Abstract
J proteins are a diverse family of co-chaperones that cooperate with heat shock protein 70 (Hsp70) to coordinate protein quality control, especially in response to cellular stress. Current models suggest that individual J proteins might play roles in recruiting Hsp70s to specific functions, such as maintaining cell wall integrity or promoting ribosome biogenesis. However, relatively few stresses have been used to test this model and, as a result, only a few specific activities have been identified. To expand our understanding of the J protein network, we used a synthetic lethal approach in which 11 Saccharomyces cerevisiae deletion strains were treated with 12 well-characterized chemical inhibitors. The results defined new roles for specific J proteins in major signaling pathways. For example, an important role for Swa2 in cell wall integrity was identified and activities of the under-explored Jjj1, Apj1, Jjj3 and Caj1 proteins were suggested. More generally, these findings support a model in which some J proteins, such as Ydj1 and Zuo1, play “generalist” roles, while others, such as Apj1 and Jjj2, are “specialists”, having roles in relatively few pathways. Together, these results provide new insight into the network of J proteins.
INTRODUCTION
The molecular chaperone heat shock protein 70 kDa (Hsp70) is a central hub of the protein quality control network in all organisms1. The Hsp70 machinery has many essential roles including nascent protein folding, complex remodeling, and protein degradation2. Hsp70 has evolved as a limited number of highly homologous isoforms, some constitutively expressed and others induced upon stress3. All Hsp70 isoforms consist of an N-terminal nucleotide-binding domain (NBD) connected by a short linker to a C-terminal substrate-binding domain (SBD) with a helical lid4. The NBD and SBD are allosterically linked and conformationally regulated by binding and hydrolysis of ATP5–7. When ATP is bound, Hsp70 has low affinity for substrate, and upon ATP hydrolysis adopts a high-affinity conformation8, 9. In Saccharomyces cerevisiae, there are two cytosolic classes of Hsp70: Ssa (Ssa1–4) and Ssb (Ssb1–2). Expression of only one of each class is required to support near wild type viability, underscoring the high structural and functional homology among Hsp70 isoforms10.
The ability of Hsp70s to engage in a wide array of cellular activities is thought to be facilitated by its cooperation with a class of co-chaperones, the J proteins (also known as Hsp40s)8, 11. J proteins share a highly conserved J domain that interacts with Hsp70 to accelerate ATP hydrolysis12–14. The J domain consists of four α-helices and requires an invariant His-Pro-Asp (HPD) motif for activity15. J proteins comprise a highly diverse class of co-chaperones that has expanded through evolution from a handful of isoforms in prokaryotes to over 20 in S. cerevisiae and over 40 in humans16. J proteins can be classified into three families based on homology to the E. coli J protein DnaJ. Class I, the most homologous to DnaJ, consists of a J domain, a glycine-phenylalanine (G/F) rich region, a zinc-finger like region (ZFLR), a barrel topology C-terminal domain, and a dimerization domain17, 18. The ZFLR region is thought to help these J proteins bind to substrates and recruit them to the Hsp70 system. Class II J proteins lack the ZFLR and have variable C-terminal domains, but are otherwise similar to Class I. Finally, Class III J proteins, the largest class, comprises the misfits; they share only a J domain and are otherwise highly diverse in structure. Recently, Kampinga and Craig suggested a reclassification of the J proteins based on function instead of structure16.
The dramatic expansion of the J proteins in higher organisms, especially compared to the relatively few Hsp70s, suggests that a larger pool of more specialized J proteins supports greater cellular complexity. Prevailing models suggest that combinations of an individual J protein with an Hsp70 might yield a complex with the ability to perform specific tasks or engage with specific subsets of substrates. In support of this idea, recent efforts have identified roles for individual J proteins in a handful of specific cellular tasks. For example, Swa2, the S. ceresvisiae ortholog of human auxilin, uncoats clathrin coated vesicles and is required for cortical ER inheritance19, 20. This activity is not readily recovered by over-expression of other J proteins, suggesting that Swa2 is specialized for this Hsp70 function. Similarly, Jjj3 is a J protein that is essential for diphthamide synthesis21. In contrast to these “specialist” J proteins, S. cerevisiae has also retained “generalists”, such as Ydj1 and Sis1, which maintain protein quality control in the cytosol by helping Hsp70 fold proteins16. A systematic study of cytosolic S. cerevisiae J proteins using deletion mutants confirmed that some J proteins play essential roles during thermal stress, while others are apparently redundant for this activity10. These observations suggest that other J proteins might be important under specific biological or environmental conditions.
Towards this goal, we envisioned that a synthetic lethal approach might enable assignment of cellular roles to individual J proteins. Using deletions of the 11 cytosolic J proteins in S. cerevisiae10, 22, 23, we tested the effects of twelve, well-characterized chemical inhibitors that act in specific cellular pathways, such as cell wall synthesis and translation (Table 1), on growth22, 24. We found that some J proteins, such as Ydj1, are multidrug resistance genes, required to buffer the cell against a diverse array of stressors. Others confer resistance to only a subset of the compounds, suggesting functions in specific pathways. Three J proteins had no synthetic lethal interactions and they may either be redundant or have specific functions in pathways not targeted by the twelve compounds. These findings define new biological roles for individual J proteins, results that could facilitate their functional classification and provide new opportunities for their use as drug targets.
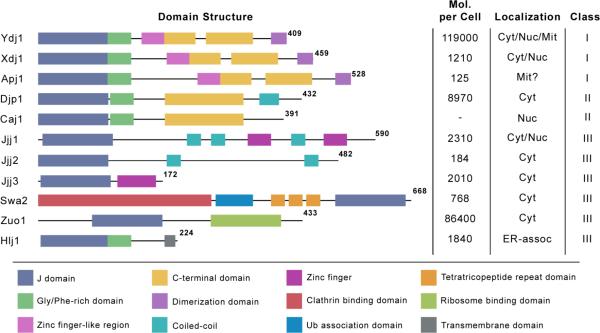
Table 1.
Eleven J Proteins were tested for synthetic lethal interactions. J proteins are depicted with domain schematics. Molecules per cell determined by Ghaemmaghami et al. 2003. Localization is in some cases predicted. Figure adapted from Kampinga and Craig, 2010.
|
RESULTS AND DISCUSSION
Selection of J proteins and chemical probes for the synthetic lethal analysis
Eleven S. cerevisiae deletion mutants of cytosolic and nuclear J proteins were selected to carry out chemical genetic investigations10 (Table 1). These J proteins represent 3 members of class I, 2 of class II and 6 of class III. In addition, twelve compounds targeting diverse cellular processes were chosen22, 23 (Table 2). Yeast strains were spotted in five-fold dilutions on rich media agar containing a sublethal concentration of each compound and grown for three days. As a secondary assay, yeast growth in liquid was also assessed using optical density (OD) values monitored for 10 hours, when all strains had reached stationary phase. Compounds that reduced growth of a J protein deletion strain in either assay were scored as synthetic lethal interactions. Of the compounds tested, only wortmannin caused growth defects in the liquid assay but not the solid media assay, likely because wortmannin is unstable in solid media.
Table 2.
Compounds with known antifungal activity used to identify synthetic lethal interactions for deletions of J proteins in S. cerevisiae.
| Compound | Target |
|---|---|
| Hygromycin B | Translation; 30S ribosomal subunit formation |
| Cycloheximide | Translation elongation |
| Rapamycin | TOR1/TOR2 signaling |
| Caffeine | TOR1 signaling; cell wall; other kinases |
| Calcofluor White | Cell wall integrity (CWI) pathway |
| Congo Red | Cell wall integrity (CWI) pathway |
| Fluconazole | Ergosterol biosynthesis (plasma membrane) |
| 17-AAG | Hsp90 |
| Camptothecin | Topoisomerase I |
| Hydroxyurea | Ribonucleotide reductase |
| FK-506 | Calcineurin |
| Wortmannin | Phosphatidylinositol kinase |
Ydj1
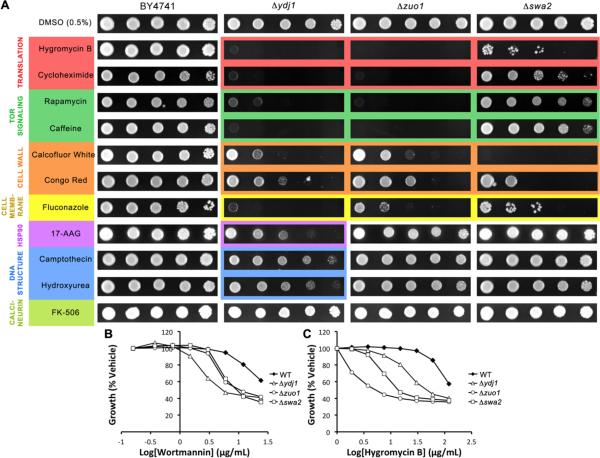
Ydj1 is an abundant J protein that partners with Hsp70s (Ssa1–4 in yeast) to carry out general protein quality control in the cytosol25, 26. Consistent with this activity, deletion of Ydj1 resulted in sensitivity to nearly every compound tested (Fig. 1A). Δydj1 cells were particularly sensitive to translation inhibitors, fluconazole, rapamycin and caffeine (Fig. 1A). Furthermore, Δydj1 was very sensitive to wortmannin, an inhibitor of PI kinases, in the liquid growth assay (Fig. 1B). Together, these results suggest that Ydj1 may play an important role in supporting the integrity of kinase signaling networks, perhaps by stabilizing the enzymes. On the other hand, Δydj1 was only moderately sensitive to cell wall stressors, suggesting that cells lacking Ydj1 can still maintain the cell wall integrity (CWI) pathway. Finally, Δydj1 was the only strain tested that was mildly sensitive to the stressors of DNA maintenance, hydroxyurea and camptothecin. This result suggests that the J proteins (and, by extension, the Hsp70 machinery) may not be responsible for stability or function in that system.
Figure 1.
Deletion of the J proteins Ydj1, Zuo1, and Swa2 confers sensitivity to many compounds. A) Five-fold dilutions of BY4741, Δydj1, Δzuo1, or Δswa2 were plated on rich medium containing DMSO or compound (see Materials and Methods for concentrations) and grown for 72 hours at 30°C. Images are representative of 3 replicates. B–D) Cultures were grown to saturation, diluted to OD600 = 0.15, and transferred to a 96-well plate containing two-fold dilutions of wortmannin (24–0 μg/mL) or hygromycin B (120–0 μg/mL) in DMSO (final concentration 2%). Plates were incubated with shaking at 30°C for 10 hours. At 10 hours, OD600 values were recorded and normalized to the OD600 with DMSO alone for each strain. The other compounds were also tested in this assay (see Fig 4) and the raw data is presented in the Supplemental Information.
Zuo1
Zuo1 is a ribosome-associated J protein that plays roles in nascent protein folding and ribosomal assembly27–29. Deletion of Zuo1 resulted in marked sensitivity to seven compounds (Fig. 1A). The sensitivity profile of Δzuo1 was similar to that of Ydj1, which is unexpected because they have markedly different localization patterns and domain structures (Table 1). For example, Δzuo1 was acutely sensitive to fluconazole, rapamycin, caffeine, and wortmannin (Fig. 1A,B) and these cells also had moderate sensitivity to cell wall perturbing compounds. It seems possible that the similar sensitivities of Δzuo1 and Δydj1 cells may reflect their activities at different stages in the same processes. For example, Zuo1 might be important in initial folding of a kinase, while Ydj1 might be important in final maturation. However, Δzuo1 cells do not share the growth defect of Δydj1 cells, so it may be that Ydj1's role is more critical to a wider range of substrates. Consistent with this idea, Δzuo1 was not sensitive to inhibitors of Hsp90, DNA-modifying enzymes or calcineurin. Another possibility is based on the observations that Zuo1 promotes expression of the drug exporter Pdr530, 31. The absence of Zuo1 may therefore allow accumulation of the specific drugs exported by Pdr5, which includes fluconazole (Fig. 1A). Finally, as expected, Δzuo1 cells were not viable in the presence of compounds that inhibit translation (Fig. 1A,C). In healthy cells, Zuo1 may confer tolerance to cycloheximide and hygromycin B by helping to disassemble arrested ribosomes and assemble new ones, and in its absence the cells cannot recover from the accumulation of arrested ribosomes.
Swa2
Swa2 functions with Hsp70s in the removal of clathrin from vesicles19 and it has essential roles in cortical ER inheritance20. In addition to its J domain and clathrin-binding domain, Swa2 includes three tetratricopeptide (TPR) motifs and a ubiquitin-binding domain32. Deletion of Swa2 caused sensitivity to 6 compounds (Fig. 1A), including weak sensitivity to caffeine and rapamycin and severe responses to calcofluor white, congo red, fluconazole, and hygromycin B. Lesser sensitivity was also observed in the presence of cycloheximide, caffeine, and rapamycin. The acute sensitivity of Δswa2 to cell wall perturbing agents suggests that Swa2 may have a role, whether direct or indirect, in cell wall assembly or integrity. CW and CR induce upregulation of the cell wall integrity (CWI) pathway, but caffeine causes cell wall defects independent of the CWI pathway33, 34. The severe sensitivity of Δswa2 to CW and CR but not caffeine thus suggests a specific defect in the cell's ability to mount a CWI response in the absence of Swa2. This may be due to impaired trafficking of cell wall components or CWI pathway members as a result of accumulated clathrin coated transport vesicles, or it could be another uncharacterized function of Swa2.
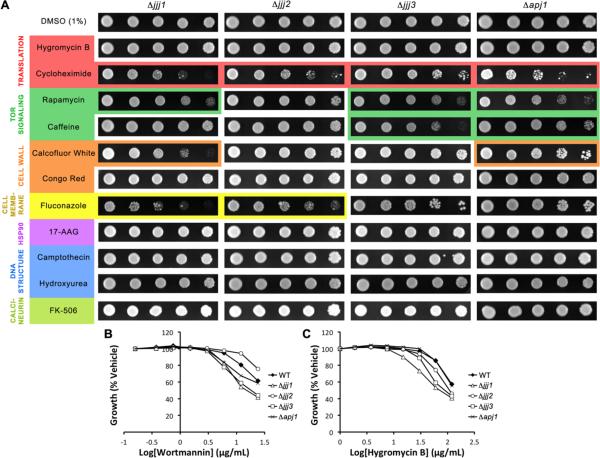
Jjj1
Jjj1 is a specialized J protein that plays a role in 60S ribosomal subunit biogenesis35, 36 and perhaps other aspects of ribosome turnover29. Deletion of this J protein conferred mild sensitivity to calcofluor white, rapamycin, cycloheximide, and fluconazole in the spot assay and hygromycin B and wortmannin in the liquid assay (Fig. 2). Previous work has demonstrated that growth defects resulting from Jjj1 deletion cannot be ameliorated by any other J protein10. On the other hand, overexpression of Jjj1 can partially rescue the slow growth of Δzuo1 cells, which is thought to be due to its ability to recruit Ssa1 (the cytosolic yeast Hsp70) to the ribosome when Zuo1 is not present to recruit Ssb1/229. Further enforcing this relationship between Jjj1 and Zuo1, their deletions confer sensitivity to nearly the same compounds in our experiments. However, loss of Jjj1 is less detrimental, as the phenotype of Δjjj1 cells is uniformly less dramatic. The differences in phenotype severity are consistent with their copy numbers; Jjj1 exists as only 2310 molecules per cell, while Zuo1 has 86400 copies and is possibly associated with every ribosome. Together, these finding are consistent with Jjj1 and Zuo1 having partially overlapping functions in ribosome function.
Figure 2.
Deletion of the J proteins Jjj1, Jjj2, Jjj3, and Apj1 confers sensitivity to a limited subset of compounds. A) Five-fold dilutions of Δjjj1, Δjjj2, Δjjj3, or Δapj1 were plated on rich medium containing DMSO or compound and grown for 72 hours at 30°C. B,C) Strains were grown in liquid YPD in the presence of wortmannin (24–0 μg/mL) or hygromycin B (120–0 μg/mL) in DMSO (final concentration 2%).
Jjj3
Jjj3 is one of the five genes required for synthesis of diphthamide (DT), a posttranslational modification of translation elongation factor 2 (EF2)21. Studies of the human ortholog of Jjj3, Dph4, found that it is an iron-binding enzyme that is capable of participating in redox processes, and it was confirmed that Jjj3 also possesses these properties37. In fact, one of the enzymes responsible for DT synthesis, Dph2, contains an iron-sulfur cluster, and it is likely that Dph4/Jjj3 cooperates with Dph237. In our synthetic lethal screen, deletion of Jjj3 conferred mild sensitivity to the TOR-targeting compounds, rapamycin and caffeine, and the translation inhibitor, cycloheximide (Fig. 2A). Mild sensitivity of Δjjj3 to hygromycin B and wortmannin in the liquid assay was also observed (Fig. 2B,C). These results clearly demonstrate that DT synthesis is not the only process in which Jjj3 is involved. Perhaps consistent with this model, Jjj3 is partially colocalized to the perinuclear site of diphthamide synthesis, but there is a significant portion of the Jjj3 pool that associates with the broader cytoskeleton38. Interestingly, protein microarray studies suggest that Jjj3 physically interacts with 8 kinases39 and it was one of only 24/4200 proteins that interacted with 8 or more kinases, including kinases involved in the PKC pathway, septin behavior, cell cycle progression, and transcriptional activation39. Together, the previous observations and these synthetic lethal studies suggest that Jjj3 may have broad roles as a redox partner.
Apj1
Deletion of Apj1 conferred mild sensitivity to caffeine, rapamycin, cyclohexmide, calcofluor white, and wortmannin (Fig. 2). The identification of phenotypes deriving from deletion of this J protein is remarkable given its extremely low abundance - there are merely 125 copies of Apj1 in the cell40. This low number suggests that this J protein may need to be localized in order to carry out its functions. Indeed, Apj1 has been identified in stringent analyses of the mitochondrial proteome41, 42. Apj1 is a class I J protein and it is highly homologous to Ydj1 (35% identity). It is therefore unsurprising that overexpression of Apj1 can rescue loss of Ydj1 in functions such as prion propagation43. However, deletion of Apj1 confers two interesting phenotypes: suppressed RNA replication of flock house virus44 and hypersensitivity to mutant huntingtin (mHtt)45. Though the phenotypes identified to date do not point to a clear single function of Apj1, it is evident that this J protein plays roles that cannot be fully compensated for by endogenous levels of other J proteins, perhaps because of its mitochondrial localization.
Jjj2
Jjj2 is another low-abundance J protein (182 copies)40, although no specific sub-cellular localization has been reported. It is a class III protein with no predicted domains other than the J domain. Deletion of Jjj2 conferred sensitivity to cycloheximide and fluconazole (Fig. 2). These are the only two compounds tested that are exported by the PDR5 transporter, suggesting that Jjj2 may support transporter expression or function, although this idea remains untested.
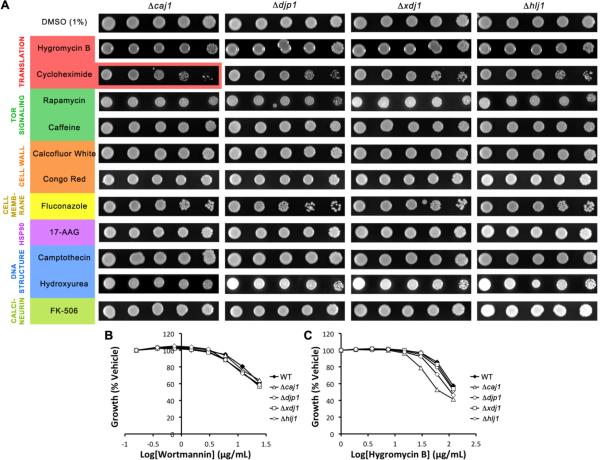
Caj1
Caj1 is thought to be located in the nucleus46, 47 but is otherwise uncharacterized. In our synthetic lethal screen, Δcaj1 cells were largely indistinguishable from wild type, with only mild phenotypes in the presence of cycloheximide in the spot assay and hygromycin B in the liquid assay (Fig. 3A, C). A previous large-scale chemical genetic screen also identified Δcaj1 as sensitive to cycloheximide24, and together these findings suggest that Caj1 may be involved in transcription.
Figure 3.
Deletion of the J proteins Caj1, Djp1, Xdj1 and Hlj1 has little or no effect in the presence of compounds tested. A) Five-fold dilutions of Δcaj1, Δdjp1, Δxdj1, or Δhlj1 were plated on rich medium containing DMSO or compound and grown for 72 hours at 30°C. B,C) Strains were grown in liquid YPD in the presence of wortmannin (24–0 μg/mL) or hygromycin B (120–0 μg/mL) in DMSO (final concentration 2%).
Djp1, Xdj1 and Hlj1
These three J proteins exhibited no phenotypes in any of the conditions tested (Fig. 3), suggesting that they either have roles in pathways not tested by these twelve compounds or that they are redundant under these conditions. It is known that Djp1 has a role in peroxisomal protein import, and it may be that this J protein is highly specialized for this role and no others48. Also, Xdj1 may not be expressed49.
Hsp70s
J proteins cooperate with Hsp70s to maintain protein quality control. When we treated Δssa1, Δssa2, or Δssa4 cells with the twelve chemical inhibitors, no apparent growth defects were observed (data not shown). These studies suggest that the Ssa proteins are redundant under these conditions. In light of this conclusion, the phenotypes resulting from deletion of individual J proteins are striking and highlight their functional diversity.
CONCLUSIONS
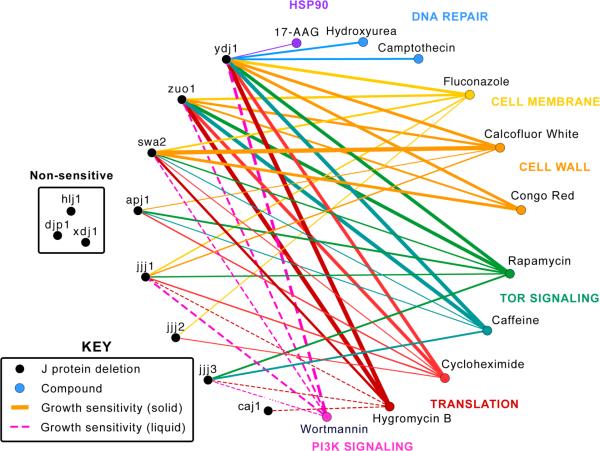
These results provide additional details to the J protein network (Fig. 4). Specifically, some J proteins, such as Ydj1 and Zuo1, have profiles that suggest broad roles in multiple cellular pathways, while others, such as Apj1 and Jjj2, seem to have focused cellular responsibilities. Although the current synthetic lethal analysis is certainly not inclusive of all possible stress conditions, these studies are supportive of a model in which J proteins have evolved to maintain a wide array of cellular processes, allowing Hsp70s to be recruited into many different types of biology.
Figure 4.
Synthetic lethal interactions identified for 11 J proteins. J proteins (black circles) are connected to compounds (colored circles) with which they exhibited a synthetic lethal interaction. Thickness of lines indicates severity of growth sensitivity phenotype. Solid lines represent interactions identified using the solid media assay and dotted lines represent interactions identified exclusively in the liquid media assay.
What do these results reveal about J proteins in disease? One interesting possibility comes from comparing these findings to the results of experiments in which proteotoxic stress (e.g. over-expression of misfolded proteins) was used to evaluate J protein activity. Specifically, Jjj3 over-expression has been shown to suppress the toxicity of polyglutamine-expanded huntingtin (Htt)50. This finding has direct relevance to our results because TOR inhibition activates autophagy and clearance of mHtt51, 52 and we found that Δjjj3 cells are sensitive to rapamycin. Similarly, we found that Apj1 is sensitive to rapamycin and this J protein is also upregulated in cells expressing mHtt50. Thus, pharmacological targeting of individual J proteins or specific Hsp70-J protein combinations53, 54 may be a compelling approach.
MATERIALS AND METHODS
Compounds
Compounds were dissolved in dimethylsulfoxide (DMSO, Sigma Aldrich) or weighed out (caffeine and hydroxyurea) and added to yeast peptone-dextrose (YPD, Teknova) rich media plates to the following final concentrations: hygromycin B (InvivoGen, 50 μg/mL), cycloheximide (Sigma Aldrich, 0.25 μg/mL), rapamycin (LC Laboratories, 0.021 μg/mL), caffeine (Sigma Aldrich, 5 mM), calcofluor white (Sigma Aldrich. 20 μg/mL), congo red (Fluka, 20 μg/mL), fluconazole (Santa Cruz, 5 μg/mL), 17-AAG (LC Laboratories, 25 μg/mL), camptothecin (Santa Cruz, 25 μg/mL), hydroxyurea (Santa Cruz, 100 mM), FK-506 (LC Laboratories, 4 μg/mL).
Yeast spot assay
Yeast knockout strains were purchased from Open Biosystems (Thermo Scientific) in the BY4741 background (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Wild type and knockout strains were grown overnight in 5 mL YPD cultures. Cells were diluted to OD600 = 0.5 and 5-fold dilutions made in YPD. A micropipette was used to spot 3 μL of cells onto plates containing DMSO (1%) or compound. Plates were inverted and incubated for 72 hours at 30°C, then imaged.
Yeast liquid assay
Compounds were dissolved in DMSO to 50 times the following final concentrations: wortmannin (24 μg/mL), hygromycin B (120 μg/mL), and calcofluor white (200 μg/mL). Two-fold serial dilutions were carried out in 96-well PCR plates (BioExpress), then 2 μL of each solution was transferred to a flat-bottom clear 96-well plate (CytoOne) using a multichannel micropipette. Yeast strains were grown overnight in 5 mL YPD cultures and diluted to OD600 = 0.15 in YPD, then 100 μL was added to each well of the plate. Plates were covered and incubated with shaking (200 RPM) at 30°C for 10 hours. OD600 values were measured using a microplate reader (Molecular Devices).
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Anuj Kumar for helpful discussions and kindly providing the yeast deletion strains and Gladis Walter for guidance and support. This work was funded by grants from the NIH (NS059690) and NSF (MCB0844512) to J.E.G. A.G. was supported by a grant from the NIH (GM008353)
REFERENCES
- 1.Mayer MP, Bukau B. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meimaridou E, Gooljar SB, Chapple JP. Journal of molecular endocrinology. 2009;42:1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 3.Daugaard M, Rohde M, Jaattela M. FEBS letters. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 4.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel M, Mayer MP, Bukau B. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- 7.Rist W, Graf C, Bukau B, Mayer MP. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 8.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 9.Young JC. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2010;88:291–300. doi: 10.1139/o09-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahi C, Craig EA. Proc Natl Acad Sci U S A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan CY, Lee S, Ren HY, Cyr DM. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene MK, Maskos K, Landry SJ. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Proc Natl Acad Sci U S A. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ER. Proc Natl Acad Sci U S A. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Protein science : a publication of the Protein Society. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kampinga HH, Craig EA. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheetham ME, Caplan AJ. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hageman J, Kampinga HH. Cell Stress Chaperones. 2009;14:1–21. doi: 10.1007/s12192-008-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gall WE, Higginbotham MA, Chen C, Ingram MF, Cyr DM, Graham TR. Curr Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- 20.Du Y, Pypaert M, Novick P, Ferro-Novick S. Mol Biol Cell. 2001;12:2614–2628. doi: 10.1091/mbc.12.9.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 23.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 24.Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A. BMC Chem Biol. 2010;10:6. doi: 10.1186/1472-6769-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caplan AJ, Douglas MG. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker J, Walter W, Yan W, Craig EA. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. Proc Natl Acad Sci U S A. 2001;98:3762–3767. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. Nature structural & molecular biology. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 29.Albanese V, Reissmann S, Frydman J. J Cell Biol. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenman HC, Craig EA. Mol Microbiol. 2004;53:335–344. doi: 10.1111/j.1365-2958.2004.04134.x. [DOI] [PubMed] [Google Scholar]
- 31.Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA. Proc Natl Acad Sci U S A. 2012;109:472–477. doi: 10.1073/pnas.1119184109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao J, Kim LS, Graham TR. Molecular Biology of the Cell. 2006;17:3281–3290. doi: 10.1091/mbc.E06-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. J Biol Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- 34.Kuranda K, Leberre V, Sokol S, Palamarczyk G, Francois J. Mol Microbiol. 2006;61:1147–1166. doi: 10.1111/j.1365-2958.2006.05300.x. [DOI] [PubMed] [Google Scholar]
- 35.Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. Proc Natl Acad Sci U S A. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer AE, Hoover LA, Craig EA. J Biol Chem. 2010;285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thakur A, Chitoor B, Goswami AV, Pareek G, Atreya HS, D'Silva P. J Biol Chem. 2012;287:13194–13205. doi: 10.1074/jbc.M112.339655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb TR, Cross SH, McKie L, Edgar R, Vizor L, Harrison J, Peters J, Jackson IJ. J Cell Sci. 2008;121:3140–3145. doi: 10.1242/jcs.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fasolo J, Sboner A, Sun MG, Yu H, Chen R, Sharon D, Kim PM, Gerstein M, Snyder M. Genes Dev. 2011;25:767–778. doi: 10.1101/gad.1998811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 41.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 42.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hines JK, Li X, Du Z, Higurashi T, Li L, Craig EA. PLoS Genet. 2011;7:e1001309. doi: 10.1371/journal.pgen.1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weeks SA, Shield WP, Sahi C, Craig EA, Rospert S, Miller DJ. J Virol. 2010;84:330–339. doi: 10.1128/JVI.01808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 46.Mukai H, Shuntoh H, Chang CD, Asami M, Ueno M, Suzuki K, Kuno T. Gene. 1994;145:125–127. doi: 10.1016/0378-1119(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 47.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I. J Cell Biol. 1998;142:421–434. doi: 10.1083/jcb.142.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarz E, Westermann B, Caplan AJ, Ludwig G, Neupert W. Gene. 1994;145:121–124. doi: 10.1016/0378-1119(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 50.Tauber E, Miller-Fleming L, Mason RP, Kwan W, Clapp J, Butler NJ, Outeiro TF, Muchowski PJ, Giorgini F. J Biol Chem. 2011;286:410–419. doi: 10.1074/jbc.M110.101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyrskyluoto A, Reijonen S, Kivinen J, Lindholm D, Korhonen L. Experimental cell research. 2012;318:33–42. doi: 10.1016/j.yexcr.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 52.Roscic A, Baldo B, Crochemore C, Marcellin D, Paganetti P. Journal of neurochemistry. 2011;119:398–407. doi: 10.1111/j.1471-4159.2011.07435.x. [DOI] [PubMed] [Google Scholar]
- 53.Evans CG, Chang L, Gestwicki JE. Journal of medicinal chemistry. 2010;53:4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patury S, Miyata Y, Gestwicki JE. Current topics in medicinal chemistry. 2009;9:1337–1351. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.