Abstract
Serum- and glucocorticoid-inducible kinase 1 (SGK1) plays a central role in epithelial sodium channel (ENaC)-dependent Na+ transport in the distal nephron. We hypothesized that SGK1 gene variants may contribute to the effect of dietary salt intake on BP in humans with hypertension, and consequentially influence renin-angiotensin-aldosterone (RAA) system activity. Our study population included 421 hypertensive Caucasian participants of the HyperPath group who had completed a dietary salt protocol with measurement of BP and RAA system activity. Three SGK1 tagging single nucleotide polymorphisms (SNPs) from the HapMap CEU population captured the genetic variation in the SGK1 region. Assuming an additive genetic model, two SNPs (rs2758151 and rs9402571) were associated with BP and plasma renin activity (PRA) effects of dietary salt intake. Major alleles were associated with higher systolic BP on high salt and decreased PRA on low salt. In contrast, low salt neutralized genotype differences. Similar, non-significant trends were observed in a normotensive population (N=152). Genotype was also associated with two salt-sensitive subtypes of hypertension. SGK1 genetic variants are associated with salt sensitivity of BP and PRA in human hypertension. Genotype status at these SGK1 variants may identify individuals prone to salt-sensitive hypertension.
Keywords: SGK1, polymorphism, renin, salt sensitivity
INTRODUCTION
Serum- and glucocorticoid-inducible kinase 1 (SGK1) is an important regulator of epithelial Na+ channel (ENaC) activity and its role in aldosterone-induced sodium reabsorption in the kidney.1 SGK1 expression is regulated by changes in intravascular volume and by mineralocorticoids.1, 2 Mice lacking the SGK1 gene are unable to appropriately increase the expression and activity of ENaC leading to impaired sodium reabsorption and hypotension on a sodium restricted diet resulting in sodium dependency to maintain blood pressure (BP).3 In humans, increases in dietary salt intake lead to increased BP in some, but not all, individuals regardless of hypertension.4,5 Variability within this BP response has been attributed in part to gene-environment interactions.6
We hypothesized that SGK1 genetic polymorphisms in humans are associated with the BP response to dietary salt intake, which in turn would result in altered renin-angiotensin-aldosterone (RAA) system activity. To test this hypothesis, we genotyped tagging single nucleotide polymorphisms (SNPs) in SGK1 in a group of hypertensive Caucasian individuals who had undergone a rigorous dietary salt protocol including assessment of BP and activity of the RAA system. We then explored whether these findings extended to a separate normotensive Caucasian group who had completed the same protocol.
METHODS
International Hypertension Pathotypes (HyperPath) Group
The data reported in this study are from the International Hypertension Pathotypes (HyperPath) Group. This group was formed in 1992 to characterize the genetic underpinnings of human hypertension through an intermediate phenotyping approach—common heritable traits within a population that define a subset while guiding candidate gene selection.7 In order to enhance the ability to detect genetic differences, normotensives in this study were included and were required to have no heritable tendency such as first-degree relatives with hypertension, coronary artery disease, or diabetes mellitus.
Study Participants and Protocol
The primary study outcome was the BP response to dietary salt intake under high and low salt conditions according to genotype status at SGK1. Data on 421 hypertensive Caucasian study volunteers and 152 normotensive Caucasian study volunteers who had participated in the International Hypertension Pathotype (HyperPath) group were analyzed. Details of this group have been described previously, but the analyses presented in this manuscript are original and not reported previously.4, 8, 9 Briefly, participants were studied on the Clinical Research Centers (CRCs) of Brigham and Women’s Hospital in Boston, Massachusetts, USA, University of Utah Medical Center in Salt Lake City, Utah, USA, Vanderbilt University in Nashville, Tennessee, USA, Hospital Broussais in Paris, France and University La Sapienza in Rome, Italy. The Institutional Review Board of each site approved the study protocol and informed consent was obtained from all study participants prior to enrollment.
As per the original HyperPath study protocol, all study participants completed a screening visit consisting of a physical examination, a medical history, and laboratory assessment.5 Study participants with a history of diabetes mellitus, any form of secondary hypertension, overt renal insufficiency (serum creatinine >114μmol/L), or any significant medical or psychiatric illnesses were excluded. Study participants with severe obesity (body mass index >34kg m−2), current tobacco or illicit drug use, or alcohol intake >12 ounces per week were also excluded. Normal laboratory values for electrolytes, thyroid and liver function tests were required. Electrocardiographic evidence of heart block, ischemia, or previous coronary events led to exclusion. Study participants had to be between 18 and 65 years of age and race was self-defined. To minimize the interference of medication with our assessment of RAA system activity, all angiotensin converting enzyme inhibitors, angiotensin receptor blockers, or mineralocorticoid receptor antagonists were discontinued 3 months before the study. If needed, study participants were placed on amlodipine or hydrochlorothiazide to control blood pressure. Beta-blockers and diuretics were withdrawn 1 month before study initiation. All antihypertensive medications were discontinued 3 weeks prior to vascular and hormonal assessment.
As per the original study design, hypertension was defined as a seated diastolic BP ≥100 mmHg off antihypertensive medication, ≥ 90mmHg while taking at least one antihypertensive medication, or treatment with at least two antihypertensive medications. Eligibility was determined while on an ad lib diet. Hypertensive individuals requiring more than 3 antihypertensive agents were excluded. Normotensive individuals had 3 repeated BP readings averaging less than 140/90 mmHg and no first-degree relative diagnosed with hypertension before 60 years of age.
As per the original HyperPath study design, participants then completed two dietary phases which varied in order: (1) high salt diet for 7 days (>200mmol/day sodium) and, (2) low salt diet for 7 days (10mmol/day sodium). In the current population, a majority of study participants (92%) underwent the high salt phase first followed by low salt. Both diets also contained 100mmol/day potassium, 20mmol/day calcium and were isocaloric. Prior to the current analyses, an examination for an order effect of dietary phase on our outcome variables was performed. We randomly chose 50 individuals who had completed the high salt phase first followed by low salt phase and 50 individuals who had completed the low salt phase first followed by high salt phase. On the final day of each diet week, study participants were admitted to CRC where they remained fasting and supine overnight. Assessment of salt balance was determined through a 24-hour urine collection for creatinine and sodium. The majority of participants completed 7 days of high salt intake while a small minority completed 5 days. We have shown previously that this variation does not affect the hormonal or vascular responsiveness.10 Regardless, all study participants were required to have attained a urine sodium concentration of > 150mmol/day sodium to be included for analysis. Low salt balance required a concentration of <30mmol/day sodium. BP and laboratory assessments were performed the following morning between the hours of 8:00-10:00AM with study participants remaining in a supine position overnight for at least 8 hours.
Laboratory Analysis and Measurement of Blood Pressure
Blood for assessment of electrolytes, plasma renin activity (PRA), and serum aldosterone were collected on ice and centrifuged for 20 minutes. Samples were kept frozen without preservatives and stored at −80 degrees centigrade until the time of assay. Both PRA and serum aldosterone were measured in a single centralized core laboratory. Radioimmunoassay techniques were used to measure aldosterone levels and renin activity.5, 11, 12 Low renin essential hypertension (LREH) was defined as PRA < 2.5ng AngI/mL/hr following 90 minutes of upright posture on low salt diet, and non-modulating (NM) hypertension was defined as an increase in serum aldosterone < 0.42 nmol L−1 after infusion of angiotensin II [3ng/kg for 60 mins]) while on a low salt diet.13, 14 The clearance of para-aminohippurate (PAH), infused as a bolus followed by a constant infusion, was used to assess renal plasma flow, as previously described.5,15 Systolic BP was measured at the time of PRA assessment and reported as the mean of 5 consecutive readings using an automated blood pressure monitor (DINAMAP; GE Healthcare, Little Chalfont, United Kingdom).
SGK1 Genotyping
Single nucleotide polymorphisms (SNPs) were selected from both CEU and YRI populations of HapMap (Phase II, November 2008) to maximize available variation using the chromosomal coordinates, chr6: 134,532,082 to 134,537,695 including the 5 kb flanking regions. Fourteen tagging SNPs–were selected when requiring an r2 > 0.8. Genotyping of these 14 SNPs was performed in the HyperPath study populations using the Illumina Bead Station GoldenGate platform (Illumina Inc, San Diego, CA). All SNPs had a genotyping completion rate of greater than 95%. (Table 1)
Table 1. Characteristics of study populations.
| Variable | HTN | NT |
|---|---|---|
| Number | 421 | 152 |
| Low Renin Essential Hypertension (%) | 81 (19%) | |
| Non-Modulating Hypertension (%) | 106 (25%) | |
| Age (years) | 48.4±8.7 | 39.5±11.0 |
| BMI (kg m−2) | 27.5±3.8 | 24.6±3.9 |
| Gender (% female) | 169(40%) | 77(51%) |
| High salt SBP (mmHg) | 146.5±19.6 | 109.9±11.3 |
| High salt DBP (mmHg) | 87.9±11.8 | 66.0±8.1 |
| Low salt SBP (mmHg) | 131.3±17.8 | 106.1±10.6 |
| Low salt DBP (mmHg) | 80.5±18.7 | 63.4±7.3 |
| High salt PRA (ng ml−1 per h) | 0.63±0.88 | 0.46±0.44 |
| Low salt PRA (ng ml−1 per h) | 2.58±2.20 | 2.96±1.94 |
| Aldosterone high salt (nmol L−1) | 0.16±0.12 | 0.11±0.09 |
| Aldosterone low salt (nmol L−1) | 0.50±0.35 | 0.52±0.32 |
| Cholesterol (mmol L−1) | 4.99±0.98 | 4.18±0.89 |
| Triglyceride (mmol L−1) | 1.69±1.02 | 1.12±0.72 |
| HDL (mmol L−1) | 1.03±0.32 | 1.20±0.48 |
| LDL (mmol L−1) | 3.13±0.81 | 2.52±0.70 |
Data presented as the mean ± standard deviation.
Statistical Analysis
Hardy-Weinberg equilibrium (HWE) was assessed for each SNP using χ2 test. The D’ and r2 measures of linkage disequilibrium (LD) were calculated using Haploview 4.1.16 All tests for association were performed using SAS version 9.1 (SAS Institute, Cary, NC). Within the HyperPath population, 7 of the 14 genotyped SNPs displayed minor allele frequencies of <10% and were not analyzed due to insufficient power. One SNP was not in HWE (rs1763527) and was also not analyzed. The remaining 6 SNPs captured 85% of the common variation in this region with an r2>0.9. These remaining 6 SNPs were represented in 3 unique blocks (ie. 3 SNPs were in LD (rs1743966, rs12663728, rs9402571) and another set of 2 SNPs were in LD (rs2758150, rs2758152)), and one SNP (rs2758151) was independent. Thus, three representative SNPs (rs2758151, rs9402571, rs2758152) were used for the present analysis as they uniquely captured all genetic variation in this region in the Caucasian HyperPath population. A generalized-estimating-equation (GEE) logistic model with an exchangeable covariance matrix was used for binary data analyses; and a mixed effect linear regression model was used for all of the continuous data analyses. GEE and mixed effect linear regression models were used to adjust for familial relatedness, as some siblings were present in the study population. All analyses were adjusted for age, gender, BMI, and study site. In the analysis of delineating the effect of different study diet sequence, PROC MIXED was used with BP and PRA as outcomes; salt intervention, experimental period and sequence were represented in the model as fixed effects; whereas study sequence was included as a random effect to allow for within-individual correlation among measurements. No indicators revealed effect of diet carryover (experimental period or study sequence) were significant for SBP(psequence= x0.58, pperiod= 0.23) or PRA(psequence = 0.12, pperiod = 0.10). Logarithmic transformations were performed for PRA concentrations to normalize the distribution of data. No evidence of population stratification was observed based on an analysis of 801 control SNPs in the study population (genomic inflation factor = 1.000).17 Bonferroni correction was applied to account for multiple comparisons. Three SNPs were evaluated in the index (hypertensive) study population, resulting in a final p-value for statistical significance of 0.5/3 or 0.017 for the primary outcome of systolic BP.
RESULTS
SGK1 Genotype Is Associated with Higher Blood Pressure in Hypertensive Study Participants on High Salt Diet
Baseline characteristics of the study populations are shown in Table 1. On a high salt diet, hypertensive study participants who were homozygous for the rs2758151 or rs9402571 major allele had significantly higher blood pressures compared with study participants carrying the minor alleles. On a low salt diet there was no association between SGK1 genotype and systolic BP (Figure 1). Consistent with these findings, SGK1 genotype was significantly associated with the change in systolic BP between dietary phases [change in systolic BP (±SEM); rs2758151: major allele homozygote 16.3 ± 1.3 mmHg; minor allele carrier 13.1 ± 1.4 mmHg; minor allele homozygote 9.1 ± 2.9 mmHg, p = 0.004; rs9402571: major allele homozygote 16.2 ± 1.2 mmHg; minor allele carrier 11.3 ± 1.5 mmHg; minor allele homozygote 8.9 ± 5.1 mmHg, p= 0.002]. SNP rs2758152 was not associated with blood pressure on either salt diet. We did not find an effect of order of the dietary intervention on blood pressure (p = 0.80). Normotensive major allele homozygotes for SGK1 showed a trend towards higher BP on a high salt diet, albeit not significant (rs2758151: 109.2 ± 3.3 mmHg; 108.8 ± 3.3 mmHg; 105.3 ± 4.0 mmHg, p = 0.26; rs9402571: 109.0 ± 3.2 mmHg; 107.6 ± 3.3 mmHg; 104.8 ± 6.3 mmHg, p = 0.32). Similar to the hypertensive population, there was no association between SGK1 genotype and BP on low salt diet (rs2758151: p = 0.67; rs9402571: p = 0.63).
Figure 1.
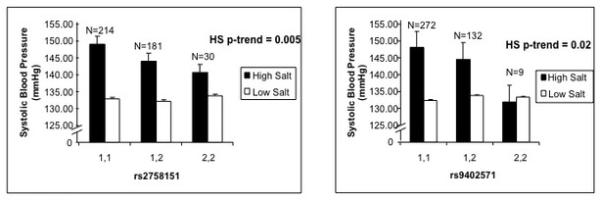
Systolic Blood Pressure on a High Salt Diet Differs by SGK1 genotype within hypertensive study participants. No difference in BP by genotype on LS diet (p=0.890, p=0.512). 1,1 represents major allele homozygotes. 2,2 represents minor allele homozygotes. Error bars represent SEM.
SGK1 Genotype and Renin-Angiotensin-Aldosterone System Activity
Consistent with BP response to dietary salt intake, SGK1 genotype at rs2758151 and rs9402571 were associated with RAA system response to dietary salt intake. On the low salt diet, PRA was significantly lower in participants homozygous for the major allele as compared with minor allele carriers (Figure 2). This difference by genotype was not observed on a high salt diet [PRA: ng/mL/hour (95% CI); rs2758151: major allele homozygote 0.42 ng/mL/hour (0.28, 0.63); minor allele carrier 0.50 ng/mL/hour (0.33, 0.75); minor allele homozygote 0.50 ng/mL/hour (0.29, 0.84), p = 0.125; rs9402571: 0.42 ng/mL/hour (0.30, 0.58); 0.45 ng/mL/hour (0.32, 0.65); 0.61 ng/mL/hour (0.28, 1.33), p = 0.264]. We did not find an effect of order of the dietary intervention on PRA (p = 0.65). No significant association was observed between SGK1 genotype and aldosterone levels on high or low salt diet (data not shown).
Figure 2.
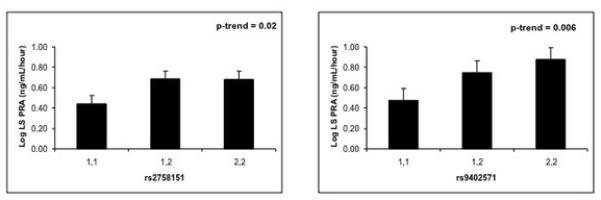
Plasma renin activity (PRA) on a Low Salt Diet differs by SGK1 genotype within hypertensive study participants. 1,1 represents major allele homozygotes. 2,2 represents minor allele homozygotes.
Exploratory analyses
SGK1 Genotype and Renal Plasma Flow
Our primary results indicate that SGK1 genotype predicts the BP and RAA system response to changes in dietary salt. We have reported altered renovascular sensitivity to salt in human hypertension.18, 19 We explored whether genotype status would, therefore, be associated with alteration in renovascular response to dietary salt intake. Renal plasma flow, measured by para-aminohippurate clearance method, significantly increased from low to high salt intake in this analysis (477.5 ± 99.0 ml/min/1.73 m2 vs. 504.3 ± 101.7 ml/min/1.73 m2, p = < 0.0001), as we have previously reported.20, 21 However, there was no significant association observed between SGK1 genotype at rs2758151and renal plasma flow on either diet (data not shown).
SGK1 Genotype in Hypertensive Subtypes
SGK1 genotype at rs2758151 did not predict increased odds for hypertension [OR 1.08 (0.98-1.19); p = 0.11], however it did identify two well-described salt-sensitive subsets within hypertension, LREH and NM hypertension Major allele homozygosity at SGK1 was associated with increased odds for low renin hypertension (OR 2.18; CI 1.36-3.49; p = 0.001) and non-modulating hypertension (OR 1.56; CI 1.02-2.39; p = 0.04).
DISCUSSION
This is the first human study of hypertension to associate SGK1 genetic variants with blood pressure and RAA system activity in response to changes in dietary salt. We demonstrated that two SNPs representing an area including the SGK1 gene are significantly associated with salt sensitivity of blood pressure in hypertensive individuals. These two SGK1 genetic variants were associated with an elevated blood pressure on a high salt diet. Further substantiating the association of these SNPs with salt intake, we demonstrated that the effects of genotype on blood pressure were abrogated when dietary salt was restricted. Within the normotensive population, there was also a trend towards higher blood pressure on high salt diet, however, these findings were not statistically significant given our limited sample size and lack of sufficient power to detect differences within the normal BP range.
Consistent with an association with salt sensitivity of blood pressure, we observed reciprocal association with PRA at the same SGK1 variants. The genotype associated with a greater blood pressure response to dietary sodium manipulation (major allele homozygote at either rs2758151 or rs9402571) also showed an association with lower PRA as detected on the low sodium diet. This was not discernible under high sodium conditions likely due to overwhelming suppression of PRA and/or assay limitations at very low PRA levels. Thus, it appears that renin is appropriately suppressed under conditions of volume expansion as represented by an increase in blood pressure observed in the major allele homozygote genotype. Given the well-described interplay between dietary salt intake, mineralocorticoid activity and SGK1 expression,22 one might anticipate a relationship between aldosterone and SGK1 genotype. However, despite differences in PRA by genotype we observed no significant association in aldosterone levels. This could be due to limitations in assay sensitivity or partial uncoupling of the normal physiologic relationship between renin and aldosterone secretion.
What might explain the association of salt sensitivity and SGK1 genotype status? We and others have described that renovascular function is dependent on dietary salt intake, whereby high salt intake results in increased renal plasma flow.23 Furthermore, loss of this relationship is seen in salt-sensitive forms of hypertension.13 Thus, we explored whether renal plasma flow response to dietary salt intake would be associated with SGK1 genotype and found no difference. This would suggest that despite an increase in BP among major allele carriers the renal vasculature is not displaying an increased blood flow---similar to what has been described in salt-sensitive hypertension in general.
Another possible mechanistic explanation for the association of SGK1 genotype with salt sensitivity of blood pressure can be found by examining renal tubular sodium handling in animal models. The SGK1 knockout mouse develops hypotension on a low sodium diet due to an inability to upregulate SGK1-dependent ENaC activity in the renal tubule and increase sodium reabsorption. Although we could not find a human analog for the SGK1 KO mouse, our results suggest that the increase in BP on high salt diet in the SGK1 major alleles may represent increased SGK1 activity. An increase in SGK1 activity could in turn increase ENaC activity, sodium reabsorption, and contribute to elevated BP.
Close to half of patients with hypertension exhibit some degree of salt sensitivity of blood pressure.24 Further, salt sensitivity is independently associated with cardiovascular events in individuals with hypertension and in individuals with normal blood pressure.25, 26 We were able to detect the association between salt-sensitivity of blood pressure and genotype in hypertensives in our group. This is consistent with a genetic predisposition to salt sensitivity. In fact, we found that genotype status was associated with increased odds for two well-described heritable salt-sensitive groups within the hypertensive population; those with low renin or those with non-modulating hypertension.14, 27, 28 Other genetic associations have been observed in these largely heritable subsets including those linked to altered sodium or volume homeostasis.29-31 Thus, our data support the importance of SGK1 as a candidate gene in identifying subsets who may benefit from therapies targeting mediators of sodium/volume homeostasis.
There are several limitations to be noted. First is the inherent inability to demonstrate causality in a retrospective or cross-sectional analytic approach. This was a tagging SNP study meant to provide polymorphic genetic coverage in a region of the human genome that includes SGK1. Thus, we can only describe associations to this region. Fine mapping or sequencing studies would be required to determine possible functional variants within this region. These analyses were performed in a modest sized population. We feel that the rigorous study conditions including a strict dietary salt protocol and medication washout further strengthen our results. Furthermore, the apparent concordance of results among the hypertensive and normotensive populations should be interpreted with caution given the smaller sample size and limited power to detect statistical significance. While we used RAA system activity as a marker of volume homeostasis, there are multiple other important contributors to intravascular volume such as the natriuretic peptide system, vasopressin, sympathetic activity and vascular tone which were not assessed in this study. Further studies are needed to directly assess SGK1 activity in renal tubules as well as other cell types.
In summary, we found that polymorphisms in the SGK1 gene are associated with blood pressure response to salt intake and RAA system activity in hypertensives. This may provide clues to the mechanisms underlying salt-sensitive hypertension and to factors that predispose to elevated blood pressure within a population. Future studies are needed to clarify this role and to determine if SGK1 genotype status can identify individuals who will respond favorably to specific non-pharmacological (salt restriction) and pharmacological approaches to managing hypertension.
Summary Table.
What is known about topic
SGK1 plays a central role in epithelial sodium channel (ENaC)-dependent Na+ transport in the distal nephron
Mice lacking the SGK1 gene are unable to appropriately increase the expression and activity of ENaC leading to impaired sodium reabsorption and hypotension on a sodium restricted diet resulting in sodium dependency to maintain blood pressure.
What this study adds
Polymorphisms in the SGK1 gene are associated with blood pressure response to salt intake and RAA system activity in hypertensives
SGK1 genotype status may provide clues to the mechanisms underlying salt-sensitive hypertension and to factors that predispose to elevated blood pressure within a population
Table 2. Minor allele frequencies and Hardy-Weinberg Equilibrium p-values: according to HapMap tagging SNPs at SGK1 in the HyperPath group.
| SNP | MAF (HTN) | MAF (NT) | HW (HTN) (p-value) |
HW (NT) (p-value) |
|---|---|---|---|---|
| rs1743966 | 19% | 23% | 0.812 | 0.319 |
| rs12663728 | 18% | 20% | 0.421 | 0.454 |
| rs2758150 | 10% | 10% | 0.273 | 0.649 |
| rs2758151 | 28% | 33% | 0.320 | 0.935 |
| rs2758152 | 9% | 12% | 0.330 | 0.898 |
| rs9402571 | 18% | 19% | 0.126 | 0.390 |
| rs1763527 | 44% | 43% | 0.000 | 0.0002 |
MAF= minor allele frequency; HTN=hypertensive group; NT=normotensive group; HW=Hardy-Weinberg equilibrium p-value (2-tailed chi2)
Acknowledgements
We would like to thank the staff of the clinical research centres at our collaborating institutions and the volunteers of this study. Sources of Funding: This project was supported in part by the following grants: U54LM008748, from the National Library of Medicine; UL1RR025758 Harvard Clinical and Translational Science Center; M01RR02634, M01RR00095, M01RR00064 from the National Center for Research Resources as well as NIH grants HL47651, HL59424, K23HL084236 (JSW), K24HL103845 (GKA) and the Specialized Center of Research (SCOR) in Molecular Genetics of Hypertension P50HL055000. ADR was supported by a NIH training grant (T32 HL007609).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Reference List
- 1.Chen S-Y, Bhargava A, Mastroberardino L, Meijer OC, Wang J, Buse P, et al. Epithelial Sodium Channel Regulated by Aldosterone-Induced Protein sgk. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(5):2514. doi: 10.1073/pnas.96.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang F, Bohmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V. (Patho)physiological Significance of the Serum- and Glucocorticoid-Inducible Kinase Isoforms. Physiological Reviews. 2006;86(4):1151. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- 3.Luft FC, Weinberger MH. Heterogeneous responses to changes in dietary salt intake: the salt-sensitivity paradigm. American Journal of Clinical Nutrition. 1997;65(2):612S. doi: 10.1093/ajcn/65.2.612S. [DOI] [PubMed] [Google Scholar]
- 4.Gumieniak O, Perlstein TS, Williams JS, Hopkins PN, Brown NJ, Raby BA, et al. Ala92 Type 2 Deiodinase Allele Increases Risk for the Development of Hypertension. Hypertension. 2007;49(3):461. doi: 10.1161/01.HYP.0000256295.72185.fd. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins PN, Lifton RP, Hollenberg NK, Jeunemaitre X, Hallouin M-C, Skuppin J, et al. Blunted renal vascular response to angiotensin II is associated with a common variant of the angiotensinogen gene and obesity. Journal of Hypertension. 1996;14:199–207. doi: 10.1097/00004872-199602000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal A, Williams GH, Fisher NDL. Genetics of human hypertension. Trends in Endocrinology and Metabolism. 2005;16(3):127–133. doi: 10.1016/j.tem.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Williams GH. Genetic approaches to understanding the pathophysiology of complex human traits. Kidney international. 1994;46(6):1550. doi: 10.1038/ki.1994.443. [DOI] [PubMed] [Google Scholar]
- 8.Chamarthi B, Williams JS, Williams GH. A mechanism for salt-sensitive hypertension: abnormal dietary sodium-mediated vascular response to angiotensin-II. Journal of Hypertension. 2010;28(5):1020. doi: 10.1097/HJH.0b013e3283375974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Underwood PC, Sun B, Williams JS, Pojoga LH, Chamarthi B, Lasky-Su J, et al. The Relationship between Peroxisome Proliferator-Activated Receptor-{gamma} and Renin: A Human Genetics Study. Journal of Clinical Endocrinology Metabolism. 2010;95(9):E75. doi: 10.1210/jc.2010-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollenberg NK, Chenitz WR, Adams DF, Williams GH. Reciprocal Influence of Salt Intake on Adrenal Glomerulosa and Renal Vascular Responses to Angiotensin II in Normal Man. Journal of Clinical Investigation. 1974;54(1):34–42. doi: 10.1172/JCI107748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Underwood RH, Williams GH. Simultaneous Measurement of Aldosterone, Cortisol, and Corticosterone in Human Peripheral Plasma by Displacement Analysis. Journal of Laboratory and Clinical Medicine. 1972;79(5):848–862. [PubMed] [Google Scholar]
- 12.Emanuel RL, Cain JP, Williams GH. DOUBLE ANTIBODY RADIOIMMUNOASSAY OF RENIN-ACTIVITY AND ANGIOTENSIN-II IN HUMAN PERIPHERAL PLASMA. Journal of Laboratory and Clinical Medicine. 1973;81(4):632–640. [PubMed] [Google Scholar]
- 13.Williams GH, Dluhy RG, Lifton RP, Moore TJ, Gleason R, Williams R, et al. Non-modulation as an intermediate phenotype in essential hypertension. Hypertension. 1992;20(6):788. doi: 10.1161/01.hyp.20.6.788. [DOI] [PubMed] [Google Scholar]
- 14.Fisher NDL, Hurwitz S, Ferri C, Jeunemaitre X, Hollenberg NK, Williams GH. Altered Adrenal Sensitivity to Angiotensin II in Low-Renin Essential Hypertension. Hypertension. 1999;34(3):388. doi: 10.1161/01.hyp.34.3.388. [DOI] [PubMed] [Google Scholar]
- 15.Raji A, Williams GH, Jeunemaitre X, Hopkins PN, Hunt SC, Hollenberg NK, et al. Insulin resistance in hypertensives: effect of salt sensitivity, renin status and sodium intake. Journal of hypertension. 2001;19(1):99. doi: 10.1097/00004872-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Moore TJ, Rich G, McKnight JA, McCullough M, Hollenberg NK. Salt sensitivity of hypertension and responses to angiotensin converting enzyme inhibition with benazepril[ast] Am J Hypertens. 1996;9(1):54–60. doi: 10.1016/0895-7061(95)00300-2. [DOI] [PubMed] [Google Scholar]
- 19.Price DA, Fisher NDL, Lansang MC, Stevanovic R, Williams GH, Hollenberg NK. Renal Perfusion in Blacks. Hypertension. 2002;40(2):186–189. doi: 10.1161/01.hyp.0000024349.85680.87. [DOI] [PubMed] [Google Scholar]
- 20.Patel TV, Williams GH, Fisher NDL. Angiotensinogen genotype predicts abnormal renal hemodynamics in young hypertensive patients. Journal of Hypertension. 2008;26(7):1353–1359. doi: 10.1097/HJH.0b013e3282ffb417. [DOI] [PubMed] [Google Scholar]
- 21.Splenser AE, Fisher NDL, Danser AHJ, Hollenberg NK. Renal plasma flow: glomerular filtration rate relationships in man during direct renin inhibition with aliskiren. Journal of the American Society of Hypertension. 2009;3(5):315–320. doi: 10.1016/j.jash.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Shibata S, Mu SY, Kawarazaki H, Muraoka K, Ishizawa K, Yoshida S, et al. Rac1 GTPase in rodent kidneys is essential for salt-sensitive hypertension via a mineralocorticoid receptor-dependent pathway. Journal of Clinical Investigation. 2011;121(8):3233–3243. doi: 10.1172/JCI43124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlstein TS, Gumieniak O, Hopkins PN, Murphey LJ, Brown NJ, Williams GH, et al. Uric acid and the state of the intrarenal renin-angiotensin system in humans. Kidney Int. 2004;66(4):1465–1470. doi: 10.1111/j.1523-1755.2004.00909.x. [DOI] [PubMed] [Google Scholar]
- 24.Williams GH, Hollenberg NK. Sodium-sensitive essential hypertension: emerging insights into an old entity. J Am Coll Nutr. 1989;8(6):490–4. doi: 10.1080/07315724.1989.10720318. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, et al. Sodium sensitivity and cardiovascular events in patients with essential hypertension. The Lancet. 1997;350(9093):1734. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 26.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt Sensitivity, Pulse Pressure, and Death in Normal and Hypertensive Humans. Hypertension. 2001;37(2):429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 27.Hurwitz S, Fisher NDL, Ferri C, Hopkins PN, Williams GH, Hollenberg NK. Controlled analysis of blood pressure sensitivity to sodium intake: interactions with hypertension type. Journal of Hypertension. 2003;21(5):951–959. doi: 10.1097/00004872-200305000-00020. [DOI] [PubMed] [Google Scholar]
- 28.Hollenberg N, Williams G. Nonmodulation and essential hypertension. Current Hypertension Reports. 2006;8(2):127–131. doi: 10.1007/s11906-006-0008-9. [DOI] [PubMed] [Google Scholar]
- 29.Pojoga L, Kolatkar NS, Williams JS, Perlstein TS, Jeunemaitre X, Brown NJ, et al. {beta}-2 Adrenergic Receptor Diplotype Defines a Subset of Salt-Sensitive Hypertension. Hypertension. 2006;48(5):892. doi: 10.1161/01.HYP.0000244688.45472.95. [DOI] [PubMed] [Google Scholar]
- 30.Kosachunhanun N, Hunt SC, Hopkins PN, Williams RR, Jeunemaitre X, Corvol P, et al. Genetic Determinants of Nonmodulating Hypertension. Hypertension. 2003;42(5):901. doi: 10.1161/01.HYP.0000095615.83724.82. [DOI] [PubMed] [Google Scholar]
- 31.Grant FD, Romero JR, Jeunemaitre X, Hunt SC, Hopkins PN, Hollenberg NH, et al. Low-Renin Hypertension, Altered Sodium Homeostasis, and an {alpha}-Adducin Polymorphism. Hypertension. 2002;39(2):191. doi: 10.1161/hy0202.104273. [DOI] [PubMed] [Google Scholar]


