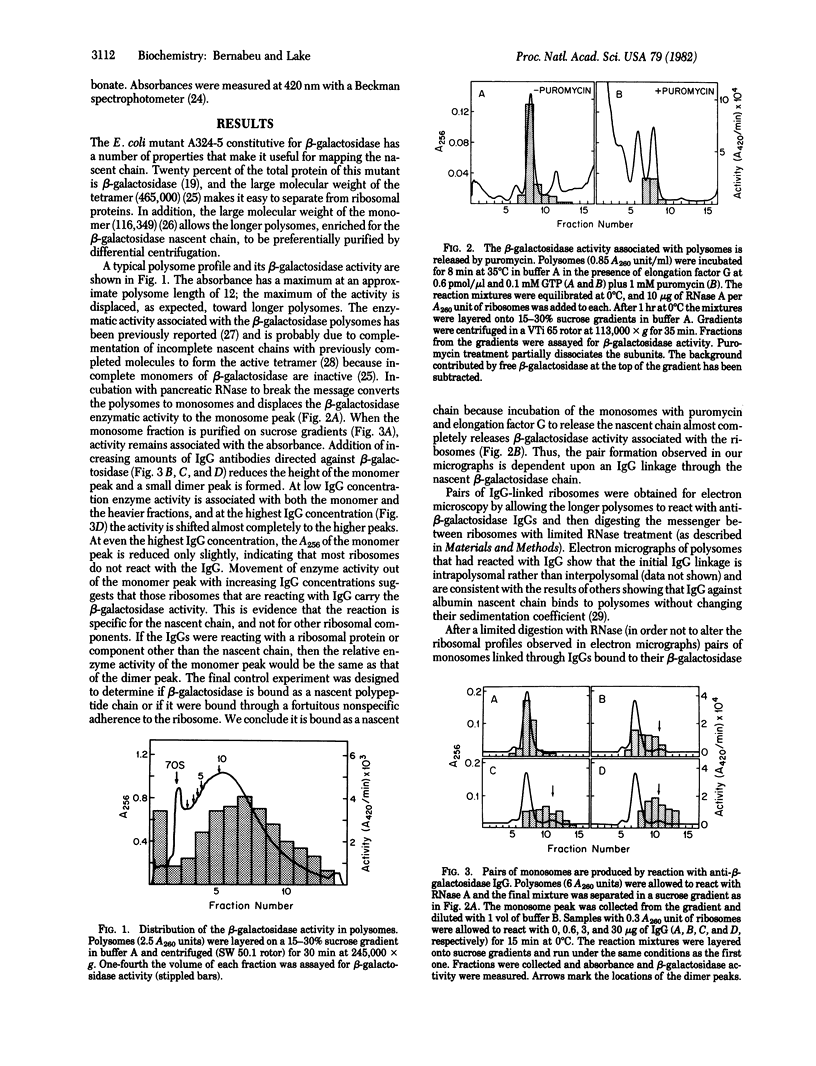
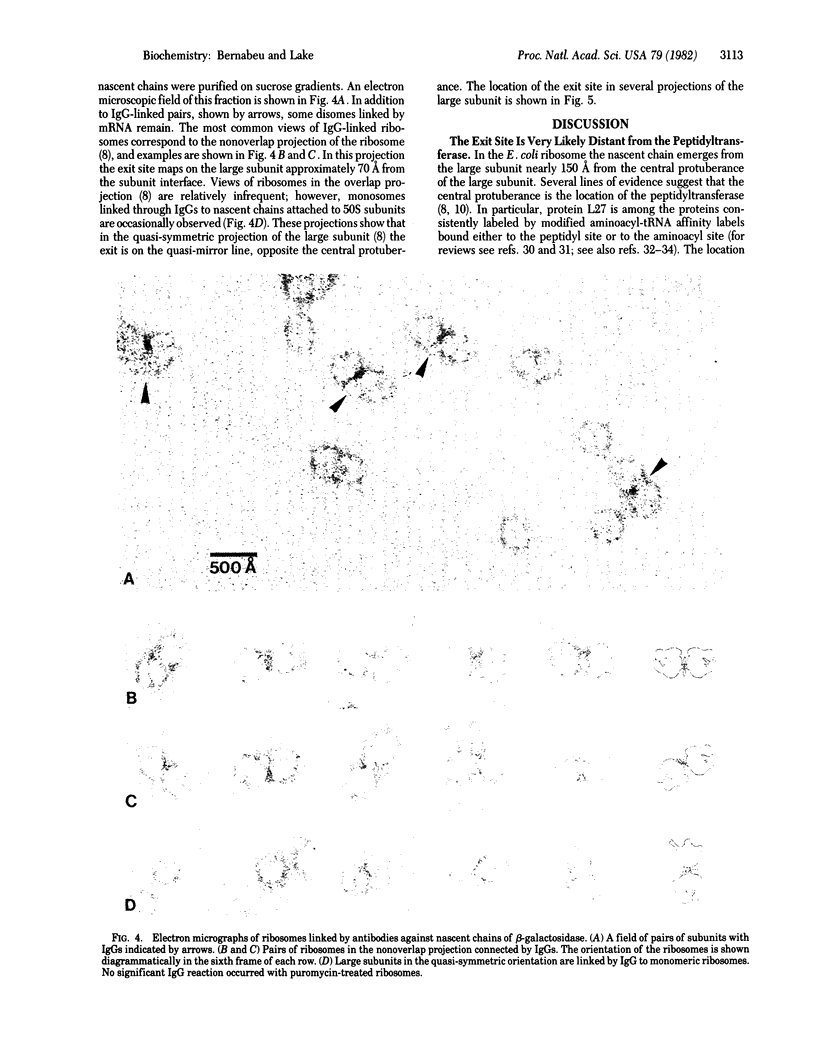
Abstract
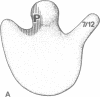
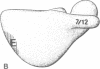
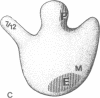
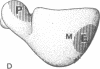
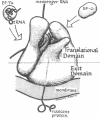
The site of the nascent polypeptide chain as it leaves the ribosome has been localized on the "exit domain" of the Escherichia coli ribosome by using IgG antibodies directed against the enzyme beta-galactosidase (EC 3.2.1.23). Thus, a functional site has been mapped on intact 70S ribosomes. The exit site is on the large subunit, approximately 70 A from the interface between subunits and nearly 150 A from the central protuberance, the likely site of peptide transfer. It is adjacent to the region corresponding to the rough endoplasmic membrane binding region of the eukaryotic ribosome but distant from ribosomal components participating in mRNA recognition and polypeptide elongation (i.e., distant from the "translational domain"). These results, together with the protease protection experiments of others, provide evidence that the nascent protein chain probably passes through the ribosome in an unfolded, fully extended conformation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman M. R., Sabatini D. D., Blobel G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973 Jan;56(1):206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta J. P.G., Montejo V., Vazquez D. Reconstitution of the 50 S ribosome subunit. Localization of activities related to the peptidyl transferase centre. FEBS Lett. 1971 Nov 15;19(1):75–78. doi: 10.1016/0014-5793(71)80609-9. [DOI] [PubMed] [Google Scholar]
- Blobel G., Sabatini D. D. Controlled proteolysis of nascent polypeptides in rat liver cell fractions. I. Location of the polypeptides within ribosomes. J Cell Biol. 1970 Apr;45(1):130–145. doi: 10.1083/jcb.45.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulik M., Hellmann W. Comparison of Artemia salina and Escherichia coli ribosome structure by electron microscopy. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2829–2833. doi: 10.1073/pnas.75.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua N. H., Blobel G., Siekevitz P., Palade G. E. Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. J Cell Biol. 1976 Nov;71(2):497–514. doi: 10.1083/jcb.71.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuilov I., Sabatini D. D., Lake J. A., Freienstein C. Localization of eukaryotic initiation factor 3 on native small ribosomal subunits. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1389–1393. doi: 10.1073/pnas.75.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. V. High-level production of -galactosidase by Escherichia coli merodiploids. J Bacteriol. 1972 Nov;112(2):856–860. doi: 10.1128/jb.112.2.856-860.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Amino acid sequence of beta-galactosidase. XI. Peptide ordering procedures and the complete sequence. J Biol Chem. 1978 Aug 10;253(15):5521–5525. [PubMed] [Google Scholar]
- Godson G. N., Sinsheimer R. L. Use of Brij lysis as a general method to prepare polyribosomes from Escherichia coli. Biochim Biophys Acta. 1967 Dec 19;149(2):489–495. doi: 10.1016/0005-2787(67)90176-1. [DOI] [PubMed] [Google Scholar]
- HERZENBERG L. A. Studies on the induction of beta-galactosidase in a cryptic strain of Escherichia coli. Biochim Biophys Acta. 1959 Feb;31(2):525–538. doi: 10.1016/0006-3002(59)90029-0. [DOI] [PubMed] [Google Scholar]
- HORIUCHI T., TOMIZAWA J. I., NOVICK A. Isolation and properties of bacteria capable of high rates of beta-galactosidase synthesis. Biochim Biophys Acta. 1962 Jan 22;55:152–163. doi: 10.1016/0006-3002(62)90941-1. [DOI] [PubMed] [Google Scholar]
- Hamlin J., Zabin I. -Galactosidase: immunological activity of ribosome-bound, growing polypeptide chains. Proc Natl Acad Sci U S A. 1972 Feb;69(2):412–416. doi: 10.1073/pnas.69.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIHO Y., RICH A. INDUCED ENZYME FORMED ON BACTERIAL POLYRIBOSOMES. Proc Natl Acad Sci U S A. 1964 Jan;51:111–118. doi: 10.1073/pnas.51.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Zur M., Boublik M., Ofengand J. Localization of the decoding region on the 30S Escherichia coli ribosomal subunit by affinity immunoelectron microscopy. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1054–1058. doi: 10.1073/pnas.76.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A., Kahan L. Ribosomal proteins S5, S11, S13 and S19 localized by electron microscopy of antibody-labeled subunits. J Mol Biol. 1975 Dec 25;99(4):631–644. doi: 10.1016/s0022-2836(75)80177-x. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Practical aspects of immune electron microscopy. Methods Enzymol. 1979;61:250–257. doi: 10.1016/0076-6879(79)61014-5. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol. 1976 Jul 25;105(1):131–139. doi: 10.1016/0022-2836(76)90200-x. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Strycharz W. A. Ribosomal proteins L1, L17 and L27 from Escherichia coli localized at single sites on the large subunit by immune electron microscopy. J Mol Biol. 1981 Dec 25;153(4):979–992. doi: 10.1016/0022-2836(81)90462-9. [DOI] [PubMed] [Google Scholar]
- Malkin L. I., Rich A. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J Mol Biol. 1967 Jun 14;26(2):329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- Nonomura Y., Blobel G., Sabatini D. Structure of liver ribosomes studied by negative staining. J Mol Biol. 1971 Sep 14;60(2):303–323. doi: 10.1016/0022-2836(71)90296-8. [DOI] [PubMed] [Google Scholar]
- Olson H. M., Glitz D. G. Ribosome structure: localization of 3' end of RNA in small subunit by immunoelectronmicroscopy. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3769–3773. doi: 10.1073/pnas.76.8.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D. D., Tashiro Y., Palade G. E. On the attachment of ribosomes to microsomal membranes. J Mol Biol. 1966 Aug;19(2):503–524. doi: 10.1016/s0022-2836(66)80019-0. [DOI] [PubMed] [Google Scholar]
- Shatsky I. N., Evstafieva A. G., Bystrova T. F., Bogdanov A. A., Vasiliev V. D. Topography of RNA in the ribosome: location of the 3'-end of 5 S RNA on the central protuberance of the 50 S subunit. FEBS Lett. 1980 Nov 17;121(1):97–100. doi: 10.1016/0014-5793(80)81274-9. [DOI] [PubMed] [Google Scholar]
- Shatsky I. N., Mochalova L. V., Kojouharova M. S., Bogdanov A. A., Vasiliev V. D. Localization of the 3' end of Escherichia coli 16 S RNA by electron microscopy of antibody-labelled subunits. J Mol Biol. 1979 Oct 9;133(4):501–515. doi: 10.1016/0022-2836(79)90404-2. [DOI] [PubMed] [Google Scholar]
- Smith W. P., Tai P. C., Davis B. D. Interaction of secreted nascent chains with surrounding membrane in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5922–5925. doi: 10.1073/pnas.75.12.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. P., Tai P. C., Davis B. D. Nascent peptide as sole attachment of polysomes to membranes in bacteria. Proc Natl Acad Sci U S A. 1978 Feb;75(2):814–817. doi: 10.1073/pnas.75.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Wilchek M., Zamir A. Mapping of Escherichia coli ribosomal components involved in peptidyl transferase activity. Proc Natl Acad Sci U S A. 1973 May;70(5):1423–1426. doi: 10.1073/pnas.70.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strycharz W. A., Nomura M., Lake J. A. Ribosomal proteins L7/L12 localized at a single region of the large subunit by immune electron microscopy. J Mol Biol. 1978 Dec 5;126(2):123–140. doi: 10.1016/0022-2836(78)90355-8. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Schimke R. T. Specific binding of albumin antibody to rat liver polysomes. J Biol Chem. 1974 Jun 10;249(11):3597–3601. [PubMed] [Google Scholar]
- Unwin P. N. Attachment of ribosome crystals to intracellular membranes. J Mol Biol. 1979 Jul 25;132(1):69–84. doi: 10.1016/0022-2836(79)90496-0. [DOI] [PubMed] [Google Scholar]
- Winkelmann D., Kahan L. The accessibility of antigenic determinants of ribosomal protein S4 in situ. J Supramol Struct. 1979;10(4):443–455. doi: 10.1002/jss.400100407. [DOI] [PubMed] [Google Scholar]