dta) files obtained from the mass spectrometric analysis and search result files (.srf) with specified search parameters were transferred to the Boise State Beowulf Cluster and submitted for search utilizing Bioworks™ 3.1 software within the TurboSEQUEST operating system. Searches were performed against NCBI (http://www.ncbi.nlm.nih.gov) nonredundant database. To minimize the number of low-scoring peptides that can give rise to false positives, all the identified peptide data were subjected to the Sequest single threshold filter of Xcorr versus charge. In brief, only peptides determined to have greater than 1.5 Xcorr for a charge state of 1+; greater than 2.0 Xcorr for a charge state of 2+; or greater than 2.5 Xcorr for a charge state of 3+, were considered for identity of a protein. Further, protein identifications were reported only if they were reproducible and found to have four or more unique peptides unless otherwise noted. The following cartilage proteins were not analyzed in our study: asporin, lubrican, lumican, versican, brevican, thrombospondin 3, CILP, cartilage derived C-type lectin, fibronectin, PRELP, osteoaderin, fibrillin, elastin, glycoprotein 39/YKL-40, matrix Gla protein, pleiotropin, chondromodulin I and II, and CD-RAP. Growth factors and cytokines which we would anticipate to be present in cartilage in low abundance were not analyzed in this study with the exception of pro-inhibin, a precursor to a member of the TGF-β superfamily, also known as semenogelin I. Trypsin autolytic fragments were removed from the data set, as were peptides corresponding to keratins, hemoglobin and histone proteins. False discovery rates were determined using decoy searches. Amino acid sequence obtained from peptide fragmentation was used to manually confirm protein identification against Swiss-Prot/TrEMBL and Entrez Protein.
2.6 STRING Query of protein interaction network
Interactions identified experimentally were evaluated with the online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to predict both direct and indirect interactions [48]. STRING integrates protein-protein association from available sources that include gene fusion, co-occurrence, experimental, database, homology and text mining data [49–56]. The combined confidence score is indicative of the probability that a pair of proteins interact. A score of 0.150 represents low confidence; 0.400 represents medium confidence; 0.700 represents high confidence; and 0.900 and higher represents the highest confidence. For the purpose of our evaluation, only associations with a score of 0.400 or higher were reported. Scores were computed as the joint probability of the probabilities from the different evidences correcting for the probability of randomly observing an interaction [57].
2.7. Surface Plasmon Resonance
SPR measurements were performed at 20°C using a Reichert SR7000 instrument. The NTD of α1(XI) collagen was immobilized to a carboxyl mixed self-assembled monolayer surface on gold sensor chip by primary amine coupling. Briefly, the chip surface was activated with a 0.4 mM N-ethyl-N-(3-diethylaminopropyl) carbodiimide, 0.1 mM N-hydroxysuccinimide solution followed by the injection of NTD of α1(XI) collagen at a concentration of 20 µg/mL in 10 mM sodium acetate buffer (pH 4.5). Unreacted N-hydroxysuccinimide ester groups were blocked with 1 M ethanolamine hydrochloride. Thrombospondin 1 (R&D Systems) solution was prepared in running buffer (phosphate buffered saline with 0.05% Tween-20), which was filtered through a 0.2 µm filter prior to use. To collect equilibrium binding data, a concentration range from 15 to 500 nM was used as analyte and pumped over the NTD ligand at a flow rate of 10 µL/min. After 100 sec, the analyte solutions were replaced with running buffer for 200 sec. The surfaces were regenerated with a 100-sec injection of running buffer containing 1 M NaCl at a flow rate of 10 µL/min. The association, dissociation, and regeneration phases were followed in real-time by monitoring changes in signal expressed in resonance units and the data displayed as response units versus time. An average of the response at equilibrium was determined for each analyte concentration and the resulting equilibrium resonance units were plotted against concentration. Data were fit to a steady-state affinity model with GraphPad Prism (GraphPad Software) using a one-site association model.
2.8. Immunofluorescence
A polyclonal antibody to the rat α1(XI) collagen α1(XI) has been previously described [9]. A monoclonal antibody to TSP1 was obtained (Calbiochem, EMD Chemicals, USA). To allow the observation of colocalization within newly synthesized pericellular matrix, cells from the Swarm rat chondrosarcoma [45] were plated at 3.5 × 104 cells/cm2 onto poly-d-lysine/laminin-coated chamber slides (BD Biosciences) and incubated in DMEM supplemented with 10% fetal bovine serum, 50 µg/ml ascorbate 2-phosphate, 50 units/ml penicillin, and 50 µg/ml streptomycin. Accumulated pericellular matrix was analyzed 48 h after plating by indirect immunofluorescence. Cells were fixed on chamber slides in −20 °C methanol for 5 min. The slides were rinsed in 50 mM Tris-buffered saline (TBS) and then permeabilized in TBS-0.5% Triton X-100 (TX) for 10 min. The slides were washed three times for 5 min in TBS-0.1% TX, followed by blocking in TBS-0.1% TX, 2% bovine serum albumin for 10 min. After incubation with the primary antibody, cells were washed five times for 5 min in TBS-0.1% TX. Secondary antibody, rhodamine (TRITC)-conjugated AffiniPure donkey anti-rabbit IgG or fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), diluted 1:100 in TBST, was applied to slides and incubated for 1 h in a humidified chamber at 37 °C. The slides were then washed three times for 5 min in TBS-0.1% TX and incubated in 3.0 µg/ml of DAPI for 10 min (Molecular Probes, Eugene, OR). The slides were washed three times 5 min in TBS-0.1% TX, rinsed in TBS, drained, mounted with Vectashield (Vector Laboratories, Burlingame, CA), and sealed. For microscopic observation and photomicrography of the fluorescently labeled cells, an Olympus BX60 fluorescence microscope equipped with a PM-10AD system was used. The fluorescent molecules were excited with a 100-watt mercury lamp. TRITC- or fluorescein isothiocyanate labeled molecules were detected with a filter set having 510–560 nm wavelength bandpass, 565 nm dichroic beam splitter, and 575-645-nm emission filters.
3. Results
3.1 Expression, extraction and purification of α1(XI) NTD
Recombinant protein was expressed, extracted and purified as previously reported for recombinant collagen α1(XI) NTD [32,45, 46]. Purity was assessed by SDS–PAGE and shown to be a homogeneous, single band migrating with an apparent molecular weight of approximately 40 kDa (Figure 3A). The NTD of collagen α1(XI) is a globular domain rich in beta sheet that extends from the surface of collagen fibrils (Figures 1 and 2). It is similar in structure to the collagen type IX NC4 domain, the LNS domain of neurexin 1 alpha, the amino terminal domain of sex hormone binding globulin, and the heparin binding domain of thrombospondin 1. Combinatorial amino acid sequence with secondary structure alignment and structural depiction of α1(XI) NTD homology models aligned with these four potential templates showing low RMSD values (0.785 Å to 8.674 Å) and E values (5.0 × 10−10 to 1.5) are included in Tables 1 and 2 and Figure 2 [18–23, 58]. These data demonstrate sequence and structural homology between the COL9A1 NC4 and COL11A1 NTD.
Figure 3.
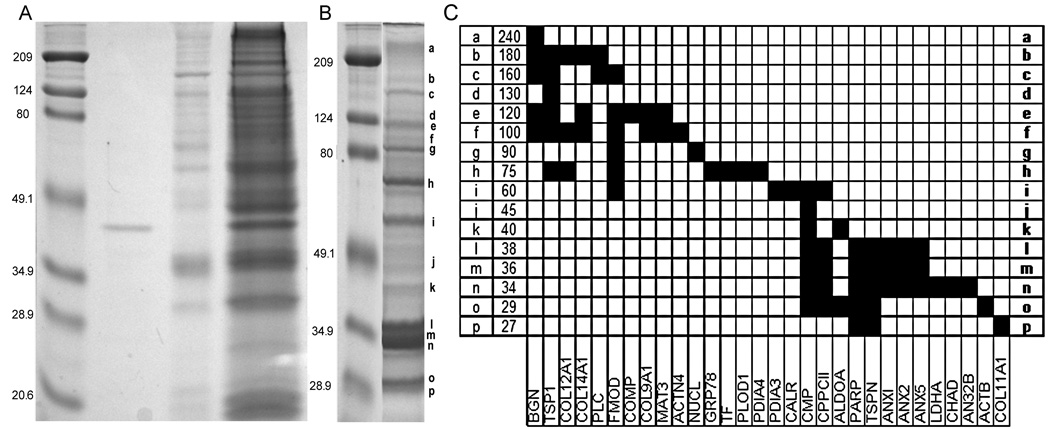
SDS-PAGE of recombinant α1(XI) NTD, cartilage extract, and affinity selected proteins. Proteins were separated by SDS-PAGE and stained with Coomassie Safe Blue. (A) Molecular weight markers (lane 1), purified recombinant α1(XI) NTD protein (lane 2), proteins bound to nonderivatized column resin indicating nonspecific binding (lane 3), and total protein extract (lane 4). Bands or regions of the gel from lane 4 were manually excised, trypsinized, and analyzed by mass spectrometry to determine identity. For each region of the gel, the following proteins were identified: In the 240/230Mr range: biglycan, collagen α1(XI), actin, aggrecan, perlecan, collagen α1(XII), decorin, collagen α1(I), epiphycan, and collagen α1(XIV); in the 210Mr range: collagen α1(XIV), collagen α1(XII), biglycan, and tenascin-C; in the 180Mr range:thrombospondin-1, biglycan, collagen α1(XIV), perlecan, biglycan, collagen α1(XII), nidogen2,tenascin-C, collagen α1(I), collagen α1(XII), and fibromodulin; in the 160Mr range: collagen α1(II), biglycan, thrombospondin-1, fibromodulin, and collagen α1(I); in the 140Mr range: biglycan and COMP; in the 130Mr range: COMP and thrombospondin-1; in the 120Mr range: COMP, collagen α1(IX), biglycan, collagen α1(XIV), fibromodulin, thrombospondin-1, and matrilin-3; in the 100Mr range: biglycan, actinin alpha-4,fibromodulin, collagen α1(IX), matrilin-3, collagen α1(XIV), thrombospondin-1, collagen α1(II), nucleolin, pyruvate kinase, ovalbumin, and elongation-factor 2; in the 75Mr range: fibromodulin, lysyl hydroxylase, biglycan, collagen α1(II), thrombospondin-1, protein disulfide isomerase A4, and heat shock protein (GRP78); in the 60Mr range: CMP, protein disulfide isomerase A4, heat shock protein (GRP78), transferrin, semenogelin/inhibin, protein disulfide isomerase A1, actin, pyruvate kinase M2, collagens, phosphoglycerate kinase, vitrin, calreticulin, chondrocalcin, protein disulfide isomerase A5, and PARP; in the 48Mr range: actin, phosphoglycerate kinase, collagen-binding protein, and vitrin; in the 45Mr range: collagen α1(XI) NTD, fructose bisphosphate aldolase, CMP, and cartilage link protein; in the 40Mr range: chondroadherin, chondrocalcin, CMP, fructose bisphosphate aldolase, cartilage link protein, actin, and collagen α1(XI) NTD; in the 38Mr range: annexin A1, annexin A2, annexin A5, chondrocalcin, chondroadherin, lactate dehydrogenase B, lactate dehydrogenase A, collagen α1(XI) NTD, PARP, CMP, thrombosponin-1, fructose bisphosphate aldolase, and actin; in the 35Mr range: chondrocalcin, chondroadherin, annexin A1, annexin A2, annexin A5, CMP, thrombospondin-1, PARP, actin, lactate dehydrogenase A, and collagen α1(XI) NTD; in the 32Mr range: chondrocalcin, chondroadherin, annexin A1, annexin A2, annexin A5, thrombospondin-1, lactate dehydrogenase A, collagen α1(XI) NTD, and ANP32B; in the 28Mr range: collagen α1(I), PARP, chondrocalcin, CMP, thrombospondin-1, and collagen α1(XI) NTD, in the 25Mr range: PARP and CMP; in the 22Mr range: keratin; in the 20Mr range: lectin, histone 2A, histone 2B, collagen α1(XI) NTD; and in the 18Mr range: hemoglobin.
(B). Cartilage proteins that interact with the NTD of collagen α1(XI). SDS-PAGE stained with Coomassie Safe Blue. Molecular weight markers (lane 1), proteins selected by affinity chromatography (lane 2). Bands or regions of the gel indicated by letters a through p on the right-hand side of gel were manually excised, trypsinized, and analyzed by mass spectrometry. Proteins identified in the specific locations a through p were as follows: a) (240Mr): biglycan; b) (180Mr) collagen α1(XIV), thrombospondin-1, perlecan, biglycan, and collagen α1(XII); c) (160Mr)collagen α1(XII), biglycan, thrombospondin-1, and fibromodulin; d) (130Mr) thrombospondin-1; e) (120Mr) COMP, collagen α1(IX), biglycan, collagen α1(XIV), fibromodulin, thrombospondin-1, and matrilin-3; f) (100Mr) biglycan, actinin alpha-4, fibromodulin, collagen α1(IX), PARP, matrilin-3, collagen α1(XIV),thrombospondin-1, and collagen α1(XII) ; g) (90Mr) nucleolin and fibromodulin; h) (75Mr) fibromodulin, lysyl hydroxylase 1, biglycan, collagen α1(XII), thrombospondin-1, protein disulfide A4, transferrin, and heat shock protein 70; i) (60Mr) CMP, protein disulfide A3, fibromodulin, chondrocalcin, PARP, and calreticulin; j) (45Mr) CMP; k) (40Mr) CMP and 1,6 fructose bisphosphate aldolase; l) (38Mr) chondrocalcin, annexin A2, annexin A1, annexin A5, CMP, PARP, thrombospondin-1, 1,6 fructose bisphosphate and actin; m) (36Mr) chondrocalcin, annexin A5, annexin A1, annexin A2, CMP. thrombospondin -1, PARP, and actin; n) (34Mr) chondrocalcin, acidic leucine-rich nuclear phosphoprotein 32 family member B, annexin A1, annexin A2, annexin A5, collagen α1(XI) NTD, chondroadherin, lactate dehydrogenase A, thrombospondin-1, PARP, and CMP; o) (29Mr) PARP, chondrocalcin, CMP, thrombospondin-1, 1,6- fructose bisphosphate aldolase, and actin; p) (27Mr): PARP, thrombospondin-1, and collagen α1(XI) NTD.
(C) Grid depicting proteins identified as a function of location on gel. Apparent molecular weight is indicated on the vertical axis and protein identity is indicated along the horizontal axis.
3.2 Cartilage protein extraction and protein identification
Cartilage tissue was harvested from the articular joints and growth plates of a fetal bovine and homogenized in 0.05 M Tris-HCl, pH 7.5, containing 1 M NaCl and protease inhibitors. Proteins from the homogenate were separated by SDS-PAGE and stained with Coomassie brilliant blue in order to define the set of cartilage proteins that were readily identifiable under these experimental conditions (Figure 3). Distinct bands or regions of the gel (lane 4 of Figure 3A) were excised and proteins contained within the gel were digested with trypsin and analyzed by mass spectrometry. Cartilage proteins identified with confidence from the total extract are listed in Table 3. These proteins represent prevalent proteins of the cartilage extracellular matrix and a subset of cellular proteins [59–68].
Table 3.
Proteins identified from total extracted proteins.
| PROTEIN NAMEa | Accession Numberb |
|---|---|
| Actin, beta | GENE ID: 280979 |
| Actinin, alpha 4 | GENE ID: 522269 |
| Aggrecan | GENE ID: 280985 |
| Aldolase A, fructose-bisphosphate | GENE ID:509566 |
| Annexin A1 | GENE ID: 327662 |
| Annexin A2 | GENE ID:282689 |
| Annexin A5 | GENE ID:281626 |
| Acidic (leucine-rich) nuclear phosphoprotein 32 family, member B | GENE ID:509685 |
| Biglycan | GENE ID:280733 |
| Calreticulin | GENE ID:281036 |
| Chondroadherin | GENE ID: 281069 |
| Collagen, type I, alpha 1 | GENE ID:282187 |
| Chondrocalcin-Carboxy propeptide of collagen type II alpha 1 | GENE ID:407142 |
| Collagen, type II, alpha 1 | GENE ID:407142 |
| Collagen, type VI, alpha 1 | GENE ID:511422 |
| Collagen, type IX, alpha 1 | GENE ID:282195 |
| Collagen, type XI, alpha 1 | GENE ID:287013 |
| Collagen, type XI, alpha 2 (PARP) | GENE ID:515435 |
| Collagen, type XII, alpha 1 | GENE ID:359712 |
| Collagen, type XIV, alpha 1 | GENE ID:7373* |
| Cartilage oligomeric matrix protein | GENE ID:281088 |
| Decorin | GENE ID:280760 |
| Epiphycan | GENE ID:281747 |
| Fibromodulin | GENE ID:281168 |
| Hyaluronan and proteoglycan link protein 1 | GENE ID: 281717 |
| Heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | GENE ID:415113 |
| Heparan sulfate proteoglycan 2 | GENE ID:444872 |
| Lactate dehydrogenase A | GENE ID:281274 |
| Matrilin 1, cartilage matrix protein | GENE ID:512059 |
| Matrilin 3 | GENE ID:540041 |
| Nidogen 2 (osteonidogen) | GENE ID:521854 |
| Nucleolin | GENE ID:497013 |
| Phosphoglycerate kinase 1 | GENE ID:507476 |
| Procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | GENE ID:281409 |
| Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | GENE ID:533642 |
| Protein disulfide isomerase family A, member 3 | GENE ID:281803 |
| Protein disulfide isomerase family A, member 4 | GENE ID:415110 |
| Protein disulfide isomerase family A, member 5 | GENE ID:511603 |
| Prolyl 4-hydroxylase | GENE ID: 281373 |
| Pyruvate kinase, muscle | GENE ID:512571 |
| Semenogelin I | GENE ID:281254 |
| Serpin peptidase inhibitor, clade H (collagen binding protein 1) | GENE ID:510850 |
| Tenascin | GENE ID:540664 |
| Serotransferrin | GENE ID:280705 |
| Thrombospondin 1 | GENE ID:281530 |
| Thrombospondin 1, Amino terminal domain | GENE ID:281530 |
| Tumor rejection antigen (gp96 or HSP90B1) | GENE ID:282646 |
| Vitrin | GENE ID:280957 |
National Center for Biotechnology Information (NCBI)
Entrez Gene Full Name Protein Accession number;
GENE ID Accession Number. Organism: Bos taurus,
By homology to human sequence; Homo sapiens
3.3 Cartilage proteins selected by association with α1(XI) NTD
Extracted proteins were applied to the α1(XI) NTD-affinity resin under conditions of moderate ionic strength as described in Materials and Methods. Under these conditions, we anticipated that protein interactions would take place. Proteins interacting with the α1(XI) NTD resin were retained while non-interacting proteins did not bind. Proteins were eluted by increasing the ionic strength of the buffer and the protein was concentrated prior to SDS-PAGE analysis. A unique banding pattern was observed in the eluate compared to total extract (compare lane 4 of Figure 3A to lane 2 of Figure 3B). For comparison, extracted proteins were applied to the affinity resin without the α1(XI) NTD derivatization (lane 3 of Figure 3A).
3.4 Identification of proteins associated with α1(XI) NTD
Bands of interest were excised, digested with trypsin and analyzed by mass spectrometry. Data were collected and subjected to a search against the complete non-redundant database acquired from the National Center for Biotechnology Information (NCBI). After the imposition of filters and sorting, fifteen extracellular matrix proteins were found to associate with the NTD of collagen α1(XI) either directly or indirectly (Figure 3C and Table 4). Collagens found to associate included the fibrillar collagens type II and type XI, and FACIT collagens types XIV, XII, and IX. Proteoglycan association was observed with the small leucine-rich repeat proteoglycans, biglycan, epiphycan, fibromodulin, and heparan sulfate proteoglycan 2/perlecan. Several noncollagenous nonproteoglycan components were identified: chondroadherin, thrombospondin-5 (COMP), thrombospondin-1, matrilin-1(CMP), and matrilin-3. Interestingly, the chondrocalcin, which is the carboxyl propeptide of collagen α1(II) and the thrombospondin-1 N-terminal domain which is proteolytically removed from the parent molecule, were both identified as interacting with the NTD of collagen α1(XI) (Table 4).
Table 4.
Identification of proteins from fetal bovine articular cartilage that associate with collagen α1(XI) NTD by affinity chromatography-mass spectrometry.
| Extracted proteins found to associate with the Collagen XI (α1) NTD | |||||
|---|---|---|---|---|---|
| Protein namea | Common Abbreviationb |
Mr Pred.c (kDa) |
Mr Experimentald (kDa) |
Peptides Matchese |
Seq.cov.(%)f |
| Extracellular Matrix Proteoglycans | |||||
| Biglycan | BGN | 41.6 | 240,140,120,100,75 | 14 | 40 |
| Fibromodulin | FMOD | 42.9 | 140,130,120,100,75 | 7 | 15 |
| Chondroadherin | CHAD | 40.8 | 35 | 12 | 37 |
| Heparan sulfate proteoglycan 2 | HSPG2 | 46.8 | 240,180 | 12 | 4 |
| Epiphycan | EPYC | 36.8 | 240,230 | 4 | 11 |
| Extracellular Matrix Structural Proteins | |||||
| Cartilage Oligomeric Matrix Protein | COMP | 139.3 | 120 | 18 | 60 |
| Thrombospondin-1 | THBS1 | 129.5 | 120 | 24 | 34 |
| Thrombospondin-Amino terminal like domain | THBS1N | 25 | 38,35,32,28,25 | 7 | 43 |
| Matrilin-1(Cartilage Matrix Protein) | MATN1 | 54 | 60, 55,38,35, 25 | 19 | 51 |
| Matrilin-3 | MATN3 | 52.8 | 120 | 4 | 12 |
| Collagen alpha 1 (II) | COL2A1 | 134.4 | 140 | 10 | 8 |
| Chondrocalcin | CPPCII | 27.4 | 75, 38,35,32,30, 25 | 9 | 40 |
| Collagen alpha 1 (IX) | COL9A1 | 92.9 | 130,120,100 | 9 | 13 |
| Collagen alpha 1 (XI) | COL11A1 | 61.7 | 32,25 | 16 | 33 |
| Collagen alpha 2(XI) amino propeptide | PARP | 20.9 | 38,32,30, 28,25, 20 | 16 | 86 |
| Collagen alpha 1(XII) | COL12A1 | 315 | 180,160,100,75 | 34 | 16 |
| Collagen alpha 1 (XIV) | COL14A1 | 202.4 | 120 | 13 | 10 |
Full name of reference sequence;
Common protein abbreviation;
Predicted Mr according to NCBI;
Experimental Mr calculated by analysis of gel images;
Number of peptide matches corresponding to the reference sequence;
Sequence coverage calculated as the number of amino acids identified of the total number of amino acids in reference sequence.
Additional proteins, shown in Table 5, were identified including several of the enzymes known to play a role in collagen biosynthesis or cartilage remodeling; procollagen lysine 2-oxoglutarate, 5-dioxygenase 1 and 2 (PLOD1 and PLOD2), protein disulfide isomerases (PDIs), and procollagen prolyl 4-hydroxylase (P4HB). PLOD and P4HB enzymes have hydroxylase activity required to hydroxylate lysine and proline residues of the procollagen, respectively and PDI activity is necessary and responsible for the formation of disulfide bonds critical to the nascent conformation of the collagen; post-translational modifications necessary in order for collagens to correctly form into a stable triple helix. PLOD1, PDIA3 and PDIA4 were found to associate with the collagen α1(XI) NTD, perhaps reflecting their role during intracellular collagen biosynthesis.
Table 5.
Identification of proteins from cell surface or cellular location with affinity for α1(XI)NTD by affinity chromatography-mass spectrometry.
| Protein namea | Common Abbreviationb |
Mr Pred.c (kDa) |
Mr Experimentald (kDa) |
Peptide Matchese | Seq.cov.(%)f |
|---|---|---|---|---|---|
| Cell Surface Location | |||||
| Annexin I | ANXA1 | 29.1 | 35, 32 | 9 | 39 |
| Annexin II | ANXA2 | 49.4 | 38,35,32 | 14 | 40 |
| Annexin V | ANXA5 | 36.1 | 35 | 16 | 62 |
| Calreticulin (GRP60) | CALR | 48 | 55 | 6 | 18 |
| Nucleolin | NUCL | 77.4 | 100 | 10 | 13 |
| Protein Fate and Modification | |||||
| Procollagen -lysine, 2-oxoglutarate 5-dioxygenase 1 | PLOD1 | 83.5 | 75 | 8 | 16 |
| Protein disulfide isomerase A3 | PDIA3 | 56.7 | 60,55 | 17 | 39 |
| Protein Disulfide Isomerase A4 | PDIA4 | 72.5 | 75 | 8 | 18 |
| Heat shock 70kDa protein 5(GRP78) | HSPA5 | 72.3 | 75 | 8 | 18 |
| Tumor rejection antigen (gp96 or HSP90B1) | HSP90B1 | 92.4 | 100 | 6 | 9 |
| Metabolic, cytoskeletal, and nuclear proteins | |||||
| Fructose Bisphophate Aldolase | ALDOA | 39.5 | 45,38 | 10 | 30 |
| Serotransferrin | TF | 77.8 | 75 | 8 | 12 |
| Lactate dehydrogenase A | LDHA | 36.6 | 32, 35 | 9 | 31 |
| Actin, beta | ACTB | 42 | 29 | 13 | 45 |
| Alpha-actinin 4 | ACTN4 | 104.8 | 100 | 9 | 15 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B | ANP32B | 29.9 | 32 | 5 | 15 |
Full name of reference sequence;
Common protein abbreviation;
Predicted Mr according to NCBI;
Experimental Mr calculated by analysis of gel images;
Number of peptide matches corresponding to the reference sequence;
Sequence coverage calculated as the number of amino acids identified of the total number of amino acids in reference sequence.
Other proteins identified have a number of different roles; calcium binding, protein fate, metabolic enzymes and cytoskeletal related proteins (Table 5). Specific proteins include the annexins, endoplasmin, calreticulin, heat shock proteins, serotransferrin, actin, actinin-α4, fructose bisphophatealdolase, and lactate dehydrogenase A.
Extracellular matrix proteins found not to interact with collagen α1(XI) NTD include aggrecan, decorin, tenascin, vitrin, nidogen 2, and collagen α2(VI).
3.5 Protein Network of Interactions with α1(XI) NTD
STRING was used to predict interactions and results were compared with those identified by our affinity chromatography-mass spectrometry approach. Our results support the predicted physical interactions by STRING with collagen type II, chondroadherin, thrombospondin 1, matrilin 1, matrilin 3, and thrombospondin 5/COMP as shown in Figure 4,where confidence is shown by the number of lines connecting the protein nodes of the network to each other and the color in the grid shown in Figure 4B. Direct interaction may take place between ANXA5 and Col11a1 NTD. Alternatively, ANXA5-Col11a1 interaction may also be mediated by Col2a1 as predicted by STRING (Figure 4). Direct interactions mediated by the triple helical domain take place with the other collagens of the heterotypic collagen fibril including Col9a1, Col9a2, Col9a3, and Col11a2.
Figure 4.
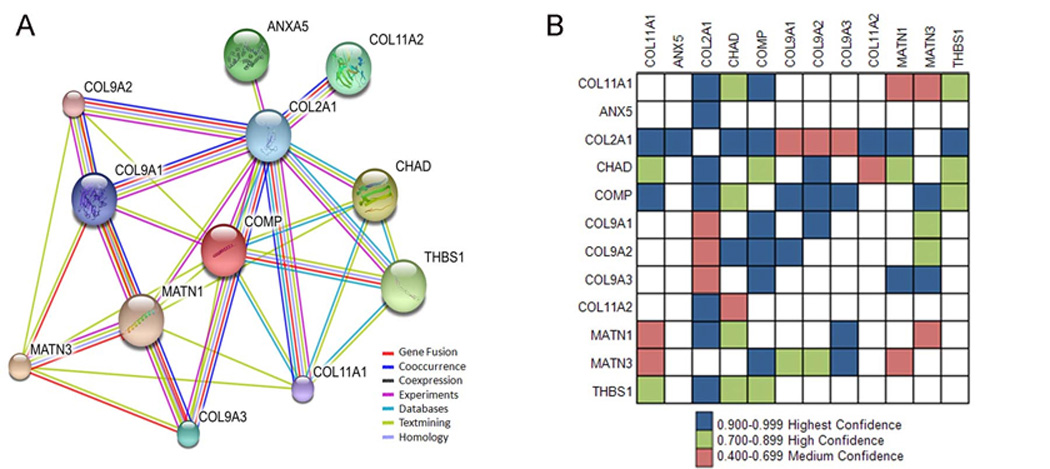
Extracellular matrix network of predicted interactions. A subset of the identified proteins are shown as a network of interacting proteins based upon this study and evidence from previously published work. (A). Col2a1, CHAD, THBS1, MATN1, MATN3, and COMP are shown within the STRING network seeded by Col11a1, where confidence is indicated by the number of lines connecting the protein nodes of the network to each other and the color in the grid shown in (B). The color of the line is indicative of the type of data supporting the predicted interaction. Purple indicates that experimental or biochemical evidence for an interaction exists in the literature, blue indicates co-occurrence evidence, purple indicates experimental evidence, yellow indicates evidence from text-mining that detected co-mentioned in PubMed abstract, light blue indicates the association in curated database, grey indicates predicted interaction based on sequence/structural homology.
Direct and indirect interactions predicted by STRING are depicted in Figure 5, in which the degree of confidence for each of the predicted interactions is indicated by thickness of the line connecting the nodes, representing proteins. Interactions with other extracellular matrix proteins were observed as well as interactions with cell surface proteins including CALR, NCL, and ANXA5, which may either be direct interactions or alternatively indirect, mediated by another ECM protein, perhaps thrombospondin 1, as indicated by the results of STRING prediction (Figure 5).
Figure 5.
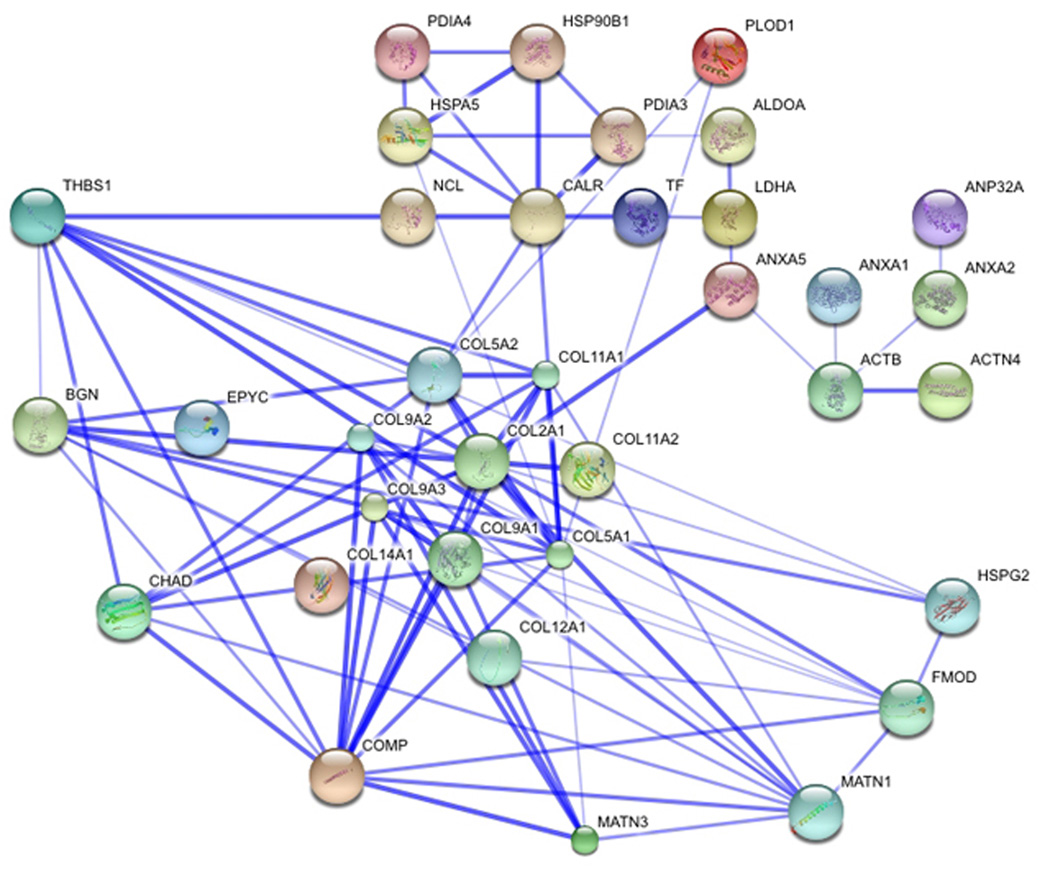
Extended network of predicted interactions. Interactions that include extracellular matrix, membrane and cellular proteins are shown to include predicted primary and secondary interactions. Each protein detected in the affinity experiment was analyzed using STRING molecular interactions. Thickness of connecting lines indicates confidence level associated with each interaction.
3.6. Confirmation of molecular interaction by SPR and co-localization by Immuno-fluorescence
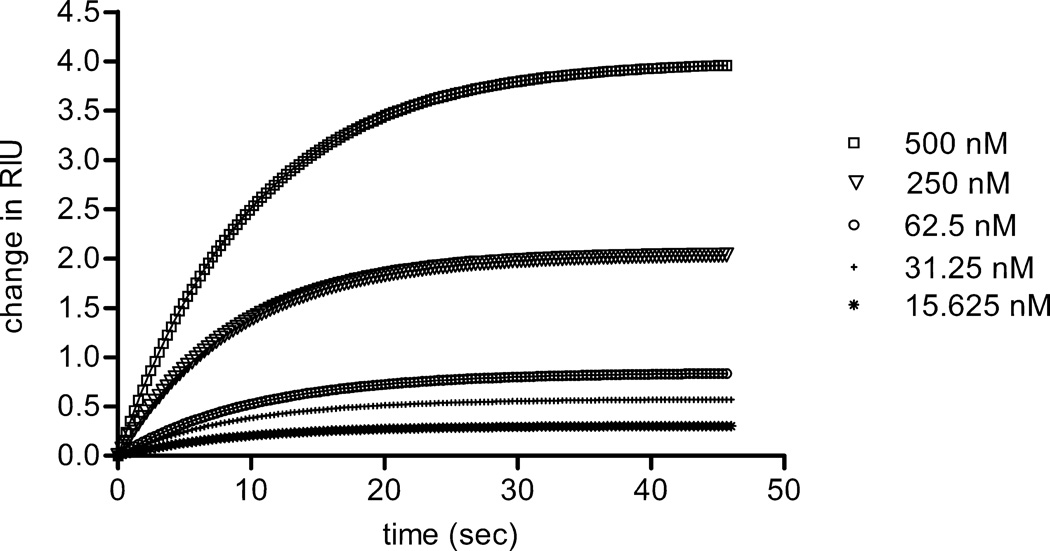
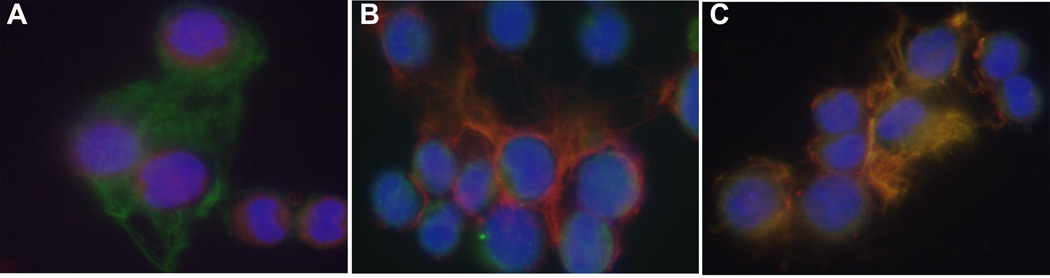
To confirm interactions for one of the proteins detected using our affinity chromatography and mass spectrometry approach, we carried out binding analysis using surface plasmon resonance (SPR) (Figure 6). Thrombospondin 1 was found to interact with the NTD of α1(XI) with a dissociation constant of 100 nM. Colocalization within newly synthesized ECM of rat chondrosarcoma cells was demonstrated by indirect immunofluorescence (Figure 7).
Figure 6.
Interaction between thrombospondin 1 and the NTD of α1(XI) collagen was analyzed by surface plasmon resonance. Collagen α1(XI) NTD was covalently coupled to the sensor chip, while thrombospondin 1 at concentrations ranging from 15 to 500 nM was allowed to bind to the Collagen α1(XI) NTD. The average of 3 runs to collect association data is shown. Using the data shown to fit to a steady state affinity model, the dissociation constant (Kd) was calculated to be 100nM for native thrombospondin 1, assuming a one-site association model.
Figure 7.
Colocalization of thrombospondin 1 and collagen type XI alpha 1 chain by immunofluorescence. A polyclonal antibody to the rat α1(XI) collagen α1(XI) and a monoclonal antibody to thrombospondin-1 were used to detect these ECM molecules within newly synthesized pericellular matrix from a cell line derived from the Swarm rat chondrosarcoma. Cells were plated at 3.5 × 104 cells/cm2 onto poly-d-lysine/laminin-coated chamber slides and accumulated pericellular matrix was analyzed after 48 hours. Primary antibodies were detected with secondary antibody, rhodamine (TRITC)-conjugated AffiniPure donkey anti-rabbit IgG or fluorescein isothiocyanate-conjugated anti-mouse IgG and counterstained with DAPI. (A) Thrombospondin-1 antibody (green), (B) Collagen α1(XI) antibody (red), (C) Thrombospondin-1 antibody (green) and Collagen α1(XI) (red) overlap resulting in yellow where colocalization occurs.
4. Discussion
The NTD of collagen α1(XI) is located on the surface of collagen fibrils and may therefore interact with other components of the ECM. This study utilized affinity chromatography and mass spectrometry to identify proteins that can interact with the NTD of α1(XI) collagen. The triple helical domain of the α1(XI) collagen is embedded in the interior of the fibril and the NTD of collagen α1(XI) is exposed on the surface. Based on these observations, we propose that the NTD of collagen α1(XI) may interact with components of the cartilage matrix to carry out the function of regulation of fibrillogenesis in concert with other ECM molecules. Additional information regarding the interactions between the proteoglycan, collagen, and noncollagenous components may explain the loss of regulated fibrillogenesis, lack of cellular organization within the growth plate, and tissue integrity observed in the absence of collagen α1(XI) in Fibrochondrogenesis, Stickler and Marshall syndrome, and in the cho/cho mouse.
Biglycan, the most prevalent of the proteoglycans detected in the proteins associating with collagen α1(XI) NTD, was identified within the relative molecular weight range from 70kDa to greater than 250kDa. This self-associating protein, consisting of a 38kDa core protein and two attached glycosaminoglycan chains (dermatan sulfate in articular cartilage) of 22kDa each, has been found to exist in both monomer-dimer equilibrium and in an associated state as a hexamer [69, 70]. Biglycan is known to provide mechanical stability through its interaction with collagen and other matrix components and for its role in cell activity regulation through binding to growth factors [71]. Matrilin-1, an ECM structural protein found to associate with α1(XI) NTD in this study, is a homotrimeric protein found specifically in the matrix surrounding chondrocytes [72–74]. Demonstrated to associate with collagen fibrils and interact with aggrecan, it has been further shown to play a role in normal matrix organization and fibril formation [75–77]. At the exposed surface of the collagen II fibril, the association of the NTD of collagen α1(XI) with biglycan and matrilin-1 could be the means by which the complex adheres, providing a connection between the pericellular environment and the ECM, and participating in the mechanism that drives normal ECM assembly.
Chondroadherin (CHAD), a member of the leucine-rich repeat family is most thoroughly described as a constituent of cartilage and tendon, and is also abundant in the territorial matrix surrounding chondrocytes [78]. Both collagen type II fibrils and CHAD bind chondrocytes by interacting with integrin α2β1. Additionally, CHAD binds to two sites on collagen type II and may be a mediator of complex formation of ECM molecules between the cell surface and the surface of the collagen fibril [79]. The presence of the NTD of collagen α1(XI) on the surface of the collagen type II fibrils may be the means by which CHAD interacts with collagen type II/IX/XI fibrils, and through this interaction may play a role in regulating fibrillogenesis.
The thrombospondin family is made up of large multimeric proteins known to be involved in interactions between cells and between the cells and matrix constituents. It has been previously shown that thrombospondin 1 interacts by direct binding with collagen type V [80, 81]. Collagens α1(V) and α1(XI) are homologous, sharing structural and functional characteristics. It is probable that the role that thrombospondin 1 plays in association with the NTD of collagen α1(XI) parallels that found in collagen α1(V) [82, 83]. While the properdin domain of thrombospondin 1 has been reported to bind the triple helix of collagen, a different domain of thrombospondin 1 may interact with the NTD α1(XI) as shown in this study.
Another member of the thrombospondin family, thrombospondin-5/COMP, capable of binding to the surface of chondrocytes through the interaction with an integrin receptor [84] is a pentameric protein made up of five identical subunits that also bind collagens. Shown to interact with collagen type IX, it has been proposed that COMP functions as a bridge between the fibrils and the ECM [85–87]. COMP was found in our investigation to associate with the NTD of collagen α1(XI) and may function as a stabilizing component in the mechanism by which the cell interacts with the ECM. It has also been shown that the expression of COMP determines whether both matrilin-3 and collagen IX are secreted and reflects the important roles that COMP, collagen IX and matrilin-3 play in the assembly of the ECM [88]. The NTD of collagen α1(XI) associated with all three components in this study and may contribute to the organization necessary for proper matrix assembly.
Annexin family members bind acidic phospholipids in a calcium dependent manner; due to their calcium-binding carboxyl termini, and specific functions within the annexin family are conferred to individual annexins by their amino termini [89]. Annexin 2, 5, and 6 are commonly associated with matrix vesicles and are critical in the initiation of the mineralization process in cartilage [90]. Annexin 5, which binds directly to collagen type II, collagen type X and the cleaved C-propeptide of collagen type II, (chondrocalcin), is suggested to provide an anchor between the matrix vesicle and its surrounding matrix and activate annexin 5 ion channel properties [91, 92]. In the cartilage matrix, chondrocalcin, which is also a calcium binding protein, is localized where mineralization occurs [93]. The demonstrated relationship that annexin 5, collagen type II and chondrocalcin have in linking matrix vesicles with the ECM during the initiation of mineralization leads us to suggest that the NTD of collagen α1(XI), in association with these components, may play a role in mineralization. This idea is further supported by the finding that collagen α1(XI) is localized adjacent to the location of perichondrial bone formation in a developing long bone [94] and our recently published finding that antisense morpholino oligonucleotides targeting the Col11a1 mRNA accelerates mineralization of osteoblast cultures [95]. Further, isolation of bone mineralization foci have identified collagen α1(XI) as a protein component of the newly mineralized bone ECM (unpublished data) and the cho/cho mouse which does not synthesize collagen α1(XI) has an increase in bone mineralization density (unpublished data).
Nucleolin was initially characterized as a multifunctional nuclear protein capable of shuttling between the nucleus and the cytoplasm [96,97]. Although not previously identified as part of the cartilage proteome, it has been demonstrated that collagen increased the localization of nucleolin to the nucleolus in fibroblasts [98]. More recently, nucleolin has been reported as a receptor located at the cell surface whose activity, associated with the actin cytoskeleton, has been linked to the internalization of components by endocytosis, and subsequent transport to the nucleus [99–103]. Further, the autolytic activity of nucleolin produces cleavage products that post-transcriptionally control the expression of at least one matrix metalloproteinase, MMP-9, known to be involved in the normal and pathophysiological degradation of the ECM. Nucleolin is a complex and multifaceted protein of special interest because expression is not only regulated by influence of the ECM components, but it may also be able to relay developmentally relevant signals between the ECM of cartilage and the nucleus of the chondrocytes.
While our study is focused on those proteins that interact, it is equally instructive to note those proteins which did not interact under conditions used in our experiment. The following proteins were identified by mass spectrometry in our study as not associating strongly with the NTD of α1(XI) collagen: collagen type I α1 chain (5 unique peptides), aggrecan (6 unique peptides), cartilage link protein (6 unique peptides), tenascin C (15 unique peptides), pro-inhibin/semenogelin-I (12 unique peptides), serpin 1/collagen binding protein (8 unique peptides), PDIA1 (16 unique peptides), PLOD2 (5 unique peptides), pyruvate kinase isozymeM2 (18 unique peptides), and phosphoglycerate kinase (10 unique peptides). Collagen type I, aggrecan, cartilage link protein, tenascin C, serpin 1, and pro-inhibin are located in the ECM while PDIA1 and PLOD2 are cellular proteins involved in collagen biosynthesis.
Concluding remarks
The implications of understanding the manner in which the components of cartilage develop and interact are far-reaching. As the majority of bone formation begins from a cartilaginous template, successful skeletal development is dependent on its structural and molecular integrity. A number of the identified proteins found to associate with the NTD of collagen α1(XI) in our study are found themselves to be candidates for the cause of a number of osteochondrodysplasias (Table 6). Continuing this work by our laboratory and others [104, 105] will provide additional information about the composition of cartilage ECM and how the ECM changes during chondrogenic differentiation. It may be that specific interactions between the NTD of collagen α1(XI) and these identified proteins, either directly or indirectly, contribute to skeletal development and understanding their association will perhaps further explain the molecular events that underlying the range of pathological skeletal phenotypes. Further investigation of the identified proteins will allow us to determine with which proteins the NTD of collagen α1(XI) directly interacts and what region(s) of the NTD they adhere to specifically. Examining the associations of these identified proteins in an isoform specific manner may lead to understanding how collagen α1(XI) aids in development of specific tissues. As the majority of the type α1(XI) collagen is embedded in the interior of the fibril with the exception of the NTD at the surface, the variable region may also interact with the matrix constituents and may do so differentially. The variable region specific to each isoform may serve as tissue specific binding domain that would allow for the possibility that collagen α1(XI) functions in regulating the formation of fibrils in a developmentally and tissue specific manner. In addition to studies of congenital skeletal defects and the onset of arthritis, results from this study may also impact the design of novel biomaterials and aid in therapeutic approaches to favor cartilage repair and regeneration.
Table 6.
Osteochondrodysplasias linked with proteins found to associate with the collagen α1(XI)NTD
| PROTEIN | SKELETAL DYSPLASIA | OMIM*a |
|---|---|---|
| COLIA1 | Ostogenesis imperfecta / Ehlers-Danlos Syndrome | 120150 |
| COL2A1 | Achondrogenesis, Hypochondrogenesis, Stickler syndrome, Kniest syndrome, Spondyloepiphyseal dysplasia | 120140 |
| COL5A1 | Ehlers-Danlos Syndrome | 120215 |
| COL9A1 | Multiple epiphyseal dysplasia, Stickler syndrome | 120210 |
| COL9A2 | Multiple epiphyseal dysplasia, Intervertebral Disc Disease | 120260 |
| COL9A3 | Multiple epiphyseal dysplasia, Intervertebral Disc Disease | 120270 |
| COL11A1 | Fibrochondrogenesis, Stickler's syndrome, Marshall's syndrome | 120280 |
| COL11A2 | Otospondylomegaepiphyseal, Non-ocular Stickler syndrome | 120290 |
| COMP | Multiple epiphyseal dysplasia, Pseudoachondroplasia | 600310 |
| MATN3 | Multiple epiphyseal dysplasia, Hand ostoarthritis | 602109 |
| HSPG2 | Dyssegmental dysplasia | 142461 |
| PLOD1 | Ehlers-Danlos Syndrome | 153454 |
| CMP | Rhematoid arthritis, Polychondritis | 115437 |
| CRTL1 | Dwarfism, Craniofacial abnormalities | 115435 |
| FMOD | Ehlers-Danlos Syndrome | 600245 |
Online Mendelian Inheritance in Man Reference number
Acknowledgments
Authors acknowledge the work of Katey Marie Anderson, Christina Blasick, Noriko Hazeki-Taylor and Linda Mercer. This work was supported in part by the Arthritis Foundation, the NIH/NIAMS Grants RO1AR47985 and KO2AR48672, NIH/NCRR Grant P20RR16454, the National Science Foundation (0619793, 0923535), M. J. Murdock Foundation, Idaho State Board of Education Higher Education Research Council, Lori and Duane Stueckle, St. Luke’s Regional Medical Center, Mountain States Tumor and Medical Research Institute, and Research Corporation Cottrell College Scholars Program.
Abbreviations
- ECM
Extracellular matrix
- NTD
Amino terminal domain
- FACIT
Fibril associated collagens with interrupted triple helices
- NC3
Noncollagenous domain 3
- NC4
Noncollagenous domain 4
- Thrombospondin-5, COMP
Cartilage oligomeric matrix protein
- MMP9
Matrix metalloproteinase 9
- ANP32B
Acidic nuclear phosphoprotein 32 family member B
- ACTB
Actin, beta
- ACTN4
Alpha actinin 4
- ANXA1, ANX1
Annexin I
- ANXA2, ANX2
Annexin II
- ANXA5, ANX5
Annexin V
- BGN
Biglycan
- GRP60, CALR
Calreticulin
- Matrilin-1, MATN1, CMP
Cartilage matrix protein
- CHAD
Chondroadherin
- Chondrocalcin, CPPCII
Carboxyl propeptide of type II collagen
- COL1A1
Collagen type I alpha 1 chain
- COL2A1
Collagen type II alpha 1 chain
- COL6A2
Collagen type VI alpha 2 chain
- COL9A1
Collagen type IX alpha 1 chain
- COL11A1
Collagen type XI alpha 1 chain
- COL11A2
Collagen type XI alpha 2
- COL12A1
Collagen type XII alpha 1 chain
- COL14A1
Collagen type XIV alpha 1 chain
- DCN
Decorin
- EPYC
Epiphycan
- FMOD
Fibromodulin
- ALDOA
Fructose bisphosphatealdolase
- HSPA5, GRP78
Glucose-regulated protein 78 (Heat shock protein 5 70 kDa
- HAPLN1
Hyaluronan and proteoglycan link protein 1
- HSP90B1
Heat shock protein 90B1
- HSPG2. PLC
Heparan sulfate proteoglycan 2/perlecan
- LDA
Lactate dehydrogenase A
- MATN3
Matrilin 3
- NCL, NUCL
Nucleolin
- PLOD1
Procollagen-lysine 1,2-oxoglutarate 5-dioxygenase 1
- PLOD2
Procollagen-lysine 2-oxoglutarate 5-dioxygenase 2
- P4HB
Prolyl 4 hydroxylase
- PARP
Proline arginine rich protein
- PDIA3
Protein disulfide isomerase family A, member 3
- PDIA4
Protein disulfide isomerase family A, member 4
- PDIA5
Protein disulfide isomerase family A, member 5
- TSP1, THBS1
Thrombospondin 1
- THBS1N, TSPN
Thrombospondin 1 amino terminal domain
- TF
Serotransferrin
- EDTA
ethylenediaminetetraacetic acid
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
Footnotes
Authors have no declared financial or commercial conflicts of interest.
References
- 1.Pogue R, Sebald E, King L, Kronstadt E, et al. A transcriptional profile of human fetal cartilage. Matrix Biology. 2004;23:299–307. doi: 10.1016/j.matbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Poole CA. In: Joint Cartilage Degradation; Basic and Clinical Aspects. Woessner JF, Howell DS, editors. New York: Marcell Dekker; 1993. [Google Scholar]
- 3.Wight TN, Heinegard DK, Hascall VC. In: Cell Biology of the Extracellular Matrix. Hay ED, editor. New York: Plenum; 1991. pp. 45–78. [Google Scholar]
- 4.Hardingham TE, Venn G. Chondrotin sulphate/dermatan sulfate proteoglycans in cartilage: aggrecan, decorin and biglycan. In: Scott JE, editor. Dermatan Sulfate Proteoglycans: Chemistry, Biology, Chemical Pathology. London, England: Portans Press; 1993. pp. 207–217. [Google Scholar]
- 5.Burgeson RE, Hollister DW. Collagen heterogeneity in human cartilage: identification of several new collagen chains. Biochem. Biophys. Res. Com. 1979;87:1124–1131. doi: 10.1016/s0006-291x(79)80024-8. [DOI] [PubMed] [Google Scholar]
- 6.Richards AJ, Yates JRW, Williams R, Payne SJ, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum. Mol. Genet. 1996;5:1339–1343. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- 7.Griffith AJ, Sprunger LK, Sirko-Osadsa DA, Tiller GE, et al. Marshall syndrome associated with a splicing defect at the COL11A1 locus. Am J Human Gen. 1998;62:816–823. doi: 10.1086/301789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tompson SW, Bacino CA, Safina NP, Bober MB, et al. Fibrochondrogenesis results from mutations in the COL11A1 Type XI collagen gene. Am J Human Gen. 2010;87:708–712. doi: 10.1016/j.ajhg.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Lacerda DA, Warman ML, Beier DR, et al. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell. 1995;80:423–430. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- 10.Greenspan DS, Cheng W, Hoffman GG. The pro-alpha 1(V) collagen chain. Complete primary structure, distribution of expression, and comparison with the pro-alpha 1(XI) collagen chain. J. Biol. Chem. 1991;266:24727–24733. [PubMed] [Google Scholar]
- 11.Nah H-D, Barembaum M, Upholt WB. The chicken alpha 1 (XI) collagen gene is widely expressed in embryonic tissues. J Biol Chem. 1992;267:22581–22586. [PubMed] [Google Scholar]
- 12.Bernard M, Yoshioka H, Rodriguez E, van der Rest M, et al. Cloning and sequencing of pro-alpha 1 (XI) collagen cDNA demonstrates that type XI belongs to the fibrillar class of collagens and reveals that the expression of the gene is not restricted to cartilaginous tissue. J. Biol. Chem. 1988;263:17159–17166. [PubMed] [Google Scholar]
- 13.Yoshioka H, Ramirez F. Pro-alpha 1(XI) collagen. Structure of the amino-terminal propeptide and expression of the gene in tumor cell lines. J Biol Chem. 1990;265:6423–6426. [PubMed] [Google Scholar]
- 14.Kimura T, Cheah KS, Chan SD, Lui VC, et al. The human alpha 2(XI) collagen (COL11A2) chain. Molecular cloning of cDNA and genomic DNA reveals characteristics of a fibrillar collagen with differences in genomic organization. J Biol Chem. 1989;264:13910–13916. [PubMed] [Google Scholar]
- 15.Zhidkova NI, Brewton RG, Mayne R. Molecular cloning of PARP (proline/arginine-rich protein) from human cartilage and subsequent demonstration that PARP is a fragment of the NH2-terminal domain of the collagen alpha 2(XI) chain. FEBS Lett. 1993;326:25–28. doi: 10.1016/0014-5793(93)81753-m. [DOI] [PubMed] [Google Scholar]
- 16.Takahara K, Sato Y, Okazawa K, Okamoto N, et al. Complete primary structure of human collagen alpha 1 (V) chain. J. Biol. Chem. 1991;266:13124–13129. [PubMed] [Google Scholar]
- 17.Imamura Y, Scott IC, Greenspan DS. The pro-alpha 3(V) collagen chain. Complete primary structure, expression domains in adult and developing tissues, and comparison to the structures and expression domains of the other types V and XI procollagen chains. J Biol Chem. 2000;275:8749–8759. doi: 10.1074/jbc.275.12.8749. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 20.Tan K, Duquette M, Liu JH, Shanmugasundaram K, Joachimiak A, Gallagher JT, Rigby AC, Wang JH, Lawler J. Heparin-induced cis- and trans-dimerization modes of the thrombospondin-1 N-terminal domain. Structure. 2006;14:33–42. doi: 10.1074/jbc.M705203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leppanen V-M, Tossavainen H, Permi P, Lehtio L, Ronnholm G, Goldman A, Kilpelainen I, Pihlajamaa T. Crystal structure of the N-terminal NC4 domain of collagen IX, a zinc binding member of the laminin-neurexin-sex hormone binding globulin (LNS) domain family. J. Biol. Chem. 2007;282:23219–23230. doi: 10.1074/jbc.M702514200. [DOI] [PubMed] [Google Scholar]
- 22.Shen KC, Kuczynska DA, Wu IJ, Murray BH, Sheckler LR, Rudenko G. Regulation of neurexin1β tertiary structure and ligand binding through alternative splicing. Structure. 2008;16:422–431. doi: 10.1016/j.str.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grishkovskaya I, Avvakumov GV, Hammond GL, Catalano MG, Muller YA. Steroid ligands bind human sex hormone-binding globulin in specific orientations and produce distinct changes in protein conformation. J. Biol. Chem. 2002;277:32086–32093. doi: 10.1074/jbc.M203999200. [DOI] [PubMed] [Google Scholar]
- 24.van der Rest M, Garrone R. Collagen family of proteins. FASEB J. 1991;5:2814–2823. [PubMed] [Google Scholar]
- 25.Shaw LM, Olsen BR. FACIT collagens: diverse molecular bridges in extracellular matrices. Trends Biochem Sci. 1991;16:191–194. doi: 10.1016/0968-0004(91)90074-6. [DOI] [PubMed] [Google Scholar]
- 26.Wälchli C, Trueb J, Kessler B, Winterhalter KH, Trueb B. Complete primary structure of chicken collagen XIV. Eur. J. Biochem. 1993;212:483–490. doi: 10.1111/j.1432-1033.1993.tb17685.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn K. In: Structure and Function of Collagen Types. Mayne R, Burgeson R, editors. New York: Academic Press; 1987. p. 2. [Google Scholar]
- 28.Keene DR, Oxford JT, Morris NP. Ultrastructural localization of collagen types II, IX, and XI in the growth plate of human rib and fetal bovine epiphyseal cartilage: type XI collagen is restricted to thin fibrils. J HistochemCytochem. 1995;43:967–979. doi: 10.1177/43.10.7560887. [DOI] [PubMed] [Google Scholar]
- 29.Holmes DF, Kadler KE. The 10+4 microfibril structure of thin cartilage fibrils. Proc. Natl. Acad. Sci. USA. 2006;103:17249–17254. doi: 10.1073/pnas.0608417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wargelius A, Fjelldal PG, Nordgarden U, Grini A, et al. Collagen type XII may be involved in the structural plasticity of the vertebral column in Atlantic salmon (Salmosala L.) J Exp. Biol. 2010;213:1207–1216. doi: 10.1242/jeb.040022. [DOI] [PubMed] [Google Scholar]
- 32.Gregory KE, Oxford JT, Chen Y, Gambee JE, et al. Structural organization of distinct domains within the non-collagenous N-terminal region of collagen type XI. J. Biol. Chem. 2000;275:11498–11506. doi: 10.1074/jbc.275.15.11498. [DOI] [PubMed] [Google Scholar]
- 33.Blaschke UK, Eikenberry EF, Hulmes DJS, Galla H-J, Bruckner P. Collagen XI nucleates self-assembly and limits lateral growth of cartilage fibrils. J. Biol. Chem. 2000;275:10370–10378. doi: 10.1074/jbc.275.14.10370. [DOI] [PubMed] [Google Scholar]
- 34.Vasios G, Nishimura I, Konomi H, van der Rest M, et al. Cartilage type IX collagen-proteoglycan contains a large amino-terminal globular domain encoded by multiple exons. J. Biol. Chem. 1988;263:2324–2329. [PubMed] [Google Scholar]
- 35.Briggs MD, Chapman KL. Pseudoachondroplasia and multiple epiphyseal dysplasia: mutation review, molecular interactions, and genotype to phenotype correlations. Hum. Mutat. 2002;19:465–478. doi: 10.1002/humu.10066. [DOI] [PubMed] [Google Scholar]
- 36.Holden P, Meadows RS, Chapman KL, Grant ME, et al. Cartilage oligomeric matrix protein interacts with type IX collagen, and disruptions to these interactions identify a pathogenetic mechanism in a bone dysplasia family. J. Biol. Chem. 2001;276:6046–6055. doi: 10.1074/jbc.M009507200. [DOI] [PubMed] [Google Scholar]
- 37.Pihlajamaa T, Lankinen H, Ylöstalo J, Valmu L, et al. Characterization of recombinant amino-terminal NC4 domain of human collagen IX: interaction with glycosaminoglycans and cartilage oligomeric matrix protein. J. Biol. Chem. 2004;279:24265–24273. doi: 10.1074/jbc.M402865200. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Nishiyama T, McDonough AM, Bruns RR, Burgeson RE. Type XII and XIV collagens mediate interactions between banded collagen fibers in vitro and may modulate extracellular matrix deformability. J. Biol.Chem. 1994;269:28193–28199. [PubMed] [Google Scholar]
- 40.Koch M, Bohrmann B, Matthison M, Haglos C, et al. Large and small splice variants of collagen XII: differential expression and ligand binding. J. Cell Biol. 1995;130:1005–1014. doi: 10.1083/jcb.130.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keene DR, Lunstrum GP, Morris NP, Stoddard DW, Burgeson RE. Two type XII-like collagens localize to the surface of banded collagen fibrils. J. Cell Biol. 1991;113:971–978. doi: 10.1083/jcb.113.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lethias C, Elefteriou F, Parsiegla G, Exposito JY, Garrone R. Identification and characterization of a conformational heparin-binding site involving two fibronectin type III modules of bovine tenascin-X. J. Biol.Chem. 2001;276:16432–16438. doi: 10.1074/jbc.M010210200. [DOI] [PubMed] [Google Scholar]
- 43.Elefteriou F, Exposito JY, Garrone R, Lethias C. Binding of tenascin-X to decorin. FEBS Lett. 2001;495:44–47. doi: 10.1016/s0014-5793(01)02361-4. [DOI] [PubMed] [Google Scholar]
- 44.Roughley PJ. Articular cartilage and changes in arthritis: Noncollagenous proteins and proteoglycans in the extracellular matrix of cartilage. Arthritis Res. 2001;3:342–347. doi: 10.1186/ar326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warner LR, Brown RJ, Yingst SMC, Oxford JT. Isoform-specific heparan sulfate binding within the amino-terminal noncollagenous domain of collagen α1(XI) J Biol Chem. 2006;281:39507–39516. doi: 10.1074/jbc.M608551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medeck RJ, Sosa S, Morris N, Oxford JT. BMP-1-mediated proteolytic processing of alternatively spliced isoforms of collagen type XI. Biochem. J. 2003;376:361–368. doi: 10.1042/BJ20030894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner LR, Blasick CM, Brown RJ, Oxford JT. Expression, purification, and refolding of recombinant collagen alpha 1(XI) amino terminal domain splice variants. PEP. 2007;52:403–409. doi: 10.1016/j.pep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarcyk D, Franceschini A, Kuhn M, Simonovic M, et al. The STRING database in 2011: functional interaction network of proteins globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bader GD, Betel D, Hogue CW. BIND: The Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xenarios I, Rice DW, Salwinski L, Baron MK, et al. DIP: The database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. http://dip.doe-mbi.ucla.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breitkreutz BJ, Stark C, Tyers M. The GRID: the General Repository for Interaction Datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. http://thebiogrid.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keshava-Prasad TS, Goel R, Kandasamy K, Keerthikumar S, et al. Human Protein Reference Database - 2009 Update. Nucleic Acids Research. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. http://www.hprd.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, et al. IntAct—an open source molecular interaction database. Nucleic Acids Research. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, et al. MINT: A Molecular INTeraction database. FEBS Letters. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. http://160.80.34.4/mint/ [DOI] [PubMed] [Google Scholar]
- 55.Schaefer CF, Anthony K, Krupa S, Buchoff J, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. http://www.genome.ad.jp/kegg/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Mering C, Jensen LJ, Snel B, Hooper SD, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The PyMOL Molecular Graphics System, Version 1.3. Schrödinger, LLC: [Google Scholar]
- 59.Neame PJ, Tapp H, Azizan A. Noncollagenous, nonproteoglycan macromolecules of cartilage. Cell. Mol. Life Sci. 1999;55:1327–1340. doi: 10.1007/s000180050373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aumailley M, Gayraud B. Structure and biological activity of the extracellular matrix. J. Mol. Med. 1998;76:253–265. doi: 10.1007/s001090050215. [DOI] [PubMed] [Google Scholar]
- 61.Heinegård D. Proteoglycans and more – from molecules to biology. Int. J. Exp. Path. 2009;90:575–586. doi: 10.1111/j.1365-2613.2009.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vincourt J-B, Lionneton F, Kratassiouk G, Guillemin F, et al. Establishment of a reliable method for direct proteome characterization of human articular cartilage. Molecular & Cellular Proteomics. 2006;5:1984–1995. doi: 10.1074/mcp.T600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Krakow D, Sebald ET, Pogue R, Rimoin LP, et al. Analysis of clones from a human cartilage cDNA library provides insight into chondrocyte gene expression and identifies novel candidate genes for the osteochondrodysplasias. Molecular Genetics and Metabolism. 2003;79:34–42. doi: 10.1016/s1096-7192(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 64.DeCeuninck F, Marcheteau E, Berger S, Caliez A, et al. Assessment of some tools for the characterization of the human osteoarthritic cartilage proteome. J. Biomol. Tech. 2005;16:256–265. [PMC free article] [PubMed] [Google Scholar]
- 65.Hocking AM, Shinomura T, McQuillan DJ. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biology. 1998;17:1–19. doi: 10.1016/s0945-053x(98)90121-4. [DOI] [PubMed] [Google Scholar]
- 66.Mayne R. Cartilage collagens. What is their function, and are they involved in articular disease? Arthritis and Rheumatism. 1989;32:241–246. doi: 10.1002/anr.1780320302. [DOI] [PubMed] [Google Scholar]
- 67.Goldring MB. The role of the chondrocyte in osteoarthritis. Arthritis & Rheumatism. 2000;43:1916–1926. doi: 10.1002/1529-0131(200009)43:9<1916::AID-ANR2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz-Romero C, Lopez-Armanda MJ, Blanco FJ. Proteomic characterization of human normal articular chondrocytes: a novel tool for the study of osteoarthritis and other rheumatic diseases. Proteomics. 2005;5:3048–3059. doi: 10.1002/pmic.200402106. [DOI] [PubMed] [Google Scholar]
- 69.Fisher LW, Termine JD, Young MF. Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J. Biol. Chem. 1989;264:4571–4576. [PubMed] [Google Scholar]
- 70.Liu J, Laue TM, Choi H, Tang L-H, Rosenberg L. The self-association of biglycan from bovine articular cartilage. J. Biol. Chem. 1994;269:28366–28373. [PubMed] [Google Scholar]
- 71.Wiberg C, Heinegård D, Wenglén C, Timpl R, Mörgelin M. Biglycan organizes collagen VI into hexagonal-like networks resembling tissue structures. J. Biol. Chem. 2002;277:49120–49126. doi: 10.1074/jbc.M206891200. [DOI] [PubMed] [Google Scholar]
- 72.Hauser N, Paulsson M. Native cartilage matrix protein (CMP) : A compact trimer of subunits assembled via a coiled-coil alpha-helix. J. Biol Chem. 1994;269:25747–25753. [PubMed] [Google Scholar]
- 73.Chen FH, Thomas AO, Hecht JT, Goldring MB, Lawler J. Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J. Biol Chem. 2005;280:32655–32661. doi: 10.1074/jbc.M504778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Q, Johnson DM, Haudenschild DR, Goetinck PF. Cartilage matrix protein: expression patterns in chicken, mouse, and human. Ann N Y Acad. Sci. 1996;785:238–240. doi: 10.1111/j.1749-6632.1996.tb56271.x. [DOI] [PubMed] [Google Scholar]
- 75.Winterbottom N, Kimata K, Line S, Strong D, et al. Cartilage matrix protein is a component of the collagen fibril of cartilage. Dev. Dyn. 1992;193:266–276. doi: 10.1002/aja.1001930307. [DOI] [PubMed] [Google Scholar]
- 76.Hauser N, Paulsson M, Heinegard D, Mörgelin M. Interaction of cartilage matrix protein with aggrecan. Increased covalent cross-linking with tissue maturation. J. Biol Chem. 1996;271:32247–32252. doi: 10.1074/jbc.271.50.32247. [DOI] [PubMed] [Google Scholar]
- 77.Huang X, Birk DE, Goetinck PF. Mice lacking matrilin-1 (cartilage matrix protein) have alterations in type II collagen fibrillogenesis and fibril organization. Develpmental Dynamics. 1999;216:434–441. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<434::AID-DVDY11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 78.Shen Z, Gantcheva S, Månsson B, Heinegård D, Sommarin Y. Chondroadherin expression changes in skeletal development. Biochem. J. 1998;330:549–557. doi: 10.1042/bj3300549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Månsson B, Wenglén C, Mörglen M, Saxne T, Heinegård D. Association of chondroadherin with collagen type II. J. Biol.Chem. 2001;276:32883–32888. doi: 10.1074/jbc.M101680200. [DOI] [PubMed] [Google Scholar]
- 80.Mumby SM, Raugi GJ, Bornstein P. Interactions of thrombospondin with extracellular matrix proteins: selective binding to type V collagen. J. Cell Biol. 1984;98:646–652. doi: 10.1083/jcb.98.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galvin NJ, Vance PM, Dixit VM, Fink B, Frazier WA. Interaction of human thrombospondin with types I-V collagen: direct binding and electron microscopy. J. Cell Biol. 1987;104:1413–1422. doi: 10.1083/jcb.104.5.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takagi J, Fujisawa T, Usui T, Aoyama T, Saito Y. A single chain 19-kDa fragment from bovine thrombospondin binds to type V collagen and heparin. J. Biol. Chem. 1993;268:15544–15549. [PubMed] [Google Scholar]
- 83.Adam JC, Lawler J. The thrombospondins. Int. J. Biochem. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Q, Johnson DM, Haudenschild DR, Tondravi MM, Goetinck PF. Cartilage matrix protein forms a type II collagen-independent filamentous network: analysis in primary cell cultures with a retrovirus expression system. Dev. Biol. 1995;172:293–306. doi: 10.1091/mbc.6.12.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mörgelin M, Heinegard D, Engel J, Paulsson M. Electron microscopy of native cartilage oligomeric matrix protein purified from the Swarm rat chondrosarcoma reveals a five-armed structure. J. Biol. Chem. 1992;276:6137–6141. [PubMed] [Google Scholar]
- 86.DiCesare PE, Carlson CS, Stollerman ES, Chen ES, et al. Expression of cartilage oligomeric matrix protein by human synovium. FEBS Lett. 1997;412:249–252. doi: 10.1016/s0014-5793(97)00789-8. [DOI] [PubMed] [Google Scholar]
- 87.Thur J, Rosenberg K, Nitsche DP, Pihlajamaa T, et al. Mutations in cartilage oligomeric matrix protein causing pseudoachondroplasia and multiple epiphyseal dysplasia affect binding of calcium and collagen I, II, and IX. J Biol Chem. 2001;276:6083–6092. doi: 10.1074/jbc.M009512200. [DOI] [PubMed] [Google Scholar]
- 88.Hecht JT, Hayes E, Haynes R, Cole WG. COMP mutations, chondrocyte function and cartilage matrix. Matrix Biology. 2005;23:525–533. doi: 10.1016/j.matbio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 89.Crompton MR, Moss SE, Crumpton MJ. Diversity in the lipocortin/calpactin family. Cell. 1988;55:1–3. doi: 10.1016/0092-8674(88)90002-5. [DOI] [PubMed] [Google Scholar]
- 90.Kirsch T, Nah H-D, Shapiro IM, Pacifici M. Regulated production of mineralization-competent matrix vesicles in hypertrophic chondrocytes. J. Cell Biol. 1997;137:1149–1160. doi: 10.1083/jcb.137.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.von der Mark K, Mollenhauer J. Annexin V interactions with collagen. J. Cell. Mol. Life Sci. 1997;53:539–545. doi: 10.1007/s000180050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kirsch T, Harrison G, Golub EE, Nah H-D. The roles of annexins and types II and X collagen in matrix vesicle-mediated mineralization of growth plate cartilage. J Biol. Chem. 2000;45:35577–35583. doi: 10.1074/jbc.M005648200. [DOI] [PubMed] [Google Scholar]
- 93.van der Rest M, Rosenberg LC, Olsen BR, Poole AR. Chondrocalcin is identical with the C-propeptide of type II procollagen. Biochem. J. 1986;237:923–925. doi: 10.1042/bj2370923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morris NP, Oxford JT, Davies GB, Smoody BF, Keene DR. Developmentally regulated alternative splicing of the alpha 1(XI) collagen chain: spatial and temporal segregation of isoforms in the cartilage of fetal rat long bones. J Histochem Cytochem. 2000;48:725–741. doi: 10.1177/002215540004800601. [DOI] [PubMed] [Google Scholar]
- 95.Kahler R, Yingst S, Krawczak D, Oxford J, Westendorf J. Collagen 11a1 is indirectly activated by Lymphocyte Enhancer-binding factor 1 (Lef1) and negatively regulates osteoblast maturation. Matrix Biology. 2008;27:330–338. doi: 10.1016/j.matbio.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jordan G. At the heart of the nucleolus. Nature. 1987;329:489–490. doi: 10.1038/329489a0. [DOI] [PubMed] [Google Scholar]
- 97.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 98.Gillery P, Georges N, Randoux A, Lefèvre F, et al. Modulation of protein synthesis by extracellular matrix: potential involvement of two nucleolar proteins, nucleolin and fibrillarin. Biochem. Biophys. Res. Comm. 1996;228:94–99. doi: 10.1006/bbrc.1996.1621. [DOI] [PubMed] [Google Scholar]
- 99.Hovenessian AG, Puvion-Dutilleul F, Nisole S, Svab J, et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp. Cell Res. 2000;261:312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- 100.Said E, Krust B, Nisole S, Svab J, et al. The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J. Biol. Chem. 2002;277:37492–37502. doi: 10.1074/jbc.M201194200. [DOI] [PubMed] [Google Scholar]
- 101.Sinclair JF, O'Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesion intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J. Biol. Chem. 2002;277:2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- 102.Turck N, Lefebvre O, Gross I, Gendry P, et al. Effect of laminin-1 on intestinal cell differentiation involves inhibition of nuclear nucleolin. J. Cell Physiol. 2006;206:545–555. doi: 10.1002/jcp.20501. [DOI] [PubMed] [Google Scholar]
- 103.Huang Y, Shi H, Zhou H, Song X, et al. Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood. 2006;107:3564–3571. doi: 10.1182/blood-2007-01-064428. [DOI] [PubMed] [Google Scholar]
- 104.Wilson R, Diseberg AF, Gordon L, Zivkovic S, et al. Comprehensive profiling of cartilage extracellular matrix formation and maturation using sequential extraction and label-free quantitative proteomics. Molecular and Cellular Proteomics. 2010;9:1296–1313. doi: 10.1074/mcp.M000014-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ji Y-H, Sun F-Y, Zeng Y-Y, He X-H, et al. Quantitative proteomics analysis of chondrogenic differentiation of C3H10T1/2 mesenchymal stem cells by iTRAQ labeling coupled with on-line two-dimensional LC/MS/MS. Molecular and Cellular Proteomics. 2010;9:550–564. doi: 10.1074/mcp.M900243-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]