Abstract
The tyrosine nitration of proteins has been observed in diverse inflammatory conditions and has been linked to the presence of reactive nitrogen species. From many in vitro experiments, it is apparent that tyrosine nitration may alter the function of proteins. A limited number of experiments under in vivo conditions also demonstrate that protein nitration is associated with altered cellular processes. To understand the association of protein nitration with the pathogenic mechanism of the disease, it is essential to identify specific protein targets of nitration with in vivo or intact tissue models. Using anti-nitrotyrosine antibodies, we demonstrated the accumulation of nitrotyrosine in a 52-kDa protein in rat kidney after lipopolysaccharide treatment. The 52-kDa protein was purified and identified with partial sequence as succinyl-CoA:3-oxoacid CoA-transferase (SCOT; EC 2.8.3.5). Western blot analysis revealed that the nitration of this mitochondrial enzyme increased in the kidneys and hearts of lipopolysaccharide-treated rats, whereas its catalytic activity decreased. These data suggest that tyrosine nitration may be a mechanism for the inhibition of SCOT activity in inflammatory conditions. SCOT is a key enzyme for ketone body utilization. Thus, tyrosine nitration of the enzyme with sepsis or inflammation may explain the altered metabolism of ketone bodies present in these disorders.
Keywords: protein nitration, lipopolysaccharide, nitric oxide
Infection and inflammation have been associated with an increased generation of reactive oxygen and nitrogen oxide species. These intermediates react with proteins, lipids, and DNA and may serve several physiological and pathophysiological functions (1). One product of the reaction of these reactive species with proteins is 3-nitrotyrosine (2). Initially, it was believed that 3-nitrotyrosine was formed almost exclusively by the reaction of tyrosine with peroxynitrite, a potent oxidant formed by the reaction of nitric oxide and superoxide anion (3). Recently, it has become recognized that other reactions, for example those involving hypochlorous acid and peroxidase enzymes, also may give a rise to nitration reactions (4). The mechanism(s) by which tyrosine is nitrated in vivo remains an area of active investigation and controversy (4–9).
Apart from the mechanism of tyrosine nitration, its biological significance is also a subject of great interest. The formation of 3-nitrotyrosine has been identified in many diverse pathological conditions such as atherosclerosis, pulmonary and heart disease, chronic rejection of transplanted organs, viral infections, and neurological disorders (for review, see ref. 10). However, many specific proteins, which undergo nitration in human disease as well as in animal and cellular models of disease, remain to be identified. In vitro studies using peroxynitrite and other nitrating agents have shown that the activity of many mammalian proteins is altered by nitration of tyrosine residue(s) (for review, see ref. 10). In addition to alterations in the structure and function of proteins, nitration of tyrosine residues may prevent tyrosine phosphorylation (11, 12). Despite the information generated from such in vitro experiments, the exact physiological relevance and functional consequences of this posttranslational protein modification remain obscure. Overall, many studies view tyrosine nitration as an incidental process with perhaps no physiologic consequence. However, a list of nitrotyrosine-containing proteins identified from in vivo studies is too limited (13–19) to draw any clear conclusion. With in vivo studies, it has been suggested that protein nitration may inhibit, activate, or have no effect on the protein's function.
Inflammation can cause a derangement of host metabolism and may lead to organ dysfunction or failure. Many of the systemic changes observed during inflammation can be duplicated by in vivo treatment of animals with lipopolysaccharide (LPS, endotoxin) from the Gram-negative bacteria outer membrane (20). In an attempt to understand whether inflammation caused tyrosine nitration of specific proteins and altered their activity, tissues from LPS-treated rats were screened for tyrosine-nitrated proteins by using Western immunoblots of tissue extracts with an anti-nitrotyrosine antibody. We observed several nitrated proteins in our screening. One of the nitrated proteins in kidney extracts was purified, partially sequenced, and identified as succinyl-CoA:3-oxoacid CoA-transferase (SCOT; EC 2.8.3.5). We demonstrate here that LPS administration enhanced SCOT nitration and decreased its catalytic activity in rat kidney and heart. These data may explain the altered ketone body metabolism during sepsis or inflammation.
Materials and Methods
Materials.
LPS (from Escherichia coli, serotype 0111:B4), tetranitromethane, soybean trypsin inhibitor, aprotinin, leupeptin, pepstatin A, antipain, phenylmethylsulfonyl fluoride, N-tosyl-L-phenylalanine chloromethyl ketone, Nα-p-tosyl-L-lysine chloromethyl ketone, 2-thiobarbituric acid, polyoxyethylene ester W-1, malonaldehyde bis(dimethyl acetal), succinyl-CoA, and acetoacetate (lithium salt) were purchased from Sigma. Ceramic hydroxyapatite and Affi-Gel Blue agarose were obtained from Bio-Rad. Sephacryl S-200 high resolution was from Amersham Pharmacia. Mouse monoclonal anti-nitrotyrosine antibody was purchased from Upstate Biotechnology (Lake Placid, NY).
Tissue Samples.
Male Wistar rats (median body weight, 250 g) were from Harlan Breeders (Indianapolis). Animals were maintained on standard rat chow and tap water ad libitum. LPS was dissolved in sterile saline and injected i.p. at a dose of 20 mg/kg body weight. Control rats received the corresponding volume of saline. Rats were killed at 6, 9, 16, or 24 h after LPS injection. The tissues were rapidly excised, immediately frozen in liquid nitrogen, and stored at −80°C until further processing.
To prepare tissue extracts, the thawed tissues were homogenized in 50 mM sodium phosphate buffer (pH 7.2)/0.15 M NaCl containing protease inhibitors (10 μg/ml soybean trypsin inhibitor/64 μM benzamidine/0.005 unit/ml aprotinin/22 μM leupeptin/15 μM pepstatin A/8 μM antipain/200 μM phenylmethylsulfonyl fluoride/500 μM N-tosyl-L-phenylalanine chloromethyl ketone/500 μM Nα-p-tosyl-L-lysine chloromethyl ketone). Homogenates were centrifuged for 10 min at 1,500 × g, and the supernatant fractions were centrifuged again for 60 min at 100,000 × g. The supernatant fractions from the high-speed centrifugation were considered to contain the total soluble protein of the tissue (i.e., cytoplasm and mitochondrial matrix). Total protein concentrations were determined by using Bio-Rad protein assay with BSA as the standard. The high-speed supernatant fractions were used for all of the following experiments.
Western Blot Analysis.
After separation by reducing SDS/PAGE in 10% gel, proteins were electrophoretically transferred to poly(vinylidene difluoride) (PVDF) membranes (Bio-Rad), which were blocked with 5% nonfat milk in 20 mM Tris⋅HCl, pH 7.2/0.3 M NaCl/0.1% Tween 20 (TBS/T). Membranes were incubated with monoclonal anti-nitrotyrosine antibodies (1:2,000 dilution, Upstate Biotechnology). After washing with TBS/T, immunoreactive proteins were detected by using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence. Control experiments for nitrotyrosine immunoreactivity were performed by preincubating (30 min at room temperature) the anti-nitrotyrosine antibodies with 4 mM free 3-nitrotyrosine or with 4 mM free L-tyrosine before incubation with PVDF membranes.
Rabbit polyclonal anti-SCOT antibody was generated against KGPKFEKRIERL peptide (BioSource International, Camarillo, CA) and used for Western blot analysis to normalize the amount of SCOT in samples assayed. Preliminary Western immunoblot experiments for SCOT also were performed with a polyclonal antibody kindly provided by T. Fukao and N. Kondo (Gifu University, Gifu, Japan).
Purification and Identification of the Nitrated 52-kDa Protein.
The high-speed supernatant fraction (1.3 ml containing 18 mg of protein) from the kidneys of rat after 6 h of LPS treatment was loaded on a Sephacryl S-200 HR column (1 × 100 cm) equilibrated with 10 mM sodium phosphate buffer (pH 7.2) containing 0.3 M NaCl and chromatographed in the same buffer at a flow rate of 10 ml/h. Collected fractions were analyzed with Western blotting by using monoclonal anti-nitrotyrosine antibodies from Upstate Biotechnology. Fractions that contained the 52-kDa nitrated protein were pooled and loaded onto a hydroxyapatite column (1 × 2 cm) equilibrated with 10 mM sodium phosphate buffer (pH 7.2). Bound proteins were eluted with a step gradient of increasing sodium phosphate concentration (50, 100, 150, and 200 mM) at a flow rate of 6 ml/h. The pool containing the 52-kDa nitrated protein was diluted with 2 vol of ice-cold deionized water and loaded onto an Affi-Gel Blue agarose column (1 × 1 cm) equilibrated with 50 mM sodium phosphate buffer (pH 7.2). After washing with 200 mM sodium phosphate buffer (pH 7.2), the bound proteins were eluted with a 1% SDS solution.
The SDS-eluted sample was resolved by SDS/PAGE, transferred to PVDF membrane, and briefly stained with Coomassie blue R-250. The 52-kDa nitrated band was excised and sent to the Protein Chemistry Core Laboratory at Baylor College of Medicine (Houston) for microsequencing analysis.
Catalytic Activity of SCOT.
SCOT activity was measured by using the assay procedure described by Williamson et al. (21). Briefly, the incubation mixture contained 50 mM Tris⋅HCl, pH 8.5/0.2 mM succinyl-CoA/0.1–10 mM acetoacetate/10 mM MgCl2/4 mM iodoacetamide, and high-speed supernatant fractions (300 μg of total protein/ml). SCOT catalytic activity was measured spectrophotometrically by following the formation of acetoacetyl-CoA (the forward direction) at 313 nm. SCOT catalytic activity was normalized to the total protein in high-speed supernatants, as Western blot analysis of these supernatants with anti-SCOT antibodies revealed similar amounts of SCOT present.
Determination of Thiobarbituric Acid-Reactive Substances (TBARS) in Mitochondria.
Mitochondria from kidney, heart, and brain were isolated by using differential centrifugation. Tissues from control rats and rats 6 h after LPS injection were minced with scissors and homogenized in a glass homogenizer with a motor-driven Teflon pestle in 10 mM phosphate buffer, pH 7.2/0.5 mM EDTA/0.25 M sucrose. After centrifugation at 750 × g for 10 min, the supernatant fractions were centrifuged at 10,000 × g for 20 min. Pellets were washed twice with 10 mM phosphate buffer, pH 7.2/0.5 mM EDTA/0.25 M sucrose. After the final wash, mitochondria were resuspended in 20 mM phosphate buffer (pH 7.2), and aliquots were quick-frozen in liquid nitrogen.
To measure TBARS, mitochondria were disrupted by four freeze-thawing cycles. Three hundred microliters of mitochondrial suspension (4 mg of total protein/ml) was mixed with 100 μl of a 10% solution of polyoxyethylene ester W-1 (nonionic detergent), 100 μl of 75% trichloroacetic acid, and 500 μl of an aqueous solution containing 0.6% 2-thiobarbituric acid. The reaction mixture was heated in boiling water for 30 min and centrifuged, and the absorbance was measured at 532 nm against a reaction mixture blank. The amounts of TBARS were calculated by using malonaldehyde bis(dimethyl acetal) as a standard.
Statistical Analysis.
Results of SCOT enzymatic activity are shown as the mean ± SEM. The results obtained from kidneys of LPS-treated rats were analyzed by ANOVA followed by Bonferroni's modified t test, whereas the other results were analyzed by unpaired t test.
Results
The 52-kDa Tyrosine Nitrated Protein in Rat Kidneys.
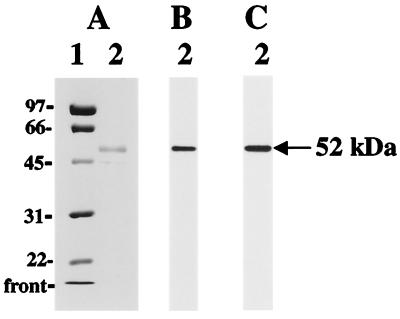
Western blot analysis of tissue extracts from kidneys of control and LPS-treated rats revealed several tyrosine-nitrated proteins. Here, we report a protein with an apparent molecular mass of 52 kDa (Fig. 1). Tyrosine-nitrated BSA (Fig. 1, lane 1) and the high-speed supernatant fraction from rat kidney after 6 h of LPS treatment (Fig. 1, lane 2) were resolved on a 10% gel by reducing SDS/PAGE, transferred to a PVDF membrane, and cut into strips. Strips were separately examined with a monoclonal antibody against nitrotyrosine (Fig. 1A), without primary antibody (Fig. 1B), and with a monoclonal antibody against nitrotyrosine that was preincubated with 4 mM free nitrotyrosine (Fig. 1C) or with 4 mM free L-tyrosine (Fig. 1D). Therefore, Fig. 1 represents a combination of negative and positive controls for this protein, which indicates the specificity of nitrotyrosine immunodetection.
Figure 1.
Western immunoblots using anti-nitrotyrosine antibodies. Lane 1, 50 ng of nitrated BSA. BSA was nitrated with 20-fold molar excess of tetranitromethane, dialyzed, and used as a positive control. Lane 2, 20 μg of a high-speed supernatant fraction from the kidneys of an LPS-treated rat (6 h after injection). After separation by reducing SDS/PAGE on 10% gel, proteins were transferred to PVDF membranes. Nitrated proteins were detected with Upstate Biotechnology monoclonal anti-nitrotyrosine antibodies (A). Control experiments for nitrotyrosine immunoreactivity were performed in the absence of primary antibody (B) or by preincubating (30 min at room temperature) the anti-nitrotyrosine antibody with 4 mM free 3-nitrotyrosine (C) or with 4 mM free L-tyrosine (D) before incubation with PVDF membranes.
To purify this 52-kDa protein, we used a combination of different protein separation methods and analyzed each separate fraction by Western blot using an anti-nitrotyrosine antibody. The procedure of purification is described in Materials and Methods in detail. Fig. 2A shows that gel filtration on the Sephacryl S-200 and chromatographies on hydroxyapatite and Affi-Gel Blue agarose result in substantial purification of the tyrosine-nitrated (Fig. 2B) 52-kDa protein. N-terminal sequencing of this protein revealed the Thr-Lys-Phe-Tyr-Thr-Asp-Pro-Val-Glu-Ala-Val sequence, which corresponds to the first 11 residues of the mature SCOT. A rabbit polyclonal anti-SCOT antibody was generated against a peptide fragment of SCOT (see Materials and Methods) and used for Western analysis to confirm that the 52-kDa nitrated protein was indeed SCOT (Fig. 2C).
Figure 2.
Purified 52-kDa nitrated protein from rat kidney 6 h after LPS injection. Lane 1, molecular mass standards; lane 2, 3 μg of 52-kDa protein after Affi-Gel Blue agarose column. The molecular masses (kDa) of protein standards are indicated on the left. Coomassie-stained SDS-polyacrylamide gel (A) and Western immunoblots with Upstate Biotechnology monoclonal anti-nitrotyrosine antibody (B) or with polyclonal anti-SCOT antibody (C) are shown.
SCOT Sequence Analysis.
SCOT is a mitochondrial matrix homodimer. The rat SCOT sequence is not available. The human (22) and pig (23) amino acid sequences possess more than 90% identity and contain 520 residues each. This includes the mitochondrial leader peptide (1–39 aa residues), which is absent in the mature enzyme (40–520 aa residues). The mature human and pig sequences contain 11 and 12 tyrosine residues, respectively. The PREDACC program (http://condor.urbb.jussieu.fr/) was used to predict the solvent accessibility of the amino acids of the human and pig enzymes. Two invariant tyrosines (Tyr-126 and Tyr-174) were predicted to be “external” with probability 73% and 74%, respectively. Although the site of nitration has not been identified, these two residues likely represent the best candidates for nitration because the major factor that determines the selectivity of protein tyrosine nitration is the solvent accessibility of tyrosine residues (24).
SCOT Nitration in Rat Tissues.
Heart and kidney are the tissues that contain the most SCOT activity in rat (21). At least 10-fold lower SCOT activities were detected in rat brain and skeletal muscle, followed by trace activities in lung and spleen. No SCOT activity has been detected in liver (21). Western immunoblot and Northern blot studies on SCOT distribution in human tissues revealed a high level of SCOT expression in heart and kidney, a reduced expression in brain and skeletal muscle, and a low level of SCOT expression in lung. No SCOT was identified in liver (25).
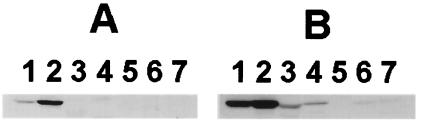
Fig. 3B shows the immunodetection of SCOT in high-speed supernatant fractions from various rat tissues 6 h after LPS injection. Our data are consistent with those previously published (21, 25) and show a high level of SCOT in heart and kidney, less amounts of SCOT in brain and skeletal muscle, trace amounts in lung and spleen, and no SCOT in liver. Fig. 3A shows the immunodetection of nitrotyrosine in the same samples. There is no detectable amount of nitrated SCOT in brain, skeletal muscle, liver, lung, or spleen. There is some nitrated SCOT in heart, and the strongest detection of nitrated SCOT was in kidney. Thus, tyrosine nitration of SCOT after LPS treatment occurs in a tissue-specific manner. Furthermore, the amounts of SCOT in heart and kidney do not differ substantially with respect to total protein, whereas tyrosine nitration was more favorable in kidney versus heart.
Figure 3.
Western immunoblots of various rat tissue extracts 6 h after LPS injection. Twenty micrograms of high-speed supernatant fractions was loaded per lane. Proteins transferred to the PVDF membrane were analyzed with Upstate Biotechnology monoclonal anti-nitrotyrosine antibody (A) or with polyclonal anti-SCOT antibody (B). Lanes 1–7 correspond to heart, kidney, brain, skeletal muscle, liver, lung, and spleen, respectively.
SCOT Catalytic Activity.
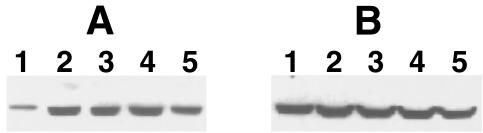
The presence of nitrated SCOT in untreated rat kidneys was observed (Fig. 4A). However, LPS treatment resulted in an increase of SCOT nitration (Fig. 4A). Fig. 4B shows that the abundance of SCOT protein was equal in all lanes. The catalytic activity of SCOT is shown in Table 1. The asterisk indicates statistically significant data. After 6 h of LPS treatment, a statistically significant decrease in SCOT activity was observed in kidney (Table 1), which was accompanied by an increase in SCOT nitration (Fig. 4A). Whereas SCOT also had increased nitration at 9, 16, and 24 h after LPS (Fig. 4A), the enzyme activity was not significantly decreased as it was at 6 h after LPS (Table 1). A statistically significant decrease in SCOT activity also was found in heart after LPS treatment (Table 1). Similar to kidney, this change also was accompanied by an increase of SCOT nitration in heart upon LPS treatment (data not shown). We did not observe SCOT nitration in brain (Fig. 3A), and no changes in SCOT catalytic activity were detected (Table 1).
Figure 4.
LPS treatment enhanced SCOT nitration in rat kidney. Forty micrograms of the high-speed supernatant fractions was loaded per lane. Proteins transferred to the PVDF membrane were analyzed with Upstate Biotechnology monoclonal anti-nitrotyrosine antibody (A) or polyclonal anti-SCOT antibody (B). Lanes 1–5 correspond to 0, 6, 9, 16, and 24 h after LPS injection, respectively.
Table 1.
LPS treatment decreases SCOT catalytic activity
| Rat tissue | Time after LPS injection, h | SCOT catalytic activity, nmol per min per mg |
|---|---|---|
| Kidney | 0 | 79.8 ± 2.4 (4) |
| 6 | 63.5 ± 4.8* (4) | |
| 9 | 65.0 ± 5.6 (4) | |
| 16 | 76.0 ± 5.4 (5) | |
| 24 | 76.7 ± 0.8 (3) | |
| Heart | 0 | 105.3 ± 6.8 (3) |
| 6 | 84.7 ± 5.3* (3) | |
| Brain | 0 | 22.4 ± 0.3 (3) |
| 6 | 25.6 ± 1.2 (3) |
The forward direction of the SCOT catalytic reaction was measured. Catalytic activity was expressed as nmol of acetoacetyl-CoA formed per min per mg of total protein. Data represent mean ± SEM. Number of animals used is shown in parentheses.
, P < 0.05.
Besides tyrosine, other amino acid residues also are susceptible to modification in inflammatory conditions. We assume that increased tyrosine nitration after LPS treatment is the primary reason for the inactivation of SCOT. However, to be sure of this assumption, the degree of SCOT nitration without alterations of other amino acids, such as tryptophan, cysteine, and methionine will have to be determined experimentally in future studies.
Mitochondrial Lipid Peroxidation.
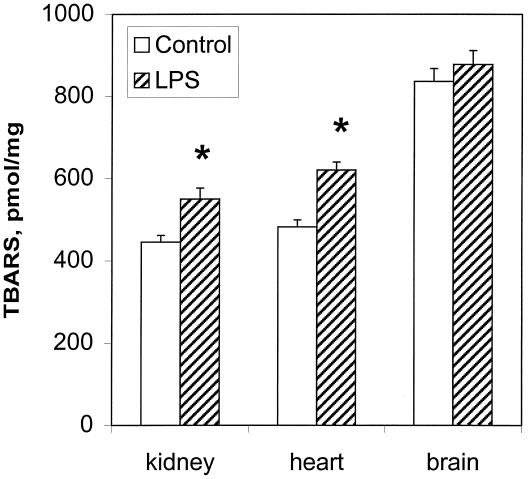
Generation of free radicals in mitochondria is well documented (26, 27). Superoxide and nitric oxide radicals react to produce peroxynitrite, which may cause protein tyrosine nitration. Peroxynitrite is a strong oxidant that also can induce membrane lipid peroxidation (28). As a measure of lipid peroxidation, the thiobarbituric acid test was used. This test estimates the amount of malondialdehyde precursors, including hydroperoxides and endoperoxides. Fig. 5 shows the formation of TBARS in kidney, heart, and brain mitochondria from control and 6-h LPS-treated rats. LPS treatment induced increases in mitochondrial lipid peroxidation in kidney and heart. No significant increase was observed in brain. The results presented in Fig. 5 are consistent with the notion that mitochondria from LPS-treated animals were exposed to an increased level of oxidative stress.
Figure 5.
Levels of lipid peroxidation in kidney, heart, and brain. TBARS were quantified in mitochondria from control rats and from rats 6 h after LPS injection and were expressed as pmol of TBARS formed per mg of total protein. Values are the mean ± SEM (n = 3 rats in each group); *, P < 0.05 compared with the corresponding control value.
Discussion
Protein tyrosine nitration in the kidney has been detected in a number of pathological conditions, such as human renal allograft rejection (13), experimental glomerulonephritis (29), diabetic nephropathy (30), LPS-induced kidney injury (31), transgenic sickle cell mice (32), IL-1β-treated mesangial cells (33), and the Goldblatt animal model for hypertension (34). However, Mn superoxide dismutase (13) and prostacyclin synthase (33) are the only previously reported proteins that are modified by nitration in kidney. In the present study, we report an additional tyrosine-nitrated protein in rat kidney, SCOT.
SCOT is localized exclusively in the mitochondrial matrix. Our findings suggest that there is an association between LPS administration, mitochondrial lipid peroxidation, SCOT nitration, and SCOT inactivation. It is important to recognize that both processes, lipid peroxidation and protein tyrosine nitration, could be a result of peroxynitrite formation. However, it should be noted that mitochondrial lipid peroxidation is not a specific indicator of mitochondrial peroxynitrite formation. Agents other than peroxynitrite can cause lipid peroxidation. Furthermore, although our data demonstrate that SCOT nitration is accompanied by SCOT inactivation, they do not necessarily establish that tyrosine nitration is the only factor responsible for the decrease in SCOT catalytic activity. Other amino acid residues in SCOT also could be modified with the conditions favorable for tyrosine nitration. Furthermore, at 9, 16, and 24 h after LPS administration, SCOT contained nitrotyrosine (Fig. 4A), whereas the enzyme activity was not significantly altered from control value (Table 1).
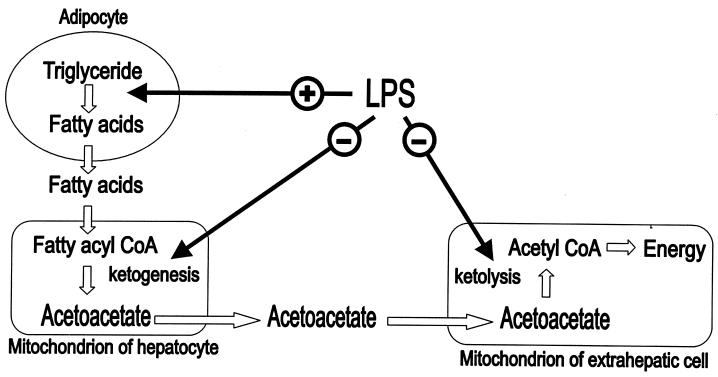
SCOT catalyzes the formation of acetoacetyl CoA from acetoacetate. This is the rate-determining step in ketolysis, which is the process of ketone body conversion into energy in the extrahepatic organs, such as heart, kidney, brain, and skeletal muscle (35). The term ketone bodies refers to three molecules. The two major ketone bodies are acetoacetate and 3-β-hydroxybutyrate, and acetone is the third one and is least abundant. The ketone body 3-β-hydroxybutyrate is formed by the reduction of acetoacetate. These two ketone bodies serve as an alternative source of energy during periods of glucose deficiency, such as starvation or prolonged exercise. Ketone body metabolism includes both ketogenesis and ketolysis (Fig. 6). Ketogenesis is the process by which free fatty acids are transformed into acetoacetate and 3-β-hydroxybutyrate in liver (35).
Figure 6.
Diagram of ketone body metabolism, which shows that inflammation induced by the administration of LPS may affect lipogenesis, ketogenesis, and ketolysis.
The putative mechanism of organ dysfunction during inflammation is a disruption of cellular energetics. It is well known that the rate of hepatic ketogenesis is diminished (Fig. 6) during an inflammatory state despite the availability of excess free fatty acids supplied to the liver (36, 37). The antiketogenic effect of inflammation results in a decrease in the plasma levels of ketone bodies. The decrease in the availability of circulating ketone bodies increases the dependence on other sources of energy and may have adverse consequences while systemic metabolism is changed by the inflammatory process.
The major observations of this study are (i) the catalytic activity of SCOT in kidney is impaired after LPS administration and (ii) this impairment is accompanied by SCOT tyrosine nitration. Overall, these data suggest that in addition to the antiketogenic effect, inflammation or endotoxin also may result in decreased efficiency of ketone body utilization (Fig. 6) through inactivation of the key enzyme of ketolysis, SCOT. Perhaps, in addition to other deleterious changes caused by inflammation or endotoxin, impaired ketone body utilization may represent another adverse contributing factor. Additional studies will be required to determine the specific tyrosine residue(s) in SCOT that is nitrated and the stoichiometry of tyrosine nitration with altered enzyme activity. As discussed above, other modifications of SCOT cannot be excluded to explain some of our observations.
Acknowledgments
This work was supported in part by National Institutes of Health Grant HL64221 (to F.M.), the John S. Dunn Foundation, G. Harold and Leila Y. Mathers Charitable Foundation, The Welch Foundation, United States Army, and the University of Texas. S.M. was supported by a fellowship from Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo (Brazil).
Abbreviations
- SCOT
succinyl-CoA:3-oxoacid CoA-transferase
- LPS
lipopolysaccharide
- TBARS
thiobarbituric acid-reactive substances
- PVDF
poly(vinylidene difluoride)
References
- 1.Davis K L, Martin E, Turko I V, Murad F. Annu Rev Pharmacol Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 2.Beckman J S. Chem Res Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 3.Koppenol W H, Moreno J J, Pryor W A, Ischiropoulos H, Beckman J S. Chem Res Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- 4.Eiserich J P, Hristova M, Cross C E, Jones A D, Freeman B A, Halliwell B, Van der Vliet A. Nature (London) 1998;391:393–397. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- 5.Pfeiffer S, Mayer B. J Biol Chem. 1998;273:27280–27285. doi: 10.1074/jbc.273.42.27280. [DOI] [PubMed] [Google Scholar]
- 6.Butler A R, Rutherford T J, Short D M, Ridd J H. Nitric Oxide. 2000;4:472–482. doi: 10.1006/niox.2000.0299. [DOI] [PubMed] [Google Scholar]
- 7.Lehnig M. Arch Biochem Biophys. 1999;368:303–318. doi: 10.1006/abbi.1999.1268. [DOI] [PubMed] [Google Scholar]
- 8.Reiter C D, Teng R-J, Beckman J S. J Biol Chem. 2000;275:32460–32466. doi: 10.1074/jbc.M910433199. [DOI] [PubMed] [Google Scholar]
- 9.Pfeiffer S, Schmidt K, Mayer B. J Biol Chem. 2000;275:6346–6352. doi: 10.1074/jbc.275.9.6346. [DOI] [PubMed] [Google Scholar]
- 10.Ischiropoulos H. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 11.Gow A, Duran D, Malcolm S, Ischiropoulos H. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 12.Kong S-K, Yim M B, Stadtman E R, Chock P B. Proc Natl Acad Sci USA. 1996;93:3377–3382. doi: 10.1073/pnas.93.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacMillan-Crow L A, Crow J P, Kerby J D, Beckman J S, Thompson J A. Proc Natl Acad Sci USA. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ara J, Przedborski S, Naini A B, Jackson-Lewis V, Trifiletti R R, Horwitz J, Ischiropoulos H. Proc Natl Acad Sci USA. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viner R I, Ferrington D A, Williams T D, Bigelow D J, Schoneich C. Biochem J. 1999;340:657–669. [PMC free article] [PubMed] [Google Scholar]
- 16.MacMillan-Crow L A, Greendorfer J S, Vickers S M, Thompson J A. Arch Biochem Biophys. 2000;377:350–356. doi: 10.1006/abbi.2000.1799. [DOI] [PubMed] [Google Scholar]
- 17.Chazotte-Aubert L, Hainaut P, Ohshima H. Biochem Biophys Res Commun. 2000;267:609–613. doi: 10.1006/bbrc.1999.2003. [DOI] [PubMed] [Google Scholar]
- 18.Gole M D, Souza J M, Choi I, Hertkorn C, Malcolm S, Foust R F, III, Finkel B, Lanken P N, Ischiropoulos H. Am J Physiol. 2000;278:L961–L967. doi: 10.1152/ajplung.2000.278.5.L961. [DOI] [PubMed] [Google Scholar]
- 19.Giasson B I, Duda J E, Murray I V J, Chen Q, Souza J M, Hurtig H I, Ischiropoulos H, Trojanowski J Q, Lee V M-Y. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 20.Raetz C R H. J Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson D H, Bates M W, Page M A, Krebs H A. Biochem J. 1971;121:41–47. doi: 10.1042/bj1210041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassovska-Bratinova S, Fukao T, Song X-Q, Duncan A M V, Chen H S, Robert M-F, Perez-Cerda C, Ugarte M, Chartrand C, Vobecky S, et al. Am J Hum Genet. 1996;59:519–528. [PMC free article] [PubMed] [Google Scholar]
- 23.Lin T, Bridger W A. J Biol Chem. 1992;267:975–978. [PubMed] [Google Scholar]
- 24.Souza J M, Daikhin E, Yudkoff M, Raman C S, Ischiropoulos H. Arch Biochem Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 25.Fukao T, Song X-Q, Mitchell G A, Yamaguchi S, Sukegawa K, Orii T, Kondo N. Pediatr Res. 1997;42:498–502. doi: 10.1203/00006450-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 26.Cadenas E, Davies K J A. Free Radical Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 27.Giulivi C, Poderoso J J, Boveris A. J Biol Chem. 1998;273:11038–11043. doi: 10.1074/jbc.273.18.11038. [DOI] [PubMed] [Google Scholar]
- 28.Radi R, Beckman J S, Bush K M, Freeman B A. Arch Biochem Biophys. 1994;288:481–487. doi: 10.1016/0003-9861(91)90224-7. [DOI] [PubMed] [Google Scholar]
- 29.Heeringa P, van Goor H, Moshage H, Klok P A, Huitema M G, Jager A D, Schep A J, Kallenberg C G M. Kidney Int. 1998;53:382–393. doi: 10.1046/j.1523-1755.1998.00780.x. [DOI] [PubMed] [Google Scholar]
- 30.Thuraisingham R C, Nott C A, Dodd S M, Yaqoob M. Kidney Int. 2000;57:1968–1972. doi: 10.1046/j.1523-1755.2000.00046.x. [DOI] [PubMed] [Google Scholar]
- 31.Bian K, Davis K, Kuret J, Binder L, Murad F. Am J Physiol. 1999;277:F33–F40. doi: 10.1152/ajprenal.1999.277.1.F33. [DOI] [PubMed] [Google Scholar]
- 32.Bank N, Kiroycheva M, Ahmed F, Anthony G M, Fabry M E, Nagel R L, Singhal P C. Kidney Int. 1998;54:1520–1528. doi: 10.1046/j.1523-1755.1998.00148.x. [DOI] [PubMed] [Google Scholar]
- 33.Zou M-H, Klein T, Pasquet J-P, Ullrich V. Biochem J. 1998;336:507–512. doi: 10.1042/bj3360507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosse H M, Bachmann S. Hypertension. 1997;30:948–952. doi: 10.1161/01.hyp.30.4.948. [DOI] [PubMed] [Google Scholar]
- 35.Laffel L. Diabetes Metab Res Rev. 1999;15:412–426. doi: 10.1002/(sici)1520-7560(199911/12)15:6<412::aid-dmrr72>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld H A, Pace J G, Kaminski M V, George D T, Jahrling P B, Wannemacher R W, Beisel W. Endocrinology. 1980;107:596–601. doi: 10.1210/endo-107-2-596. [DOI] [PubMed] [Google Scholar]
- 37.Memon R A, Feingold K R, Moser A H, Doerrler W, Adi S, Dinarello C A, Grunfeld C. Am J Physiol. 1992;263:E301–E309. doi: 10.1152/ajpendo.1992.263.2.E301. [DOI] [PubMed] [Google Scholar]